Abstract
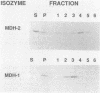
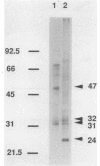
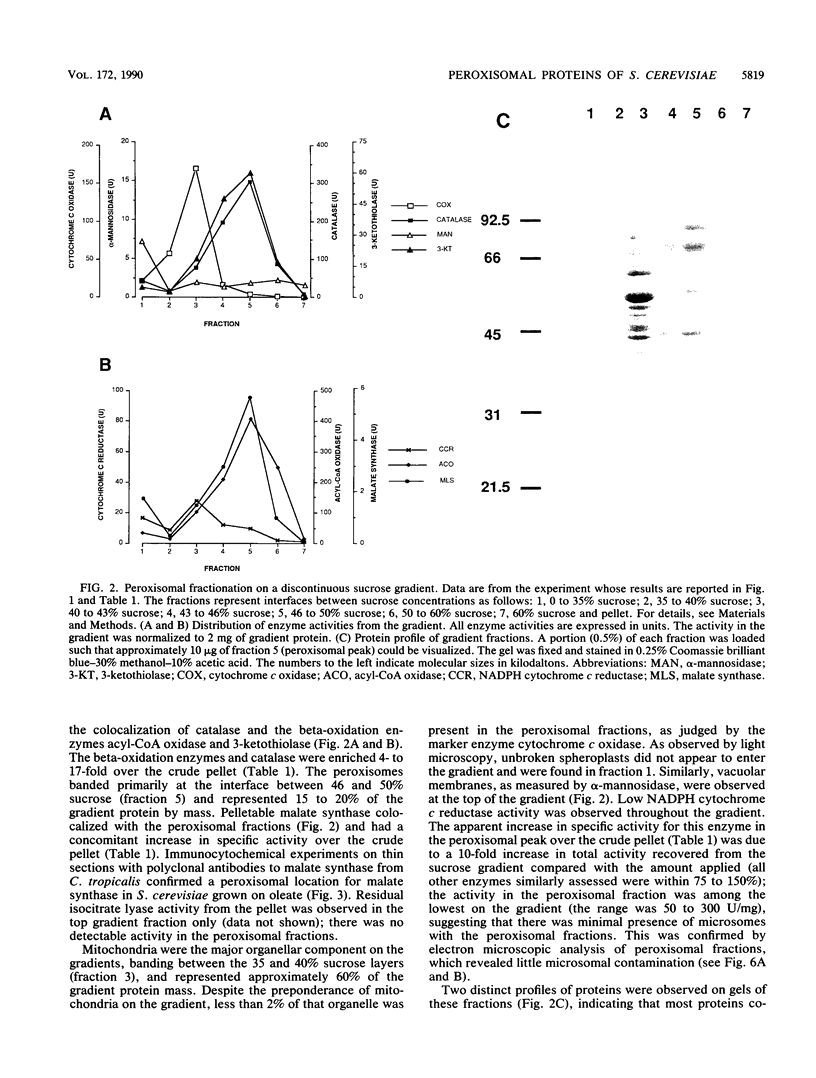
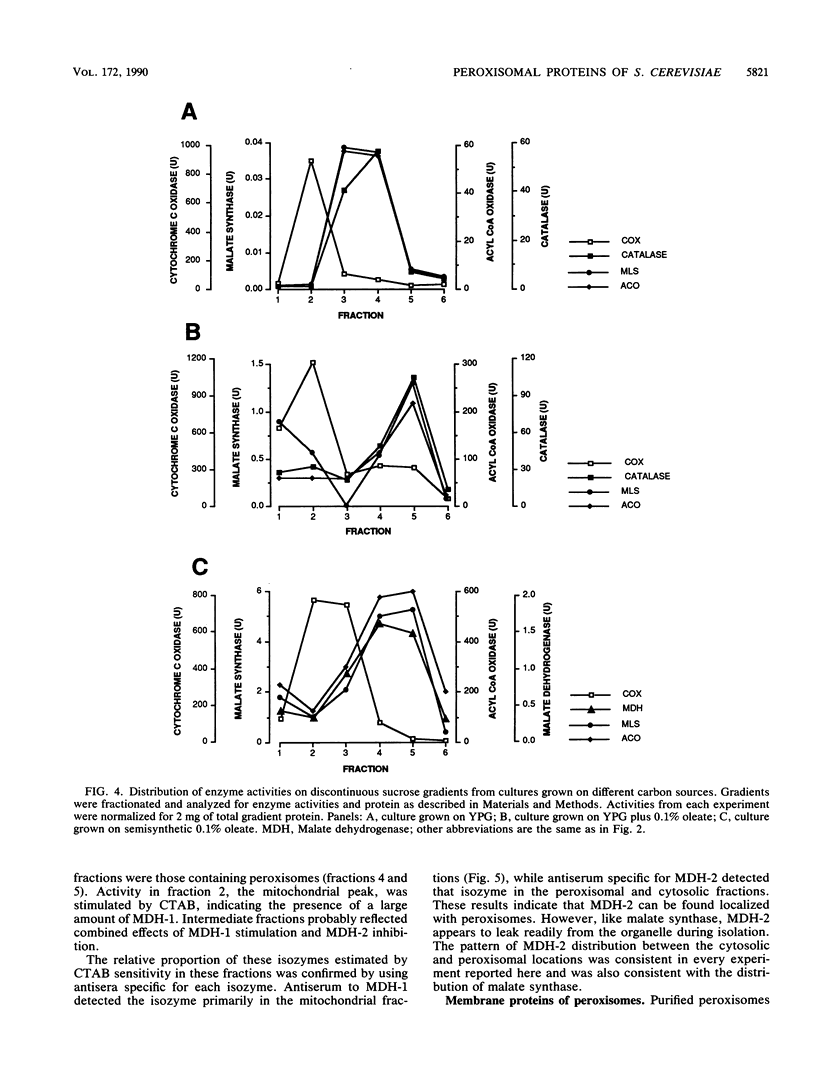
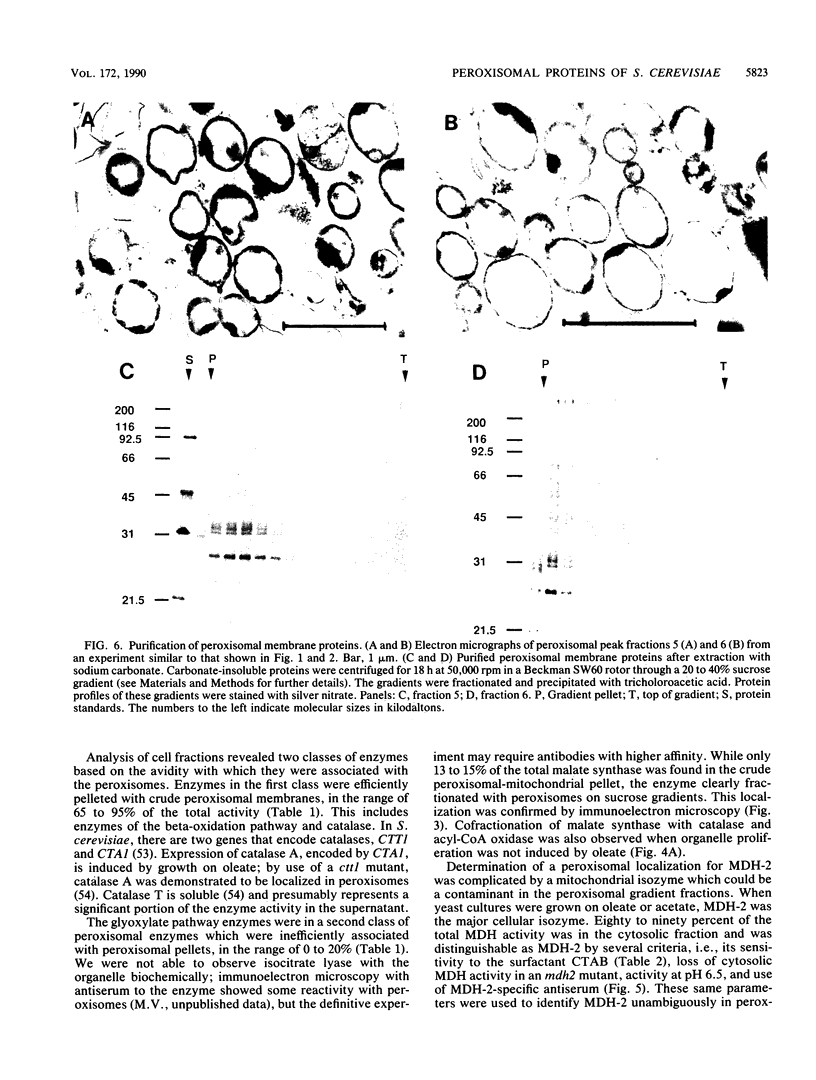
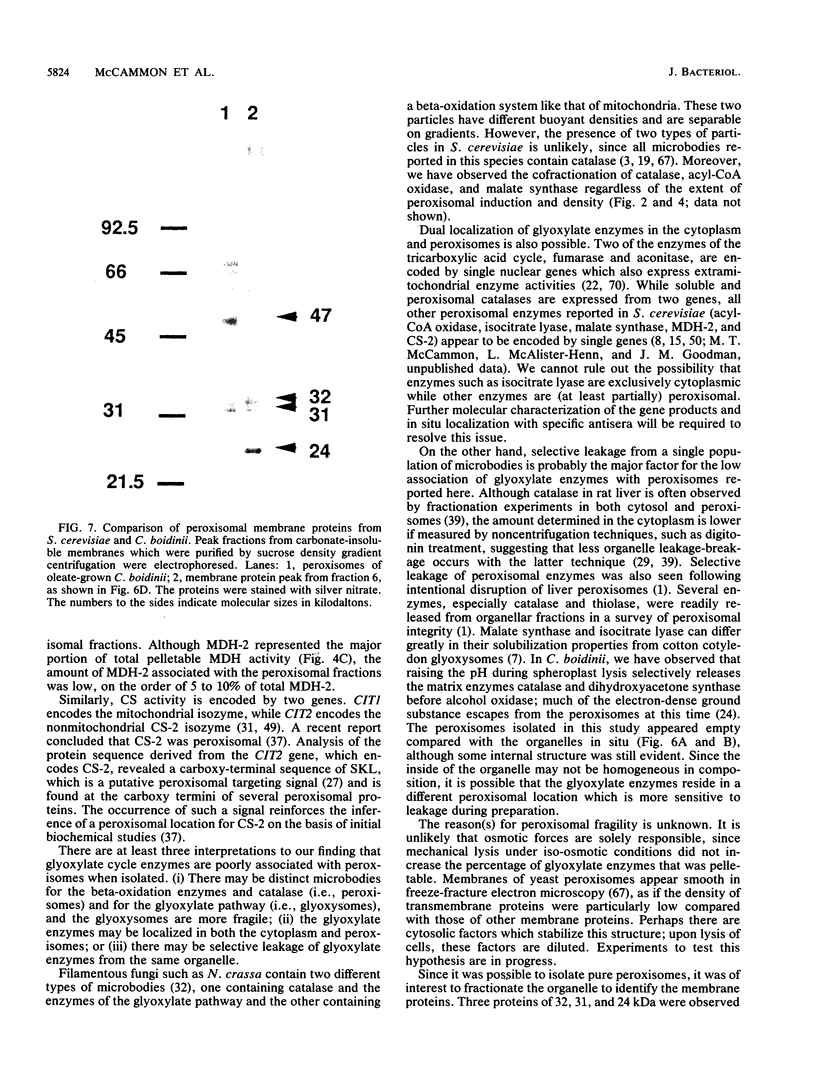
Although peroxisomes are difficult to identify in Saccharomyces cerevisiae under ordinary growth conditions, they proliferate when cells are cultured on oleic acid. We used this finding to study the protein composition of these organelles in detail. Peroxisomes from oleic acid-grown cells were purified on a discontinuous sucrose gradient; they migrated to the 46 to 50% (wt/wt) sucrose interface. The peroxisomal fraction was identified morphologically and by the presence of all of the enzymes of the peroxisomal beta-oxidation pathway. These organelles also contained a significant but minor fraction of two enzymes of the glyoxylate pathway, malate synthase and malate dehydrogenase-2. The localization of malate synthase in peroxisomes was confirmed by immunoelectron microscopy. It is postulated that glyoxylate pathway enzymes are readily and preferentially released from peroxisomes upon cell lysis, accounting for their incomplete recovery from isolated organelles. Small uninduced peroxisomes from glycerol-grown cultures were detected on sucrose gradients by marker enzymes. Under these conditions, catalase, acyl-coenzyme A oxidase, and malate synthase cofractionated at equilibrium close to the mitochondrial peak, indicating smaller, less dense organelles than those from cells grown on oleic acid. Peroxisomal membranes from oleate cultures were purified by buoyant density centrifugation. Three abundant proteins of 24, 31, and 32 kilodaltons were observed.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexson S. E., Fujiki Y., Shio H., Lazarow P. B. Partial disassembly of peroxisomes. J Cell Biol. 1985 Jul;101(1):294–304. doi: 10.1083/jcb.101.