Abstract
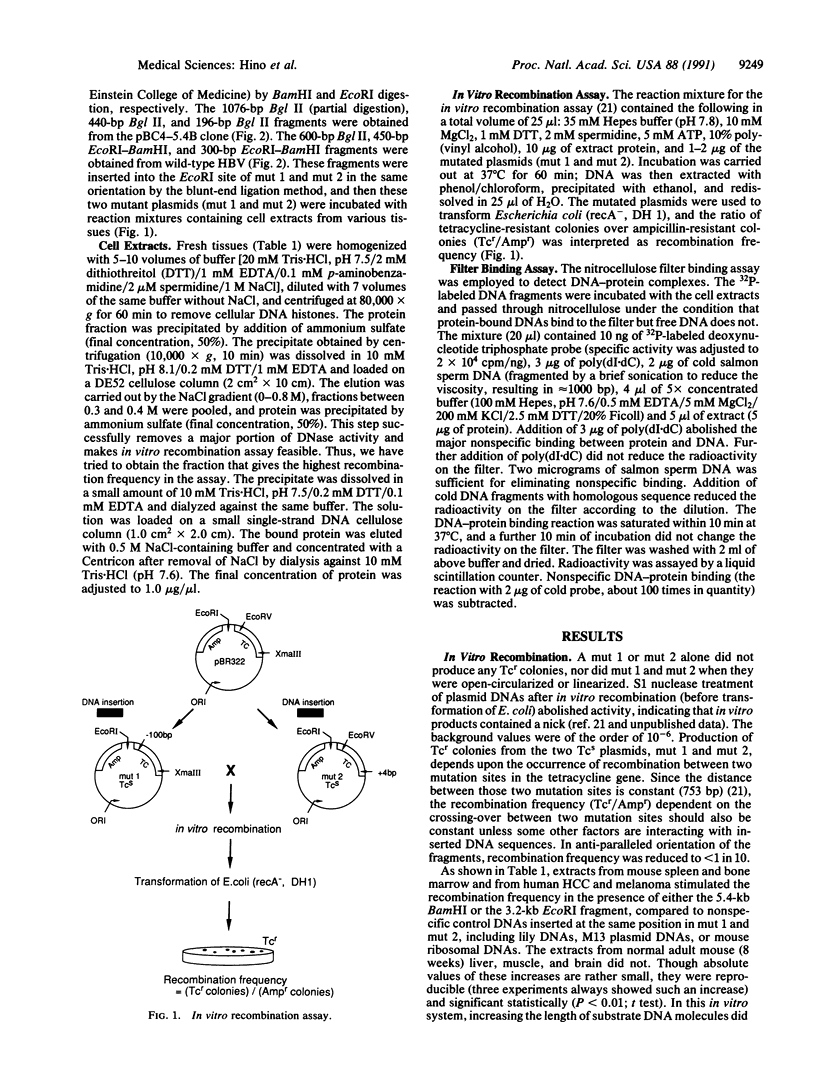
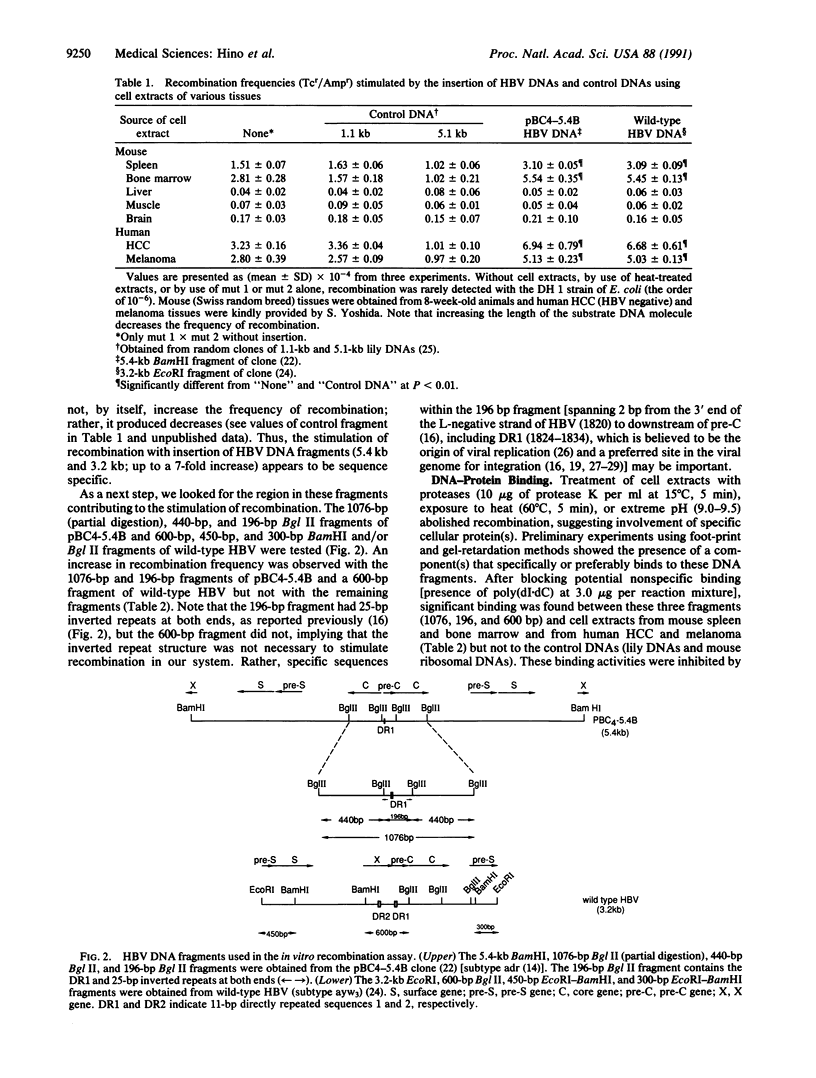
Chromosomal translocation, deletion, and inversion/duplication directly linked to hepatitis B virus (HBV) DNA integration occur frequently in host DNA of human hepatocellular carcinomas. To test the possible recombinogenic effect of HBV DNA, we have utilized an in vitro recombination assay. Fragments containing the region spanning DR1, which is believed to be the origin of viral replication and a preferred site in the viral genome for integration, increased the recombination events reproducibly in the presence of extracts from actively dividing cells (e.g., hepatocellular carcinoma) but not resting cells (e.g., normal liver). Moreover, in these extracts we have found a protein(s) that specifically binds to these HBV DNA fragments. These results support the notion that in some instances integrated HBV DNAs cause further genomic instability, possibly involving specific cellular protein(s). The fact that extracts from nondividing, normal liver did not increase recombination events suggests that genomic instability depends upon active cellular growth, a feature more commonly found subsequent to HBV-induced hepatocellular injury than in healthy liver. Our results offer an explanation for the high incidence of liver cancer that accompanies chronic hepatitis and add HBV to the list of agents that can cause genetic recombination.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brand N., Petkovich M., Krust A., Chambon P., de Thé H., Marchio A., Tiollais P., Dejean A. Identification of a second human retinoic acid receptor. Nature. 1988 Apr 28;332(6167):850–853. doi: 10.1038/332850a0. [DOI] [PubMed] [Google Scholar]
- Buetow K. H., Murray J. C., Israel J. L., London W. T., Smith M., Kew M., Blanquet V., Brechot C., Redeker A., Govindarajah S. Loss of heterozygosity suggests tumor suppressor gene responsible for primary hepatocellular carcinoma. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8852–8856. doi: 10.1073/pnas.86.22.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisari F. V., Klopchin K., Moriyama T., Pasquinelli C., Dunsford H. A., Sell S., Pinkert C. A., Brinster R. L., Palmiter R. D. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell. 1989 Dec 22;59(6):1145–1156. doi: 10.1016/0092-8674(89)90770-8. [DOI] [PubMed] [Google Scholar]
