Abstract
Δ4-3-oxosteroid 5β-reductase is member D1 of the aldo-keto reductase family 1 (AKR1D1), which catalyzes 5β-reduction of molecules with a 3-oxo-4-ene structure. Bile acid intermediates and most of the steroid hormones carry the 3-oxo-4-ene structure. Therefore, AKR1D1 plays critical roles in both bile acid synthesis and steroid hormone metabolism. Currently our understanding on transcriptional regulation of AKR1D1 under physiological and pathological conditions is very limited. In this study, we investigated the regulatory effects of primary bile acids, chenodeoxycholic acid (CDCA) and cholic acid (CA), on AKR1D1 expression. The expression levels of AKR1D1 mRNA and protein in vitro and in vivo following bile acid treatments were determined by real-time PCR and Western blotting. We found that CDCA markedly repressed AKR1D1 expression in vitro in human hepatoma HepG2 cells and in vivo in mice. On the contrary, CA significantly upregulated AKR1D1 expression in HepG2 cells and in mice. Further mechanistic investigations revealed that the farnesoid x receptor (FXR) signaling pathway was not involved in regulating AKR1D1 by bile acids. Instead, CDCA and CA regulated AKR1D1 through the mitogen-activated protein kinases/c-Jun N-terminal kinases (MAPK/JNK) signaling pathway. Inhibition of the MAPK/JNK pathway effectively abolished CDCA and CA-mediated regulation of AKR1D1. It was thus determined that AKR1D1 expression was regulated by CDCA and CA through modulating the MAPK/JNK signaling pathway. In conclusion, AKR1D1 expression was differentially regulated by primary bile acids through negative and positive feedback mechanisms. The findings indicated that both bile acid concentrations and compositions play important roles in regulating AKR1D1 expression, and consequently bile acid synthesis and steroid hormone metabolism.
Introduction
Bile acids are the ultimate metabolites of cholesterol. Studies in the past decade uncovered a broad spectrum of functions associated with bile acids as hormone-like signaling molecules through various nuclear receptors, notably farnesoid x receptor (FXR) and G protein-coupled bile acid receptor 5 (TGR5) [1–3]. Both FXR and TGR5 signaling pathways play critical roles in regulating bile acids, cholesterol, lipids and glucose homeostasis [4–7]. Bile acid homeostasis is maintained through tightly regulated bile acid synthesis and enterohepatic circulation. Cholesterol 7α-hydroxylase (CYP7A1) is the rate limiting enzyme in classical bile acid synthesis while sterol 12α-hydroxylase (CYP8B1) is the determinant enzyme for the production of CA [8–11]. Canalicular secretion of bile acids through bile salt export pump (BSEP) is the rate-limiting step in the enterohepatic circulation of bile acids [12, 13]. Activation of FXR by bile acids represses CYP7A1 while induces BSEP expression to maintain hepatic bile acid homeostasis [14, 15].
Aldo-keto reductase family 1, member D1 (AKR1D1) is a Δ4-3-oxosteroid 5β-reductase, which catalyzes the reduction of molecules with a 3-oxo-4-ene structure in the presence of nicotinamide adenine dinucleotide phosphate (NADPH) as a hydride donor [16–18]. AKR1D1 is required for cholesterol metabolism into bile acids. In the bile acid synthesis pathway, AKR1D1 catalyzes a reaction downstream of the reaction mediated by CYP7A1. It reduces the double bond in the A ring of the bile acid intermediates to eventually produce primary bile acids [19–21]. Consistent with its vital role in bile acid synthesis, deficiency in AKR1D1 expression or activity has been directly linked to severe cholestatic liver disease in infancy, with characteristic appearance of unsaturated 3-oxo-4-ene-bile acids [22–27]. In addition to its critical role in bile acid synthesis, AKR1D1 is also involved in steroid hormone metabolism and clearance [28, 29]. Most of the steroid hormones including testosterones, progesterones, mineralocorticoids and glucocorticoids carry the 3-oxo-4-ene structure. Since 5β-reduction is a common transformation and major deactivation pathway for steroid hormones, and AKR1D1 is the only enzyme in humans capable of catalyzing a 5β-reduction in those steroids, AKR1D1 plays a critical role in regulating and maintaining the homeostasis of steroid hormones [19, 22, 28, 29]. Taken together, AKR1D1 is sitting at the interface between the two important biological pathways, bile acid synthesis and steroid hormone metabolism, both of which have a broad spectrum of impacts on metabolisms and energy expenditure. However, our current understanding on the regulation of AKR1D1 expression under physiological and pathological conditions is very limited.
In this study, we investigated the regulatory effects of bile acids on AKR1D1 expression. Chenodeoxycholic acid (CDCA) and cholic acid (CA) are the two primary bile acids in human. We found that CDCA markedly repressed AKR1D1 expression in vitro in human hepatoma HepG2 cells and in vivo in mice. In contrast, CA significantly upregulated AKR1D1 expression in HepG2 cells and in mice. Further mechanistic studies revealed that the FXR signaling pathway was not involved in regulating AKR1D1 by bile acids. Instead, CDCA and CA regulated AKR1D1 expression through the mitogen-activated protein kinases/c-Jun N-terminal kinases (MAPK/JNK) signaling pathway. Inhibition of the MAPK/JNK pathway effectively abolished CDCA and CA-mediated regulatory effects on AKR1D1. It was thus concluded that AKR1D1 expression was differentially regulated by CDCA and CA through modulating the MAPK/JNK signaling pathway.
Materials and Methods
Chemicals and reagents
CDCA and CA were purchased from Sigma-Aldrich. FXR agonist GW4064 was obtained from Tocris Biosciences. Cell culture reagents Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), TaqMan master-mix and probes were purchased from Life Technologies. RNA Bee for RNA isolation, dimethyl sulfoxide (DMSO), and propanediol were obtained from Fisher Scientific. Primary and secondary antibodies for Western blotting were purchased from Santa Cruz Biotechnologies. All Western blotting gels, buffers, and markers were purchased through BioRad Laboratories. Complementary DNA synthesis kit was purchased through Promega. Protease inhibitor, Halt, and bovine serum albumin quantification reagents were purchased from Thermo Scientific.
Treatment of HepG2 cells
Human hepatoma HepG2 cells (ATCC, HB-8065) were seeded in 12-well plates at a cell density of 4.0 x 105 in 1.0 mL DMEM supplemented with 10% (v/v) FBS, 1% (v/v) penicillin/streptomycin and 1% (v/v) non-essential amino acids (NEAA) and cultured in a 5% CO2 incubator overnight at 37°C before treatment. After overnight incubation, cells were treated with 25 μM CDCA, 25 μM CA, 1 μM GW4064 or vehicle DMSO for 30 hours. Total mRNA and protein lysates were prepared for real-time PCR and Western blot. HepG2 cells seeded in 12-well plates were transfected with FXRα1 or FXRα2 expression plasmid (1μg/well) [30], followed by treatment of the transfected cells with FXR agonist GW4064 for 30 h. Total cellular RNAs were isolated for real-time PCR analyses. HepG2 cells were treated with CDCA (25μM), CDCA+MAPK/JNK inhibitor SP600125 (1μM), CDCA+MAPK/ERK1/2 inhibitor PD98059 (5μM), CA (50μM), CA+SP600125, CA+PD98059, or vehicle for 30 hours, followed by preparation of total RNA for real-time PCR and cell lysates for Western blot analysis.
Treatment of mice
Twenty four male and female CD-1 mice (mus musculus) were bred in-house and randomly divided into 4 groups with 6 mice per group at ages of 6–8 weeks. The mice had free access to food and water and were on a 12-hour dark/light cycle. The mice were treated with CDCA (5mg/kg), CA (5mg/kg), GW4064 (1mg/kg) or vehicle propanediol through intraperitoneal injection twice a day for 3 days. Twelve hours after the last injection, mice were euthanized by CO2 euthanasia method and liver tissues were harvested and processed for mRNA and protein analyses. In addition, a group of wild type (Wt) and FXR-knockout mice (FXR-/-) were euthanized with CO2 euthanasia method and liver tissues were collected for gene expression analysis. The Institutional Animal Care and Use Committee (IACUC) at the University of Rhode Island approved all animal studies with a protocol number of AN09-02-008.