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa H., Takiguchi M., Amaya Y., Nagata S., Hayashi H., Mori M. cDNA-derived amino acid sequence of rat mitochondrial 3-oxoacyl-CoA thiolase with no transient presequence: structural relationship with peroxisomal isozyme. EMBO J. 1987 May;6(5):1361–1366. doi: 10.1002/j.1460-2075.1987.tb02376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avers C. J., Federman M. The occurrence in yeast of cytoplasmic granules which resemble microbodies. J Cell Biol. 1968 May;37(2):555–559. doi: 10.1083/jcb.37.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellion E., Goodman J. M. Proton ionophores prevent assembly of a peroxisomal protein. Cell. 1987 Jan 16;48(1):165–173. doi: 10.1016/0092-8674(87)90367-9. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chapman K. D., Turley R. B., Trelease R. N. Relationship between Cottonseed Malate Synthase Aggregation Behavior and Suborganellar Location in Glyoxysomes and Endoplasmic Reticulum. Plant Physiol. 1989 Jan;89(1):352–359. doi: 10.1104/pp.89.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriacy M. Isolation and characterization of yeast mutants defective in intermediary carbon metabolism and in carbon catabolite derepression. Mol Gen Genet. 1977 Jul 20;154(2):213–220. doi: 10.1007/BF00330840. [DOI] [PubMed] [Google Scholar]
- Cohen G., Rapatz W., Ruis H. Sequence of the Saccharomyces cerevisiae CTA1 gene and amino acid sequence of catalase A derived from it. Eur J Biochem. 1988 Sep 1;176(1):159–163. doi: 10.1111/j.1432-1033.1988.tb14263.x. [DOI] [PubMed] [Google Scholar]
- Crane D. I., Hemsley A. C., Masters C. J. Purification of peroxisomes from livers of normal and clofibrate-treated mice. Anal Biochem. 1985 Aug 1;148(2):436–445. doi: 10.1016/0003-2697(85)90250-7. [DOI] [PubMed] [Google Scholar]
- DIXON G. H., KORNBERG H. L., LUND P. Purification and properties of malate synthetase. Biochim Biophys Acta. 1960 Jul 1;41:217–233. doi: 10.1016/0006-3002(60)90004-4. [DOI] [PubMed] [Google Scholar]
- Desel H., Zimmermann R., Janes M., Miller F., Neupert W. Biosynthesis of glyoxysomal enzymes in Neurospora crassa. Ann N Y Acad Sci. 1982;386:377–393. doi: 10.1111/j.1749-6632.1982.tb21429.x. [DOI] [PubMed] [Google Scholar]
