Abstract
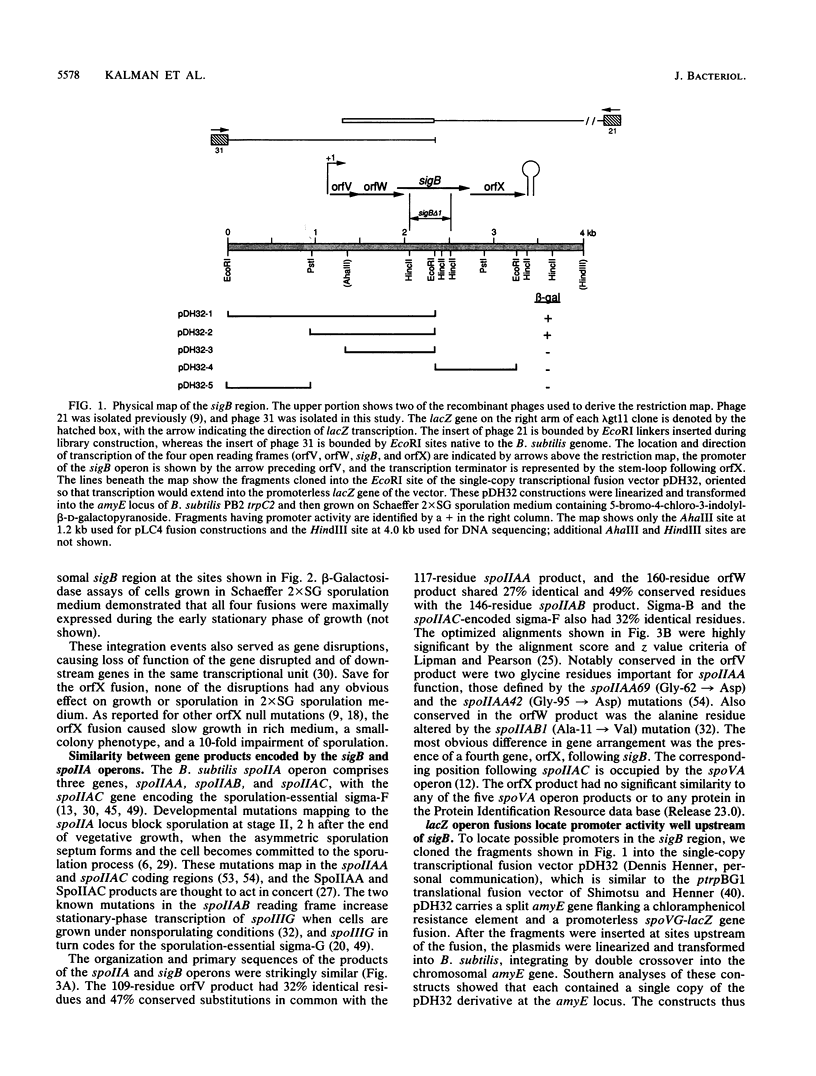
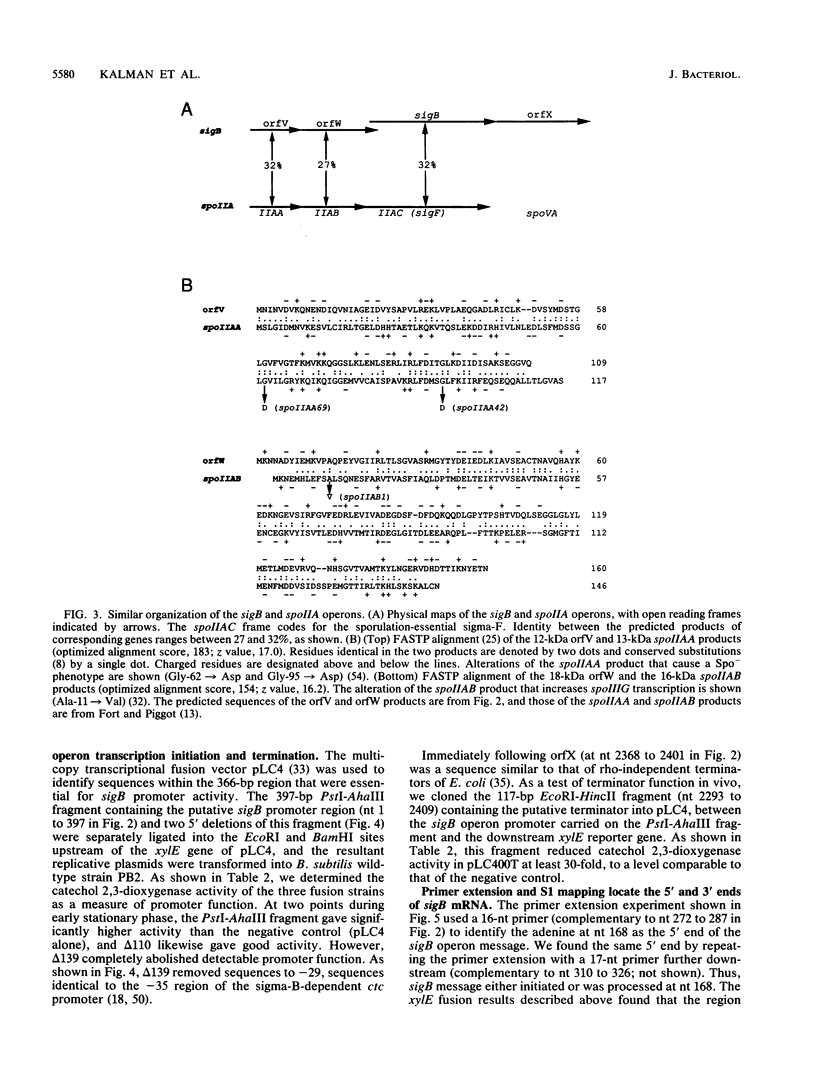
Bacillus subtilis sigma-B is an alternate sigma factor implicated in controlling stationary-phase gene expression. We characterized the genetic organization and regulation of the region containing the sigma-B structural gene (sigB) to learn which metabolic signals and protein factors govern sigma-B function. sigB lay in an operon with four open reading frames (orfs) in the order orfV-orfW-sigB-orfX, and lacZ gene fusions showed that all four frames were translated in vivo. Experiments with primer extension, S1 nuclease mapping, and lacZ transcriptional fusions found that sigB operon transcription initiated early in stationary phase from a site 32 nucleotides upstream of orfV and terminated 34 nucleotides downstream of orfX. Fusion expression was abolished in a strain carrying an in-frame deletion in sigB, suggesting that sigma-B positively regulated its own synthesis, and deletions in the sigB promoter region showed that sequences identical to the sigma-B-dependent ctc promoter were essential for promoter activity. Fusion expression was greatly enhanced in a strain carrying an insertion mutation in orfX, suggesting that the 22-kilodalton (kDa) orfX product was a negative effector of sigma-B expression or activity. Notably, the genetic organization of the sigB operon was strikingly similar to that of the B. subtilis spoIIA operon, which has the gene order spoIIAA-spoIIAB-spoIIAC, with spoIIAC encoding the sporulation-essential sigma-F. The predicted sequence of the 12-kDa orfV product was 32% identical to that of the 13-kDa SpoIIAA protein, and the 18-kDa orfW product was 27% identical to the 16-kDa SpoIIAB protein. On the basis of this clear evolutionary conservation, we speculate these protein pairs regulate their respective sigma factors by a similar molecular mechanism and that the spoIIA and sigB operons might control divergent branches of stationary-phase gene expression.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnosti D. N., Chamberlin M. J. Secondary sigma factor controls transcription of flagellar and chemotaxis genes in Escherichia coli. Proc Natl Acad Sci U S A. 1989 Feb;86(3):830–834. doi: 10.1073/pnas.86.3.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnie C., Lampe M., Losick R. Gene encoding the sigma 37 species of RNA polymerase sigma factor from Bacillus subtilis. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5943–5947. doi: 10.1073/pnas.83.16.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan S. A., Chun K. T., Edson B. A., Price C. W. Early-blocked sporulation mutations alter expression of enzymes under carbon control in Bacillus subtilis. Mol Gen Genet. 1988 May;212(2):271–280. doi: 10.1007/BF00334696. [DOI] [PubMed] [Google Scholar]
