Abstract
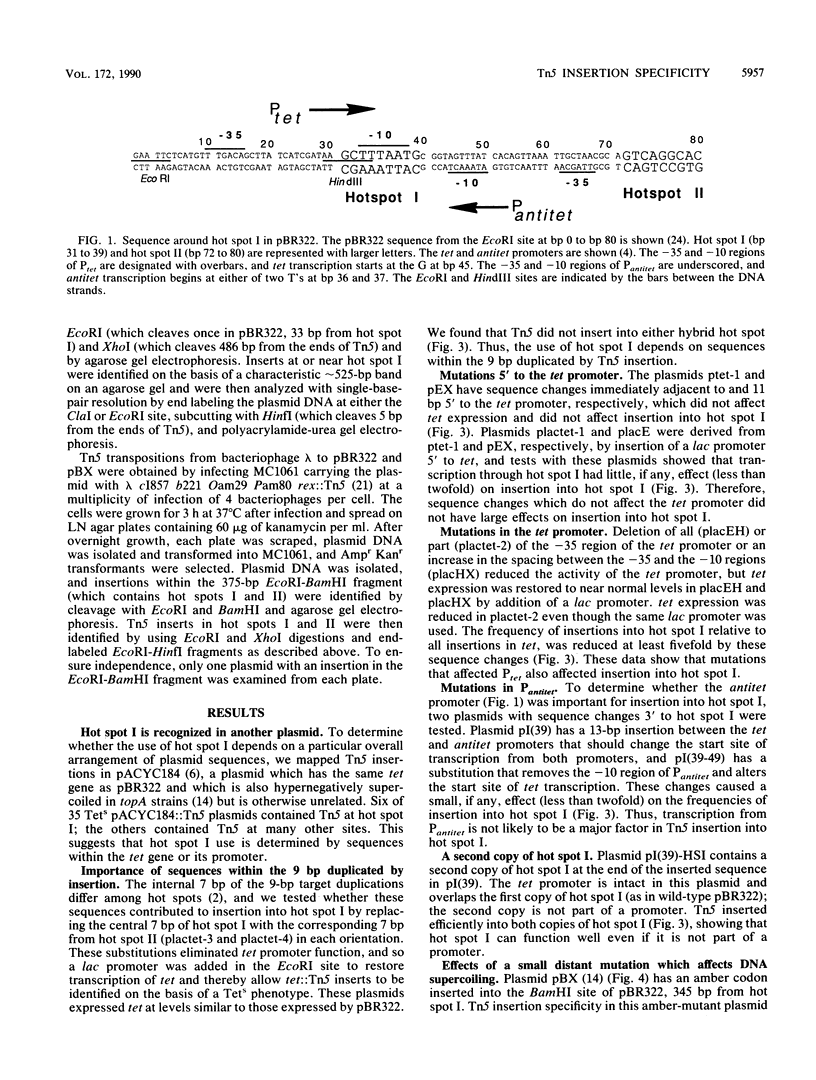
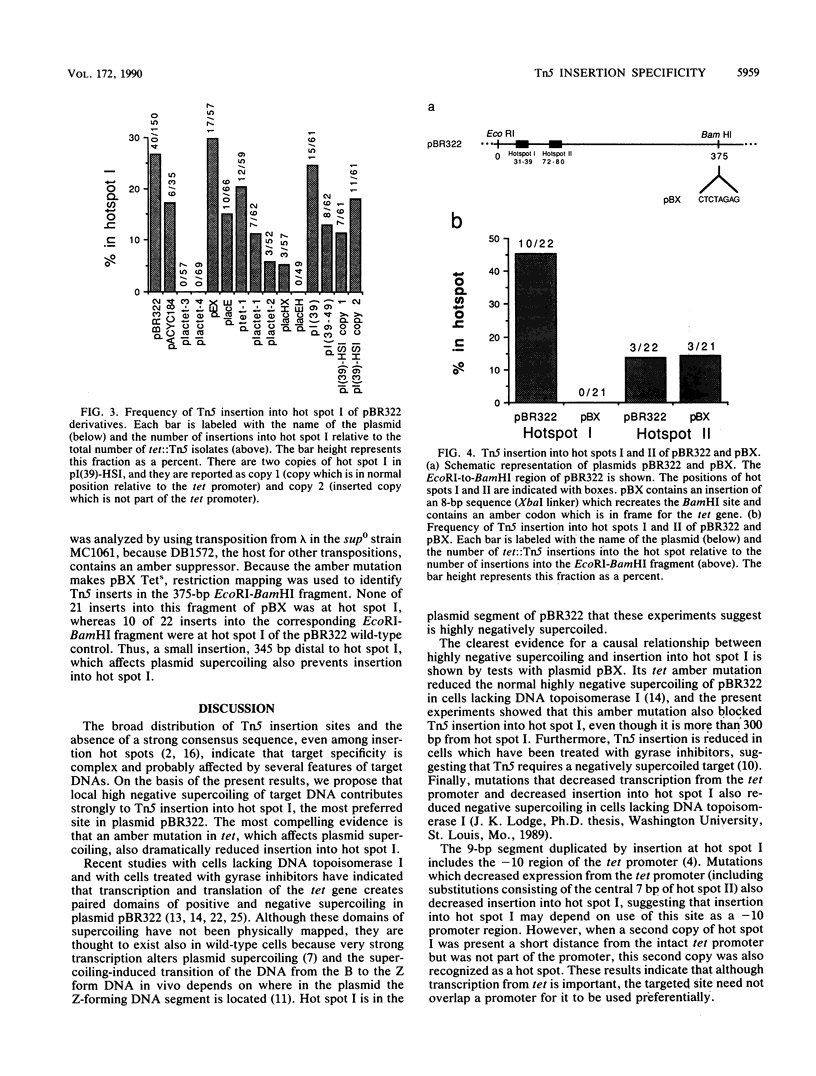
The major hot spot of transposon Tn5 insertion in plasmid pBR322 (hot spot I) is in the promoter for the tetracycline resistance gene (tet). We made a series of pBR322 derivatives with mutations in and around this promoter and assayed their effects on insertion of Tn5 into hot spot I. Those mutations which reduced transcription from the tet promoter also reduced the frequency of insertion into hot spot I. Transcription and translation of tet are thought to cause the formation of paired domains of negative and positive supercoiling in pBR322. An amber codon in tet, 345 base pairs from hot spot I, decreases the negative supercoiling of the DNA segment containing hot spot I because it terminates translation of tet prematurely. We report here that this amber mutation also reduces insertion into hot spot I. These results suggest that the ability of Tn5 to insert into its major hot spot in pBR322 depends directly on negative supercoiling of the target DNA.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balbás P., Soberón X., Merino E., Zurita M., Lomeli H., Valle F., Flores N., Bolivar F. Plasmid vector pBR322 and its special-purpose derivatives--a review. Gene. 1986;50(1-3):3–40. doi: 10.1016/0378-1119(86)90307-0. [DOI] [PubMed] [Google Scholar]
- Berg D. E., Schmandt M. A., Lowe J. B. Specificity of transposon Tn5 insertion. Genetics. 1983 Dec;105(4):813–828. doi: 10.1093/genetics/105.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogosian G., Kane J. F. Nucleotide sequence of the EcoRI fragment from pLJ3 bearing two tandem lacUV5 promoters. Nucleic Acids Res. 1987 Sep 11;15(17):7185–7185. doi: 10.1093/nar/15.17.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Cate R. L., Perlmutter A. P. Precise location of two promoters for the beta-lactamase gene of pBR322. S1 mapping of ribonucleic acid isolated from Escherichia coli or synthesized in vitro. J Biol Chem. 1982 Aug 10;257(15):9205–9210. [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa N., Bossi L. Transcription induces gyration of the DNA template in Escherichia coli. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9416–9420. doi: 10.1073/pnas.85.24.9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas P., Chandler M. G., Prentki P., Galas D. J. Escherichia coli integration host factor binds specifically to the ends of the insertion sequence IS1 and to its major insertion hot-spot in pBR322. J Mol Biol. 1987 May 20;195(2):261–272. doi: 10.1016/0022-2836(87)90648-6. [DOI] [PubMed] [Google Scholar]
- Halling S. M., Kleckner N. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell. 1982 Jan;28(1):155–163. doi: 10.1016/0092-8674(82)90385-3. [DOI] [PubMed] [Google Scholar]
