SUMMARY
The suprachiasmatic nucleus (SCN) is the central circadian clock in mammals. It is entrained by light, but resistant to temperature shifts that entrain peripheral clocks [1–5]. The SCN expresses many functionally important neuropeptides including vasoactive intestinal peptide (VIP), which drives light entrainment, synchrony, and amplitude of SCN cellular clocks, and organizes circadian behavior [5–16]. The transcription factor LHX1 drives SCN Vip expression, and cellular desynchrony in Lhx1-deficient SCN largely results from Vip loss [17,18]. LHX1 regulates many genes other than Vip, yet activity rhythms in Lhx1-deficient mice are similar to Vip−/− mice under light-dark cycles, and only somewhat worse in constant conditions. We suspected that LHX1 targets other than Vip have circadian functions overlooked in previous studies. In this study, we compared circadian sleep and temperature rhythms of Lhx1 and Vip-deficient mice, and found loss of acute light control of sleep in Lhx1 but not Vip mutants. We also found loss of circadian resistance to fever in Lhx1 but not Vip mice, which was partially recapitulated by heat application to cultured Lhx1-deficient SCN. Having identified VIP-independent functions of LHX1, we mapped the VIP-independent transcriptional network downstream of LHX1, and a largely separable VIP-dependent transcriptional network. The VIP-independent network does not affect core clock amplitude and synchrony, unlike the VIP-dependent network. . These studies identify Lhx1 as the first gene required for temperature resistance of the SCN clockworks, and demonstrate that acute light control of sleep is routed through the SCN and its immediate output regions.
Results and Discussion
Lhx1 in the SCN/vSPZ Mediates Light and Circadian Control of Sleep/Wake
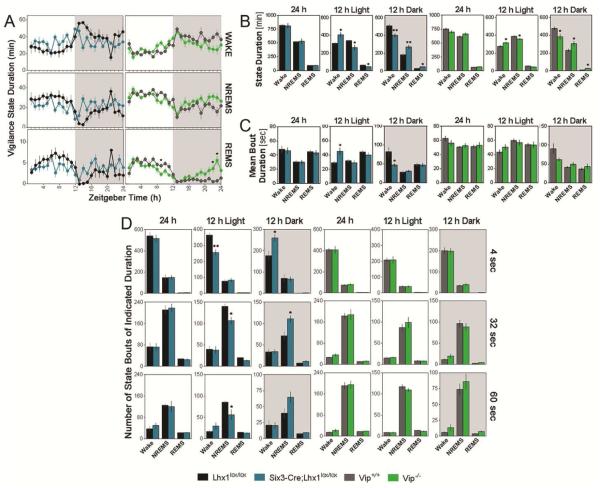
Rodents show three distinct vigilance states detectable by electroencephalography and electromyography (EEG/EMG): wake, NREMS (non-rapid-eye movement sleep), and REMS (rapid-eye movement sleep). Six3-Cre;Lhx1lox/lox sleep/wake distribution in 12:12 LD lacks significant temporal regulation of wake, NREMS, or REMS (p=0.17, 0.16, 0.24; Figure 1A; Supplemental Data Set). Six3-Cre;Lhx1lox/lox waking is higher during the day and lower at night than controls, and NREMS and REMS show the opposite pattern (p<0.05 or p<0.01); time in each state over the full 24-hour cycle is unchanged (Figure 1B). However, this does not reflect a flat distribution of vigilance; lack of circadian and light-dependent regulation of sleep in Lhx1 mutants unmasks rapid oscillations of sleep/wake state, which coalesce into ultradian rhythms in some animals (2/5 mice with one or more p<0.05 ultradian rhythm by JTK-Cycle; Figure S1A; Supplemental Data Set). Parameters of sleep consolidation were also altered in Six3-Cre;Lhx1lox/lox mice in LD. Wake bout duration was longer during daytime and shorter at night in Lhx1 mutants compared to controls (p<0.05), though day and night bout duration of REMS and NREMS were unchanged. The full 24-hour cycle bout duration of all states were similar in Six3-Cre;Lhx1lox/lox and control mice (Figure 1C). Lhx1 mutants also had fewer short (4 sec) wake bouts during daytime (p<0.01) and more at night (p<0.05) compared to controls, and had fewer longer 32 and 60 second bouts of NREMS during daytime, and more 32 second bouts at night (p<0.05; Figure 1D). Importantly, these changes in sleep structure (especially selective lengthening of mean wake bout duration due to loss of very short, abortive wake bouts during daytime, and cases of significant ultradian rhythmicity unmasked by profound lack of all sources of higher-order temporal structure) support profound loss of both circadian and light control of sleep in Lhx1-deficient mice. Unsurprisingly, sleep/wake was also profoundly disorganized in Six3-Cre;Lhx1lox/lox mice in DD (Figure S1B).
Figure 1. Lhx1 Regulates Both Light and Circadian Control of Sleep Timing.
(A) Line graphs showing vigilance state duration in Lhx1lox/lox (black) and Six3-Cre;Lhx1lox/lox (blue) mice (left), and Vip+/+ (gray) and Vip−/− (green) mice (right) across a 24-hour cycle in 12:12 LD (n=5,5,8,8; mean +/− SEM). Significance of temporal regulation of sleep/wake within each population was assessed by Greenhouse-Geisser corrected ANOVA. Significance of individual animals’ ultradian rhythmicity was assessed by Benjamini-Hochberg corrected JTK-Cycle analyses of each vigilance state (n=72). See Supplemental Data Set for details.
(B) Bar graphs showing vigilance state duration across the full 24-hour cycle (left), day (middle) and night (right), in Lhx1lox/lox (black) and Six3-Cre;Lhx1lox/lox (blue) mice, and Vip+/+ (gray) and Vip−/− (green) mice (n=5,5,8,8; two-tailed t-tests; *p<0.05; **p<0.01; mean +/− SEM).
(C) Bar graphs showing vigilance bout duration across the full 24-hour cycle (left), day (middle) and night (right), in Lhx1lox/lox (black) and Six3-Cre;Lhx1lox/lox (blue) mice, and Vip+/+ (gray) and Vip−/− (green) mice (n=5,5,8,8; two-tailed t-tests; *p<0.05; mean +/− SEM).
