Abstract
Gene therapy platforms offer a variety of potentially effective solutions for development of targeted agents that can be exploited for cancer treatment. The physicochemical properties of nanocarriers can be tuned to enhance their localization in tumors, and cell specificity can also be increased by appropriate selection of gene targets. A relatively underexploited approach to enhance therapeutic selectivity in cancer tissues is the use of nanocarriers whose nuclear targeting and uptake are triggered by the altered expression of specific endomembrane trafficking proteins in cancer cells. Previously, we showed that histone 3 (H3) peptide-targeted DNA polyplexes traffic to the nucleus efficiently through caveolar endocytosis followed by transfer through the Golgi and endoplasmic reticulum (ER). We hypothesized that these polyplexes would exhibit enhanced activity in inflammatory breast cancer (IBC) cells, which overexpress caveolin-1 as part of their invasive phenotype, and we also posited that this targeting effect could be exploited to facilitate IBC-specific transfection and prodrug conversion in the presence of normal breast epithelial cells. Using cellular transfection experiments, function-blocking assays, and confocal imaging in both IBC SUM149 cell monocultures and IBC SUM149 co-cultures with MCF10A normal breast epithelial cells, we found that our H3-targeted polyplexes selectively transfected IBC SUM149 cells at a 4-fold higher level than normal breast epithelial cells. This selectivity and increased transfection were caused by a 2.2-fold overexpression of caveolin-1 in IBC SUM149 cells, which led to increased polyplex trafficking to the nucleus through the Golgi and ER. We also saw similar enhancements in cell selectivity and transfection when cells were transfected with a suicide gene/prodrug combination, as the increased expression of the suicide gene in IBC SUM149 cells led to a 55% decrease in viability in IBC SUM149 cells as compared to a 25% decrease in MCF10A cells. These findings demonstrate that differences in the expression of the endocytic membrane protein caveolin-1 can be exploited for cell-selective gene delivery, and ultimately, these gene-based targeting approaches may be useful in potential treatments for aggressive cancer types.
Introduction
Research and development in the field of cancer nanotherapeutics has grown at an exponential rate since the early 2000s and offers a number of exciting potential approaches to improve drug efficacy (Amer 2014). Nanomedicines are ideal in oncology because they have the ability to encapsulate cytotoxic drugs and deliver them more selectively to targeted cells. For instance, one of the first FDA approved nanomedicines, Doxil®, approved in 1995, is used for the treatment of Kaposi sarcoma. This liposomal formulation of doxorubicin passively targets tumors through the enhanced permeability and retention (EPR) effect and then releases the drug (Barenholz 2012). However, passively targeted nanocarriers enhance the concentration of the drug in the tumor interstitial fluid, as well as in other sites of passive nanocarrier accumulation, such as the liver and spleen. As a result, there is a significant probability of toxicity in non-target tissues before the drug concentration in tumors reaches the therapeutic level (Akhtar 2006). Recently, the first actively targeted nanoparticles have entered human clinical trials (Kamaly et al. 2012). These nanocarriers offer the ability to increase cell specificity, yet receptor-targeting approaches continue to exhibit off-target effects due to the expression of surface markers in healthy tissues. Multi-modal nanostructures that combine receptor targeting with selectivity for specific endomembrane trafficking pathways may provide a compelling alternative with significantly increased cell/tissue specificity, and incorporation of endocytic targeting is also ideal for DNA nanotherapeutics and other nanomedicines destined for specific cellular organelles (Bertrand et al. 2014).
Approaches to target specific endocytic pathways to achieve increased cell or tissue specificity and/or enhanced gene transfer have previously shown promise. Over the past decade, several groups have sought to create targeted therapies by harnessing specific endomembrane markers within different organs. For example, Schnitzer and colleagues developed a new proteomic mapping and imaging approach to discover targets residing within lung endothelial caveolae, and they were able to use these targets to generate new antibody probes that selectively targeted lung caveolae in vivo (Schnitzer 2001). A key finding was that the molecular heterogeneity of the endothelium permitted vascular targeting to achieve selective transcytosis only into lung tissues (McIntosh et al. 2002). Moreover, they showed that targeting caveolae allowed active pumping of antibodies across the endothelial cell layer, leading to rapid accumulation in the surrounding lung tissue with minimal off-target delivery (Oh et al. 2007). Other approaches have sought to explore whether differences in cell physiology alter both the internalization pathways and gene transfer efficacy of non-viral carriers. One study utilized three different cell types to directly link cell line-dependent differences in transfection efficiency to cell-specific internalization mechanisms and subsequent internal trafficking of the gene carriers (Douglas et al. 2008).
A major challenge in implementing such intracellular targeting approaches for gene delivery is the need to target pathways that exhibit both the desired cell/tissue specificity and also support efficient gene transfer into the nucleus. Caveolae represent an intriguing target that may meet these needs in oncology-related applications due to their relevance to the invasive phenotypes in several aggressive cancers (Martinez-Outschoorn et al. 2015) and their correlation to increased gene transfer efficacy (Rejman et al. 2005). In particular, caveolin-1, a structural component of caveolae, is overexpressed in tumor-associated endothelial cells, metastatic and androgen-resistant prostate cancer, and inflammatory breast cancer (IBC) (Van den Eynden et al. 2006). IBC is a distinct and particularly aggressive subtype of locally advanced breast cancer, with less than 50% of IBC patients surviving more than 5 years (Kleer et al. 2000), and the underlying invasive mechanism of IBC is directly related to the overexpression of caveolin-1 (Van den Eynden et al. 2006). Hypomethylation of the caveolin-1 and -2 promoters leads to increased expression of caveolin-1 and -2 both at the mRNA and protein level in IBC (Van den Eynden et al. 2006). Normal breast endothelial cells express caveolin-1 (Hnasko and Lisanti 2003); however, caveolin-1 expression is substantially increased in IBC cells, imparting the potential capacity to exploit caveolar endocytic routes in order to gain cell specificity (Van den Eynden et al. 2006). These findings motivated our interest in caveolin-1 as a therapeutic target for IBC.
The potential role for caveolae in gene transfer has also been documented. Caveolae constitute an alternative endocytic pathway to clathrin-coated pits. They support receptor-mediated endocytosis of viruses such as SV40 (Pelkmans et al. 2001) as well as other nanostructures including cholera toxin, tetanus toxin, and albumin-gold complexes (Montesano et al. 1982). Caveolar uptake of these structures leads to transfer to intracellular compartments including the ER, Golgi, endosomes, and lysosomes (Shin and Abraham 2001). Caveolar targeting has increasingly been examined as a route for gene delivery, as multiple pieces of literature suggest that caveolar targeting can be an efficient pathway for reaching the nucleus in several different types of cells (Fichter et al. 2013; Munsell et al. 2015; Rejman et al. 2005; Schnitzer 2001). For example, the Reineke group has demonstrated that cellular internalization of poly(glycoamidoamine) polyplexes in HeLa cells occurs through a multifaceted internalization mechanism primarily involving caveolae, and efficient nuclear delivery and transgene expression appeared to depend directly upon caveolar uptake (McLendon et al. 2010). Another study in HeLa cells also demonstrated that targeting polyethylenimine complexes to the caveolar pathway with folic acid yielded enhanced gene delivery, and that a contributing factor to the success of this targeting approach was the avoidance of lysosomes (Gabrielson and Pack 2009).
Our recent studies with histone H3-targeted polyplexes provided additional support for the roles of caveolar uptake and retrograde trafficking in efficient gene transfer (Reilly et al. 2012b; Ross et al. 2015). Polyplexes are a widely-used class of gene carriers comprised of positively charged polymers that bind cooperatively with DNA to form compact nanostructures (Midoux et al. 2008). Polyplexes have several advantages in gene delivery, including the capacity to protect DNA against enzymatic degradation, and polyplexes also have sizes and properties that promote cellular uptake; however, the gene transfer activity of most cationic polymers is directly correlated with increased cytotoxicity (Grandinetti et al. 2012). We recently demonstrated a new class of H3-targeted polyplexes with significantly improved gene expression and high cell viability in several cell types (Reilly et al. 2012a), and the inclusion of the H3 tail peptide was specifically shown to improve gene delivery by enhancing trafficking via caveolae-mediated endocytosis and retrograde routing through the Golgi and ER (Reilly et al. 2012b). These H3-targeted polyplexes enhanced plasmid transport to the nucleus by evading endolysosomal trafficking routes and harnessing histone effectors and the Rab6 GTPase to target perinuclear compartments and ultimately accumulate within the nucleus (Ross et al. 2015).