- Isberg R. R., Syvanen M. DNA gyrase is a host factor required for transposition of Tn5. Cell. 1982 Aug;30(1):9–18. doi: 10.1016/0092-8674(82)90006-x. [DOI] [PubMed] [Google Scholar]
- Jaworski A., Blaho J. A., Larson J. E., Shimizu M., Wells R. D. Tetracycline promoter mutations decrease non-B DNA structural transitions, negative linking differences and deletions in recombinant plasmids in Escherichia coli. J Mol Biol. 1989 Jun 5;207(3):513–526. doi: 10.1016/0022-2836(89)90461-0. [DOI] [PubMed] [Google Scholar]
- Klaer R., Kühn S., Fritz H. J., Tillmann E., Saint-Girons I., Habermann P., Pfeifer D., Starlinger P. Studies on transposition mechanisms and specificity of IS4. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):215–224. doi: 10.1101/sqb.1981.045.01.034. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge J. K., Kazic T., Berg D. E. Formation of supercoiling domains in plasmid pBR322. J Bacteriol. 1989 Apr;171(4):2181–2187. doi: 10.1128/jb.171.4.2181-2187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge J. K., Weston-Hafer K., Berg D. E. Transposon Tn5 target specificity: preference for insertion at G/C pairs. Genetics. 1988 Nov;120(3):645–650. doi: 10.1093/genetics/120.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandecki W. Oligonucleotide-directed double-strand break repair in plasmids of Escherichia coli: a method for site-specific mutagenesis. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7177–7181. doi: 10.1073/pnas.83.19.7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKinnon R. D., Waye J. S., Bautista D. S., Graham F. L. Nonrandom insertion of Tn5 into cloned human adenovirus DNA. Gene. 1985;40(1):31–38. doi: 10.1016/0378-1119(85)90021-6. [DOI] [PubMed] [Google Scholar]
- Noll M. Internal structure of the nucleosome: DNA folding in the conserved 140-base-pair core particle. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):77–85. doi: 10.1101/sqb.1978.042.01.009. [DOI] [PubMed] [Google Scholar]
- Phadnis S. H., Berg D. E. Identification of base pairs in the outside end of insertion sequence IS50 that are needed for IS50 and Tn5 transposition. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9118–9122. doi: 10.1073/pnas.84.24.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss G. J., Drlica K. Topoisomerase I mutants: the gene on pBR322 that encodes resistance to tetracycline affects plasmid DNA supercoiling. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8952–8956. doi: 10.1073/pnas.83.23.8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Y., Shyy S. H., Wang J. C., Liu L. F. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988 May 6;53(3):433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]