(D) Bar graphs showing the number of short (top), intermediate (middle), or long (bottom) vigilance state bouts across the full 24-hour cycle (left), day (middle) and night (right), in Lhx1lox/lox (black) and Six3-Cre;Lhx1lox/lox (blue) mice, and Vip+/+ (gray) and Vip−/− (green) mice (n=5,5,8,8; two-tailed t-tests; *p<0.05; **p<0.01; mean +/− SEM).
See also Figure S1 and Supplemental Data Set.
Vip−/− mice reportedly retain damped rhythms in wake, NREMS, and REMS under LD, in marked contrast to Lhx1 mutants [19]. We repeated this study, and confirmed that Vip loss alone cannot explain Six3Cre;Lhx1lox/lox LD sleep/wake phenotypes. Vip−/− mice had milder day-night changes in state duration relative to controls that, unlike in Lhx1 mutants, left Vip−/− mice with significant temporal regulation of vigilance (p<0.01 for REMS; genotype/time interaction p>0.05 obviated genotype-specific post-analysis for wake and NREMS; Figure 1A-B; Supplemental Data Set). Vip−/− mice also lacked differences in mean bout duration and number of bouts of any length relative to controls (Figure 1C-D), and no Vip−/− mice exhibited significant ultradian rhythms (Supplemental Data Set). Our sleep timing results were similar to those previously reported; however, we observed no significant loss of daytime REMS in Vip−/− mice, leading to increased instead of decreased total REMS in our Vip−/− mice compared to controls (p<0.05; Figure 1B) [19] .
SCN-lesioned animals in LD more closely mirror the sleep/wake phenotype of Six3-Cre;Lhx1lox/lox mice, exhibiting profound loss of all temporal control over the 24-hour cycle that unmasks ultradian rhythms [20–22]. However, retinohypothalamic tract damage in these lesion studies left unanswered to what extent their sleep/wake phenotypes reflected lost retinal input. Retinal input is intact in Six3-Cre;Lhx1lox/lox SCN [17]. Moreover, pupillary light reflexes are preserved in Six3-Cre;Lhx1lox/lox mice, indicating that retinal ipRGCs (which innervate both the SCN and pupil-controlling OPN) are functional [17]. However, ventral subparaventricular zone (vSPZ) Lhx1 expression is also lost in Six3-Cre;Lhx1lox/lox mice. vSPZ preferentially conveys circadian information about sleep/wake cycles (likely contributing to worse sleep/wake disruption in Six3-Cre;Lhx1lox/lox mutants compared to other rhythmic behaviors) and can cause ultradian sleep phenotypes when lesioned selectively [17,23]. Thus, Lhx1 loss in both the SCN and vSPZ may contribute to Six3-Cre;Lhx1lox/lox sleep phenotypes.
Our data show that the SCN/vSPZ complex is essential for direct induction of sleep by light. Comparatively worse impairment of Six3-Cre;Lhx1lox/lox sleep/wake rhythms compared to their wheel-running behavior support findings that light-dependent masking of activity and sleep/wake rely upon at least partially non-overlapping circuitry[17,24] . Importantly, our results extend to temporal organization of sleep findings by others that SCN-relayed light inputs can clock-independently synchronize peripheral oscillators [25,26], and provide important clues to the SCN circuitry underlying light control of this process.
Lhx1 in the SCN Mediates Resistance of Circadian Behavior to Temperature
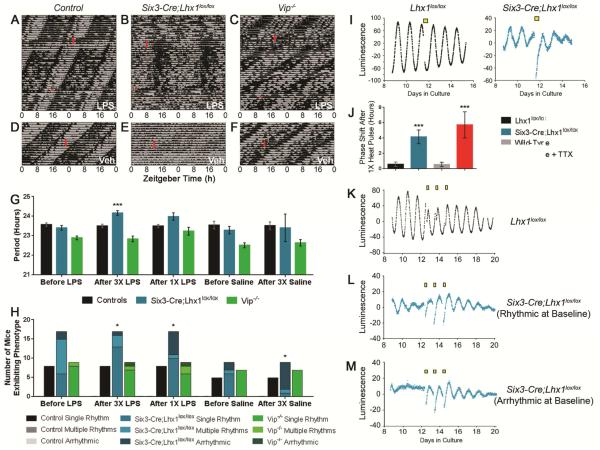
SCN network properties are essential for SCN resistance to thermal perturbation [2]. Since Lhx1 regulates SCN signaling so broadly, we hypothesized that Six3-Cre;Lhx1lox/lox circadian behavior could be modified by thermal cues. To test this, we induced fever by injecting lipopolysaccharide (LPS) while recording core body temperature (CBT) rhythms by telemetry. Three daily injections (3X) of LPS consolidated CBT rhythms of Six3-Cre;Lhx1lox/lox mice (Figure 2B), shifting the distribution of mutants with single, multiple, or no rhythms significantly toward single rhythms (hereafter referred to as consolidation) (p<0.05; Figure 2H; Supplemental Data Set). Mean period also lengthened to approximate the injection interval of 24 hours (p<0.001 vs. baseline) (Figure 2G). These effects were durable, persisting up to three weeks. After a single (1X) LPS pulse, period became indistinguishable from baseline (p>0.05), but consolidation was unchanged from 3X LPS (p<0.01 vs. baseline; p=0.11 vs. 3X LPS) (Figure 2G-H). 1X LPS thus “reset” (reverted to baseline) period but not consolidation phenotypes induced by 3X LPS. Wheel-running behavior after LPS largely recapitulated period effects in Six3-Cre;Lhx1lox/lox mice, demonstrating behavioral multimodality of LPS circadian effects (p<0.01 baseline vs 3X LPS but not 1X LPS; Figure S2A-F). However, Six3-Cre;Lhx1lox/lox wheel-running was not consolidated by LPS, likely reflecting interference by the consolidating effects of running wheels [27].
Figure 2. Six3-Cre;Lhx1lox/lox Circadian Behavior and SCN Clock Are Susceptible to Heat.
(A-C) Sample CBT recordings from control (A), Six3-Cre;Lhx1lox/lox (B), and Vip−/− (C) mice in DD, before and after 3X (long red rectangle), and 1X (red square) LPS injections.