- Dmochowska A., Dignard D., Maleszka R., Thomas D. Y. Structure and transcriptional control of the Saccharomyces cerevisiae POX1 gene encoding acyl-coenzyme A oxidase. Gene. 1990 Apr 16;88(2):247–252. doi: 10.1016/0378-1119(90)90038-s. [DOI] [PubMed] [Google Scholar]
- Dommes V., Baumgart C., Kunau W. H. Degradation of unsaturated fatty acids in peroxisomes. Existence of a 2,4-dienoyl-CoA reductase pathway. J Biol Chem. 1981 Aug 25;256(16):8259–8262. [PubMed] [Google Scholar]
- Duntze W., Neumann D., Gancedo J. M., Atzpodien W., Holzer H. Studies on the regulation and localization of the glyoxylate cycle enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1969 Aug;10(1):83–89. doi: 10.1111/j.1432-1033.1969.tb00658.x. [DOI] [PubMed] [Google Scholar]
- Erdmann R., Veenhuis M., Mertens D., Kunau W. H. Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5419–5423. doi: 10.1073/pnas.86.14.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y., Fowler S., Shio H., Hubbard A. L., Lazarow P. B. Polypeptide and phospholipid composition of the membrane of rat liver peroxisomes: comparison with endoplasmic reticulum and mitochondrial membranes. J Cell Biol. 1982 Apr;93(1):103–110. doi: 10.1083/jcb.93.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S. P., Marguet D., Lauquin G. J. Molecular cloning of the yeast mitochondrial aconitase gene (ACO1) and evidence of a synergistic regulation of expression by glucose plus glutamate. Mol Cell Biol. 1990 Jul;10(7):3551–3561. doi: 10.1128/mcb.10.7.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrard L. J., Goodman J. M. Two genes encode the major membrane-associated protein of methanol-induced peroxisomes from Candida boidinii. J Biol Chem. 1989 Aug 15;264(23):13929–13937. [PubMed] [Google Scholar]
- Goodman J. M. Dihydroxyacetone synthase is an abundant constituent of the methanol-induced peroxisome of Candida boidinii. J Biol Chem. 1985 Jun 10;260(11):7108–7113. [PubMed] [Google Scholar]
- Goodman J. M., Scott C. W., Donahue P. N., Atherton J. P. Alcohol oxidase assembles post-translationally into the peroxisome of Candida boidinii. J Biol Chem. 1984 Jul 10;259(13):8485–8493. [PubMed] [Google Scholar]