Quantitative real-time PCR
Total RNA isolation from HepG2 cells or mouse liver tissues and subsequent TaqMan real-time PCR were carried out as described [31, 32]. Transcript levels of AKR1D1, CYP7A1, CYP8B1 and BSEP were quantified by real-time PCR with gene-specific probes. The expression levels of target mRNA were normalized against the mRNA levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Western blotting
Cell lysates were made from HepG2 or mouse liver tissues as described [33, 34]. Thirty micrograms of total protein were loaded into each well. Membranes were blotted with antibodies against human or mouse AKR1D1, CYP7A1, CYP8B1 or BSEP (Santa Cruz Biotechnologies). The same membranes were stripped and re-blotted with antibodies against GAPDH (Sigma-Aldrich). Quantifications of AKR1D1, CYP7A1, CYP8B1 and BSEP protein levels were performed with all the samples in each individual groups or treatments. Representative images of Western blot from each individual groups or treatments were presented. The expression levels of GAPDH were used to normalize the expression of target genes [35].
Signal Transduction 45-Pathway Reporter Array
The Cignal 45-Pathway Reporter Array was purchased from Qiagen (Cat#: CCA-901L). The array assays were essentially carried out according to the protocol recommended by the manufacture. Reverse transfection was performed using HepG2 cells (8x104 cells/well in 98-well plates). Sixteen hours post-transfection, transfected cells were treated with CDCA (25 μM), CA (25 μM) or vehicle DMSO for 30 hours, followed by detection of luciferase activities by the Dual Luciferase Assay [36].
Statistical analysis
Student’s t-test was applied to pair-wise comparison for normally distributed data. One-way ANOVA was applied to analyze data with multiple groups, followed by Tukey post-hoc test for multiple comparisons. Non-parametric Mann-Whitney test was used for pair-wise comparison for non-normally distributed data. A p value of 0.05 or less was considered statistically significant.
Results
Bile acid CDCA markedly repressed AKR1D1 expression in human hepatoma HepG2 cells
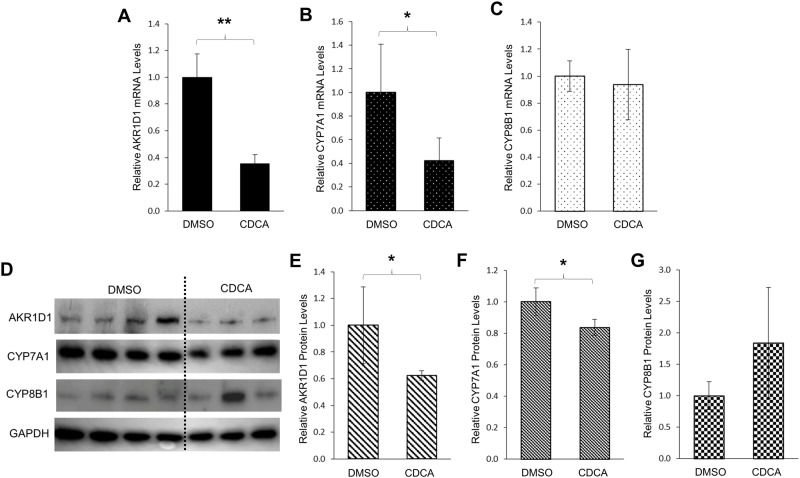
CDCA is one of the major primary bile acids in human. AKR1D1 is an enzyme involved in both bile acid synthesis and steroid hormone metabolism. To investigate whether AKR1D1 expression is regulated by its ultimate product CDCA, the effects of CDCA treatment on AKR1D1 expression were determined in human hepatoma HepG2 cells. As shown in Fig 1A, AKR1D1 mRNA expression levels were markedly decreased by 65% in cells treated with CDCA in comparison with the levels in cells treated with vehicle. It is well established that CDCA down-regulates CYP7A1 through a negative feedback loop [8, 9]. Indeed, CDCA treatment reduced CYP7A1 mRNA levels by 58% (Fig 1B). However, the expression levels of CYP8B1 mRNA were not significantly changed (Fig 1C). Consistent with the altered mRNA levels, CDCA treatment significantly reduced the protein levels of AKR1D1 (Fig 1D and 1E) by 40%. As expected, CYP7A1 protein levels were also significantly decreased (Fig 1D and 1F). On the other hand, CYP8B1 protein levels were slightly increased without reaching a statistical significance (Fig 1D and 1G). The data thus demonstrated that CDCA repressed AKR1D1 expression in human HepG2 cells.
Fig 1. CDCA markedly repressed AKR1D1 expression in human hepatoma HepG2 cells.
HepG2 cells were treated with CDCA (25μM) or vehicle DMSO (0.1%) for 30h, followed by detection of the mRNA levels of (A) AKR1D1, (B) CYP7A1 and (C) CYP8B1 by real-time PCR, and (D) protein levels of AKR1D1, CYP7A1 and CYP8B1 by Western blotting. (E) quantification of AKR1D1, (F) CYP7A1, and (G) CYP8B1 protein levels in (D). The data are presented as mean ± SD of at least three separate experiments or treatments. The Student’s t-test was applied to pair-wise comparison. *p<0.05, **p<0.01.
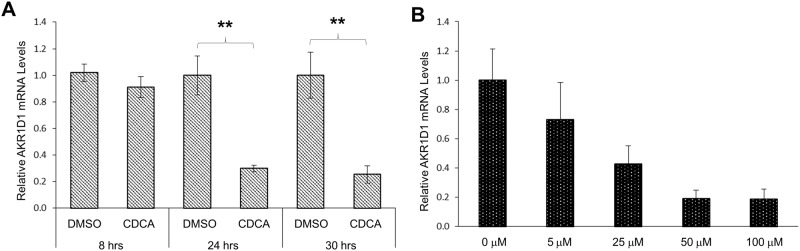
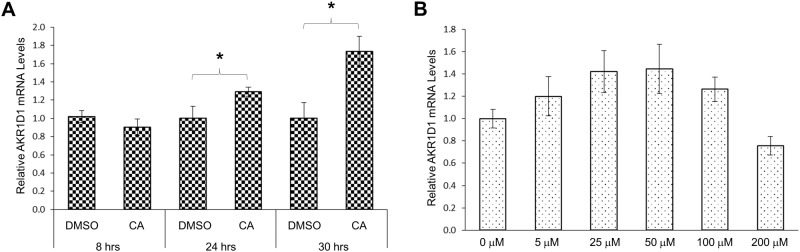
A time course study revealed that no significant repression of AKR1D1 expression by CDCA was detected 8 hours (hrs) post-treatment. However, marked repressions of AKR1D1 (70% and 75% reduction) were detected 24 and 30 hrs post-treatment of CDCA (Fig 2A). A dose-response study showed that CDCA dose-dependently repressed AKR1D1 expression (Fig 2B). AKR1D1 expression levels were decreased by 27%, 57%, 81% and 81% in cells treated with 5, 25, 50 and 100 μM CDCA, respectively. Taken together, the results demonstrated that AKR1D1 expression was repressed by CDCA in time and dose-dependent manners.
Fig 2. CDCA repressed AKR1D1 expression in time and dose-dependent manners in HepG2 cells.
(A) HepG2 cells were treated with either CDCA (25 μM) or vehicle DMSO for 8, 24 or 30 hrs, followed by detection of AKR1D1 mRNA with real-time PCR. (B) HepG2 cells were treated with various concentrations of CDCA (0, 5, 25, 50, 100 μM) for 30 hrs, followed by detection of AKR1D1 mRNA expression with real-time PCR. **p<0.01 with the Student’s t-test for pair-wise comparison.