- Boylan S. A., Suh J. W., Thomas S. M., Price C. W. Gene encoding the alpha core subunit of Bacillus subtilis RNA polymerase is cotranscribed with the genes for initiation factor 1 and ribosomal proteins B, S13, S11, and L17. J Bacteriol. 1989 May;171(5):2553–2562. doi: 10.1128/jb.171.5.2553-2562.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater K. F., Bruton C. J., Plaskitt K. A., Buttner M. J., Méndez C., Helmann J. D. The developmental fate of S. coelicolor hyphae depends upon a gene product homologous with the motility sigma factor of B. subtilis. Cell. 1989 Oct 6;59(1):133–143. doi: 10.1016/0092-8674(89)90876-3. [DOI] [PubMed] [Google Scholar]
- Cooney P. H., Whiteman P. F., Freese E. Media dependence of commitment in Bacillus subtilis. J Bacteriol. 1977 Feb;129(2):901–907. doi: 10.1128/jb.129.2.901-907.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan M. L., Kalman S. S., Thomas S. M., Price C. W. Gene encoding the 37,000-dalton minor sigma factor of Bacillus subtilis RNA polymerase: isolation, nucleotide sequence, chromosomal locus, and cryptic function. J Bacteriol. 1987 Feb;169(2):771–778. doi: 10.1128/jb.169.2.771-778.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J., Mandelstam J. Use of a lacZ gene fusion to determine the dependence pattern of sporulation operon spoIIA in spo mutants of Bacillus subtilis. J Gen Microbiol. 1986 Nov;132(11):2967–2976. doi: 10.1099/00221287-132-11-2967. [DOI] [PubMed] [Google Scholar]
- Ferrari F. A., Trach K., Hoch J. A. Sequence analysis of the spo0B locus reveals a polycistronic transcription unit. J Bacteriol. 1985 Feb;161(2):556–562. doi: 10.1128/jb.161.2.556-562.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort P., Errington J. Nucleotide sequence and complementation analysis of a polycistronic sporulation operon, spoVA, in Bacillus subtilis. J Gen Microbiol. 1985 May;131(5):1091–1105. doi: 10.1099/00221287-131-5-1091. [DOI] [PubMed] [Google Scholar]
- Grossman A. D., Erickson J. W., Gross C. A. The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell. 1984 Sep;38(2):383–390. doi: 10.1016/0092-8674(84)90493-8. [DOI] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Helmann J. D., Masiarz F. R., Chamberlin M. J. Isolation and characterization of the Bacillus subtilis sigma 28 factor. J Bacteriol. 1988 Apr;170(4):1560–1567. doi: 10.1128/jb.170.4.1560-1567.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol. 1982 May;150(2):804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo M. M., Losick R. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J Mol Biol. 1986 Oct 20;191(4):615–624. doi: 10.1016/0022-2836(86)90449-3. [DOI] [PubMed] [Google Scholar]
- Igo M., Lampe M., Ray C., Schafer W., Moran C. P., Jr, Losick R. Genetic studies of a secondary RNA polymerase sigma factor in Bacillus subtilis. J Bacteriol. 1987 Aug;169(8):3464–3469. doi: 10.1128/jb.169.8.3464-3469.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmazyn-Campelli C., Bonamy C., Savelli B., Stragier P. Tandem genes encoding sigma-factors for consecutive steps of development in Bacillus subtilis. Genes Dev. 1989 Feb;3(2):150–157. doi: 10.1101/gad.3.2.150. [DOI] [PubMed] [Google Scholar]