- Gould S. J., Keller G. A., Subramani S. Identification of peroxisomal targeting signals located at the carboxy terminus of four peroxisomal proteins. J Cell Biol. 1988 Sep;107(3):897–905. doi: 10.1083/jcb.107.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes R. S., Masters C. J. Species specific features of the distribution and multiplicity of mammalian liver catalase. Arch Biochem Biophys. 1972 Jan;148(1):217–223. doi: 10.1016/0003-9861(72)90134-8. [DOI] [PubMed] [Google Scholar]
- Kamijo K., Taketani S., Yokota S., Osumi T., Hashimoto T. The 70-kDa peroxisomal membrane protein is a member of the Mdr (P-glycoprotein)-related ATP-binding protein superfamily. J Biol Chem. 1990 Mar 15;265(8):4534–4540. [PubMed] [Google Scholar]
- Kim K. S., Rosenkrantz M. S., Guarente L. Saccharomyces cerevisiae contains two functional citrate synthase genes. Mol Cell Biol. 1986 Jun;6(6):1936–1942. doi: 10.1128/mcb.6.6.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kionka C., Kunau W. H. Inducible beta-oxidation pathway in Neurospora crassa. J Bacteriol. 1985 Jan;161(1):153–157. doi: 10.1128/jb.161.1.153-157.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer R., Palmieri F. Molecular aspects of isolated and reconstituted carrier proteins from animal mitochondria. Biochim Biophys Acta. 1989 Apr 17;974(1):1–23. doi: 10.1016/s0005-2728(89)80160-4. [DOI] [PubMed] [Google Scholar]
- Labarca P., Wolff D., Soto U., Necochea C., Leighton F. Large cation-selective pores from rat liver peroxisomal membranes incorporated to planar lipid bilayers. J Membr Biol. 1986;94(3):285–291. doi: 10.1007/BF01869724. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lemmens M., Verheyden K., Van Veldhoven P., Vereecke J., Mannaerts G. P., Carmeliet E. Single-channel analysis of a large conductance channel in peroxisomes from rat liver. Biochim Biophys Acta. 1989 Sep 18;984(3):351–359. doi: 10.1016/0005-2736(89)90302-7. [DOI] [PubMed] [Google Scholar]