(D-E) Sample CBT recordings from control (D), Six3-Cre;Lhx1lox/lox (E), and Vip−/− (F) mice in DD, before and after 3X saline injections (red rectangle).
(G) Bar graph showing period of control (black), Six3-Cre;Lhx1lox/lox (blue), and Vip−/− (green) mice before and after 3X LPS, 1X LPS, and 3X saline (Supplemental Data Set; two-way ANOVA with Tukey post-hoc tests; ***p<0.001; mean +/− SEM; only within-genotype comparisons to baseline are shown).
(H) Bar graphs showing the number of control (black), Six3-Cre;Lhx1lox/lox (blue), and Vip−/− (green) mice that had singly rhythmic, multiply rhythmic, or arrhythmic CBTs before and after 3X LPS, 1X LPS, and 3X saline (Supplemental Data Set; 2x3 Fisher’s exact test; *p<0.05; only within-genotype comparisons to baseline are shown).
(I) Background-subtracted Per2::Luc bioluminescence traces from Lhx1lox/lox (black) or Six3-Cre;Lhx1lox/lox (blue) SCN, before and after a single 3-hour 38.5°C heat pulse (yellow square).
(J) Phase shifts of Per2::Luc rhythms after a single 3-hour 38.5°C heat pulse in Lhx1lox/lox (black), Six3-Cre;Lhx1lox/lox (blue), wild type (gray), and wild type + 1mM TTX (red) SCN (n=7,4,7,6; one way ANOVA with Tukey post-hoc; ***p<0.001; mean +/− SEM).
(K-M) Background-subtracted Per2::Luc bioluminescence traces of Lhx1lox/lox (black, K), rhythmic Six3-Cre;Lhx1lox/lox (blue, L), or arrhythmic Six3-Cre;Lhx1lox/lox (blue, M) SCNs. SCNs received 3X 3 hour, 38.5°C heat pulses (yellow squares). This treatment merely phase-shifted Six3-Cre;Lhx1lox/lox slices rhythmic at baseline (L), but was able to induce rhythms in Six3-Cre;Lhx1lox/lox slices arrhythmic at baseline (M).
See also Figure S2 and Supplemental Data Set.
3X saline increased the number of Six3-Cre;Lhx1lox/lox mice with arrhythmic CBTs (p<0.05; Figure 2H; Supplemental Data Set), with no effect on period (p>0.05; Figure 2G). Thus, the effects of 3X LPS in Lhx1 mutants are unrelated to vehicle. Period and consolidation were unaffected by LPS or saline in control mice (p>0.05; Figure 2A,D,G-H). Vip−/− mice lacked changes in population consolidation or period after LPS or saline (p>0.05; Figure 2C,F-H), though individuals show period scatter, especially after 1X LPS (Figure S2I). This mirrors phase scatter in wild-type SCNs subjected to both Vipr2 antagonist and a heat pulse [2].
To assess the likelihood that fever drives LPS effects on Lhx1 mutant behavior, we subjected Six3-Cre;Lhx1lox/lox;Per2:Luc/+ untreated, Per2:Luc/+ tetrodotoxin (TTX)-treated, Lhx1lox/lox;Per2:Luc/+ untreated, and Per2:Luc/+ untreated SCN slice cultures to 1X heat pulse. Lhx1-deficient SCNs lacked resistance to this thermal resetting stimulus, reflected in phase shifts much larger than untreated controls and identical to SCN slices in TTX (p<0.001 vs. untreated, p>0.05 vs. TTX; Figure 2I-J). Poor Six3-Cre;Lhx1lox/lox baseline behavioral rhythms forbade phase shift analysis in vivo, but phase resetting in culture recalls the “resetting” of period and consolidation phenotypes in Lhx1 mutants after 1X LPS.
We then tested whether Six3-Cre;Lhx1lox/lox SCN Per2:Luc rhythms could be entrained by 3X heat pulses. Control slices never showed amplitude or period changes after 3X heat (Figure 2K). 3X heat only very weakly tended to lengthen Six3-Cre;Lhx1lox/lox; Per2:Luc/+ period, perhaps due to plateauing of their already long baseline period compared to controls (p<0.05; Figure S2L). Lhx1-deficient slices rhythmic at baseline also lacked amplitude changes, though they did phase reset to 3X heat (Figures 2L and S2M; 4/7 slices; p<0.001). However, 3X heat induced rhythms in all Six3-Cre; Lhx1lox/lox;Per2:Luc/+ SCNs arrhythmic before treatment (indicated by Fourier transform in 3/7 slices; Figure 2M), suggesting a mechanism for LPS consolidation of circadian behavior.
Our in slice studies strongly support fever as the LPS response that consolidates and resets behavioral rhythms in Six3-Cre;Lhx1lox/lox mice after 3X LPS, and to which Vip−/− mice remain resistant. Thus, though VIP is sufficient to modify SCN thermal responsiveness [1], our LPS data supports previous VIPR2 antagonist studies showing that VIP is not necessary for circadian resistance to temperature [2]. Our study identifies Lhx1 as the first gene necessary for temperature resistance of the SCN clock, and provides the first direct evidence that the SCN is the substrate of temperature resistance of circadian behavior.
Mapping Vip-Dependent and Vip-Independent Lhx1 Transcriptional Networks
The SCN signals underlying both light control of sleep and SCN temperature resistance are very poorly understood, making elucidation of the LHX1 transcriptional network impacting these processes particularly mechanistically interesting. Vip expression is controlled by LHX1 and mediates some of LHX1’s other circadian effects (such as synchronization of SCN oscillators) [17,18]. We thus mapped VIP-dependent and VIP-independent LHX1 transcriptional networks by RNA-sequencing Six3-Cre;Lhx1lox/lox, Vip−/−, and control SCNs collected at mid-day, to identify candidate mediators of circadian behaviors dependent upon LHX1 but not VIP.