- Kroos L., Kunkel B., Losick R. Switch protein alters specificity of RNA polymerase containing a compartment-specific sigma factor. Science. 1989 Jan 27;243(4890):526–529. doi: 10.1126/science.2492118. [DOI] [PubMed] [Google Scholar]
- Kustu S., Santero E., Keener J., Popham D., Weiss D. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989 Sep;53(3):367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landick R., Vaughn V., Lau E. T., VanBogelen R. A., Erickson J. W., Neidhardt F. C. Nucleotide sequence of the heat shock regulatory gene of E. coli suggests its protein product may be a transcription factor. Cell. 1984 Aug;38(1):175–182. doi: 10.1016/0092-8674(84)90538-5. [DOI] [PubMed] [Google Scholar]
- Leighton T. New types of RNA polymerase mutations causing temperature-sensitive sporulation in bacillus subtilis. J Biol Chem. 1977 Jan 10;252(1):268–272. [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Losick R., Pero J. Cascades of Sigma factors. Cell. 1981 Sep;25(3):582–584. doi: 10.1016/0092-8674(81)90164-1. [DOI] [PubMed] [Google Scholar]
- Losick R., Youngman P., Piggot P. J. Genetics of endospore formation in Bacillus subtilis. Annu Rev Genet. 1986;20:625–669. doi: 10.1146/annurev.ge.20.120186.003205. [DOI] [PubMed] [Google Scholar]
- Piggot P. J., Coote J. G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976 Dec;40(4):908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot P. J., Curtis C. A., de Lencastre H. Use of integrational plasmid vectors to demonstrate the polycistronic nature of a transcriptional unit (spoIIA) required for sporulation of Bacillus subtilis. J Gen Microbiol. 1984 Aug;130(8):2123–2136. doi: 10.1099/00221287-130-8-2123. [DOI] [PubMed] [Google Scholar]
- Price C. W., Doi R. H. Genetic mapping of rpoD implicates the major sigma factor of Bacillus subtilis RNA polymerase in sporulation initiation. Mol Gen Genet. 1985;201(1):88–95. doi: 10.1007/BF00397991. [DOI] [PubMed] [Google Scholar]
- Rather P. N., Coppolecchia R., DeGrazia H., Moran C. P., Jr Negative regulator of sigma G-controlled gene expression in stationary-phase Bacillus subtilis. J Bacteriol. 1990 Feb;172(2):709–715. doi: 10.1128/jb.172.2.709-715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray C., Hay R. E., Carter H. L., Moran C. P., Jr Mutations that affect utilization of a promoter in stationary-phase Bacillus subtilis. J Bacteriol. 1985 Aug;163(2):610–614. doi: 10.1128/jb.163.2.610-614.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M., Solomon L., Zipkas D. Relationship between gene function and gene location in Escherichia coli. J Mol Evol. 1978 May 12;11(1):47–56. doi: 10.1007/BF01768024. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sala-Trepat J. M., Evans W. C. The meta cleavage of catechol by Azotobacter species. 4-Oxalocrotonate pathway. Eur J Biochem. 1971 Jun 11;20(3):400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savva D., Mandelstam J. Synthesis of spoIIA and spoVA mRNA in Bacillus subtilis. J Gen Microbiol. 1986 Nov;132(11):3005–3011. doi: 10.1099/00221287-132-11-3005. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Shimotsu H., Henner D. J. Construction of a single-copy integration vector and its use in analysis of regulation of the trp operon of Bacillus subtilis. Gene. 1986;43(1-2):85–94. doi: 10.1016/0378-1119(86)90011-9. [DOI] [PubMed] [Google Scholar]