- Dejean A., Sonigo P., Wain-Hobson S., Tiollais P. Specific hepatitis B virus integration in hepatocellular carcinoma DNA through a viral 11-base-pair direct repeat. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5350–5354. doi: 10.1073/pnas.81.17.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori M., Tokino T., Hino O., Kitagawa T., Imamura T., Okamoto E., Mitsunobu M., Ishikawa T., Nakagama H., Harada H. Allelotype study of primary hepatocellular carcinoma. Cancer Res. 1991 Jan 1;51(1):89–93. [PubMed] [Google Scholar]
- Henderson A. S., Ripley S., Hino O., Rogler C. E. Identification of a chromosomal aberration associated with a hepatitis B DNA integration site in human cells. Cancer Genet Cytogenet. 1988 Feb;30(2):269–275. doi: 10.1016/0165-4608(88)90194-x. [DOI] [PubMed] [Google Scholar]
- Higashitani A., Tabata S., Ogawa T., Ogawa H., Shibata M., Hotta Y. ATP-independent strand transfer protein from murine spermatocytes, spermatids, and spermatozoa. Exp Cell Res. 1990 Feb;186(2):317–323. doi: 10.1016/0014-4827(90)90311-w. [DOI] [PubMed] [Google Scholar]
- Hino O., Nomura K., Ohtake K., Kawaguchi T., Sugano H., Kitagawa T. Instability of integrated hepatitis B virus DNA with inverted repeat structure in a transgenic mouse. Cancer Genet Cytogenet. 1989 Feb;37(2):273–278. doi: 10.1016/0165-4608(89)90059-9. [DOI] [PubMed] [Google Scholar]
- Hino O., Ohtake K., Rogler C. E. Features of two hepatitis B virus (HBV) DNA integrations suggest mechanisms of HBV integration. J Virol. 1989 Jun;63(6):2638–2643. doi: 10.1128/jvi.63.6.2638-2643.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino O., Shows T. B., Rogler C. E. Hepatitis B virus integration site in hepatocellular carcinoma at chromosome 17;18 translocation. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8338–8342. doi: 10.1073/pnas.83.21.8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta Y., Tabata S., Bouchard R. A., Piñon R., Stern H. General recombination mechanisms in extracts of meiotic cells. Chromosoma. 1985;93(2):140–151. doi: 10.1007/BF00293161. [DOI] [PubMed] [Google Scholar]
- Hotta Y., Tabata S., Stubbs L., Stern H. Meiosis-specific transcripts of a DNA component replicated during chromosome pairing: homology across the phylogenetic spectrum. Cell. 1985 Apr;40(4):785–793. doi: 10.1016/0092-8674(85)90338-1. [DOI] [PubMed] [Google Scholar]