In this study, we hypothesized that H3-targeted polyplexes would have increased gene transfer efficiency and selectivity in IBC cells vs. normal breast epithelial cells due to the increased expression of caveolin-1. Accordingly, using IBC SUM149 cells and MCF10A normal breast epithelial cells, we detailed the role of caveolin-1 in H3-targeted polyplex trafficking, nuclear accumulation, and transfection efficiency. We demonstrated significantly enhanced transfection for the H3-targeted polyplexes in IBC SUM149 cells, as compared with MCF10A cells, whereas untargeted polyplexes and polyplexes formed with a scrambled H3 (sH3) sequence exhibited no cell specificity. We also showed that the improved transfection in IBC SUM149 cells was related to increased caveolar trafficking and polyplex transfer to the perinuclear region and ER, based on colocalization studies with caveolin-1 and Rab-GTPases. Gene silencing studies demonstrated the extent to which transfection depended upon polyplex trafficking via caveolin-1, as transfection in IBC SUM149 cells, but not MCF10A cells, was significantly decreased when caveolin-1 was silenced. When we delivered our H3-targeted polyplexes to IBC SUM149/MCF10A co-cultures that were an in vitro mimic of the cell types present in IBC patients, gene transfer occurred efficiently in the IBC SUM149 cells but minimally in the MCF10A cells, demonstrating the potential capacity for cell-selective gene transfer in more complex, multicellular tissues. Finally, when we transfected each cell line with polyplexes formed with plasmids encoding the prodrug converting enzyme cytosine deaminase (CD) we found a significant decrease in viability in IBC cells when the prodrug 5-fluorocytosine (5-FC) was delivered to cells transfected with the H3-targeted polyplexes, whereas there was only a minor effect on viability when 5-FC was delivered to transfected MCF10A cells. Collectively, these data demonstrate that differences in the expression of the membrane scaffolding protein caveolin-1 can be exploited for high efficiency and cell-selective gene delivery. These findings may enable new and safer treatment approaches for aggressive cancers such as IBC.
Results
Caveolin-1 is required for H3-targeted polyplex transfection in IBC SUM149 cells
Our prior studies demonstrated the selective utilization of caveolar uptake pathways for efficient gene delivery by H3-targeted polyplexes in several cell types (Reilly et al. 2012b). Herein, we sought to determine whether H3-targeted polyplexes would also associate with caveolar vesicles in IBC and non-IBC cells, and also whether the increased expression of caveolin-1 in IBC would correlate with increased polyplex association. Accordingly, IBC SUM149 cells and MCF10A normal breast epithelial cells were transfected with H3-targeted polyplexes, untargeted polyplexes, or polyplexes formed with a scrambled H3 sequence (sH3). Using immunocytochemical staining and confocal microscopy, we quantified colocalization of polyplexes and caveolin-1 in both cell lines at various times following transfection. We found that a significantly larger fraction of the H3-targeted polyplexes colocalized with caveolin-1 in SUM149 cells vs. MCF10A cells, with approximately 60% more colocalization in the SUM149 cells at all time points (Figure 1). The association between polyplexes and caveolin-1 in IBC SUM149 cells was substantially enhanced by inclusion of the H3 targeting peptide, with 3.3-fold and 10-fold higher association levels between caveolin-1 and the H3-targeted polyplexes as compared with untargeted polyplexes or sH3 polyplexes, respectively (Figure 1). Colocalization between the H3-targeted polyplexes and caveolin-1 decreased gradually over a 2 h period in IBC SUM149 cells, whereas colocalization in the MCF10A cells was maximal at 1 h post-transfection and then declined. Caveolin-1 colocalization with the untargeted polyplexes and the sH3 polyplexes remained low at all time points and did not change significantly as a function of time. These experiments also verified the overexpression of caveolin-1 in IBC SUM149 cells, with 2.2-fold higher caveolin-1 expression in IBC SUM149 cells as compared with the MCF10A cells based on ImageJ analysis and western blotting (Figure S1).
Figure 1.
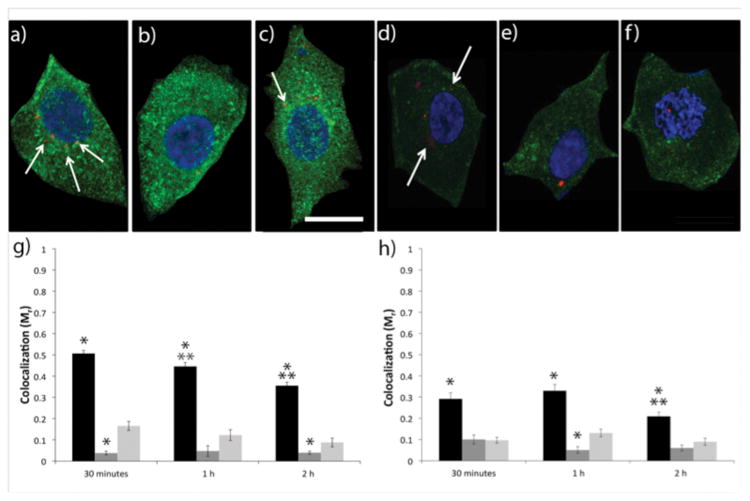
Colocalization of fluorescently labeled pDNA polyplexes (red) with caveolin-1 (green) in IBC SUM149 and MCF10A cells. Representative confocal microscopy z-slice images of IBC cells (a-c) and MCF10A cells (d–f) with nuclei stained with DAPI (blue) following a pulse transfection with (a, d) H3-targeted polyplexes (b,e) sH3 polyplexes or (c,f) untargeted PEI polyplexes. The scale bar (shown in c) = 10 μm. Cells were imaged at 2 h post-transfection. Quantification of colocalization between caveolin-1 and H3-targeted polyplexes (black), sH3 polyplexes (dark gray), or untargeted polyplexes (light gray) at different times post-transfection in IBC SUM149 cells (g) and MCF10A cells (h), performed using Volocity Image Analysis software. Each data point represents the mean ± SE for a minimum of 100 polyplexes, with ~10 images analyzed per colocalization replicate. *Indicates a statistically significant difference from PEI polyplexes at the same time point (P < 0.05). **Indicates a statistically significant difference from the previous time point for the given polyplex (P < 0.05).
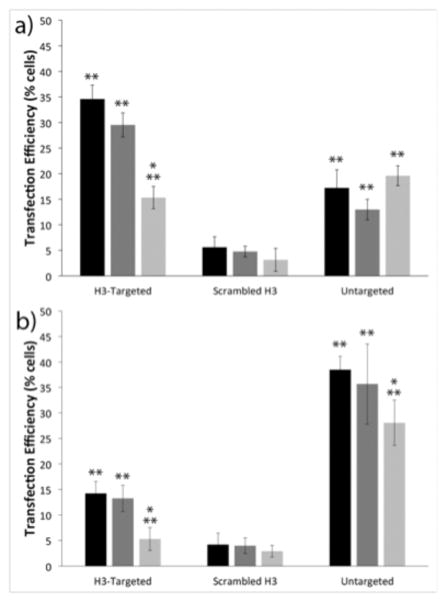
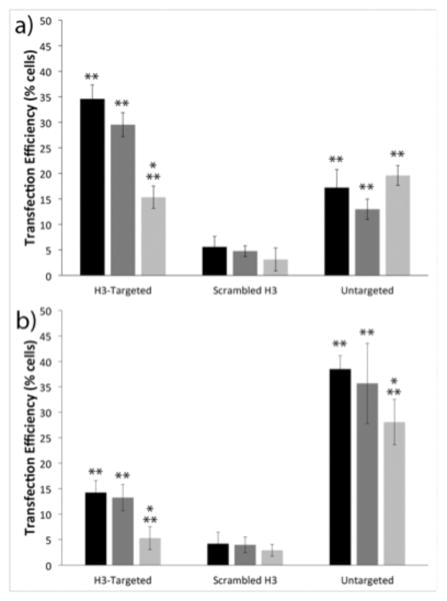
To determine whether increased caveolin-1 association by the H3-targeted polyplexes conferred enhancements in transfection efficiency and targeting specificity in IBC SUM149 cells, we next investigated cell-line dependent differences in transfection. IBC SUM149 cells and MCF10A cells were transfected with H3-targeted polyplexes, untargeted polyplexes, or sH3 polyplexes formed with green fluorescent protein (GFP)-encoding pDNA, and transfection efficiency was measured using flow cytometry. As shown in Figure 2, there were clear differences in transfection in the two cell lines, and these transfection differences were directly related to polyplex type. Specifically, in the IBC SUM149 cells, the H3-targeted polyplexes exhibited 2- and 7-fold higher transfection efficiencies as compared to the untargeted PEI polyplexes and sH3 polyplexes, respectively, with 35% of cells transfected by the H3-targeted polyplexes, but only 18% transfected by the untargeted polyplexes and 5% transfected by the sH3 polyplexes. These trends were reversed in the MCF10A cells, in which the untargeted polyplexes exhibited more than twice the transfection efficiency of the H3-targeted polyplexes, with 38% of cells transfected by the untargeted polyplexes vs. 14% of cells transfected by the H3-targeted polyplexes; only 4% of cells were transfected by the sH3 polyplexes.