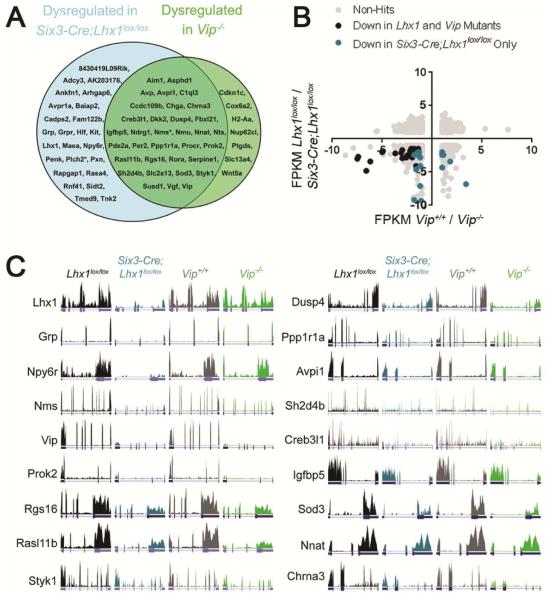
Many transcripts were reduced by similar amounts compared to controls in both Vip−/− and Six3-Cre;Lhx1lox/lox SCN, suggesting that LHX1 control of their expression is fully epistatic to VIP (Figures 3C; 4A; S3I-T). These included genes with known circadian effects, including neuropeptides (e.g. Avp, Prok2), signal transducers (e.g. Dusp4, Rgs16), and core clock genes (e.g. Per2, Rora). Nms was the only transcript expressed in Vip−/− SCN at levels intermediate between control and Six3-Cre;Lhx1lox/lox SCN (Figures 3C; S3W,AA; and [17]).
Figure 3. Elucidation of Two Distinct Lhx1-Regulated Transcriptional Networks.
(A) Venn diagram of transcripts dysregulated in SCN of Six3-Cre;Lhx1lox/lox SCN (blue), Vip−/− SCN (green), or both (overlap) at mid-day. * indicates conflicting RNA-Seq and ISH result.
(B) XY plot of unique transcripts, with mean FPKM fold-change in Vip−/− vs. Vip+/+ SCN on the x-axis and in Six3-Cre;Lhx1lox/lox vs. Lhx1lox/lox SCN on the y-axis. Gray transcripts are false positives, black transcripts are down in both mutants, and blue transcripts are down in Six3-Cre;Lhx1lox/lox SCN. Vip and several false positives with extreme FPKMs were excluded from this plot.
(C) Sample RNA-Seq gene expression traces visualized on the UCSC browser. Data from Lhx1lox/lox (black), Six3-Cre;Lhx1lox/lox (blue), Vip+/+ (gray), and Vip−/− (green) replicates are shown.
See also Supplemental Data Set.
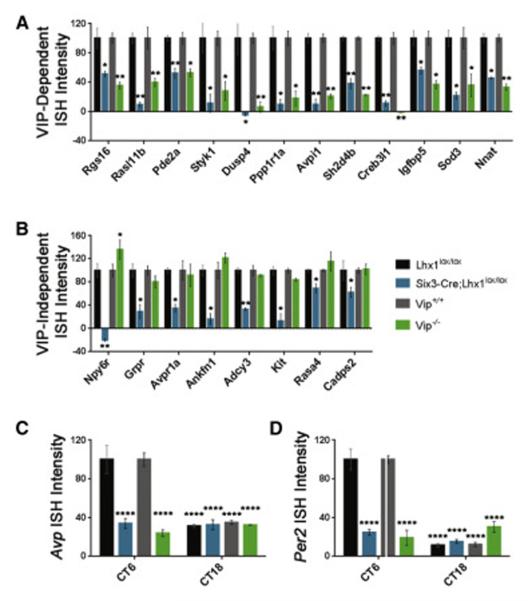
Figure 4. Effects of Lhx1 and Vip on SCN Gene Expression.
(A) Densitometry showed similarly decreased expression in Six3-Cre;Lhx1lox/lox and Vip−/− SCN for transcripts J-U (n=3, two-tailed paired t-tests, *p<0.05; **p<0.01, mean +/− SEM).
(B) Densitometry showed decreased expression in Six3-Cre;Lhx1lox/lox but not Vip−/− SCN for transcripts A-H. It also showed selectively increased Npy6r in Vip−/− SCN (n=3, two-tailed paired t-tests, *p<0.05; **p<0.01, mean +/− SEM). Tissue in (A-B) collected in a window from ~ZT6-9.
(C-D) Densitometry of SCN ISH showed decreased Avp and Per2 expression at CT6, normally a circadian time of peak or near-peak expression for these transcripts, to trough levels similar to control expression at CT18 in both Six3-Cre;Lhx1lox/lox and Vip−/− mutants (n=3, two-way ANOVA with post-hoc Tukey tests, ****p<0.0001, graph depicts mean +/− SEM).
See also Figure S3 and Supplemental Data Set.
Most VIP-dependent SCN transcripts (73.5%) are clock-controlled, often robustly, and are often expressed in the dorsomedial shell, which is enriched for robustly rhythmic genes more generally (Supplemental Data Set; Figure S3I-T,X-AA,DD). Further, many of these transcripts provide direct input to the core clock, often in ways that bridge intercellular and intracellular signaling. RGS16 drives cAMP rhythms required for normal phasing of SCN subdomains [28]. CREB3L1 couples the core clock to Avp expression downstream of cAMP signaling [29,30]. And DUSP4 gates pERK responses to light input, likely mediating lost gating of light responses in Vip−/− mice [16,31]. Other VIP-driven transcripts like Ppp1r1a are promising SCN signal transduction candidates. PPP1R1A inhibits PP1 (which regulates clock gene phosphorylation in Drosophila) downstream of cAMP-PKA signaling, in a manner gated by [Ca2+i] through both PKC and calcineurin [32–34]. Thus, PPP1R1A is a potential integrator of cAMP- and PLC-mediated signaling in the SCN [35–37]. Other intriguing hits include Avpi1, a MAPK activator regulated by cAMP-PKA signaling[38,39]; Nnat, a regulator of [Ca2+i] [40]; and the GTPase Rasl11b. Elucidation of this transcriptional network illuminates how the cell-autonomous amplitude-increasing and intercellular synchrony-promoting effects of VIP interact with each other at the cellular level [7,9,11,12,41].