- Kim C. M., Koike K., Saito I., Miyamura T., Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991 May 23;351(6324):317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- Knudson A. G., Jr Genetics of human cancer. Annu Rev Genet. 1986;20:231–251. doi: 10.1146/annurev.ge.20.120186.001311. [DOI] [PubMed] [Google Scholar]
- Li Y., Togashi Y., Sato S., Emoto T., Kang J. H., Takeichi N., Kobayashi H., Kojima Y., Une Y., Uchino J. Spontaneous hepatic copper accumulation in Long-Evans Cinnamon rats with hereditary hepatitis. A model of Wilson's disease. J Clin Invest. 1991 May;87(5):1858–1861. doi: 10.1172/JCI115208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizusawa H., Taira M., Yaginuma K., Kobayashi M., Yoshida E., Koike K. Inversely repeating integrated hepatitis B virus DNA and cellular flanking sequences in the human hepatoma-derived cell line huSP. Proc Natl Acad Sci U S A. 1985 Jan;82(1):208–212. doi: 10.1073/pnas.82.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. A., Kitagawa T. Hepatocarcinogenesis in the rat: the effect of promoters and carcinogens in vivo and in vitro. Int Rev Cytol. 1986;101:125–173. doi: 10.1016/s0074-7696(08)60248-x. [DOI] [PubMed] [Google Scholar]
- Nagaya T., Nakamura T., Tokino T., Tsurimoto T., Imai M., Mayumi T., Kamino K., Yamamura K., Matsubara K. The mode of hepatitis B virus DNA integration in chromosomes of human hepatocellular carcinoma. Genes Dev. 1987 Oct;1(8):773–782. doi: 10.1101/gad.1.8.773. [DOI] [PubMed] [Google Scholar]
- Pasek M., Goto T., Gilbert W., Zink B., Schaller H., MacKay P., Leadbetter G., Murray K. Hepatitis B virus genes and their expression in E. coli. Nature. 1979 Dec 6;282(5739):575–579. doi: 10.1038/282575a0. [DOI] [PubMed] [Google Scholar]
- Preston-Martin S., Pike M. C., Ross R. K., Jones P. A., Henderson B. E. Increased cell division as a cause of human cancer. Cancer Res. 1990 Dec 1;50(23):7415–7421. [PubMed] [Google Scholar]
- Rogler C. E., Sherman M., Su C. Y., Shafritz D. A., Summers J., Shows T. B., Henderson A., Kew M. Deletion in chromosome 11p associated with a hepatitis B integration site in hepatocellular carcinoma. Science. 1985 Oct 18;230(4723):319–322. doi: 10.1126/science.2996131. [DOI] [PubMed] [Google Scholar]
- Seeger C., Ganem D., Varmus H. E. Biochemical and genetic evidence for the hepatitis B virus replication strategy. Science. 1986 Apr 25;232(4749):477–484. doi: 10.1126/science.3961490. [DOI] [PubMed] [Google Scholar]