Figure 2.
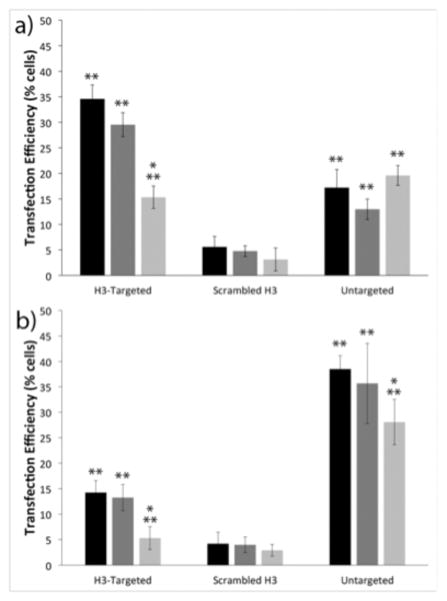
Flow cytometry analyses of cell transfection. Transfection efficiencies of the indicated polyplexes in (a) IBC SUM149 cells and (b) MCF10A cells. Transfection with no treatment control (black), scrambled siRNA (dark gray), and caveolin-1 siRNA (light gray). Each data point represents the mean ± standard deviation for a total of at least four separately prepared samples. * indicates statistically significant difference from the no treatment control (P < 0.05). ** indicates statistically significant difference between cell lines for each treatment (P < 0.05).
To assess the extent to which transfection depended upon polyplex trafficking via caveolin-1, these experiments were also performed in the presence of caveolin-1-specific siRNAs. IBC SUM149 cells or MCF10A cells were pre-transfected with caveolin-1 siRNA using RNAiMAX, and the cells were subsequently transfected with polyplexes. Western blots were performed to confirm and quantify the extent of caveolin-1 silencing in each cell line (Figure S1). These experiments demonstrated similar reductions in caveolin-1 protein levels in the two cell types; with a 73% decrease in caveolin-1 expression in the IBC SUM149 cells and a 67% decrease in the MCF10A cells. When caveolin-1 was silenced in the IBC cell line, the transfection efficiency of the H3-targeted polyplexes decreased by over 50%, whereas the transfection levels achieved with the untargeted polyplexes and sH3 polyplexes remained unchanged. These results demonstrated a clear dependence on caveolar trafficking by the H3-targeted polyplexes in IBC SUM149 cells. Similar results were also found in the MCF10A cell line, in which the levels of transfection with the H3-targeted polyplexes dropped by almost 65% when caveolin-1 was silenced. MCF10A cell transfection with the untargeted polyplexes was reduced only slightly, with a 27% reduction when caveolin-1 was silenced. Silencing caveolin-1 had no effect on transfection efficiency with the sH3 polyplexes in MCF10A cells.
H3-targeted polyplexes selectively traffic through Rab6-linked ER vesicles in IBC SUM149 cells
In order to further probe the trafficking mechanisms leading to transfection, we next sought to determine whether enhanced transfection following caveolar uptake in IBC cells was related to increased polyplex transfer to the perinuclear region and ER. One of the major classes of proteins regulating transfer of cargo between endomembrane organelles is the Rab family of small GTPases (Stenmark 2009). Rab proteins are well-known biomarkers because they have compartment-specific localization profiles and interact with key effectors that define the molecular identity of the organelles to which they are bound (Sandin et al. 2012). Our prior studies in CHO cells demonstrated that H3-targeting enhanced gene transfer by increasing polyplex transport through Rab6-linked caveolar pathways (Ross et al. 2015). The H3-targeted polyplexes accumulated in Rab6 vesicles until mitosis, when they were transferred into the nucleus. In contrast, a majority of untargeted PEI polyplexes were internalized through clathrin-mediated endocytosis in CHO cells (Reilly et al. 2012b), and a large fraction of these polyplexes were subsequently shuttled to Rab11-linked recycling endosomes (Ross et al. 2015). Hence, we sought to explore the roles of various Rab-GFPs in polyplex trafficking in IBC SUM149 cells and MCF10A cells. Cells were pre-transfected with Rab-GFP plasmids, and subsequently, the cells were synchronized and pulse transfected (Ross et al. 2015) with AlexaFluor555-labeled H3-targeted polyplexes. Polyplex colocalization with the Rab-GFPs was quantified as a function of time by using the Manders’ correlation coefficient (Mr).
We initially examined polyplex colocalization with Rab6, which regulates a native transport pathway from the Golgi to the ER (White et al. 1999) that is co-opted by pathogens including Shiga toxin B-fragment and herpes simplex virus 1 during transport to the nucleus (Johns et al. 2014). Colocalization between polyplexes and Rab6 reached a value of 0.73 at 4 h in IBC SUM149 cells, with a significantly higher amount (3.4-fold) of polyplex-Rab6 colocalization in IBC SUM149 cells as compared with MCF10A cells at this time (Figure 3a). Colocalization with Rab6 dropped significantly after mitosis in IBC SUM149 cells, consistent with the behavior documented previously in CHO-K1 cells (Ross et al. 2015). In MCF10A cells, colocalization with Rab6 peaked at 2 h and then declined; there was no significant decrease in post-mitotic polyplex-Rab6 colocalization in MCF10A cells. Colocalization levels with untargeted polyplexes and Rab6 remained below 20% for each cell line; however, both cell lines showed a significant decrease with mitosis (Figure 3c). These results suggest an accumulation of H3-targeted polyplexes within Golgi/ER vesicles through a Rab6-dependent pathway, with increased use of Rab6-linked trafficking in IBC cells due to their overexpression of caveolin-1. These results also suggest that mitosis may not play a large role in trafficking H3-targeted polyplexes into the nucleus of MCF10A cells.
Figure 3.
Quantification of colocalization of H3-targeted (a,b) and untargeted (c,d) polyplexes with (a,c) Rab6 and (b,d) Rab11 at four different time points following a pulse transfection in MCF10A (light gray) and IBC SUM149 (dark gray) cells. Quantification of colocalization from confocal microscopy images was performed with Volocity Image Analysis software. Each data point represents the mean ± SE for a minimum of 100 polyplexes, with ~10 images analyzed per colocalization replicate. Gray box indicates mitosis. * indicates statistically significant difference between cell line (P < 0.05). ** indicates statistically significant difference from previous time point (P < 0.05).
