Abstract
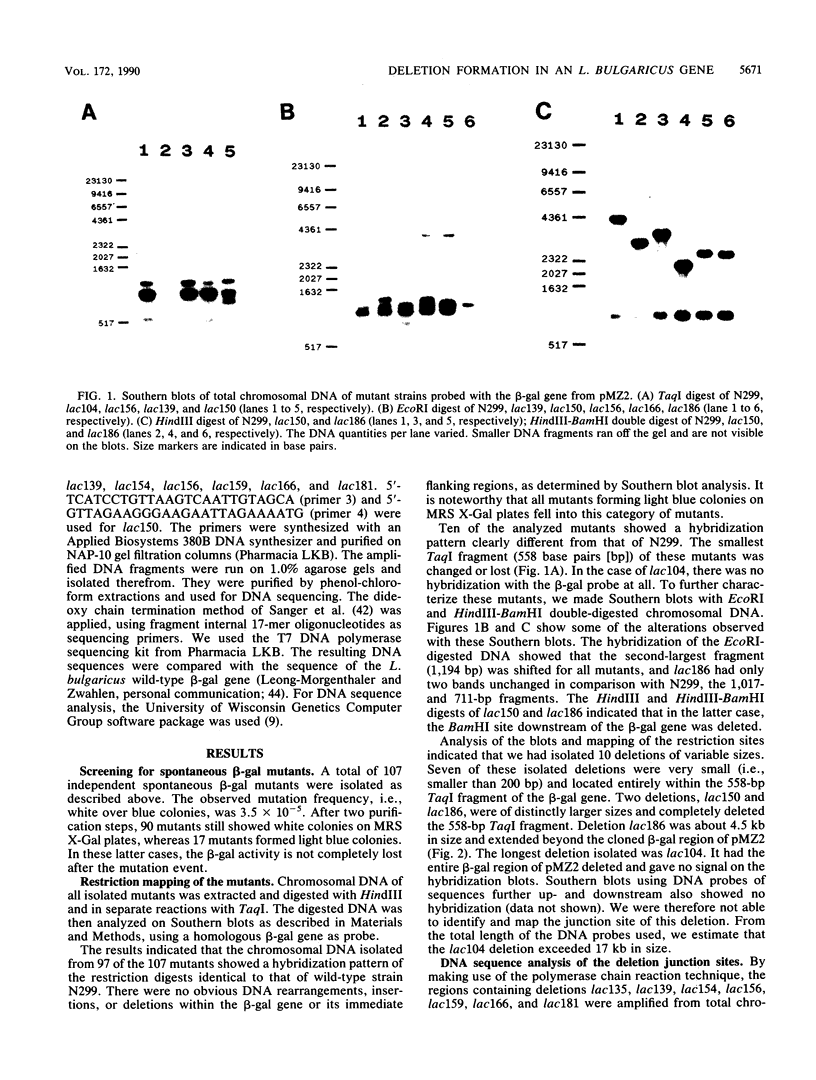
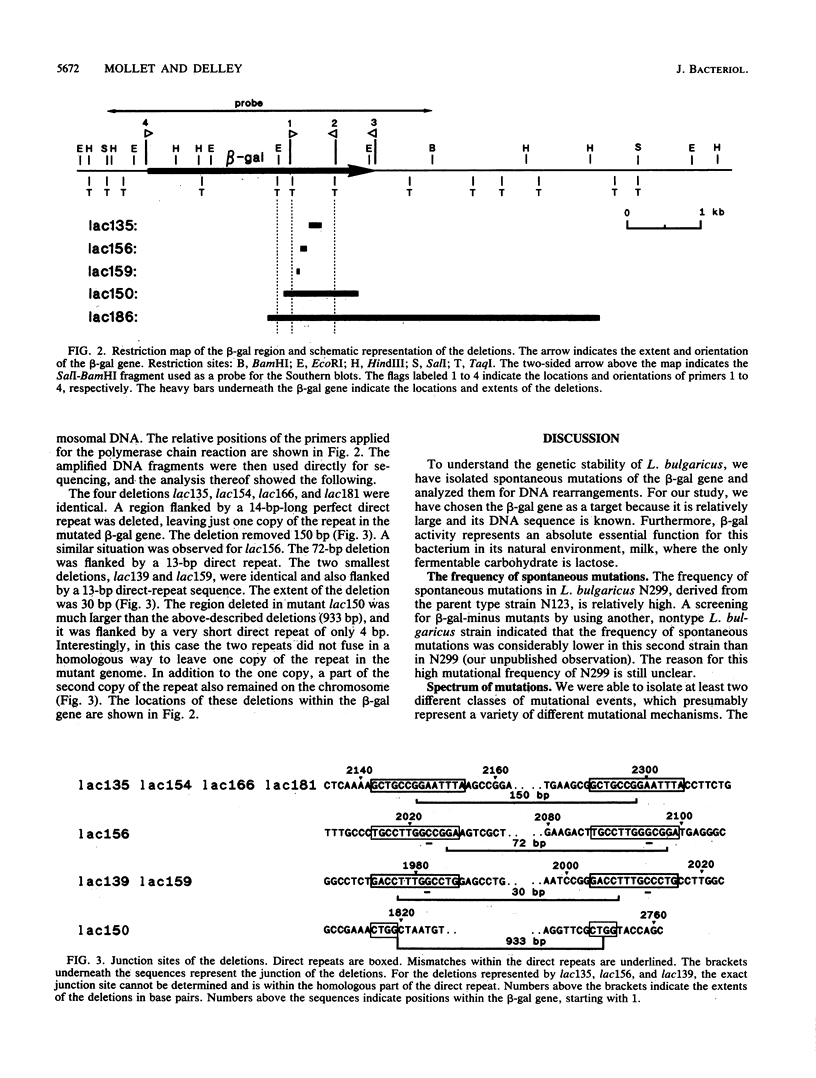
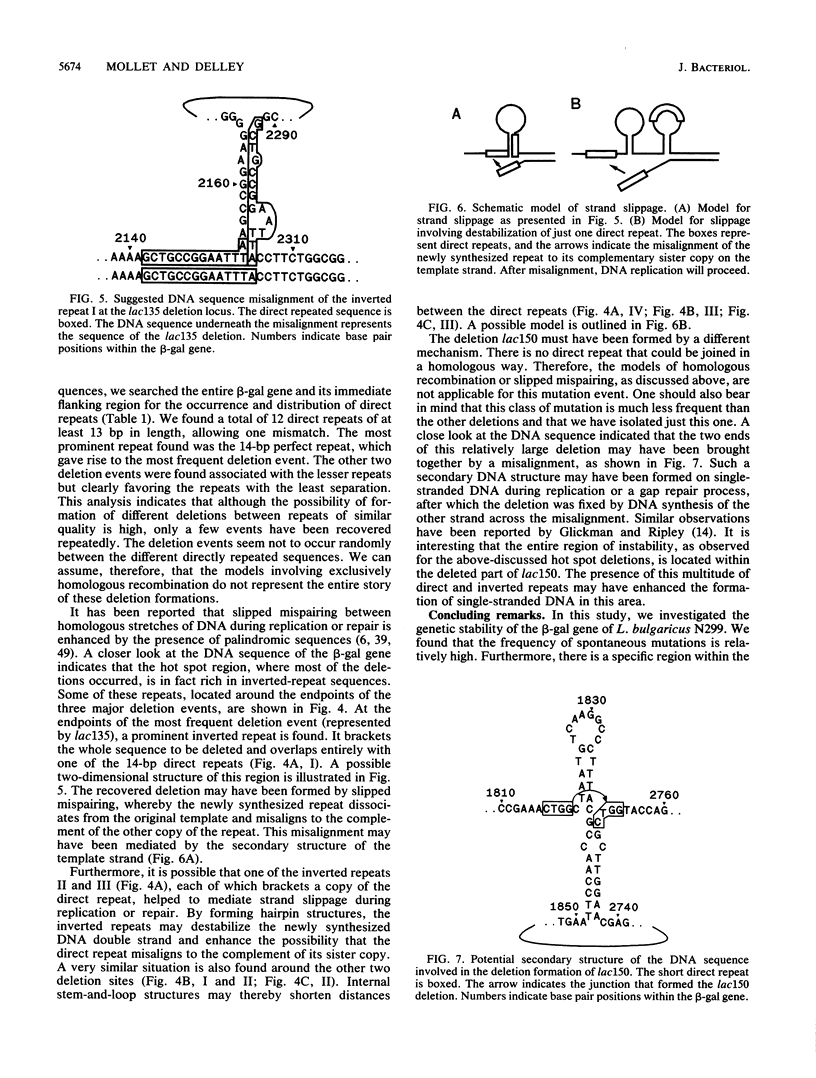
To investigate the genetic stability of the dairy organism Lactobacillus bulgaricus, we have analyzed 107 spontaneous mutations of the beta-galactosidase gene of this organism. Ten of these mutations were DNA rearrangements giving rise to different deletions, located predominantly within a small hot spot area. The DNA sequences of the different deletion junctions have been determined. The analysis showed that the deletions can be divided into two classes, depending on the presence of short direct-repeat sequences at the deletion endpoints and on the length of the deleted sequences. Possible mechanisms of these deletion formations and the involvement of inverted-repeat sequences that may enhance slipped DNA mispairing are discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Boizet B., Flickinger J. L., Chassy B. M. Transfection of Lactobacillus bulgaricus protoplasts by bacteriophage DNA. Appl Environ Microbiol. 1988 Dec;54(12):3014–3018. doi: 10.1128/aem.54.12.3014-3018.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunier D., Peeters B. P., Bron S., Ehrlich S. D. Breakage--reunion and copy choice mechanisms of recombination between short homologous sequences. EMBO J. 1989 Oct;8(10):3127–3133. doi: 10.1002/j.1460-2075.1989.tb08465.x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Casadesus J., Roth J. R. Transcriptional occlusion of transposon targets. Mol Gen Genet. 1989 Apr;216(2-3):204–209. doi: 10.1007/BF00334357. [DOI] [PubMed] [Google Scholar]
- Coukell M. B., Yanofsky C. Increased frequency of deletions in DNA polymerase mutants of Escherichia coli. Nature. 1970 Nov 14;228(5272):633–635. doi: 10.1038/228633a0. [DOI] [PubMed] [Google Scholar]
- DasGupta U., Weston-Hafer K., Berg D. E. Local DNA sequence control of deletion formation in Escherichia coli plasmid pBR322. Genetics. 1987 Jan;115(1):41–49. doi: 10.1093/genetics/115.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delley M., Mollet B., Hottinger H. DNA Probe for Lactobacillus delbrueckii. Appl Environ Microbiol. 1990 Jun;56(6):1967–1970. doi: 10.1128/aem.56.6.1967-1970.