Our SCN RNA-Seq identified few Six3-Cre;Lhx1lox/lox -specific hits (Figure 3B; Supplemental Data Set), which mostly lacked known or probable circadian functions, aside from the neuropeptide Grp and receptor Npy6r (which represents a novel mechanism linking LHX1 to Vip expression, given that Npy6r drives SCN Vip transcription [42], is co-regulated with Lhx1 and Vip in SCN by light [18], and is selectively up-regulated in Vip−/− SCN, in a manner suggesting homeostatic compensation for Vip loss (Figure 3B)). However, some known LHX1-regulated neuropeptides (Avp, Penk) fell short of our criteria, likely due to contamination from nearby regions that highly express these transcripts [17,43]. Pathway analysis of our RNA-Seq revealed that SCN signaling is even more broadly regulated by LHX1 than previously thought, and also indicated a possible role for LHX1 in SCN cytoskeletal dynamics (Supplemental Data Set). We conducted ISH studies guided by these analyses and found more transcripts important for signaling that are regulated by LHX1 independently of VIP, including Adcy3, Ankfn1, Avpr1a, Cadps2, Grpr, Kit, and Rasa4 (p<0.01 or p<0.05; Figures 4B; S3B-H). We also found VIP-dependent regulation of Pde2a and Nts (p<0.01 or p<0.05; Figures 4A; S3K,W,DD). Finally, we validated our RNA-Seq for 18/19 hits tested (p<0.05 or p<0.01; Figures 4A-B; S3). The only hit not confirmed by ISH, Plch2, trended down in Lhx1 mutant SCN (p>0.05; data not shown). The VIP-independent LHX1 transcriptional network identified by these studies is only modestly clock-controlled, with only 36% of SCN transcripts significantly rhythmic and even many of these having only marginal rhythms (Supplemental Data Set). Many of these transcripts are also located in the core and central SCN, where less robust oscillators are common (Figure S3A-H,AA-CC). Finally, many of the VIP-independent network’s components couple the core clock to its input or outputs, rather than modulating the core clock per se (Supplemental Data Set) [37,42,44–46]. Other network components like Grp and Grpr, while supportive of core clock functions like synchronization [12,47], are non-essential for behavioral rhythmicity and primarily mediate photoentrainment [48,49].
To directly test whether the VIP-independent LHX1 transcriptional network meaningfully interacts with the core clock separately from the VIP-dependent network, we did ISH for Per2 and Avp in Vip−/−, Six3-Cre;Lhx1lox/lox, and control SCNs, at high and low circadian times (CT6,18). Per2 and Avp in both mutants was statistically indistinguishable from trough expression in controls regardless of circadian time (p>0.05; Figure 4C-D,S3U-V), and down by similar amounts from peak expression in their respective controls (p<0.001). Thus, the VIP-independent LHX1 transcriptional network exerts its circadian effects mostly through SCN signaling that does not impinge on the core clock, consistent with a key role in regulating SCN input and output. Surprisingly, this implies that unlike the strong relationship between core clock amplitude and SCN resistance to photic input, core clock amplitude is largely dispensable for SCN resistance to thermal input [50–52].
Finally, this study identified a promising cluster of signaling pathways that likely mediate VIP-independent functions of LHX1. Components of this transcriptional module like Ankfn1 (the murine homolog of Drosophila wide awake) and Nms (a member of the sleep-regulating neuromedin family) may contribute to light control of sleep [44,53,54]. Furthermore, localization of these and many other VIP-independent, LHX1-regulated transcripts in the SCN core and central domains give insight into the SCN circuitry underlying regulation of sleep timing by light.
Finally, GRP, NMS, and AVP signaling are all regulated by LHX1 independently of Vip at the neuropeptide and/or receptor level (Figures 3,4,S3), and directly impact SCN [Ca2+i]. Since Ca2+-fluxing Trp channels and Ca2+-regulated effectors like SOLH protease and HSF1 convey heat input to the molecular clock, the AVP/GRP/NMS signaling cluster is a candidate substrate of SCN thermal resistance [2–4,55–57]. These findings give considerable insight that can guide the large-scale loss-of-function and gain-of-function studies needed to further plumb the signaling pathways underlying these phenomena going forward.
Experimental Procedures
Animal Housing, Care, and Genotyping
Six3-Cre;Lhx1lox/lox and Lhx1lox/lox mice, and Vip−/− and Vip+/+ mice, were used for most studies. Six3-Cre;Lhx1lox/lox;Per2:Luc/+ and control Lhx1lox/lox;Per2:Luc/+ mice were used for luciferase studies. Adult (2-8 month) males were used for behavior; other studies used adult sex-matched pairs of both sexes. Lhx1lox/lox was originally on mixed C57BL6/Sv129 background; other alleles were pure C57BL6. Thus, littermate controls were always used for Six3-Cre;Lhx1lox/lox mice, while age-matched non-littermate controls were sometimes substituted for Vip−/− mice. Except where noted, mice were on 15:9 hr light:dark (LD) cycle. Procedures and care were approved by Johns Hopkins IACUC, consistent with NIH guidelines. Transnetyx conducted most genotyping. Cre was detected by presence of a ~300 bp band amplified using 5′-TTCCCGCAGAACCTGAAGAT-3′ and 5′-CCCCAGAAATGCCAGATTAC-3′.
RNA-Sequencing
Brains were dissected from ~ZT6-9 in 15:9 LD and SCNs were collected as described [58]. Five punches were pooled for each of three biological replicates. cDNA libraries were generated from extracted RNA and sequenced to a paired-end read depth of 100 cycles. Alignment and fold-change was computed in Galaxy, and data was processed through a labor-intensive custom analysis pipeline to minimize false-positives. See Supplement for details.
In Situ Hybridization
Brains were prepared in a window of ~ZT6-9, as described [17]. Densitometry was done in ImageJ, using rolling-ball correction and subtracting cell-poor signal to compensate for background. Control intensity was normalized to 100, expressing mutants as % control signal. Some probes were hydrolyzed to improve signal-to-noise. Probes listed in Supplemental Data Set.
Electroencephalography/Electromyography (EEG/EMG)
Anesthetized mice were implanted with EEG/EMG headmounts secured by anterior 0.1” and posterior 0.12” surgical screws (Pinnacle Technology) located at approximately +2 mm AP, ±1.3 mm ML and −3.5 mm AP, ±1.3 mm ML from Bregma. These were connected to the headmount contacts with conductive silver epoxy and protected with a skull cap of Jet Denture Repair cement (Lang Dental). Recordings were done with a 3-channel acquisition system and Sirenia software (Pinnacle Technology), and analyzed as described [59–61].