Akr1d1 expression was severely repressed by CDCA in vivo in mice
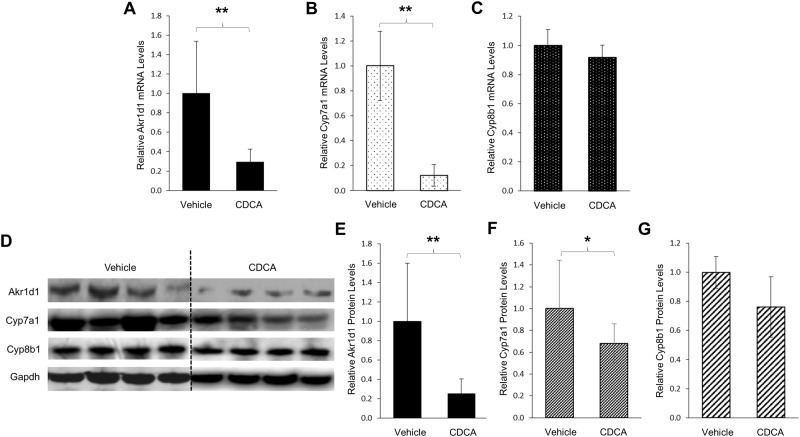
To determine whether CDCA-mediated regulation of AKR1D1 in HepG2 cells is conserved in vivo in mice, we carried out a mouse study to determine the effects of CDCA on Akr1d1 expression. As shown in Fig 3A, Akr1d1 mRNA levels were severely repressed by CDCA. Compared with vehicle control, Akr1d1 mRNA levels were decreased by 70%. As expected, CDCA treatment markedly decreased Cyp7a1 mRNA levels by 88% (Fig 3B). However, Cyp8b1 mRNA levels were minimally changed by CDCA treatment (Fig 3C). Consistently, Akr1d1 protein levels were decreased by 75% in mice treated with CDCA when compared with vehicle-treated mice (Fig 3D and 3E). Similarly, the Cyp7a1 protein levels were significantly decreased by CDCA treatment (Fig 3D and 3F). On the other hand, no significant changes in CYP8B1 protein levels were detected (Fig 3D and 3G). Taken together, the data demonstrated that consistent with the results from human HepG2 cells, CDCA down-regulated AKR1D1 expression in vivo in mice. It was thus concluded that similar to CYP7A1, AKR1D1 was regulated by CDCA in a negative feedback manner.
Fig 3. Akr1d1 expression was severely repressed by CDCA in vivo in mice.
Two groups of mice (n = 6/group) were treated with CDCA (5mg/kg) or vehicle propanediol through intraperitoneal injection twice a day for 3 days. Twelve hours after the last injection, mice were euthanized and liver tissues were harvested and processed for mRNA and protein analyses. (A) the effects of CDCA treatment on Akr1d1, (B) Cyp7a1 and (C) Cyp8b1 mRNA levels detected by real-time PCR. (D) the effects of CDCA treatment on Akr1d1, Cyp7a1 and Cyp8b1 protein expression detected by Western blotting, and (E) quantification of Akr1d1, (F) Cyp7a1 and (G) Cyp8b1 protein levels in (). The data are presented as mean ± SD of the groups. The Student’s t-test was applied to pair-wise comparison. *p<0.05, **p<0.01.
Bile acid CA significantly upregulated AKR1D1 expression in human HepG2 cells
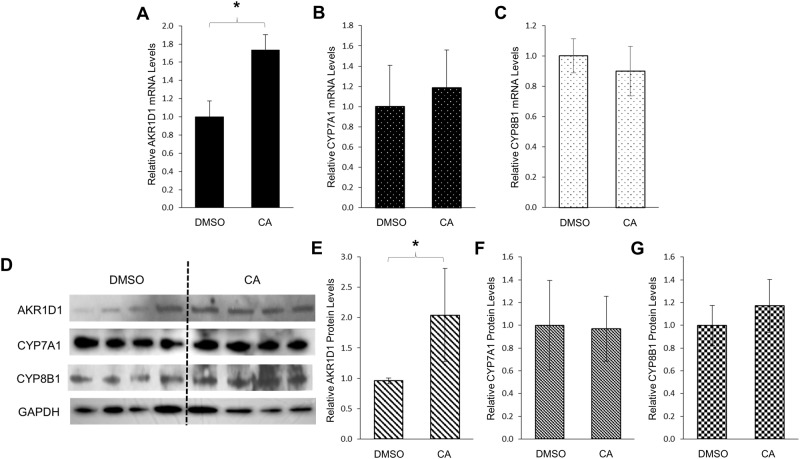
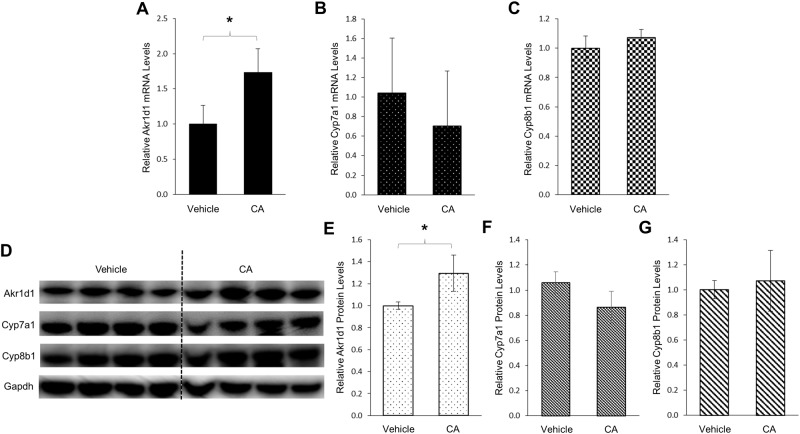
CA is the other primary bile acid in human. To determine whether CA acts similarly as CDCA in modulating AKR1D1 expression, the effects of CA treatment on AKR1D1 expression was investigated in HepG2 cells. As shown in Fig 4A, in contrast to CDCA, CA treatment significantly increased AKR1D1 mRNA levels by 74% when compared with vehicle. On the other hand, the expression levels of CYP7A1 and CYP8B1 mRNA were not significantly altered by CA treatment (Fig 4B and 4C). In line with mRNA levels, AKR1D1 protein levels were increased by 1.1 fold in cells treated with CA (Fig 4D and 4E) while CYP7A1 and CYP8B1 protein levels were minimally changed. (Fig 4D, 4F and 4G).
Fig 4. CA significantly upregulated AKR1D1 expression in human HepG2 cells.
HepG2 cells were treated with CA (25μM) or vehicle DMSO (0.1%) for 30h, followed by detection of the mRNA levels of (A) AKR1D1, (B) CYP7A1 and (C) CYP8B1 by real-time PCR, and the protein levels of (D) AKR1D1, CYP7A1 and CYP8B1 detected by Western blotting. (E) quantification of AKR1D1, (F) CPY7A1 and (G) CYP8B1 protein levels in (D). The data are presented as mean ± SD of at least three separate experiments or treatments. The Student’s t-test was applied to pair-wise comparison. *p<0.05.
A time course study showed that minimal changes in AKR1D1 expression were detected in cells treated with CA for 8 hrs. A significant 30% increase in AKR1D1 expression was detected in cells treated with CA for 24 hrs with additional increases up to 74% being detected in cells treated with CA for 30 hrs (Fig 5A). A dosing study revealed an atypical response in AKR1D1 expression following treatments with various concentrations of CA (Fig 5B). From the dose range of 0 to 50 μM, AKR1D1 expression levels were gradually increased as CA concentrations increased. However, from the dose range of 50 to 200 μM, AKR1D1 expression levels were gradually decreased as CA concentrations increased (Fig 5B). Therefore, CA-mediated modulating effects on AKR1D1 expression were dose-dependent and binary.
Fig 5. CA modulated AKR1D1 expression in time and dose-dependent manners in HepG2 cells.
(A) HepG2 cells were treated with either CA (25 μM) or vehicle DMSO for 8, 24 or 30 hrs, followed by detection of AKR1D1 mRNA with real-time PCR. (B) HepG2 cells were treated with various concentrations of CA (0, 5, 25, 50, 100, 200 μM) for 30 hrs, followed by detection of AKR1D1 mRNA expression with real-time PCR. *p<0.05 with the Student’s t-test for pair-wise comparison.
Akr1d1 expression was induced by CA in vivo in mice
To confirm the findings that AKR1D1 expression was upregulated by CA in HepG2 cells, we performed a mouse study to determine the effects of CA on Akr1d1 expression in vivo. As shown in Fig 6A, CA treatment significantly increased the expression levels of Akr1d1 mRNA by 73%. On the other hand, Cyp7a1 expression was slightly decreased by CA treatment without reaching statistical significance (Fig 6B). Minimal changes in Cyp8b1 mRNA levels were detected in mice treated with CA (Fig 6C). Consistent with the results of mRNA levels, Akr1d1 protein levels were significantly increased by 36% following CA treatment (Fig 6D and 6E) while a slight decrease in Cyp7a1 and minimal change in Cyp8b1 protein levels were detected (Fig 6F and 6G). The data thus demonstrated that in contrast to CDCA, CA induced AKR1D1 expression in vitro in human HepG2 cells and in vivo in mice. Taken together, the results revealed that bile acids CDCA and CA differentially down- and upregulated AKR1D1 expression.