Rab11 is associated with endocytic recycling downstream of clathrin-mediated endocytosis (Ullrich et al. 1996). In both IBC SUM149 cells and MCF10A cells, a small portion of the H3-targeted polyplexes colocalized with Rab11, suggesting that some polyplexes were non-specifically transported back to the plasma membrane via late endosomes in both cell types (Figure 3b). This finding was consistent with our previous studies in CHO cells, in which colocalization with Rab11 reached a maximum at 1 h post-transfection and then decreased significantly after 2 h. Overall, both the IBC SUM149 and MCF10A cells had approximately half of the polyplexes associated with Rab11 as compared with CHO cells (Ross et al. 2015), indicating that fewer polyplexes were recycled in the cells with breast epithelial origin. However, there was a significantly larger fraction of polyplexes that were colocalized with Rab11 in MCF10A cells versus IBC SUM149 cells, suggesting that more H3-targeted polyplexes used clathrin-mediated trafficking in cells with lower levels of caveolin-1 expression. Colocalization between untargeted polyplexes and Rab11 significantly increased until 2 h post-transfection in both cell types (Figure 3d), reaching a higher value in IBC SUM149 cells at the 4 h time point. There was also a significantly larger amount of untargeted polyplex colocalization with Rab11 in IBC SUM149 cells as compared with the MCF10A cells at the 30-minute time points. These data showed that untargeted polyplexes were actively shuttled back to the plasma membrane, as untargeted polyplexes colocalized significantly more with Rab11 as compared with Rab6 in both cell types.
Increased nuclear localization in IBC SUM149 cells occurs during mitosis
Because the decrease in H3-targeted polyplex colocalization with Rab6 occurred during mitosis, we hypothesized that the Rab6/ER-associated polyplexes were redistributed into the nucleus during cell division. Hence, we quantified nuclear localization over similar time frames using confocal imaging (Figure 4). Consistent with our previous studies in CHO cells, we saw moderate increases in nuclear localization of the H3-targeted polyplexes within the SUM149 cells prior to mitosis, and larger increases coincident with mitosis. This led to a high level (~74%) of nuclear localization by the H3-targeted polyplexes during cell division. These results suggested that pre-mitotic accumulation in Rab6-linked compartments led to H3-targeted transfection in SUM149 cells. Polyplex delivery demonstrated a similar pattern in MCF10A cells, although fewer H3-targeted polyplexes localized within the nucleus at all times. Specifically, the levels of post-mitotic nuclear accumulation were 40% lower at 11 h post-transfection in the MCF10A cells as compared to IBC SUM149 cells. These data coincide with the lower transfection efficiencies in MCF10A cells, and are also consistent with the decreased amount of polyplex accumulation within Rab6/ER vesicles leading to decreased nuclear import.
Figure 4.
Localization of H3-targeted polyplexes within the nucleus at five different time points following H3-targeted (a-d), untargeted (e), and sH3 (f) polyplex transfection in MCF10A (light gray) and IBC SUM149 (dark gray) cells. (a-c) Representative confocal microscopy z-slice images of IBC cells with the nuclei stained with DAPI (4',6-diamidino-2-phenylindole) (blue) following a pulse transfection with H3-targeted PEI polyplexes (red) (a) 1 h, (b) 4 h, and (c) 11 h post-transfection. The scale bar (shown in a) = 5 μm. The cell borders were outlined in white by comparison with the corresponding phase images by using Zen software. (d-f) Quantification of colocalization from confocal microscopy images was performed with Volocity Image Analysis software. Each data point represents the mean ± SE for a minimum of 100 polyplexes, with ~10 images analyzed per colocalization replicate. Gray box indicates mitosis. * indicates statistically significant difference between cell line for given time point (P < 0.05). ** indicates statistically significant difference between previous time point for given cell line (P < 0.05).
In contrast, nuclear localization with untargeted polyplexes only reached a level of 35% in IBC cells at 11 h and 32% in MCF10A cells at 9 h, with no significant pattern of increase through mitosis. Nuclear localization was even lower with sH3 polyplexes, reaching a maximum value of 22% in IBC SUM149 cells at 4 h, and 19% in MCF10A cells at 7 h, also with no significant increase through the measured time points.
IBC SUM149 and MCF10A co-cultures demonstrate selective transfection of IBC SUM149 cells
In order to investigate the capacity for IBC cell-specific transfection within the in vivo environment, we initially examined an in vitro mimic comprised of co-cultured IBC SUM149 cells together with MCF10A cells. To enable colorimetric detection of each cell type in the mixed cultures, we labeled each cell line with a different Cell Tracker dye, combined the labeled cells, and transfected the co-cultured cells with either the H3-targeted polyplexes or untargeted polyplexes. Cell-selective gene delivery occurred in co-cultures transfected with the H3-targeted polyplexes, whereas delivery occurred non-specifically in co-cultures treated with the untargeted polyplexes (Figure 5). In particular, the data in Figure 5 show the percentage of total transfected cells that were MCF10A cells and the percentage of total transfected cells that were SUM149 cells based upon flow cytometry, using both the untargeted and H3-targeted polyplexes. When H3-targeted polyplexes were delivered, these analyses demonstrated that 80% of the transfected cells were IBC SUM149 cells, whereas only 20% of the transfected cells were MCF10A cells. In contrast, in co-cultures treated with untargeted polyplexes, there was no selectivity in transfection. Fifty-two percent of the transfected cells in these samples were MCF10A cells and 43% were IBC SUM149 cells. These results clearly demonstrate selectivity in transfection, with H3-targeted polyplexes favoring cells that overexpress caveolin-1. The transfection efficiencies for each cell type in co-culture (Figure S2) were consistent with the transfection efficiency data for each cell type in mono-culture (Figure 2).
Figure 5.
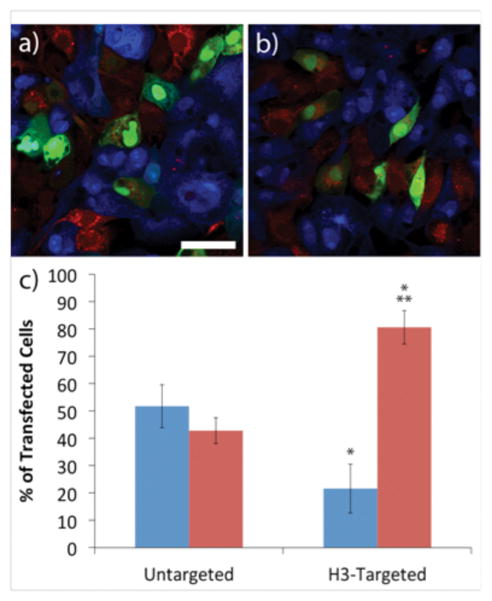
Transfection experiments with co-cultures of MCF10A cells and IBC SUM149 cells. Representative images of co-culture transfections with (a) untargeted polyplexes and (b) H3-targeted polyplexes. MCF10A cells (blue) and IBC SUM149 cells (red) were stained using Cell Tracker dyes. Green indicates GFP expression. The scale bar (shown in a) = 25 μm. (c) Summary of flow cytometry analyses of cell transfection. MCF10A cells (blue) or IBC SUM149 cells (red). Each data point represents the mean ± standard deviation for a total of at least four separately prepared samples. * indicates statistically significant difference from untargeted polyplexes for given cell type (P < 0.05). ** indicates statistically significant difference between cell type for given polyplex (P < 0.05).
H3-targeted polyplexes delivering a suicide gene efficiently decrease IBC SUM149 cell viability
The CD gene is one of the first and most widely studied suicide genes for treating cancer (Moolten 1994). CD catalyzes the deamination of cytosine to uracil, and thus can induce efficient conversion of the prodrug 5-FC into the highly cytotoxic metabolite 5-fluorouracil (5-FU) (Moolten 1994). Hence, we assembled H3-targeted polyplexes and untargeted polyplexes containing DNA encoding for the CD gene to determine whether H3-targeted polyplexes could selectively reduce cell viability in IBC SUM149 cells when the cells were treated with the 5-FC prodrug. Each cell type was transfected with polyplexes formed with yeast CD-encoded plasmid DNA (yCD-pDNA), and the cells were subsequently treated with 5-FC and analyzed by flow cytometry. These data demonstrated a 55% decrease in cell viability after the H3-targeted CD polyplexes were delivered to IBC SUM149 cells, whereas the untargeted CD polyplexes produced only a 33% decrease in viability (Figure 6). The trends were reversed in MCF10A cells, which exhibited only a 25% decrease in cell viability when transfected with H3-targeted polyplexes, but a 54% decrease when treated with untargeted polyplexes. We note that cell viability in all samples transfected with H3-targeted polyplexes or sH3 polyplexes, but lacking the prodrug, was high, indicating that polyplex treatment itself did not significantly detract from cell health.