- Shih C., Burke K., Chou M. J., Zeldis J. B., Yang C. S., Lee C. S., Isselbacher K. J., Wands J. R., Goodman H. M. Tight clustering of human hepatitis B virus integration sites in hepatomas near a triple-stranded region. J Virol. 1987 Nov;61(11):3491–3498. doi: 10.1128/jvi.61.11.3491-3498.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slagle B. L., Zhou Y. Z., Butel J. S. Hepatitis B virus integration event in human chromosome 17p near the p53 gene identifies the region of the chromosome commonly deleted in virus-positive hepatocellular carcinomas. Cancer Res. 1991 Jan 1;51(1):49–54. [PubMed] [Google Scholar]
- Tiollais P., Buendia M. A. Hepatitis B virus. Sci Am. 1991 Apr;264(4):116–123. doi: 10.1038/scientificamerican0491-116. [DOI] [PubMed] [Google Scholar]
- Tokino T., Fukushige S., Nakamura T., Nagaya T., Murotsu T., Shiga K., Aoki N., Matsubara K. Chromosomal translocation and inverted duplication associated with integrated hepatitis B virus in hepatocellular carcinomas. J Virol. 1987 Dec;61(12):3848–3854. doi: 10.1128/jvi.61.12.3848-3854.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda H., Zhang W. D., Shimosato Y., Yokota J., Terada M., Sugimura T., Miyamura T., Hirohashi S. Allele loss on chromosome 16 associated with progression of human hepatocellular carcinoma. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6791–6794. doi: 10.1073/pnas.87.17.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano Y., Watanabe K., Lin C. C., Hino O., Tamaoki T. Interstitial chromosomal deletion within 4q11-q13 in a human hepatoma cell line. Cancer Res. 1991 Mar 1;51(5):1460–1464. [PubMed] [Google Scholar]
- Wang H. P., Rogler C. E. Deletions in human chromosome arms 11p and 13q in primary hepatocellular carcinomas. Cytogenet Cell Genet. 1988;48(2):72–78. doi: 10.1159/000132593. [DOI] [PubMed] [Google Scholar]
- Wang H. P., Rogler C. E. Topoisomerase I-mediated integration of hepadnavirus DNA in vitro. J Virol. 1991 May;65(5):2381–2392. doi: 10.1128/jvi.65.5.2381-2392.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Chenivesse X., Henglein B., Bréchot C. Hepatitis B virus integration in a cyclin A gene in a hepatocellular carcinoma. Nature. 1990 Feb 8;343(6258):555–557. doi: 10.1038/343555a0. [DOI] [PubMed] [Google Scholar]