- Sripati C. E., Warner J. R. Isolation, characterization, and translation of mRNA from yeast. Methods Cell Biol. 1978;20:61–81. doi: 10.1016/s0091-679x(08)62009-9. [DOI] [PubMed] [Google Scholar]
- Stahl M. L., Ferrari E. Replacement of the Bacillus subtilis subtilisin structural gene with an In vitro-derived deletion mutation. J Bacteriol. 1984 May;158(2):411–418. doi: 10.1128/jb.158.2.411-418.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989 Dec;53(4):450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P. Comment on 'Duplicated sporulation genes in bacteria' by J. Errington, P. Fort and J. Mandelstam. FEBS Lett. 1986 Jan 20;195(1-2):9–11. doi: 10.1016/0014-5793(86)80119-3. [DOI] [PubMed] [Google Scholar]
- Stragier P., Kunkel B., Kroos L., Losick R. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science. 1989 Jan 27;243(4890):507–512. doi: 10.1126/science.2536191. [DOI] [PubMed] [Google Scholar]
- Straus D. B., Walter W. A., Gross C. A. The heat shock response of E. coli is regulated by changes in the concentration of sigma 32. Nature. 1987 Sep 24;329(6137):348–351. doi: 10.1038/329348a0. [DOI] [PubMed] [Google Scholar]
- Suh J. W., Boylan S. A., Price C. W. Gene for the alpha subunit of Bacillus subtilis RNA polymerase maps in the ribosomal protein gene cluster. J Bacteriol. 1986 Oct;168(1):65–71. doi: 10.1128/jb.168.1.65-71.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D. X., Stragier P., Setlow P. Identification of a new sigma-factor involved in compartmentalized gene expression during sporulation of Bacillus subtilis. Genes Dev. 1989 Feb;3(2):141–149. doi: 10.1101/gad.3.2.141. [DOI] [PubMed] [Google Scholar]
- Tatti K. M., Moran C. P., Jr Promoter recognition by sigma-37 RNA polymerase from Bacillus subtilis. J Mol Biol. 1984 May 25;175(3):285–297. doi: 10.1016/0022-2836(84)90349-8. [DOI] [PubMed] [Google Scholar]
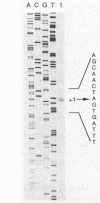
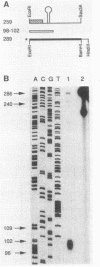
- Wu J. J., Howard M. G., Piggot P. J. Regulation of transcription of the Bacillus subtilis spoIIA locus. J Bacteriol. 1989 Feb;171(2):692–698. doi: 10.1128/jb.171.2.692-698.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Yeast RNA polymerase II genes: isolation with antibody probes. Science. 1983 Nov 18;222(4625):778–782. doi: 10.1126/science.6356359. [DOI] [PubMed] [Google Scholar]
- Yudkin M. D., Jarvis K. A., Raven S. E., Fort P. Effects of transition mutations in the regulatory locus spoIIA on the incidence of sporulation in Bacillus subtilis. J Gen Microbiol. 1985 Apr;131(4):959–962. doi: 10.1099/00221287-131-4-959. [DOI] [PubMed] [Google Scholar]
- Yudkin M. D. Structure and function in a Bacillus subtilis sporulation-specific sigma factor: molecular nature of mutations in spoIIAC. J Gen Microbiol. 1987 Mar;133(3):475–481. doi: 10.1099/00221287-133-3-475. [DOI] [PubMed] [Google Scholar]