Figure 6.
Flow cytometry analyses of cell viability following polyplex transfection with CD and application of the prodrug 5-FC. (a) IBC SUM149 and (b) MCF10A cell viability following transfection. Each cell line was transfected either with polyplexes containing GFP-pDNA as a control (dark gray) or with polyplexes containing yCD-pDNA (dark gray). Each data point represents the mean ± standard deviation for a total of at least three separately prepared samples. * indicates a statistically significant difference from control transfection (P < 0.05).
Discussion
A major limitation in gene therapy is the ability of vehicles to both target specific tissues and transfer DNA payloads efficiently into the nuclei of the targeted cells. Generalized receptor-mediated delivery approaches are often ineffective for highly invasive cancers such as IBC, which has an unusual metastatic signature and aberrant gene expression patterns (Kleer et al. 2000). Simultaneously, the ability to efficiently transport genes into the nucleus depends strongly upon a nanocarrier’s propensity to interact with the targeted cell’s membrane and harness its endomembrane trafficking network (Munsell et al. 2015; Reilly et al. 2012b). Our data demonstrate a new approach to achieve both IBC selectivity and efficient gene transfer by targeting the endomembrane marker caveolin-1 that is overexpressed during IBC metastasis (Van den Eynden et al. 2006). In particular, our studies support the efficacy of using endocytic targeting in IBC as a novel approach to induce efficient and more cell selective nuclear delivery and gene expression.
We anticipated that caveolin-1 overexpression in IBC SUM149 cells would increase transfection efficiency with our H3-targeted polyplexes, which were previously shown to use caveolar uptake routes to initiate delivery to the nucleus (Ross et al. 2015). We found that caveolin-1 overexpression indeed increased transfection efficiency in IBC SUM149 cells. Colocalization between H3-targeted polyplexes and Rab6 was found in both cell lines; however, our studies show the overexpression of caveolin-1 in IBC SUM149 cells led to increased colocalization with Rab6, which also increased nuclear accumulation. These results supported our previous studies showing that Rab6 colocalization led to increased nuclear colocalization with H3-targeted polyplexes after caveolar uptake in CHO cells (Reilly et al. 2012b; Ross et al. 2015). More generally, these data demonstrate that the H3-mediated targeting of caveolar uptake may represent a more general approach to route nanostructures to the nucleus via perinuclear accumulation in Golgi vesicles.
IBC accounts for up to 10% of all breast cancer mortality because of treatment resistance and the propensity for early metastasis. To date, no IBC-specific therapies exist, in part because of the lack of models that faithfully recapitulate the clinical features of the disease (Lacerda and Woodward 2012). Co-culture experiments performed in this study allowed us to explore an in vitro mimic of cancerous tissue. In particular, whereas monoculture systems cannot evaluate competitive interactions between polyplexes and cancerous vs. non-cancerous cells, this co-culture system provided us with an opportunity to evaluate simplified aspects of the microenvironment in breast cancer. We demonstrated that culturing IBC SUM149 cells together with non-cancerous breast epithelial MCF10A cells magnified the significance of our results, with 4-fold more IBC SUM149 cells were transfected in co-culture models when H3-targeted polyplexes were employed. Given the large differences in caveolin-1 expression in the two cell types (Figure 1), we suspect that the selectivity for IBC SUM149 cells vs. MCF10A cells could be even further increased through changes in polyplex surface chemistry. In particular, while H3-targeting substantially increased the fraction of polyplexes routed through caveolar pathways in breast epithelial cells (Figure 1) as well as other cell types (Reilly et al. 2012b), a fraction of the H3-targeted polyplex population shuttled through non-caveolar uptake routes. This lack of selectivity is most likely due to the positive net charge of the polyplexes (Reilly et al. 2012a), which is well known to promote non-specific interactions with the cell surface.
Gene therapy can facilitate chemotherapeutic treatment of tumor cells by delivering a cytotoxic gene to the tumor. For example, genes encoding enzymes with the capacity to convert non-cytotoxic prodrugs into lethal compounds have been identified in viruses and bacteria, and when expressed in mammalian cells, these gene products can be used in combination with their prodrug substrates to selectively kill cancer cells. In particular, CD is an enzyme that is found in many bacteria and fungi but not in mammalian cells (Mullen et al. 1992). This system has been studied in retroviral, adenoviral, and liposomal mediated gene transfer strategies (Chaszczewska-Markowska et al. 2008; Khil et al. 1996). CD converts the non-cytotoxic prodrug 5-FC into 5-FU, a chemotherapeutic agent that is commonly used in the treatment of various tumors. Selective expression of CD within targeted tumors combined with 5-FC administration would theoretically result in locally high concentrations of 5-FU while minimizing systemic 5-FU toxicity.
Our prodrug results were consistent with the transfection efficiencies displayed in both cell types, with,a large fraction (52%) of IBC SUM149 cells undergoing apoptosis after CD transfection with H3-targeted polyplexes and prodrug treatment. On the other hand, 24% of MCF10A cells went through prodrug-mediated apoptosis. The differences between transfection efficiency and viability may be due to the bystander effect that occurs with suicide gene therapy (Kuriyama et al. 1998), where tumor cells not actually expressing a suicide gene, but in proximity to suicide gene-transduced cells, are killed after exposure to the prodrug. Specifically, the CD/5-FC pair is known to have a significant bystander effect that does not require direct cell contact, as 5-FU can readily move into and out of cells by spontaneous diffusion (Huber et al. 1994).
In contrast to H3-targeted polyplexes, untargeted and sH3 polyplexes exhibited fundamentally different transfection efficiencies depending on cell line. Untargeted polyplexes exhibited higher transfection in MCF10A cells, coinciding with the different trafficking patterns of untargeted polyplexes, and their increased use of clathrin versus caveolar uptake pathways. Silencing caveolin-1 also had little effect on the transfection efficiency of untargeted polyplexes. These results are consistent with previous studies demonstrating that untargeted polyplexes utilize a different trafficking pathway to enter the nucleus (Ross et al. 2015). In these experiments, we note that we opted to use an N:P ratio of 10 for the untargeted polyplexes to keep the N:P ratios consistent between the untargeted and H3-targeted samples. While we have tested lower charge ratios of the PEI polyplexes and shown that these formulations exhibit more minimal cytotoxicity, PEI polyplexes at these lower charge ratios do not transfect SUM149 or MCF10A cells effectively.
sH3 polyplexes were even less affected by the expression of caveolin-1, as transfection efficiency remained unchanged after caveolin-1 silencing in both cell lines, which demonstrates sH3 polyplexes are utilizing a different endocytic pathway to enter cells. These results agree with our previous findings with sH3 polyplexes, which showed low cellular uptake and minimal transfection efficiency in CHO cells (Ross et al. 2015). This demonstrates the sequence-specificity of the H3 peptide and its impacts on polyplex interactions with the plasma membrane to alter endocytic trafficking.
In this study, we observed that H3 targeting enhanced polyplex transfection by harnessing the overexpression of caveolin-1 in IBC SUM149 cells. These results are consistent with previous studies, which show that caveolar trafficking efficiently delivers these polyplexes, as well as other types of polyplexes, to the nucleus. Although effective identification of cancer cells will ultimately need even greater cell/tissue selectivity, what is clear from these results is that a simple, endocytic targeting strategy can offer promising increases in cell specificity that could be combined with additional ligand-mediated targeting approaches. Indeed, multi-modal targeting has already demonstrated promising benefits in several preclinical cancer models (Bazak et al. 2015; Massey and Schnitzer 2010). Furthermore, our study demonstrates the versatility of histone-targeted polyplex transfection by showing its efficacy for selective expression of two different genes including a suicide gene with translational potential in various cancers. Thus, these findings demonstrate a potential strategy for the treatment of IBC and other types of cancers that possess composite biomolecular or phenotypic characteristics which distinguishes them from normal tissues, and which can be exploited for gene therapy against a variety of gene targets.