Fig 6. Akr1d1 expression was induced by CA in vivo in mice.
Two groups of mice (n = 6/group) were treated with CA (5mg/kg) or vehicle propanediol through intraperitoneal injection twice a day for 3 days. Twelve hours after the last injection, mice were euthanized and liver tissues were harvested and processed for mRNA and protein analyses. (A) the effects of CA treatment on Akr1d1, (B) Cyp7a1 and (C) Cyp8b1 mRNA levels detected by real-time PCR. (D) the effects of CA treatment on Akr1d1, Cyp7a1 and Cyp8b1 protein expression detected by Western blotting, and (E) quantification of Akr1d1, (F) Cyp7a1 and (G) Cyp8b1 protein levels in (D). The data are presented as mean ± SD of the groups. The Student’s t-test was applied to pair-wise comparison. *p<0.05.
FXR signaling was not involved in regulating AKR1D1
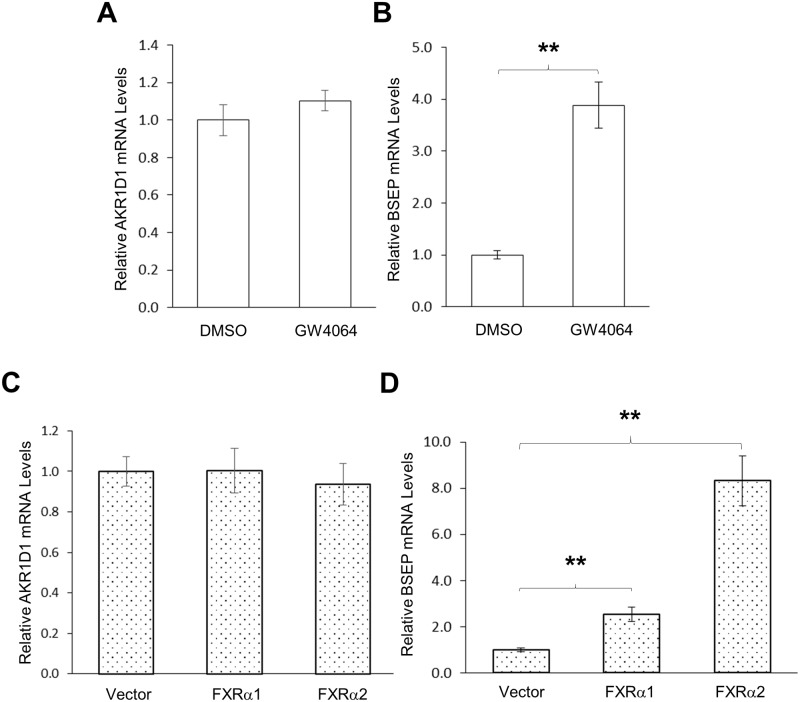
FXR is the master regulator of bile acid homeostasis [1, 2], and bile acids including CDCA and CA are endogenous ligands for FXR with different agonistic activities. To determine whether FXR is involved in CDCA and CA-mediated regulation of AKR1D1, we performed a series of experiments. First, we determined whether activation of FXR by a synthetic agonist GW4064 has any effects on AKR1D1 expression in vitro in HepG2 cells. As shown in Fig 7A, activation of FXR by GW4064 had minimal effects on AKR1D1 expression while FXR target gene BSEP was markedly induced by GW4064 in HepG2 cells (Fig 7B). Next, we evaluated the effects of FXR over-expression on AKR1D1 expression. As shown in Fig 7C, over-expression of FXRα1 or FXRα2 had minimal effects on AKR1D1 expression in HepG2 cells. On the other hand, as expected, over-expression of FXRα1 or FXRα2 significantly upregulated BSEP expression (Fig 7D). The results indicated that FXR signaling was not involved in regulating AKR1D1 in human HepG2 cells.
Fig 7. FXR signaling was not involved in regulating AKR1D1 by bile acids in HepG2 cells.
(A) HepG2 cells were treated with FXR agonist GW4064 (1μM) or vehicle DMSO (0.1%) for 30h, followed by detection of AKR1D1 and BSEP mRNA levels by real-time PCR. (B) HepG2 cells were transfected with FXRα1, FXRα2 or vector, followed by treatment with GW4064 (1μM) for 30h. The expression levels of AKR1D1 and BSEP were detected by real-time PCR. The Student’s t-test was applied to pair-wise comparison. One-way ANOVA was applied to analyze data with multiple groups, followed by Tukey post-hoc test for multiple comparisons. ** p<0.01.
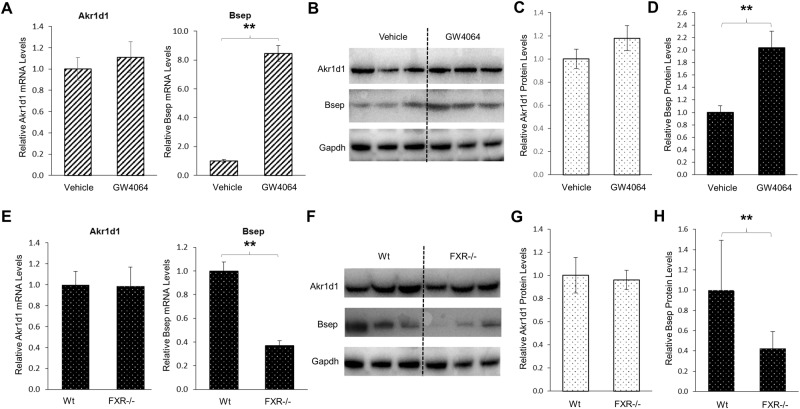
To confirm the findings, we carried out a mouse study to determine the effects of FXR activation on Akr1d1 expression in vivo. Consistent with the results from HepG2 cells, activation of FXR by its agonist GW4064 in vivo in mice did not significantly alter Akr1d1 expression at both mRNA and protein levels (Fig 8A, 8B and 8C) while it did markedly induced FXR target Bsep expression at both mRNA and protein levels (Fig 8A, 8B and 8D). Finally, we compared Akr1d1 expression levels in wt and FXR knockout (FXR-/-) mice. No significant differences in the expression levels of Akr1d1 at both mRNA and protein levels were detected between the wt and FXR-/- mice (Fig 8E, 8F and 8G). On the other hand, as expected, Bsep expressions at both mRNA and protein levels were significantly reduced in FXR-/- mice (Fig 8E, 8F and 8H). The consistent data from those series of in vitro and in vivo experiments concluded that FXR signaling was not involved in regulating AKR1D1 expression, thus excluded the possibility that CDCA and CA differentially regulated AKR1D1 through the FXR signaling pathway.
Fig 8. FXR signaling was not involved in regulating AKR1D1 by bile acids in vivo in mice.
(A) two groups of mice (n = 6/group) were treated with GW4064 (5mg/kg) or vehicle propanediol through intraperitoneal injection twice a day for 3 days. The expression levels of Akr1d1 and Bsep were quantified by real-time PCR and (B) Western blot. (C) quantification of Akr1d1 and (D) Bsep protein levels in (B). (E) the expression levels of Akr1d1 and Bsep in wt and FXR-knockout (FXR-/-) mice were determined by real-time PCR and (F) Western blot. (G) quantification of Akr1d1 and (H) Bsep protein levels in (F). The data are presented as mean ± SD of the groups of mice. The Student’s t-test was applied to pair-wise comparison. ** p<0.01.