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner C., Berg D. E. Excision of transposon Tn5 is dependent on the inverted repeats but not on the transposase function of Tn5. Proc Natl Acad Sci U S A. 1981 Jan;78(1):459–463. doi: 10.1073/pnas.78.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Foster T. J., Lundblad V., Hanley-Way S., Halling S. M., Kleckner N. Three Tn10-associated excision events: relationship to transposition and role of direct and inverted repeats. Cell. 1981 Jan;23(1):215–227. doi: 10.1016/0092-8674(81)90286-5. [DOI] [PubMed] [Google Scholar]
- Galas D. J. An analysis of sequence repeats in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):858–863. doi: 10.1016/0022-2836(78)90024-4. [DOI] [PubMed] [Google Scholar]
- Glickman B. W., Ripley L. S. Structural intermediates of deletion mutagenesis: a role for palindromic DNA. Proc Natl Acad Sci U S A. 1984 Jan;81(2):512–516. doi: 10.1073/pnas.81.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J., Dubnau D. Analysis of plasmid deletional instability in Bacillus subtilis. J Bacteriol. 1985 Jun;162(3):1014–1023. doi: 10.1128/jb.162.3.1014-1023.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones I. M., Primrose S. B., Ehrlich S. D. Recombination between short direct repeats in a recA host. Mol Gen Genet. 1982;188(3):486–489. doi: 10.1007/BF00330053. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Konrad E. B. Method for the isolation of Escherichia coli mutants with enhanced recombination between chromosomal duplications. J Bacteriol. 1977 Apr;130(1):167–172. doi: 10.1128/jb.130.1.167-172.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. The mutational specificity of DNA polymerase-beta during in vitro DNA synthesis. Production of frameshift, base substitution, and deletion mutations. J Biol Chem. 1985 May 10;260(9):5787–5796. [PubMed] [Google Scholar]
- Kupsch J., Alonso J. C., Trautner T. A. Analysis of structural and biological parameters affecting plasmid deletion formation in Bacillus subtilis. Mol Gen Genet. 1989 Sep;218(3):402–408. doi: 10.1007/BF00332402. [DOI] [PubMed] [Google Scholar]
- Langella P., Chopin A. Conjugal transfer of plasmid pIP501 from Lactococcus lactis to Lactobacillus delbrückii subsp. bulgaricus and Lactobacillus helveticus. FEMS Microbiol Lett. 1989 Jul 15;51(1):149–152. doi: 10.1016/0378-1097(89)90498-9. [DOI] [PubMed] [Google Scholar]