Materials & Methods
Materials
H3 tail peptides comprised of the mammalian N-terminal H3 residues 1–25 (ARTKQTARKSTGGKAPRKQLATKAA-CONH2), and ethidium homodimer-1 (EthD-1), were purchased from Anaspec (Fremont, CA) at ≥95% purity. The sH3 peptide sequence was designed to incorporate residues 1-25 of the N-terminal tail of the H3 protein in a randomized sequence (LSAATPRTAKGARQTKRQKAKGTAK-CONH2). The peptide was synthesized by solid phase peptide synthesis using previous protocols (Ross et al. 2015). The gWIZ mammalian expression plasmid encoding GFP was obtained from Genlantis (San Diego, CA) and Rab-GFP plasmids were purchased from Addgene (ID# 12674, 31733). The fcy1 gene from yeast was inserted into the pCDNA3.1(+) vector backbone (Invitrogen, Carlsbad, CA), at NheI and ApaI restriction sites by polymerase chain reaction techniques. This fcy1 gene contains three mutations for thermostabilityA23L/V108I/I140L. All plasmids were amplified in DH5α Escherichia coli in Lysogeny Broth, and purified with a QIAGEN Plasmid Mega Kit (QIAGEN, Valencia, CA) following the manufacturer’s protocols. Alexa Fluor 555-labeled PNA clamps were custom-synthesized and purified to >90% by Panagene (Daejeon, Korea). Cell culture reagents were purchased from Fisher Scientific (Pittsburgh, PA). Primary antibodies reactive with caveolin-1 (sc-894) and actin (sc1616R), and caveolin-1 siRNA (sc-29942), which consists of pools of three to five target-specific 19–25 nt siRNAs, were obtained from Santa Cruz Biotechnology (Dallas, TX). Secondary antibody (AlexaFluor488 anti-rabbit IgG), RNAiMAX reagent, and Cell Tracker dyes Deep Red (C34552) and Blue CMAC (C2110) were obtained from Invitrogen. Branched PEI (25 kDa) and all other reagents were purchased at analytical grade from Sigma (St. Louis, MO).
Polyplex formation and transfection
H3-targeted PEI polyplexes or untargeted PEI polyplexes were formed in 20 mmol/l 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) at a pH of 6 (Reilly et al. 2012a). For all experiments, polyplexes were formed at an N:P ratio of 10. For polyplexes formed with a mixture of H3 and PEI, an N:P ratio of 6/4 was used, where the total N:P ratio was 10, with N = 6 from H3 and N = 4 from PEI. This corresponds to ~90% (w/w) H3 and 10% PEI in the polycation solution used for pDNA complexation. The structures of the polyplexes were analyzed in our prior work by gel electrophoresis experiments as well as dynamic light scattering and zeta potential experiments (Larsen et al. 2012; Reilly et al. 2012a). The amount of peptide within the polyplex was also quantified previously (Larsen et al. 2012). For all colocalization studies, gWIZ plasmid DNA (pDNA) was fluorescently labeled prior to polyplex formation with PNA-AlexaFluor555 at a ratio of 1:20 (DNA:PNA), as previously described (Millili et al. 2010).
For all polyplex transfections, cells were rinsed with PBS, and subsequently, serum-free Opti-MEM was added to the cells prior to polyplex addition. We used similar transfection conditions for consistency with the literature and for consistency with our earlier experiments with the H3-targeted polyplexes (Ross et al. 2015). Such procedures, e.g. cell maintenance in complete medium and the use of low serum during polyplex exposure, are typically used because they enable both robust cell proliferation/polyplex uptake and maximal polyplex activity.
Cell culture and synchronization
IBC SUM149 cells were a gift from Kenneth van Golen’s laboratory (Department of Biological Sciences, University of Delaware). These cells were originally established from primary IBC patient samples and authenticated by the Johns Hopkins Genomics Resource Center as described (Lehman et al. 2012). MCF10A cells were obtained from American Type Culture Collection (ATCC). Both cell lines were cultured according to previously established methods (Van den Eynden et al. 2006; van Golen et al. 1999). Briefly, IBC SUM149 cells were grown in Ham’s F12 medium supplemented with 5% FBS, 1% (v/v) penicillin/streptomycin, 1% (v/v) L-glutamine, 1% (v/v) mycoplasma antibiotic supplement, insulin (5μg/mL), transferrin (2.5 μg/mL), selenium (200 ng/mL), and hydrocortisone (1 μg/mL). All cells were grown at 37°C in 5% CO2. MCF10A cells were grown in 50/50 DMEM/Ham’s F12 medium supplemented with 5% FBS, 1% (v/v) penicillin/streptomycin, bovine pituitary extract (50 μg/mL), insulin (10 μg/mL), hydrocortisone (0.5 μg/mL), cholera toxin (100 ng/mL), and epidermal growth factor (20 ng/mL).
For cell synchronization, cells were plated at ~7,200 cells/cm2 in complete growth medium. Twenty-four hours after plating, medium without serum was added to cells for 72 h. In order to resume the cell division cycle, the serum-free medium was removed, the cells were washed with phosphate buffered saline (PBS), and fresh complete medium was added to cells. Cells were then incubated for an additional 12 h so that transfection could take place during the S-phase of cell division.
Immunocytochemistry analysis of caveolin-1 expression
For colocalization studies with caveolin-1, cells were plated, synchronized, and 15-minute pulse transfected, as previously described (Ross et al. 2015), with 50 μl of polyplex solution (either H3-targeted polyplexes, sH3 polyplexes, or untargeted PEI polyplexes) in 150 μl of Opti-MEM containing 1 μg pDNA labeled with PNA-AlexaFluor555. At the specified time points, cells were rinsed (“chased”) with PBS, washed with 10 μg/ml heparin, washed again with PBS, and fixed with 4% paraformaldehyde in PBS for 15 minutes. Cells were subsequently permeabilized with 0.1% saponin in PBS (Sap) and blocked with 0.5% bovine serum albumin in 0.1% Sap. A 1:500 solution of primary antibody was incubated with the cells overnight at 4 °C in the blocking buffer. Cells were subsequently rinsed three times with PBS and incubated with the secondary antibody in blocking buffer for 1 h at room temperature. Following secondary antibody treatment, cells were rinsed three more times with PBS and stored at 4 °C prior to imaging.
siRNA silencing experiments
Cells were transfected with 10 μM caveolin-1 siRNA for 48 h using the RNAiMAX reagent. Twenty-four hours after siRNA treatment, cells were transfected with 250 μl of polyplex solution (either H3-targeted polyplexes, sH3 polyplexes, or untargeted PEI polyplexes) in 2 ml of Opti-MEM containing 5 μg of pDNA labeled with PNA-AlexaFluor555 for 2 h, according to previously published procedures (Reilly et al. 2012a). GFP expression was quantified on an BD Accuri C6 flow cytometer (BD Biosciences, San Jose, CA). For cytometry analyses, cells were collected after imaging and prepared for analysis by standard trypsin-mediated collection protocols. Cells were resuspended in PBS, filtered through a 35 μm nylon mesh to remove aggregates, and stored at 4 °C until analysis. Scattering plots were gated for quantification purposes, and a total of 10,000 cells were analyzed for each cell sample. Dead cells were excluded from the analyses of transfection efficiency.
Rab-GFP transfection experiments
For Rab-GFP colocalization analyses, cells were plated in 8-well glass bottom plates from LabTek (Thermo Fisher Scientific, Waltham, MA), cultured for 24 h, and synchronized. Subsequently, the cells were transfected with 1 μg of the specified Rab-GFP fusion plasmid in 2 μl Lipofectamine and 100 μl of Opti-MEM for 2 h, washed with PBS, and cultured for an additional 24 h. Rab-GFP transfection was confirmed by fluorescence microscopy and flow cytometry to ensure that expression patterns were appropriate and that GFP was visible in a majority (~90%) of cells. Next, each well was transfected with 50 μl of polyplex solution in 150 μl of Opti-MEM containing 1 μg of pDNA labeled with PNA-AlexaFluor555. After a 15-minute pulse-transfection with polyplexes, cells were rinsed with PBS and then incubated in complete medium. At the specified time points, cells were fixed with 4% paraformaldehyde according to routine procedures.