CDCA and CA differentially modulated the MAPK/JNK signaling pathway
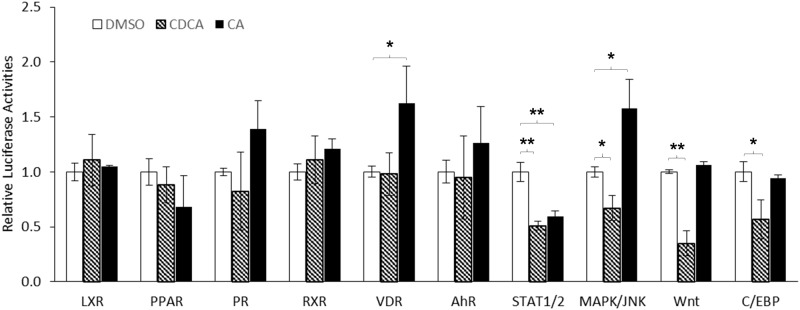
To identify potential signaling pathways through which CDCA and CA differentially regulate AKR1D1 expression, we screened 45 signaling pathways using Signal Transduction 45-Pathway Reporter Array. CDCA and CA did not show significant effects on most of the signaling pathways (data not shown). A set of representative data including the signaling pathways altered by CDCA, CA or both were presented in Fig 9. CDCA treatment had no significant effects on various nuclear receptor pathways including liver x receptor (LXR), peroxisome proliferator-activated receptor (PPAR), progesterone receptor (PR), retinoid X receptor (RXR), vitamin D receptor (VDR) and aryl hydrocarbon receptor (AhR). However, CDCA significantly down-regulated the following signaling pathways including the signal transducer and activator of transcription 1 and 2 (STAT1/2), mitogen-activated protein kinases/c-Jun N-terminal kinases (MAPK/JNK), Wnt and CCAAT-enhancer-binding proteins (C/EBP). For CA treatment, minimal effects on the following signaling pathways were detected including LXR, PPAR, PR, RXR, AhR, Wnt and C/EBP. However, the VDR and MAPK/JNK signaling were significantly elevated by CA treatment. On the other hand, CA down-regulated STAT1/2 signaling pathways. Taken together, among those signaling pathways, CDCA and CA differentially down- and upregulated MAPK/JNK signaling pathway, which exhibits the similar regulatory pattern of AKR1D1 following CDCA and CA treatments. Therefore, the MAPK/JNK pathway was the candidate pathway through which CDCA and CA differentially regulated AKR1D1.
Fig 9. CDCA and CA differentially modulated the MAPK/JNK signaling pathway.
HepG2 cells were reversely transfected with 45 signaling pathway element reporter plasmids, followed by treatment of transfected cells with CDCA (25μM), CA (25μM) or vehicle DMSO (0.1%) for 30 h. The luciferase activities were detected with the Dual Luciferase Assays. The data are presented as mean ± SD of at least three separate experiments. One-way ANOVA was applied to analyze data with multiple groups, followed by Tukey post-hoc test for multiple comparisons. * p<0.05 and ** p<0.01.
Inhibition of MAPK/JNK signaling pathway abolished CDCA and CA-mediated regulatory effects on AKR1D1
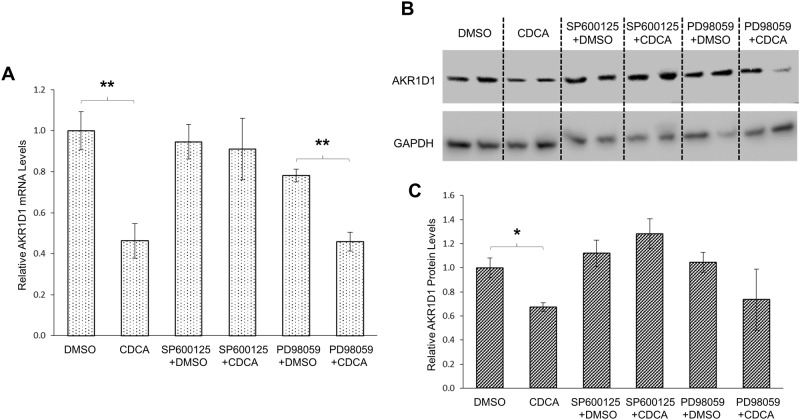
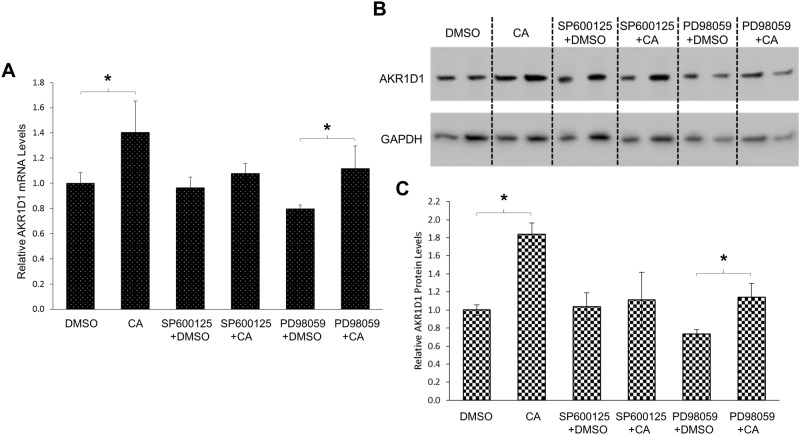
To determine whether CDCA and CA regulated AKR1D1 expression through the MAPK/JNK signaling pathway, the effects of MAPK/JNK inhibition on CDCA or CA-mediated regulation of AKR1D1 were evaluated in HepG2 cells. The AKR1D1 expression levels were detected in the absence or presence of a selective MAPK/JNK inhibitor, SP600125. A selective MAPK/extracellular signal-regulated kinases 1/2 (ERK1/2) inhibitor, PD98059, was included in the experiments as a negative control. As shown in Fig 10A, CDCA significantly decreased AKR1D1 mRNA expression by 54% while SP600125 had minimal effects. However, such CDCA-mediated decrease in AKR1D1 mRNA expression was completely abolished in the presence of MAPK/JNK inhibitor SP600125. On the other hand, the ability of CDCA to repress AKR1D1 mRNA expression remained intact in the presence of MAPK/ERK1/2 inhibitor PD98059. AKR1D1 mRNA expression was slightly decreased by PD98059 alone and was further significantly reduced by CDCA. Consistent with the results of AKR1D1 mRNA levels, AKR1D1 protein expression was significantly repressed by CDCA (Fig 10B and 10C). However, such CDCA-mediated repression of AKR1D1 protein expression was totally abolished in the presence of MAPK/JNK inhibitor SP600125. In contrast, AKR1D1 protein levels were slightly increased in cells treated with SP600125 and CDCA. On the other hand, CDCA treatment resulted in decreased AKR1D1 protein expression by 27% in the presence of PD98059. The consistent results of CDCA’s effects on both AKR1D1 mRNA and protein expression thus established that MAPK/JNK signaling pathway was involved in CDCA-mediated downregulation of AKR1D1 expression. Similar treatments were applied to determine whether MAPK/JNK signaling is involved in CA-mediated regulation of AKR1D1. As shown in Fig 11A, CA treatment increased AKR1D1 mRNA expression by 40%. However such CA-mediated induction of AKR1D1 mRNA expression was completely abolished by MAPK/JNK inhibitor SP600125 but not MAPK/ERK1/2 inhibitor PD98059. Consistent modulating effects of CA on AKR1D1 protein levels were detected (Fig 11B and 11C) in the absence and presence of MAPK/JNK inhibitor. CA treatment induced AKR1D1 protein expression by 84%. However, such induction was blunted in the presence of SP600125. Treatment with PD98059 reduced AKR1D1 expression. However, addition of CA significantly elevated AKR1D1 protein levels by 54%. The results thus demonstrated that MAPK/JNK signaling was involved in CA-mediated upregulation of AKR1D1. Taken together, the data revealed that MAPK/JNK signaling was involved in CDCA and CA-mediated differential regulation of AKR1D1 expression. It was thus concluded that CDCA and CA differentially regulated AKR1D1 expression through modulating the MAPK/JNK signaling pathway.
Fig 10. Inhibition of MAPK/JNK signaling pathway abolished CDCA-mediated regulation of AKR1D1.
(A) HepG2 cells were treated with CDCA (25μM) in the absence or presence of MAPK/JNK inhibitor SP600125 (1μM), MAPK/ERK1/2 inhibitor PD98059 (5μM) or vehicle for 30 hrs, followed by detection of AKR1D1 mRNA by real-time PCR and (B) AKR1D1 protein by Western blot. (C) quantification of AKR1D1 protein levels in (B). One-way ANOVA was applied to analyze the data, followed by Tukey post-hoc test for multiple comparisons. * p<0.05 and ** p<0.01.
Fig 11. Inhibition of MAPK/JNK signaling pathway abolished CA-mediated regulation of AKR1D1.
(A) HepG2 cells were treated with CA (50μM) in the absence or presence of MAPK/JNK inhibitor SP600125 (1μM), MAPK/ERK1/2 inhibitor PD98059 (5μM) or vehicle for 30 hrs, followed by detection of AKR1D1 mRNA by real-time PCR and (B) AKR1D1 protein by Western blot. (C) quantification of AKR1D1 protein levels in (B). One-way ANOVA was applied to analyze the data, followed by Tukey post-hoc test for multiple comparisons. * p<0.05.