- Lopez P., Espinosa M., Greenberg B., Lacks S. A. Generation of deletions in pneumococcal mal genes cloned in Bacillus subtilis. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5189–5193. doi: 10.1073/pnas.81.16.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V., Kleckner N. Mismatch repair mutations of Escherichia coli K12 enhance transposon excision. Genetics. 1985 Jan;109(1):3–19. doi: 10.1093/genetics/109.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., D'Alençon E., Ehrlich S. D. Deletion hot spots in chimeric Escherichia coli plasmids. J Bacteriol. 1989 Apr;171(4):1846–1853. doi: 10.1128/jb.171.4.1846-1853.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Illegitimate recombination at the replication origin of bacteriophage M13. Proc Natl Acad Sci U S A. 1986 May;83(10):3386–3390. doi: 10.1073/pnas.83.10.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbantoglu J., Hartley D., Phear G., Tear G., Meuth M. Spontaneous deletion formation at the aprt locus of hamster cells: the presence of short sequence homologies and dyad symmetries at deletion termini. EMBO J. 1986 Jun;5(6):1199–1204. doi: 10.1002/j.1460-2075.1986.tb04347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbantoglu J., Phear G., Meuth M. DNA sequence analysis of spontaneous mutations at the aprt locus of hamster cells. Mol Cell Biol. 1987 Apr;7(4):1445–1449. doi: 10.1128/mcb.7.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters B. P., de Boer J. H., Bron S., Venema G. Structural plasmid instability in Bacillus subtilis: effect of direct and inverted repeats. Mol Gen Genet. 1988 Jun;212(3):450–458. doi: 10.1007/BF00330849. [DOI] [PubMed] [Google Scholar]
- Pierce J. C., Masker W. Genetic deletions between directly repeated sequences in bacteriophage T7. Mol Gen Genet. 1989 Jun;217(2-3):215–222. doi: 10.1007/BF02464884. [DOI] [PubMed] [Google Scholar]
- Pittet A. C., Hottinger H. A Lactobacillus bulgaricus DNA fragment containing a 5S RNA gene adjacent to a pentameric tRNA gene cluster. Nucleic Acids Res. 1989 Jun 26;17(12):4874–4874. [PMC free article] [PubMed] [Google Scholar]