Confocal microscopy and quantification of polyplex colocalization
Cell imaging was performed with a 40× water immersion objective (NA = 0.55) on an LSM 710 microscope (Carl Zeiss, Thornwood, NY) equipped with appropriate lasers and filters for the selected fluorescent dyes. Volocity Imaging Software (PerkinElmer) was used for image analysis and quantification of colocalization, where the locations of polyplexes and caveolin-1 or Rab proteins were determined from measurement statistics associated with individual voxel intensities. The fraction of polyplexes (red voxels) that colocalized with the vesicle or organelle of interest (green voxels) was analyzed by calculation of the Mr, which represents the sum of the colocalized red intensity divided by the sum of the total red intensity. Mr values range from 0 (no colocalization) to 1 (complete colocalization of red voxels with green voxels).(Manders et al. 1993) Volocity software automatically determined minimum values for red and green intensities, and these minimum values were set as the threshold to distinguish signal from background. Statistical analyses of Mr values were performed using a two-tailed Student’s t-test and the SE reported represents the population of polyplexes analyzed. A range of 80–100 polyplexes per data point in each colocalization study were analyzed to obtain these values.
Co-culture of IBC SUM149 and MCF10A cells
Each cell line was plated individually in a well of a 12 well plate. Twenty-four hours after plating, cells were incubated for 30 minutes with Cell Tracker dyes in serum-free medium (Deep Red at 1 μM or Blue CMAC at 10 μM); subsequently, the dye containing solutions were replaced with fresh complete medium. Twelve hours later, cells were trypsinized and plated together in a 6 well plate, in a 50:50 mixture of SUM149 medium and MCF10A medium. Cells were subsequently transfected with polyplexes for 2 h, 24 h after co-culturing. Polyplexes were then removed, and the cells were rinsed with PBS and allowed to grow in complete medium. Flow cytometry analyses of GFP expression and the presence of the Cell Tracker dyes were performed 24 h after transfection using a BD FACSCalibur Flow Cytometer (San Jose, CA). A total of 10,000 cells were analyzed for each cell sample.
Prodrug treatments and cell viability
For prodrug treatment, cells were seeded in 6 well plates, and allowed to grow for 24 h. Cells were rinsed with PBS and subsequently transfected in serum-free medium with 250 μl of polyplex solution formed with 5 μg of pDNA containing the yCD gene in a total of 2 ml of Opti-MEM. After 2 h, cells were rinsed with PBS and the medium was replaced with fresh complete medium. Twenty-four hours post-transfection, cells were incubated for 24 h with medium containing 100 μg/ml 5-FC. Dead cells were visualized by fluorescence microscopy. Solutions containing 4 μM EthD-1 were formulated in PBS and incubated with cells for 10 min. Subsequently, cells were washed and imaged with a Leica 6000 fluorescent microscope (Wetzler, Germany). The fraction of viable cells was quantified via flow cytometry analysis. Live/dead cell gating was based on forward and side scatter patterns characteristic of dead cells, and verified based on overlap with EthD-1 staining. Scatter plots were analyzed for both untransfected cells and cells transfected with polyplexes containing the gWiz GFP expression vector as controls. A total of 10,000 cells were analyzed for each cell sample.
Supplementary Material
Figure S1: Western blots for caveolin-1 siRNA
Figure S2: Transfection efficiencies for MCF10A/IBC SUM149 co-cultures
Acknowledgments
This work was supported by the National Science Foundation under Grant No. DMR-0746458. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. This work was also partially supported by the National Institutes of Health under Grant No. R01 EB017766. We acknowledge the Delaware Biotechnology Institute Bioimaging Facility for use of their confocal microscopes. We thank Jeffrey L Caplan and Michael Moore for training and continuing guidance with the equipment and software used for all cellular imaging and analysis. We thank the Van Golen lab for their generous donation of IBC SUM149 and MCF10A cell lines. We thank the Chen lab, and Rebecca Chen, for their generous donation of the yCD plasmid. We also thank Lynn Opdenaker, at the Center for Translational Cancer Research, for use of the flow cytometer.
References
- Akhtar S. Non-viral cancer gene therapy: beyond delivery. Gene Ther. 2006;13(9):739–40. doi: 10.1038/sj.gt.3302692. [DOI] [PubMed] [Google Scholar]
- Amer MH. Gene therapy for cancer: present status and future perspective. Molecular and Cellular Therapies. 2014;2:27. doi: 10.1186/2052-8426-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenholz Y. Doxil(R)--the first FDA-approved nano-drug: lessons learned. J Control Release. 2012;160(2):117–34. doi: 10.1016/j.jconrel.2012.03.020. [DOI] [PubMed] [Google Scholar]
- Bazak R, Houri M, El Achy S, Kamel S, Refaat T. Cancer active targeting by nanoparticles: a comprehensive review of literature. J Cancer Res Clin Oncol. 2015;141(5):769–84. doi: 10.1007/s00432-014-1767-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC. Cancer nanotechnology: the impact of passive and active targeting in the era of modern cancer biology. Adv Drug Deliv Rev. 2014;66:2–25. doi: 10.1016/j.addr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaszczewska-Markowska M, Stebelska K, Sikorski A, Madej J, Opolski A, Ugorski M. Liposomal formulation of 5-fluorocytosine in suicide gene therapy with cytosine deaminase--for colorectal cancer. Cancer Lett. 2008;262(2):164–72. doi: 10.1016/j.canlet.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Douglas KL, Piccirillo CA, Tabrizian M. Cell line-dependent internalization pathways and intracellular trafficking determine transfection efficiency of nanoparticle vectors. Eur J Pharm Biopharm. 2008;68(3):676–87. doi: 10.1016/j.ejpb.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Fichter KM, Ingle NP, McLendon PM, Reineke TM. Polymeric nucleic acid vehicles exploit active interorganelle trafficking mechanisms. ACS Nano. 2013;7(1):347–64. doi: 10.1021/nn304218q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielson NP, Pack DW. Efficient polyethylenimine-mediated gene delivery proceeds via a caveolar pathway in HeLa cells. J Control Release. 2009;136(1):54–61. doi: 10.1016/j.jconrel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Grandinetti G, Smith AE, Reineke TM. Membrane and nuclear permeabilization by polymeric pDNA vehicles: efficient method for gene delivery or mechanism of cytotoxicity? Mol Pharm. 2012;9(3):523–38. doi: 10.1021/mp200368p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnasko R, Lisanti MP. The biology of caveolae: lessons from caveolin knockout mice and implications for human disease. Mol Interv. 2003;3(8):445–64. doi: 10.1124/mi.3.8.445. [DOI] [PubMed] [Google Scholar]
- Huber BE, Austin EA, Richards CA, Davis ST, Good SS. Metabolism of 5-fluorocytosine to 5-fluorouracil in human colorectal tumor cells transduced with the cytosine deaminase gene: significant antitumor effects when only a small percentage of tumor cells express cytosine deaminase. Proc Natl Acad Sci U S A. 1994;91(17):8302–6. doi: 10.1073/pnas.91.17.8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns HL, Gonzalez-Lopez C, Sayers CL, Hollinshead M, Elliott G. Rab6 dependent post-Golgi trafficking of HSV1 envelope proteins to sites of virus envelopment. Traffic. 2014;15(2):157–78. doi: 10.1111/tra.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamaly N, Xiao Z, Valencia PM, Radovic-Moreno AF, Farokhzad OC. Targeted polymeric therapeutic nanoparticles: design, development and clinical translation. Chem Soc Rev. 2012;41(7):2971–3010. doi: 10.1039/c2cs15344k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil MS, Kim JH, Mullen CA, Kim SH, Freytag SO. Radiosensitization by 5-fluorocytosine of human colorectal carcinoma cells in culture transduced with cytosine deaminase gene. Clin Cancer Res. 1996;2(1):53–7. [PubMed] [Google Scholar]