- Lewin A. S., Hines V., Small G. M. Citrate synthase encoded by the CIT2 gene of Saccharomyces cerevisiae is peroxisomal. Mol Cell Biol. 1990 Apr;10(4):1399–1405. doi: 10.1128/mcb.10.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister-Henn L., Thompson L. M. Isolation and expression of the gene encoding yeast mitochondrial malate dehydrogenase. J Bacteriol. 1987 Nov;169(11):5157–5166. doi: 10.1128/jb.169.11.5157-5166.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton B. 3-Ketoacyl-CoA thiolases of mammalian tissues. Methods Enzymol. 1975;35:128–136. doi: 10.1016/0076-6879(75)35148-3. [DOI] [PubMed] [Google Scholar]
- Nuttley W. M., Aitchison J. D., Rachubinski R. A. cDNA cloning and primary structure determination of the peroxisomal trifunctional enzyme hydratase-dehydrogenase-epimerase from the yeast Candida tropicalis pK233. Gene. 1988 Sep 30;69(2):171–180. doi: 10.1016/0378-1119(88)90428-3. [DOI] [PubMed] [Google Scholar]
- Nuttley W. M., Bodnar A. G., Mangroo D., Rachubinski R. A. Isolation and characterization of membranes from oleic acid-induced peroxisomes of Candida tropicalis. J Cell Sci. 1990 Mar;95(Pt 3):463–470. doi: 10.1242/jcs.95.3.463. [DOI] [PubMed] [Google Scholar]
- Okada H., Ueda M., Sugaya T., Atomi H., Mozaffar S., Hishida T., Teranishi Y., Okazaki K., Takechi T., Kamiryo T. Catalase gene of the yeast Candida tropicalis. Sequence analysis and comparison with peroxisomal and cytosolic catalases from other sources. Eur J Biochem. 1987 Dec 30;170(1-2):105–110. doi: 10.1111/j.1432-1033.1987.tb13673.x. [DOI] [PubMed] [Google Scholar]
- Okazaki K., Takechi T., Kambara N., Fukui S., Kubota I., Kamiryo T. Two acyl-coenzyme A oxidases in peroxisomes of the yeast Candida tropicalis: primary structures deduced from genomic DNA sequence. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1232–1236. doi: 10.1073/pnas.83.5.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperdoes F. R., Baudhuin P., Coppens I., De Roe C., Edwards S. W., Weijers P. J., Misset O. Purification, morphometric analysis, and characterization of the glycosomes (microbodies) of the protozoan hemoflagellate Trypanosoma brucei. J Cell Biol. 1984 Apr;98(4):1178–1184. doi: 10.1083/jcb.98.4.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish R. W. The isolation and characterization of peroxisomes (microbodies) from baker's yeast, Saccharomyces cerevisiae. Arch Microbiol. 1975 Nov 7;105(3):187–192. doi: 10.1007/BF00447136. [DOI] [PubMed] [Google Scholar]