Discussion
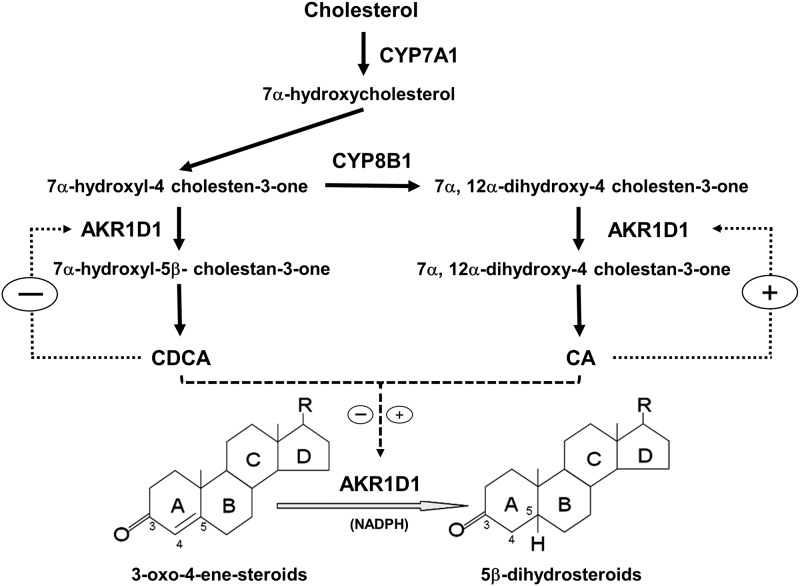
In the bile acid synthesis pathway, AKR1D1 catalyzes the 5β-reduction of bile acid intermediates to eventually produce the primary bile acids CDCA and CA. It is well established that CYP7A1-mediated 7α-hydroxylation is the rate-limiting step in bile acid synthesis and bile acid homeostasis is maintained by regulating CYP7A1 expression through several negative feedback mechanisms [8–10]. In this study, we found that AKR1D1 was differentially regulated by the two primary bile acids. Similar to the effects on CYP7A1, CDCA markedly repressed AKR1D1 expression (Figs 1 and 3). On the other hand, CA induced AKR1D1 expression while CYP7A1 expression was minimally affected by CA (Figs 4 and 6). Our new findings indicate that bile acid synthesis is regulated at multiple steps in the bile acid synthesis pathway. In addition to CYP7A1, AKR1D1-mediated 5β-reduction of bile acid intermediates is also regulated by bile acids. CDCA-mediated repression of AKR1D1 expression represents a negative feedback mechanism to restrain bile acid production while CA-mediated upregulation of AKR1D1 represents a feed-forward regulation loop to promote bile acid production (Fig 12). The data thus suggest that both bile acid concentration and composition have an impact on controlling bile acid synthesis.
Fig 12. Functions and regulation of AKR1D1 in bile acid synthesis and steroid hormone metabolism.
In the bile acid synthesis pathway, bile acid intermediate 7α-hydroxy-4-cholesten-3-one can take one of the two routes in subsequent steps. If the intermediate is acted upon by AKR1D1, the ultimate product is CDCA. If the intermediate is acted upon by CYP8B1, followed by AKR1D1, the ultimate product is CA. In this study, we demonstrated that CDCA and CA regulated AKR1D1 through a negative and positive feedback mechanism, respectively. In addition to bile acid synthesis, AKR1D1 is also involved in steroid hormone metabolism. 5β-reduction by AKR1D1 is a common transformation and major deactivation pathway for many steroid hormones. Therefore, AKR1D1 plays a critical role in regulating and maintaining the homeostasis of steroid hormones. Thus bile acids crosstalk with steroid hormone signaling pathways through modulating AKR1D1 expression. Plus (+) and minus (-) indicated positive and negative feedback regulation, respectively.
Various bile acid species exhibited diverse or even opposite biochemical and physiological properties. For example, CA promotes while CDCA inhibits intestinal cholesterol adsorption [37–40]. Alterations in bile acid compositions are associated with various pathological conditions [41–44]. It is therefore critical to maintain a normal bile acid composition. Bile acid composition is mainly determined by the relative concentrations of the two primary bile acids CDCA and CA or the ratio of CA/CDCA. CYP8B1 is the determining factor for the production of CA and its expression levels dictate the bile acid composition or the ratio of CA/CDCA [11, 45]. However, as shown in Fig 8, in the bile acid synthesis pathway, bile acid intermediate 7α-hydroxy-4-cholesten-3-one can take one of the two routes in subsequent steps. If the intermediate is acted upon by AKR1D1, the ultimate product is CDCA. If the intermediate is acted upon by CYP8B1, followed by AKR1D1, the ultimate product is CA (Fig 12). Therefore, the relative CA and CDCA production or the CA/CDCA ratio, is determined by both CYP8B1 and AKR1D1. It is speculated that AKR1D1 works together with CYP8B1 to coordinately maintain the relative production of CDCA and CA under physiological condition. Repression of AKR1D1 by CDCA is a typical negative feedback mechanism to control the production of CDCA. On the other hand, upregulation of AKR1D1 by CA represents a feed-forward regulation of CA production (Fig 12).
In addition to bile acid synthesis, AKR1D1 is involved in steroid hormone metabolism [28, 29]. 5β-reduction by AKR1D1 is a common transformation and major deactivation pathway for many steroid hormones. Therefore, AKR1D1 plays a critical role in regulating and maintaining the homeostasis of steroid hormones [19, 22, 28, 29]. In this investigation, we revealed that AKR1D1 expression was regulated by bile acids. It was thus speculated that bile acids may interplay with steroid hormone signaling pathways through modulating AKR1D1 expression (Fig 12). Indeed, bile acids were reported to modulate glucocorticoid metabolism and had an impact on the hypothalamic-pituitary-adrenal axis [46]. On the other hand, steroid hormones such as glucocorticoids are capable of directly or indirectly regulating bile acid homeostasis [47]. It remains to be determined how bile acid and steroid hormone signaling pathways crosstalk each other, especially through AKR1D1.
It is well established that bile acids down-regulate CYP7A1 expression through activating FXR [8–11]. Activation of FXR by bile acids induce the expression of small heterodimer partner (SHP) in the liver and fibroblast growth factor 19 (FGF19) in the intestine, which in turn repress CYP7A1 expression. In the current study, it was surprisingly found that FXR signaling pathway was not involved in bile acids-mediated regulation of AKR1D1. Activation of FXR by synthetic FXR agonist GW4064 in vitro and in vivo or knockout of FXR in vivo had minimal effects on AKR1D1 expression. Instead, we discovered that bile acids-mediated regulation of AKR1D1 involved the MAPK/JNK signaling pathway. Blockage of the MAPK/JNK signaling pathway totally abolished CDCA and CA-mediated regulatory effects on AKR1D1. The connection between bile acids and MAPK signaling pathways have long been recognized [48–53]. Previous studies showed that MAPK/JNK and MAPK/ERK1/2 signaling pathways also contribute to the down-regulation of CYP7A1 by bile acids in addition to the FXR signaling pathway [48, 49, 54, 55]. The MAPK signal transduction pathway is one of the most important regulatory mechanisms in eukaryotic cells that transduce environmental and developmental signals into adaptive and programmed responses including cell proliferation, differentiation and death, and inflammation. The JNKs were initially identified as the stress-activated protein kinases and are mainly activated in response to stress and proinflammatory cytokines [56]. Bile acids-mediated regulation of AKR1D1 and CYP7A1 through MAPK/JNK signaling pathways may represent a mechanism for the cells to cope with cellular stress and inflammation while bile acids-mediated feedback repression of CYP7A1 through FXR signaling pathway represents the main mechanism for hepatocytes to maintain bile acid homeostasis.