Lipopolysaccharide (LPS) Telemetry and Wheel Running Studies
For telemetry, G2 emitters (Starr Life Sciences) were implanted into the IP cavity of anesthetized mice. CBT was recorded in Vitalview (Respironics) and analyzed in Clocklab (Actimetrics). Wheel running was assayed as described [17]. After pre-conditioning with 1X LPS in 12:12 LD during entrainment, mice were moved to DD for 2-3 weeks, then given 3X LPS, followed by 1X LPS 2-3 weeks later. See Supplement for LPS protocol details.
Per2:Luc Temperature Studies
Experiments were conducted as described[2] . See Supplemental Methods for details.
Supplementary Material
Acknowledgements
We thank the Johns Hopkins Deep Sequencing and Microarray Core for library prep and sequencing; Hong Wang for genotyping; Shirley Zhang and Gaétan Poirier for help with analysis; and Amita Sehgal and Wendy Yap for comments. This work was supported by the Johns Hopkins Brain Science Institute. JLB was supported by Wilmer Eye Institute VNTP and NSF GRFP fellowships. VM is supported by the Fonds de recherche du Québec-Santé and Canadian Institutes of Health Research. EB and RVG were supported by NIH EY001730 and an unrestricted grant from Research to Prevent Blindness. SB was a W.M. Keck Distinguished Young Scholar in Medical Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributions
JLB, SH, and SB conceived the study and wrote the manuscript. JLB and JL did RNA-Seq studies. JLB and AB did all ISH and LPS studies, and maintained all mice used. TAL and BB did EEG/EMG recordings. VM did LD EEG/EMG analysis. TAL did DD EEG/EMG analysis. EB did Per2:Luc studies. All authors contributed intellectually.
References
- An S, Harang R, Meeker K, Granados-Fuentes D, Tsai CA, Mazuski C, Kim J, Doyle FJ, 3rd, Petzhold LR, Herzog ED. A neuropeptide speeds circadian entrainment by reducing intercellular synchrony. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E4355–61. doi: 10.1073/pnas.1307088110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr ED, Yoo S-HH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330:379–85. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd P, Young M, Siggia E. Temperature compensation and temperature sensation in the circadian clock. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E6284–92. doi: 10.1073/pnas.1511215112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini C, Morf J, Stratmann M, Gos P, Schibler U. Simulated body temperature rhythms reveal the phase-shifting behavior and plasticity of mammalian circadian oscillators. Genes Dev. 2012;26:567–80. doi: 10.1101/gad.183251.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattar S, Liao HW, Takao M, Berson DM, Yau KW. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science. 2002;295:1065–70. doi: 10.1126/science.1069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS, Michel S, Itri J, Rodriguez W, Tam J, Lelievre V, Hu Z, Liu X, Waschek JA. Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003;285:R939–49. doi: 10.1152/ajpregu.00200.2003. [DOI] [PubMed] [Google Scholar]
- Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat. Neurosci. 2005;8:476–83. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh DH, Dragich JM, Kudo T, Schroeder AM, Nakamura TJ, Waschek JA, Block GD, Colwell CS. Effects of vasoactive intestinal peptide genotype on circadian gene expression in the suprachiasmatic nucleus and peripheral organs. J. Biol. Rhythms. 2011;26:200–9. doi: 10.1177/0748730411401740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TM, Colwell CS, Waschek JA, Piggins HD. Disrupted neuronal activity rhythms in the suprachiasmatic nuclei of vasoactive intestinal polypeptide-deficient mice. J. Neurophysiol. 2007;97:2553–8. doi: 10.1152/jn.01206.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthasubramaniam B, Herzog ED, Herzel H. Timing of neuropeptide coupling determines synchrony and entrainment in the mammalian circadian clock. PLoS Comput. Biol. 2014;10:e1003565. doi: 10.1371/journal.pcbi.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, et al. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell. 2002;109:497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- Maywood ES, Chesham JE, O’Brien JA, Hastings MH. A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc. Natl. Acad. Sci. U.S.A. 2011;108:14306–11. doi: 10.1073/pnas.1101767108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosko A, Diepen HC van, Kuljis D, Chiu AM, Heyer D, Terra H, Carpenter E, Michel S, Meijer JH, Colwell CS. Role of vasoactive intestinal peptide in the light input to the circadian system. Eur. J. Neurosci. 2015;42:1839–48. doi: 10.1111/ejn.12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman SJ, Silver R, Sauter J Le, Bult-Ito A, McMahon DG. Phase resetting light pulses induce Per1 and persistent spike activity in a subpopulation of biological clock neurons. J. Neurosci. 2003;23:1441–50. doi: 10.1523/JNEUROSCI.23-04-01441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916:172–91. doi: 10.1016/s0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- Dragich JM, Loh DH, Wang LM, Vosko AM, Kudo T, Nakamura TJ, Odom IH, Tateyama S, Hagopian S, Waschek JA, et al. The role of the neuropeptides PACAP and VIP in the photic regulation of gene expression in the suprachiasmatic nucleus. Eur. J. Neurosci. 2010;31:864–75. doi: 10.1111/j.1460-9568.2010.07119.x. [DOI] [PubMed] [Google Scholar]