- Perlman P. S., Mahler H. R. Intracellular localization of enzymes in yeast. Arch Biochem Biophys. 1970 Jan;136(1):245–259. doi: 10.1016/0003-9861(70)90348-6. [DOI] [PubMed] [Google Scholar]
- Rickey T. M., Lewin A. S. Extramitochondrial citrate synthase activity in bakers' yeast. Mol Cell Biol. 1986 Feb;6(2):488–493. doi: 10.1128/mcb.6.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenkrantz M., Alam T., Kim K. S., Clark B. J., Srere P. A., Guarente L. P. Mitochondrial and nonmitochondrial citrate synthases in Saccharomyces cerevisiae are encoded by distinct homologous genes. Mol Cell Biol. 1986 Dec;6(12):4509–4515. doi: 10.1128/mcb.6.12.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHATZ G., KLIMA J. TRIPHOSPHOPYRIDINE NUCLEOTIDE: CYTOCHROME C REDUCTASE OF SACCHAROMYCES CEREVISIAE: A "MICROSOMAL" ENZYME. Biochim Biophys Acta. 1964 Mar 9;81:448–461. doi: 10.1016/0926-6569(64)90130-0. [DOI] [PubMed] [Google Scholar]
- Schutgens R. B., Wanders R. J., Heymans H. S., Schram A. W., Tager J. M., Schrakamp G., van den Bosch H. Zellweger syndrome: biochemical procedures in diagnosis, prevention and treatment. J Inherit Metab Dis. 1987;10 (Suppl 1):33–45. doi: 10.1007/BF01812845. [DOI] [PubMed] [Google Scholar]
- Seah T. C., Bhatti A. R., Kaplan J. G. Novel catalatic proteins of bakers' yeast. I. An atypical catalase. Can J Biochem. 1973 Nov;51(11):1551–1555. doi: 10.1139/o73-208. [DOI] [PubMed] [Google Scholar]
- Skoneczny M., Chełstowska A., Rytka J. Study of the coinduction by fatty acids of catalase A and acyl-CoA oxidase in standard and mutant Saccharomyces cerevisiae strains. Eur J Biochem. 1988 Jun 1;174(2):297–302. doi: 10.1111/j.1432-1033.1988.tb14097.x. [DOI] [PubMed] [Google Scholar]
- Smith K., Sundaram T. K. Action of surfactants on porcine heart malate dehydrogenase isoenzymes and a simple method for the differential assay of these isoenzymes. Biochim Biophys Acta. 1986 Oct 29;884(1):109–118. doi: 10.1016/0304-4165(86)90233-3. [DOI] [PubMed] [Google Scholar]