It was well documented that different bile acid species exhibit various regulatory effects on their target genes through modulating the MAPK signaling pathways. CA was identified as the most potent bile acid to activate MAPK/JNK while CDCA exhibited a much less potency in rat primary hepatocytes [55]. Deoxycholic acid (DCA) was found to be the most potent activator of MAPK/ERK signaling when compared with other bile acids [57]. DCA and UDCA exhibited opposite effects on colon cancer progression by differentially modulating MAPK pathways [58]. On the other hand, the same bile acids exhibited different or even opposite effects on various target genes by modulating the MAPK pathways. For examples, CDCA down-regulated CYP7A1 and acetyl-coenzyme A carboxylase-α (ACCα) while upregulated early growth response factor-1 (EGR-1) and low-density lipoprotein receptor (LDLR) through differentially modulating the MAPK signaling pathways [48–55]. In this investigation, CDCA and CA regulated AKR1D1 through the MAPK/JNK pathway with opposite effects. The underlying mechanisms for such differential effects of CDCA and CA on AKR1D1 expression remain to be determined. One possible explanation is that CDCA and CA act upon different upstream stress or inflammation-related signals in the MAPK/JNK cascade. Supporting such speculation is our data that CDCA-mediated down-regulation of AKR1D1 in HepG2 cells was consistently detected regardless cell sources and passage history. In contrast, CA-mediated upregulation of AKR1D1 was cell status-dependent (the source and passage history of the HepG2 cells). In addition, CDCA exhibited a linear (Fig 2B) while CA showed a binary dose-response in modulating AKR1D1 expression in HepG2 cells (Fig 5B). Taken together, AKR1D1 expression was differentially regulated by CDCA and CA, and such feedback regulation was mediated through the MAPK/JNK signaling pathway.
Acknowledgments
This work was supported by the National Institutes of Health Grant R01DK087755. C.N. is partially supported by the pre-doctoral fellowship from American Foundation for Pharmaceutical Education. We thank Dr. Bingfang Yan, University of Rhode Island, for sharing instruments and reagents in his laboratory with us. Instrumental supports from the RI-INBRE Core Facility in the College of Pharmacy, University of Rhode Island are greatly appreciated, which is supported by an Institutional Development Award (IDeA) from the National Institutes of General Medical Sciences of the National Institutes of Health under grant P20 GM103430-13.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by the National Institutes of Health Grant R01DK087755 (RD). CN is partially supported by the pre-doctoral fellowship from American Foundation for Pharmaceutical Education. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, et al. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999; 284:1365–1368. [DOI] [PubMed] [Google Scholar]
- 2.Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, et al. Identification of a nuclear receptor for bile acids. Science. 1999; 284:1362–1365. [DOI] [PubMed] [Google Scholar]
- 3.Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003; 278:9435–9440. 10.1074/jbc.M209706200 [DOI] [PubMed] [Google Scholar]
- 4.Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000; 102:731–744. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, Heyman RA, et al. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004; 113:1408–1418. 10.1172/JCI21025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009; 10:167–177. 10.1016/j.cmet.2009.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houten SM, Watanabe M, Auwerx J. Endocrine functions of bile acids. Embo J. 2006; 25:1419–1425. 10.1038/sj.emboj.7601049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, et al. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000; 6:507–515. [DOI] [PubMed] [Google Scholar]
- 9.Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000; 6:517–526. [DOI] [PubMed] [Google Scholar]
- 10.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005; 2:217–225. 10.1016/j.cmet.2005.09.001 [DOI] [PubMed] [Google Scholar]
- 11.Pandak WM, Bohdan P, Franklund C, Mallonee DH, Eggertsen G, Björkhem I, et al. Expression of sterol 12alpha-hydroxylase alters bile acid pool composition in primary rat hepatocytes and in vivo. Gastroenterology. 2001; 120:1801–1809. [DOI] [PubMed] [Google Scholar]
- 12.Meier PJ, Stieger B. Bile salt transporters. Annu Rev Physiol. 2002; 64:635–661. 10.1146/annurev.physiol.64.082201.100300 [DOI] [PubMed] [Google Scholar]
- 13.Kullak-Ublick GA, Stieger B, Meier PJ. Enterohepatic bile salt transporters in normal physiology and liver disease. Gastroenterology. 2004; 126:322–342. [DOI] [PubMed] [Google Scholar]
- 14.Ananthanarayanan M, Balasubramanian N, Makishima M, Mangelsdorf DJ, Suchy FJ. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J Biol Chem. 2001; 276:28857–28865. 10.1074/jbc.M011610200 [DOI] [PubMed] [Google Scholar]
- 15.Plass JR, Mol O, Heegsma J, Geuken M, Faber KN, Jansen PL, et al. Farnesoid X receptor and bile salts are involved in transcriptional regulation of the gene encoding the human bile salt export pump. Hepatology. 2002; 35:589–596. 10.1053/jhep.2002.31724 [DOI] [PubMed] [Google Scholar]
- 16.Okuda A, Okuda K. Purification and characterization of delta 4-3-ketosteroid 5 beta-reductase. J Biol Chem. 1984; 259:7519–7524. [PubMed] [Google Scholar]
- 17.Onishi Y, Noshiro M, Shimosato T, Okuda K. Molecular cloning and sequence analysis of cDNA encoding delta 4-3-ketosteroid 5 beta-reductase of rat liver. FEBS Lett. 1991; 283:215–218. [DOI] [PubMed] [Google Scholar]
- 18.Kondo KH, Kai MH, Setoguchi Y, Eggertsen G, Sjöblom P, Setoguchi T, et al. Cloning and expression of cDNA of human delta 4-3-oxosteroid 5 beta-reductase and substrate specificity of the expressed enzyme. Eur J Biochem. 1994; 219:357–363. [DOI] [PubMed] [Google Scholar]
- 19.Jin Y, Chen M, Penning TM. Rate of steroid double-bond reduction catalysed by the human steroid 5β-reductase (AKR1D1) is sensitive to steroid structure: implications for steroid metabolism and bile acid synthesis. Biochem J. 2014; 462:163–171. 10.1042/BJ20140220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen M, Penning TM. 5β-Reduced steroids and human Δ(4)-3-ketosteroid 5β-reductase (AKR1D1). Steroids. 2014; 83:17–26. 10.1016/j.steroids.2014.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003; 72:137–174. 10.1146/annurev.biochem.72.121801.161712 [DOI] [PubMed] [Google Scholar]
- 22.Palermo M, Marazzi MG, Hughes BA, Stewart PM, Clayton PT, Shackleton CH. Human Δ4-3-oxosteroid 5beta-reductase (AKR1D1) deficiency and steroid metabolism. Steroids. 2008; 73:417–423. 10.1016/j.steroids.2007.12.001 [DOI] [PubMed] [Google Scholar]
- 23.Clayton PT, Patel E, Lawson AM, Carruthers RA, Tanner MS, Strandvik B, et al. 3-Oxo-delta 4 bile acids in liver disease. Lancet. 1988; 1:1283–1284. [DOI] [PubMed] [Google Scholar]
- 24.Setchell KD, Suchy FJ, Welsh MB, Zimmer-Nechemias L, Heubi J, Balistreri WF. Delta 4-3-oxosteroid 5 beta-reductase deficiency described in identical twins with neonatal hepatitis. A new inborn error in bile acid synthesis. J Clin Invest. 1988; 82:2148–2157. 10.1172/JCI113837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shneider BL, Setchell KD, Whitington PF, Neilson KA, Suchy FJ. Delta 4-3-oxosteroid 5 beta-reductase deficiency causing neonatal liver failure and hemochromatosis. J Pediatr. 1994; 124:234–238. [DOI] [PubMed] [Google Scholar]
- 26.Lemonde HA, Custard EJ, Bouquet J, Duran M, Overmars H, Scambler PJ, et al. Mutations in SRD5B1 (AKR1D1), the gene encoding delta(4)-3-oxosteroid 5beta-reductase, in hepatitis and liver failure in infancy. Gut. 2003; 52:1494–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daugherty CC, Setchell KD, Heubi JE, Balistreri WF. Resolution of liver biopsy alterations in three siblings with bile acid treatment of an inborn error of bile acid metabolism (delta 4-3-oxosteroid 5 beta-reductase deficiency). Hepatology. 1993; 18:1096–1101. [PubMed] [Google Scholar]