- Bedont JL, LeGates TA, Slat EA, Byerly MS, Wang H, Hu J, Rupp AC, Qian J, Wong GW, Herzog ED, et al. Lhx1 controls terminal differentiation and circadian function of the suprachiasmatic nucleus. Cell Rep. 2014;7:609–22. doi: 10.1016/j.celrep.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori M, Gill S, Mure L, Goulding M, O’Leary D, Panda S. Lhx1 maintains synchrony among circadian oscillator neurons of the SCN. eLife. 2014;3:e03357. doi: 10.7554/eLife.03357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WP, Li JD, Colwell CS, Zhou QY. Decreased REM sleep and altered circadian sleep regulation in mice lacking vasoactive intestinal polypeptide. Sleep. 2011;34:49–56. doi: 10.1093/sleep/34.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker FC, Angara C, Szymusiak R, McGinty D. Persistence of sleep-temperature coupling after suprachiasmatic nuclei lesions in rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005;289:R827–38. doi: 10.1152/ajpregu.00093.2005. [DOI] [PubMed] [Google Scholar]
- Ibuka N, Inouye SI, Kawamura H. Analysis of sleep-wakefulness rhythms in male rats after suprachiasmatic nucleus lesions and ocular enucleation. Brain Res. 1977;122:33–47. doi: 10.1016/0006-8993(77)90660-6. [DOI] [PubMed] [Google Scholar]
- Eastman CI, Mistlberger RE, Rechtschaffen A. Suprachiasmatic nuclei lesions eliminate circadian temperature and sleep rhythms in the rat. Physiol. Behav. 1984;32:357–68. doi: 10.1016/0031-9384(84)90248-8. [DOI] [PubMed] [Google Scholar]
- Lu J, Zhang YH, Chou TC, Gaus SE, Elmquist JK, Shiromani P, Saper CB. Contrasting effects of ibotenate lesions of the paraventricular nucleus and subparaventricular zone on sleep-wake cycle and temperature regulation. J. Neurosci. 2001;21:4864–74. doi: 10.1523/JNEUROSCI.21-13-04864.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altimus CM, Güler AD, Villa KL, McNeill DS, Legates TA, Hattar S. Rods-cones and melanopsin detect light and dark to modulate sleep independent of image formation. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19998–20003. doi: 10.1073/pnas.0808312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husse J, Leliavski A, Tsang AH, Oster H, Eichele G. The light-dark cycle controls peripheral rhythmicity in mice with a genetically ablated suprachiasmatic nucleus clock. FASEB J. 2014;28:4950–60. doi: 10.1096/fj.14-256594. [DOI] [PubMed] [Google Scholar]
- Izumo M, Pejchal M, Schook A, Lange R, Walisser J, Sato T, Wang X, Bradfield CA, Takahashi JS. Differential effects of light and feeding on circadian organization of peripheral clocks in a forebrain Bmal1 mutant. eLife. 2014;3 doi: 10.7554/eLife.04617. doi: 10.7554/eLife.04617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power A, Hughes AT, Samuels RE, Piggins HD. Rhythm-promoting actions of exercise in mice with deficient neuropeptide signaling. J. Biol. Rhythms. 2010;25:235–46. doi: 10.1177/0748730410374446. [DOI] [PubMed] [Google Scholar]
- Doi M, Ishida A, Miyake A, Sato M, Komatsu R, Yamazaki F, Kimura I, Tsuchiya S, Kori H, Seo K, et al. Circadian regulation of intracellular G-protein signalling mediates intercellular synchrony and rhythmicity in the suprachiasmatic nucleus. Nat. Commun. 2011;2:327. doi: 10.1038/ncomms1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood M, Greenwood M, Mecawi A, Loh S, Rodrigues J, Paton J, Murphy D. Transcription factor CREB3L1 mediates cAMP and glucocorticoid regulation of arginine vasopressin gene transcription in the rat hypothalamus. Molecular Brain. 2015;8:68. doi: 10.1186/s13041-015-0159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood M, Bordieri L, Greenwood M, Melo M, Colombari D, Colombari E, et al. Transcription Factor CREB3L1 Regulates Vasopressin Gene Expression in the Rat Hypothalamus. J. Neurosci. 2014;34:3810–3820. doi: 10.1523/JNEUROSCI.4343-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher G, Lee B, Obrietan K. Temporal Regulation of Light-Induced Extracellular Signal-Regulated Kinase Activation in the Suprachiasmatic Nucleus. J. Neurophysiol. 2003;90:3854–3863. doi: 10.1152/jn.00524.2003. [DOI] [PubMed] [Google Scholar]
- El-Armouche A, Bednorz A, Pamminger T, Ditz D, Didié M, Dobrev D, Eschenhagen T. Role of calcineurin and protein phosphatase-2A in the regulation of phosphatase inhibitor-1 in cardiac myocytes. Biochem. Biophys. Res. Commun. 2006;346:700–6. doi: 10.1016/j.bbrc.2006.05.182. [DOI] [PubMed] [Google Scholar]
- Sahin B, Shu H, Fernandez J, El-Armouche A, Molkentin J, Nairn A, Bibb JA. Phosphorylation of protein phosphatase inhibitor-1 by protein kinase C. J. Biol. Chem. 2006;281:24322–35. doi: 10.1074/jbc.M603282200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Sathyanarayanan S, Sehgal A. Post-translational regulation of the Drosophila circadian clock requires protein phosphatase 1 (PP1) Genes Dev. 2007;21:1506–18. doi: 10.1101/gad.1541607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio M, Maywood ES, Chesham JE, Loudon AS, Hastings MH. A Gq-Ca2+ axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron. 2013;78:714–28. doi: 10.1016/j.neuron.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton SJ, Huettner JE, Straume M, Herzog ED. GABA and Gi/o differentially control circadian rhythms and synchrony in clock neurons. Proc. Natl. Acad. Sci. U.S.A. 2006;103:19188–93. doi: 10.1073/pnas.0607466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S, Irwin RP, Allen CN, Tsai C, Herzog ED. Vasoactive intestinal polypeptide requires parallel changes in adenylate cyclase and phospholipase C to entrain circadian rhythms to a predictable phase. J. Neurophysiol. 2011;105:2289–96. doi: 10.1152/jn.00966.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicod M, Michlig S, Flahaut M, Salinas M, Jaeger N, Horisberger J, Rossier BC, Firsov D. A novel vasopressin-induced transcript promotes MAP kinase activation and ENaC downregulation. EMBO J. 2002;21:5109–5117. doi: 10.1093/emboj/cdf509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CP, Loftus RW, Liu KZ. AVP-induced VIT32 gene expression in collecting duct cells occurs via trans-activation of a CRE in the 5’-flanking region of the VIT32 gene. A.J.P.: Renal Physiology. 2004;287:F460–F468. doi: 10.1152/ajprenal.00107.2004. [DOI] [PubMed] [Google Scholar]