- Pittet A. C., Hottinger H. Sequence of a hexameric tRNA gene cluster associated with rRNA genes in Lactobacillus bulgaricus. Nucleic Acids Res. 1989 Jun 26;17(12):4873–4873. [PMC free article] [PubMed] [Google Scholar]
- Pribnow D., Sigurdson D. C., Gold L., Singer B. S., Napoli C., Brosius J., Dull T. J., Noller H. F. rII cistrons of bacteriophage T4. DNA sequence around the intercistronic divide and positions of genetic landmarks. J Mol Biol. 1981 Jul 5;149(3):337–376. doi: 10.1016/0022-2836(81)90477-0. [DOI] [PubMed] [Google Scholar]
- Ripley L. S., Glickman B. W. Unique self-complementarity of palindromic sequences provides DNA structural intermediates for mutation. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):851–861. doi: 10.1101/sqb.1983.047.01.097. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper R. M., Danforth B. N., Glickman B. W. Mechanisms of spontaneous mutagenesis: an analysis of the spectrum of spontaneous mutation in the Escherichia coli lacI gene. J Mol Biol. 1986 May 20;189(2):273–284. doi: 10.1016/0022-2836(86)90509-7. [DOI] [PubMed] [Google Scholar]
- Schmidt B. F., Adams R. M., Requadt C., Power S., Mainzer S. E. Expression and nucleotide sequence of the Lactobacillus bulgaricus beta-galactosidase gene cloned in Escherichia coli. J Bacteriol. 1989 Feb;171(2):625–635. doi: 10.1128/jb.171.2.625-635.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B. S., Westlye J. Deletion formation in bacteriophage T4. J Mol Biol. 1988 Jul 20;202(2):233–243. doi: 10.1016/0022-2836(88)90454-8. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Simon M. N., Dunn J. J. Genetic and physical mapping in the early region of bacteriophage T7 DNA. J Mol Biol. 1979 Dec 25;135(4):917–937. doi: 10.1016/0022-2836(79)90520-5. [DOI] [PubMed] [Google Scholar]
- Taylor J. M., Illmensee R., Summers J. Efficeint transcription of RNA into DNA by avian sarcoma virus polymerase. Biochim Biophys Acta. 1976 Sep 6;442(3):324–330. doi: 10.1016/0005-2787(76)90307-5. [DOI] [PubMed] [Google Scholar]
- Weston-Hafer K., Berg D. E. Palindromy and the location of deletion endpoints in Escherichia coli. Genetics. 1989 Apr;121(4):651–658. doi: 10.1093/genetics/121.4.651. [DOI] [PMC free article] [PubMed] [Google Scholar]