- 28.Rižner TL, Penning TM. Role of aldo-keto reductase family 1 (AKR1) enzymes in human steroid metabolism. Steroids. 2014; 79:49–63. 10.1016/j.steroids.2013.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bauman DR, Steckelbroeck S, Penning TM. The roles of aldo-keto reductases in steroid hormone action. Drug News Perspect. 2004; 17:563–578. [DOI] [PubMed] [Google Scholar]
- 30.Song X, Chen Y, Valanejad L, Kaimal R, Yan B, Stoner M, et al. Mechanistic insights into isoform-dependent and species-specific regulation of bile salt export pump by farnesoid X receptor. J Lipid Res. 2013; 54:3030–3044. 10.1194/jlr.M038323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng R, Yang D, Radke A, Yang J, Yan B. The hypolipidemic agent guggulsterone regulates the expression of human bile salt export pump: dominance of transactivation over farsenoid X receptor-mediated antagonism. J Pharmacol Exp Ther. 2007; 320:1153–1162. 10.1124/jpet.106.113837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song X, Kaimal R, Yan B, Deng R. Liver receptor homolog 1 transcriptionally regulates human bile salt export pump expression. J Lipid Res. 2008; 49:973–984. 10.1194/jlr.M700417-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Song X, Valanejad L, Vasilenko A, More V, Qiu X, et al. Bile salt export pump is dysregulated with altered farnesoid X receptor isoform expression in patients with hepatocellular carcinoma. Hepatology. 2013; 57:1530–1541. 10.1002/hep.26187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Vasilenko A, Song X, Valanejad L, Verma R, You S, et al. Estrogen and Estrogen Receptor-α-Mediated Transrepression of Bile Salt Export Pump. Mol Endocrinol. 2015; 29:613–626. 10.1210/me.2015-1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song X, Vasilenko A, Chen Y, Valanejad L, Verma R, Yan B, et al. Transcriptional dynamics of bile salt export pump during pregnancy: mechanisms and implications in intrahepatic cholestasis of pregnancy. Hepatology. 2014; 60:1993–2007. 10.1002/hep.27171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng R, Yang D, Yang J, Yan B. Oxysterol 22(R)-hydroxycholesterol induces the expression of the bile salt export pump through nuclear receptor farsenoid X receptor but not liver X receptor. J Pharmacol Exp Ther. 2006; 317:317–325. 10.1124/jpet.105.097758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woollett LA, Buckley DD, Yao L, Jones PJ, Granholm NA, Tolley EA, et al. Cholic acid supplementation enhances cholesterol absorption in humans. Gastroenterology. 2004; 126: 724–731. [DOI] [PubMed] [Google Scholar]
- 38.Leiss O, von Bergmann K, Streicher U, Strotkoetter H. Effect of three different dihydroxy bile acids on intestinal cholesterol absorption in normal volunteers. Gastroenterology. 1984; 87: 144–149. [PubMed] [Google Scholar]
- 39.Ponz de Leon M, Carulli N, Loria P, Iori R, Zironi F. The effect of chenodeoxycholic acid (CDCA) on cholesterol absorption. Gastroenterology. 1979; 77:223–230. [PubMed] [Google Scholar]
- 40.Reynier MO, Montet JC, Gerolami A, Marteau C, Crotte C, Montet AM, et al. Comparative effects of cholic, chenodeoxycholic, and ursodeoxycholic acids on micellar solubilization and intestinal absorption of cholesterol. J Lipid Res. 1981; 22:467–473. [PubMed] [Google Scholar]
- 41.Azer SA, Coverdale SA, Byth K, Farrell GC, Stacey NH. Sequential changes in serum levels of individual bile acids in patients with chronic cholestatic liver disease. J Gastroenterol Hepatol. 1996; 11:208–215. [DOI] [PubMed] [Google Scholar]
- 42.de Leon M. P., Ferenderes R, and Carulli N. Bile lipid composition and bile acid pool size in diabetes. Am J Dig Dis. 1978; 23:710–716. [DOI] [PubMed] [Google Scholar]
- 43.Haeusler RA, Astiarraga B, Camastra S, Accili D, Ferrannini E. Human insulin resistance is associated with increased plasma levels of 12α-hydroxylated bile acids. Diabetes. 2013; 62:4184–4191. 10.2337/db13-0639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okuda H, Obata H, Nakanishi T, Hisamitsu T, Matsubara K, Watanabe H. Quantification of individual serum bile acids in patients with liver diseases using high-performance liquid chromatography. Hepatogastroenterology. 1984; 31:168–171. [PubMed] [Google Scholar]
- 45.Vlahcevic ZR, Eggertsen G, Björkhem I, Hylemon PB, Redford K, Pandak WM. Regulation of sterol 12alpha-hydroxylase and cholic acid biosynthesis in the rat. Gastroenterology. 2000; 118:599–607. [DOI] [PubMed] [Google Scholar]
- 46.McNeilly AD, Macfarlane DP, O'Flaherty E, Livingstone DE, Mitić T, McConnell KM, et al. Bile acids modulate glucocorticoid metabolism and the hypothalamic-pituitary-adrenal axis in obstructive jaundice. J Hepatol. 2010; 52:705–711. 10.1016/j.jhep.2009.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosales R, Romero MR, Vaquero J, Monte MJ, Requena P, Martinez-Augustin O, et al. FXR-dependent and -independent interaction of glucocorticoids with the regulatory pathways involved in the control of bile acid handling by the liver. Biochem Pharmacol. 2013; 85:829–938. 10.1016/j.bcp.2013.01.001 [DOI] [PubMed] [Google Scholar]
- 48.Li T, Jahan A, Chiang JY. Bile acids and cytokines inhibit the human cholesterol 7 alpha-hydroxylase gene via the JNK/c-jun pathway in human liver cells. Hepatology. 2006; 43:1202–1210. 10.1002/hep.21183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Fabiani E, Mitro N, Anzulovich AC, Pinelli A, Galli G, Crestani M. The negative effects of bile acids and tumor necrosis factor-alpha on the transcription of cholesterol 7alpha-hydroxylase gene (CYP7A1) converge to hepatic nuclear factor-4: a novel mechanism of feedback regulation of bile acid synthesis mediated by nuclear receptors. J Biol Chem. 2001; 276:30708–30716. 10.1074/jbc.M103270200 [DOI] [PubMed] [Google Scholar]
- 50.Talukdar S, Bhatnagar S, Dridi S, Hillgartner FB. Chenodeoxycholic acid suppresses the activation of acetyl-coenzyme A carboxylase-alpha gene transcription by the liver X receptor agonist T0-901317. J Lipid Res. 2007; 48:2647–2663. 10.1194/jlr.M700189-JLR200 [DOI] [PubMed] [Google Scholar]
- 51.Allen K, Kim ND, Moon JO, Copple BL. Upregulation of early growth response factor-1 by bile acids requires mitogen-activated protein kinase signaling. Toxicol Appl Pharmacol. 2009; 243:63–67. 10.1016/j.taap.2009.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakahara M, Fujii H, Maloney PR, Shimizu M, Sato R. Bile acids enhance low density lipoprotein receptor gene expression via a MAPK cascade-mediated stabilization of mRNA. J Biol Chem. 2002; 277:37229–37234. 10.1074/jbc.M206749200 [DOI] [PubMed] [Google Scholar]
- 53.Yashiro T, Yokoi Y, Shimizu M, Inoue J, Sato R. Chenodeoxycholic acid stabilization of LDL receptor mRNA depends on 3'-untranslated region and AU-rich element-binding protein. Biochem Biophys Res Commun. 2011; 409:155–159. 10.1016/j.bbrc.2011.04.006 [DOI] [PubMed] [Google Scholar]
- 54.Song KH, Li T, Owsley E, Strom S, Chiang JY. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression. Hepatology. 2009; 49:297–305. 10.1002/hep.22627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta S, Stravitz RT, Dent P, Hylemon PB. Down-regulation of cholesterol 7alpha-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-Jun N-terminal kinase pathway. J Biol Chem. 2001; 276:15816–15822. 10.1074/jbc.M010878200 [DOI] [PubMed] [Google Scholar]
- 56.Bogoyevitch MA, Kobe B. Uses for JNK: the many and varied substrates of the c-Jun N-terminal kinases. Microbiol Mol Biol Rev. 2006; 70:1061–1095. 10.1128/MMBR.00025-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rao YP, Studer EJ, Stravitz RT, Gupta S, Qiao L, Dent P, Hylemon PB. Activation of the Raf-1/MEK/ERK cascade by bile acids occurs via the epidermal growth factor receptor in primary rat hepatocytes. Hepatology. 2002; 35:307–314. 10.1053/jhep.2002.31104 [DOI] [PubMed] [Google Scholar]
- 58.Centuori SM, Martinez JD. Differential regulation of EGFR-MAPK signaling by deoxycholic acid (DCA) and ursodeoxycholic acid (UDCA) in colon cancer. Dig Dis Sci. 2014; 59:2367–2380. 10.1007/s10620-014-3190-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.