- Kleer CG, van Golen KL, Merajver SD. Molecular biology of breast cancer metastasis. Inflammatory breast cancer: clinical syndrome and molecular determinants. Breast Cancer Res. 2000;2(6):423–9. doi: 10.1186/bcr89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama S, Masui K, Sakamoto T, Nakatani T, Kikukawa M, Tsujinoue H, Mitoro A, Yamazaki M, Yoshiji H, Fukui H, et al. Bystander effect caused by cytosine deaminase gene and 5-fluorocytosine in vitro is substantially mediated by generated 5-fluorouracil. Anticancer Res. 1998;18(5A):3399–406. [PubMed] [Google Scholar]
- Lacerda L, Woodward WA. Models of Inflammatory Breast Cancer. In: Ueno NT, Fink T, editors. Inflammatory Breast Cancer: An Update. New York: Springer; 2012. pp. 139–150. [Google Scholar]
- Larsen JD, Reilly MJ, Sullivan MO. Using the epigenetic code to promote the unpackaging and transcriptional activation of DNA polyplexes for gene delivery. Mol Pharm. 2012;9(5):1041–51. doi: 10.1021/mp200373p. [DOI] [PubMed] [Google Scholar]
- Lehman HL, Van Laere SJ, van Golen CM, Vermeulen PB, Dirix LY, van Golen KL. Regulation of inflammatory breast cancer cell invasion through Akt1/PKBalpha phosphorylation of RhoC GTPase. Mol Cancer Res. 2012;10(10):1306–18. doi: 10.1158/1541-7786.MCR-12-0173. [DOI] [PubMed] [Google Scholar]
- Manders E, Verbeek F, Aten J. Measurement of colocalization of objects in dual-color confocal images. Journal of Microscopy-Oxford. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Caveolae and signalling in cancer. Nat Rev Cancer. 2015;15(4):225–37. doi: 10.1038/nrc3915. [DOI] [PubMed] [Google Scholar]
- Massey KA, Schnitzer JE. Caveolae and cancer. Recent Results Cancer Res. 2010;180:217–31. doi: 10.1007/978-3-540-78281-0_13. [DOI] [PubMed] [Google Scholar]
- McIntosh DP, Tan XY, Oh P, Schnitzer JE. Targeting endothelium and its dynamic caveolae for tissue-specific transcytosis in vivo: a pathway to overcome cell barriers to drug and gene delivery. Proc Natl Acad Sci U S A. 2002;99(4):1996–2001. doi: 10.1073/pnas.251662398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLendon PM, Fichter KM, Reineke TM. Poly(glycoamidoamine) vehicles promote pDNA uptake through multiple routes and efficient gene expression via caveolae-mediated endocytosis. Mol Pharm. 2010;7(3):738–50. doi: 10.1021/mp900282e. [DOI] [PubMed] [Google Scholar]
- Midoux P, Breuzard G, Gomez J, Pichon C. Polymer-Based Gene Delivery: A Current Review on the Uptake and Intracellular Trafficking of Polyplexes. Curr Gene Ther. 2008;8(5):335–352. doi: 10.2174/156652308786071014. [DOI] [PubMed] [Google Scholar]
- Millili PG, Yin DH, Fan H, Naik UP, Sullivan MO. Formulation of a Peptide nucleic Acid based nucleic Acid delivery construct. Bioconjug Chem. 2010;21(3):445–55. doi: 10.1021/bc900328j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesano R, Roth J, Robert A, Orci L. Non-coated membrane invaginations are involved in binding and internalization of cholera and tetanus toxins. Nature. 1982;296(5858):651–3. doi: 10.1038/296651a0. [DOI] [PubMed] [Google Scholar]
- Moolten FL. Drug sensitivity ("suicide") genes for selective cancer chemotherapy. Cancer Gene Ther. 1994;1(4):279–87. [PubMed] [Google Scholar]
- Mullen CA, Kilstrup M, Blaese RM. Transfer of the bacterial gene for cytosine deaminase to mammalian cells confers lethal sensitivity to 5-fluorocytosine: a negative selection system. Proc Natl Acad Sci U S A. 1992;89(1):33–7. doi: 10.1073/pnas.89.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsell EV, Ross NL, Sullivan MO. Journey to the Center of the Cell: Current Nanocarrier Design Strategies Targeting Biopharmaceuticals to the Cytoplasm and Nucleus. Curr Pharm Des. 2015 doi: 10.2174/1381612822666151216151420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh P, Borgström P, Witkiewicz H, Li Y, Borgström BJ, Chrastina A, Iwata K, Zinn KR, Baldwin R, Testa JE, et al. Live dynamic imaging of caveolae pumping targeted antibody rapidly and specifically across endothelium in the lung. Nat Biotechnol. 2007;25(3):327–337. doi: 10.1038/nbt1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkmans L, Kartenbeck J, Helenius A. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat Cell Biol. 2001;3(5):473–83. doi: 10.1038/35074539. [DOI] [PubMed] [Google Scholar]
- Reilly MJ, Larsen JD, Sullivan MO. Histone H3 tail peptides and poly(ethylenimine) have synergistic effects for gene delivery. Mol Pharm. 2012a;9(5):1031–40. doi: 10.1021/mp200372s. [DOI] [PubMed] [Google Scholar]
- Reilly MJ, Larsen JD, Sullivan MO. Polyplexes traffic through caveolae to the Golgi and endoplasmic reticulum en route to the nucleus. Mol Pharm. 2012b;9(5):1280–90. doi: 10.1021/mp200583d. [DOI] [PubMed] [Google Scholar]
- Rejman J, Bragonzi A, Conese M. Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol Ther. 2005;12(3):468–74. doi: 10.1016/j.ymthe.2005.03.038. [DOI] [PubMed] [Google Scholar]
- Ross NL, Munsell EV, Sabanayagam C, Sullivan MO. Histone-targeted Polyplexes Avoid Endosomal Escape and Enter the Nucleus During Postmitotic Redistribution of ER Membranes. Molecular Therapy—Nucleic Acids. 2015;4(2):e226. doi: 10.1038/mtna.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandin P, Fitzpatrick LW, Simpson JC, Dawson KA. High-speed imaging of Rab family small GTPases reveals rare events in nanoparticle trafficking in living cells. ACS Nano. 2012;6(2):1513–21. doi: 10.1021/nn204448x. [DOI] [PubMed] [Google Scholar]
- Schnitzer JE. Caveolae: from basic trafficking mechanisms to targeting transcytosis for tissue-specific drug and gene delivery in vivo. Adv Drug Deliv Rev. 2001;49(3):265–80. doi: 10.1016/s0169-409x(01)00141-7. [DOI] [PubMed] [Google Scholar]
- Shin JS, Abraham SN. Cell biology. Caveolae--not just craters in the cellular landscape. Science. 2001;293(5534):1447–8. doi: 10.1126/science.1061079. [DOI] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nature Reviews Molecular Cell Biology. 2009;10(8):513–25. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Ullrich O, Reinsch S, Urbé S, Zerial M, Parton RG. Rab11 regulates recycling through the pericentriolar recycling endosome. The Journal of Cell Biology. 1996;135(4):913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Eynden GG, Van Laere SJ, Van der Auwera I, Merajver SD, Van Marck EA, van Dam P, Vermeulen PB, Dirix LY, van Golen KL. Overexpression of caveolin-1 and -2 in cell lines and in human samples of inflammatory breast cancer. Breast Cancer Res Treat. 2006;95(3):219–28. doi: 10.1007/s10549-005-9002-1. [DOI] [PubMed] [Google Scholar]
- van Golen KL, Davies S, Wu ZF, Wang Y, Bucana CD, Root H, Chandrasekharappa S, Strawderman M, Ethier SP, Merajver SD. A novel putative low-affinity insulin-like growth factor-binding protein, LIBC (lost in inflammatory breast cancer), and RhoC GTPase correlate with the inflammatory breast cancer phenotype. Clin Cancer Res. 1999;5(9):2511–9. [PubMed] [Google Scholar]
- White J, Johannes L, Mallard F, Girod A, Grill S, Reinsch S, Keller P, Tzschaschel B, Echard A, Goud B, et al. Rab6 coordinates a novel Golgi to ER retrograde transport pathway in live cells. J Cell Biol. 1999;147(4):743–60. doi: 10.1083/jcb.147.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Western blots for caveolin-1 siRNA
Figure S2: Transfection efficiencies for MCF10A/IBC SUM149 co-cultures