- Steinman H. M., Hill R. L. Bovine liver crotonase (enoyl coenzyme A hydratase). EC 4.2.1.17 L-3-hydroxyacyl-CoA hydrolyase. Methods Enzymol. 1975;35:136–151. doi: 10.1016/0076-6879(75)35149-5. [DOI] [PubMed] [Google Scholar]
- Susani M., Zimniak P., Fessl F., Ruis H. Localization of catalase A in vacuoles of Saccharomyces cerevisiae: evidence for the vacuolar nature of isolated "yeast peroxisomes". Hoppe Seylers Z Physiol Chem. 1976 Jul;357(7):961–970. doi: 10.1515/bchm2.1976.357.2.961. [DOI] [PubMed] [Google Scholar]
- Tolbert N. E. Isolation of subcellular organelles of metabolism on isopycnic sucrose gradients. Methods Enzymol. 1974;31:734–746. doi: 10.1016/0076-6879(74)31077-4. [DOI] [PubMed] [Google Scholar]
- Tolbert N. E. Metabolic pathways in peroxisomes and glyoxysomes. Annu Rev Biochem. 1981;50:133–157. doi: 10.1146/annurev.bi.50.070181.001025. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Veldhoven P. P., Just W. W., Mannaerts G. P. Permeability of the peroxisomal membrane to cofactors of beta-oxidation. Evidence for the presence of a pore-forming protein. J Biol Chem. 1987 Mar 25;262(9):4310–4318. [PubMed] [Google Scholar]
- Veenhuis M., Mateblowski M., Kunau W. H., Harder W. Proliferation of microbodies in Saccharomyces cerevisiae. Yeast. 1987 Jun;3(2):77–84. doi: 10.1002/yea.320030204. [DOI] [PubMed] [Google Scholar]
- Wu M., Tzagoloff A. Mitochondrial and cytoplasmic fumarases in Saccharomyces cerevisiae are encoded by a single nuclear gene FUM1. J Biol Chem. 1987 Sep 5;262(25):12275–12282. [PubMed] [Google Scholar]
- van Dijken J. P., Otto R., Harder W. Growth of Hansenula polymorpha in a methanol-limited chemostat. Physiological responses due to the involvement of methanol oxidase as a key enzyme in methanol metabolism. Arch Microbiol. 1976 Dec 1;111(1-2):137–144. doi: 10.1007/BF00446560. [DOI] [PubMed] [Google Scholar]
- van der Klei I., Veenhuis M., van der Ley I., Harder W. Heterologous expression of alcohol oxidase in Saccharomyces cerevisiae: properties of the enzyme and implications for microbody development. FEMS Microbiol Lett. 1989 Jan 15;48(2):133–137. doi: 10.1111/j.1574-6968.1989.tb03287.x. [DOI] [PubMed] [Google Scholar]