- Lin H, Bell E, Uwanogho D, Perfect L, Noristani H, Bates T, Snetkov V, Price J, Sun YM. Neuronatin Promotes Neural Lineage in ESCs via Ca2+ Signaling. Stem Cells. 2010;28:1950–1960. doi: 10.1002/stem.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Reddy AB, Wong GK, O’Neill JS, O’Brien JA, McMahon DG, Hamar AJ, Okamura H, Hastings MH. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr. Biol. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Yulyaningsih E, Loh K, Lin S, Lau J, Zhang L, Shi Y, Berning BA, Enriquez R, Driessler F, Macia L, et al. Pancreatic polypeptide controls energy homeostasis via Npy6r signaling in the suprachiasmatic nucleus in mice. Cell Metab. 2014;19:58–72. doi: 10.1016/j.cmet.2013.11.019. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Bugoski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–76. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Liu S, Lamaze A, Liu Q, Tabuchi M, Yang Y, Fowler M, Bharadwaj R, Zhang J, Bedont JL, Blackshaw S, et al. WIDE AWAKE mediates the circadian timing of sleep onset. Neuron. 2014;82:151–66. doi: 10.1016/j.neuron.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Suzuki T, Mizoro Y, Kori H, Okada K, Chen Y, Fustin JM, Yamazaki F, Mizuguchi N, Zhang J, et al. Mice Genetically Deficient in Vasopressin V1a and V1b Receptors Are Resistant to Jet Lag. Science. 2013;342:85–90. doi: 10.1126/science.1238599. [DOI] [PubMed] [Google Scholar]
- Li J-D, Burton K, Zhang C, Hu S-B, Zhou Q-Y. Vasopressin receptor V1a regulates circadian rhythms of locomotor activity and expression of clock-controlled genes in the suprachiasmatic nuclei. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009;296:R824–R830. doi: 10.1152/ajpregu.90463.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TM, Hughes AT, Piggins HD. Gastrin-releasing peptide promotes suprachiasmatic nuclei cellular rhythmicity in the absence of vasoactive intestinal polypeptide-VPAC2 receptor signaling. J. Neurosci. 2005;25:11155–64. doi: 10.1523/JNEUROSCI.3821-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida R, Moriya T, Araki M, Akiyama M, Wada K, Wada E, Shibata S. Gastrin-releasing peptide mediates photic entrainable signals to dorsal subsets of suprachiasmatic nucleus via induction of period gene in mice. Mol. Pharmacol. 2002;61:26–34. doi: 10.1124/mol.61.1.26. [DOI] [PubMed] [Google Scholar]
- Gamble KL, Allen GC, Zhou T, McMahon DG. Gastrin-releasing peptide mediates light-like resetting of the suprachiasmatic nucleus circadian pacemaker through cAMP response element-binding protein and Per1 activation. J. Neurosci. 2007;27:12078–87. doi: 10.1523/JNEUROSCI.1109-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulivarthy S, Tanaka N, Welsh D, Haro L, Verma I, Panda S. Reciprocity between phase shifts and amplitude changes in the mammalian circadian clock. Proc. Natl. Acad. Sci. U.S.A. 2007;104:20356–61. doi: 10.1073/pnas.0708877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Silver R. Differential induction and localization of mPer1 and mPer2 during advancing and delaying phase shifts. Eur. J. Neurosci. 2002;16:1531–40. doi: 10.1046/j.1460-9568.2002.02224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer JH, Watanabe K, Schaap J, Albus H, Détári L. Light responsiveness of the suprachiasmatic nucleus: long-term multiunit and single-unit recordings in freely moving rats. J. Neurosci. 1998;18:9078–87. doi: 10.1523/JNEUROSCI.18-21-09078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CN, Rihel J, Lee DA, Singh C, Mosser EA, Chen S, Sapin V, Pham U, Engle J, Niles BJ, et al. A zebrafish genetic screen identifies neuromedin U as a regulator of sleep/wake states. Neuron. 2016;89:842–56. doi: 10.1016/j.neuron.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahnaou A, Drinkenburg WH. Neuromedin U(2) receptor signaling mediates alteration of sleep-wake architecture in rats. Neuropeptides. 2011;45:165–74. doi: 10.1016/j.npep.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Tataroglu O, Zhao X, Busza A, Ling J, O’Neill JS, Emery P. Calcium and SOL protease mediate temperature resetting of circadian clocks. Cell. 2015;163:1214–24. doi: 10.1016/j.cell.2015.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lee Y, Montell C. Drosophila TRPA1 functions in temperature control of circadian rhythm in pacemaker neurons. J. Neurosci. 2013;33:6716–25. doi: 10.1523/JNEUROSCI.4237-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang W, Simoni A, Gentile C, Stanewsky R. The Pyrexia transient receptor potential channel mediates circadian clock synchronization to low temperature cycles in Drosophila melanogaster. Proc. Biol. Sci. 2013;280:20130959. doi: 10.1098/rspb.2013.0959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MJ, Brancaccio M, Sethi S, Maywood ES, Satija R, Edwards JK, Jagannath A, Couch Y, Finelli MJ, Smyllie NJ, et al. The regulatory factor ZFHX3 modifies circadian function in SCN via an AT motif-driven axis. Cell. 2015;162:607–21. doi: 10.1016/j.cell.2015.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helou J, Bélanger-Nelson E, Freyburger M, Dorsaz S, Curie T, La Spada F, Gaudreault PO, Beaumont E, Pouliot P, Lesage F, et al. Neuroligin-1 links neuronal activity to sleep-wake regulation. Proc. Natl. Acad. Sci. U.S.A. 2013;110:9974–9. doi: 10.1073/pnas.1221381110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massart R, Freyburger M, Suderman M, Paquet J, El Helou J, Belanger-Nelson E, Rachalski A, Koumar OC, Carrier J, Szyf M. The genome-wide landscape of DNA methylation and hydroxymethylation in response to sleep deprivation impacts on synaptic plasticity genes. Translational Psychiatry. 2014;4:e347. doi: 10.1038/tp.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyburger M, Pierre A, Paquette G, Bélanger-Nelson E, Bedont J, Gaudreault PO, Drolet G, Laforest S, Blackshaw S, Cermakian N, et al. EphA4 is involved in sleep regulation but not in the electrophysiological response to sleep deprivation. Sleep. 2016;39:613–24. doi: 10.5665/sleep.5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.