Abstract
Bone mineral density (BMD) is a highly heritable predictor of osteoporotic fracture. Genome-wide association studies (GWAS) for BMD have identified dozens of associations; yet, the genes responsible for most associations remain elusive. Here, we used a bone co-expression network to predict causal genes at BMD GWAS loci based on the premise that genes underlying a disease are often functionally related and functionally related genes are often co-expressed. By mapping genes implicated by BMD GWAS onto a bone co-expression network, we predicted and inferred the function of causal genes for 30 of 64 GWAS loci. We experimentally confirmed that two of the genes predicted to be causal, SPTBN1 and MARK3, are potentially responsible for the effects of GWAS loci on Chromosomes 2p16.2 and 14q32.32, respectively. This approach provides a roadmap for the dissection of additional BMD GWAS associations. Furthermore, it should be applicable to GWAS data for a wide range of diseases.
Graphical abstract
INTRODUCTION
Osteoporosis is a metabolic disease characterized by decreased bone mineral density (BMD) and an increased risk of fracture (Black and Rosen, 2016). In the U.S., osteoporosis affects over 12 million individuals and is directly responsible for 1.5 million fractures annually at a cost of ~$17 billion (Gass and Dawson-Hughes, 2006). Osteoporosis is, in large-part, influenced by genetic variation with fracture-related traits, such as BMD, being among the most heritable disease-associated quantitative traits (h2>0.50) (Liu et al., 2012b; Ralston and de Crombrugghe, 2006; Ralston and Uitterlinden, 2010; Zheng et al., 2011). As a result, a comprehensive understanding of the genetic basis of osteoporosis is critical for the development of approaches for its treatment and prevention.
Over the last decade, genome-wide association studies (GWAS) have revolutionized the genetic analysis of complex diseases by discovering thousands of loci for hundreds of diseases and disease-related quantitative traits, including BMD (Frazer et al., 2009). In 2012, the GEnetic Factors for OSteoporosis Consortium (GEFOSII) conducted a meta-analysis of lumbar spine (LSBMD) and femoral neck (FNBMD) BMD in ~80 K individuals and identified 64 independent BMD associations (Estrada et al., 2012). The loci identified by GEFOSII represent a wealth of information with the potential to reveal novel genes and pathways that play important roles in bone biology and inform drug discovery (Nelson et al., 2015). Yet, “unlocking” this information requires an intimate understanding of the causal variants and genes underlying each locus and, to date, there has not been a systematic functional characterization of BMD GWAS loci.
One approach successfully used for causal gene prediction is the integration of GWAS data and biological networks (Jia and Zhao, 2014; Leiserson et al., 2013). A number of studies have used biological networks (e.g. protein-protein interaction and co-expression networks) to both predict causal genes at GWAS loci and use GWAS data to pinpoint key network modules in disease (as examples (Califano et al., 2012; Farber, 2010; 2013; Gustafsson et al., 2015; Huan et al., 2015; Mäkinen et al., 2014)). The idea behind this approach is that groups of genes influencing a complex disease are often functionally related and participate in similar pathways or processes (e.g. the function of bone-forming osteoblasts or bone-resorbing osteoclasts) and functionally related genes are often co-expressed (Goh et al., 2007). Biological networks provide a framework to reconstruct pathways in an unbiased manner and by mapping genes located in GWAS regions onto a biological network it is possible to extract disease relevant pathways and causal genes. Co-expression networks are particularly useful for this purpose, because when constructed using disease relevant expression profiles they can capture the tissue and cell-type specific nature of disease (Nayak et al., 2009; Zhang and Horvath, 2005).
The goal of this study was to use a bone co-expression network to inform BMD GWAS. We used a co-expression network constructed from mouse cortical bone expression profiles due to the lack of similar (i.e. bone minus marrow) population-scale transcriptomic data on human bone. By mapping mouse homologs of human genes located in GWAS regions onto network modules, we identified an Osteoblast Functional Module containing 33 genes implicated by GWAS. These genes are candidates for 30 of the 64 GEFOSII BMD GWAS regions. Importantly, by characterizing specific network connections, we were also able to predict how these genes influenced BMD. As a proof-of-principle, we determined if genes we predicted to be causal at two separate BMD GWAS loci were involved in the regulation of BMD. We confirmed our predictions using in vitro and in vivo approaches and at both loci the data support the causality of the investigated genes. We expect this approach will be useful to interrogate GWAS data for other complex diseases.
RESULTS
Identification of the Osteoblast Functional Module
Defining a list of genes implicated by BMD GWAS
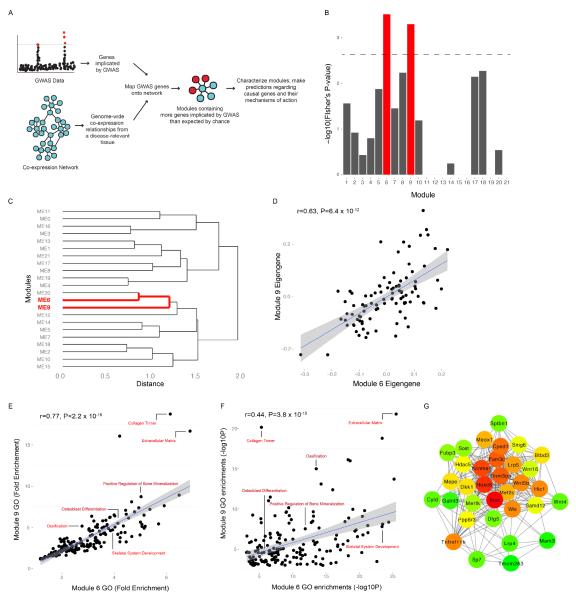
An overview of our strategy to inform BMD GWAS is shown in Figure 1A. We began by generating a list of genes located within BMD GWAS loci. As a set of loci, we used the 64 independent associations (P<5.0 × 10−8) for FNBMD and/or LSBMD identified by the GEFOSII GWAS meta-analysis (discovery and replication N~83,000) (Table S1) (Estrada et al., 2012). We included all RefSeq genes that were located within or overlapped with the region defined by linkage disequilibrium (LD; r2≥0.7) for each of the 64 lead GWAS SNPs. If a region did not contain or overlap a gene, we included the genes closest up- and downstream. The resulting BMD GWAS Implicated Gene list (BGIG) contained 167 genes (Table S2). The number of genes per association ranged from 2 to 16, with a mean of 2.8±1.9. The BGIG was enriched for gene ontology (GO) terms such as “ossification” (P=9.2 × 10−11), “skeletal system development” (P=2.2 × 10−8), “bone remodeling” (P=1.8 × 10−7) and “osteoblast differentiation” (P=6.6 × 10−7) (full list in Table S3), suggesting it contained many truly causal BMD GWAS genes.
Figure 1.
Identification of the Osteoblast Functional Module (OFM). A). Overview of the approach used to predict genes responsible for BMD GWAS associations. B). Enrichment of genes located in BMD GWAS regions in network modules 6 and 9. C). Eigengenes for modules 6 and 9 cluster. D). Module 6 and 9 eigengenes are highly correlated. E). Gene ontology fold enrichments and F) significance are correlated for GO terms shared between modules 6 and 9. G). The OFM is a cohesive, highly interconnected functional module. OFM genes with a topological overlap measure (TOM) of 0.05 are connected. A darker red color indicates increased number of connections with other OFM genes.
Identifying BGIG genes co-expressed in bone
BGIG genes were identified based solely on their proximity to GWAS variants; therefore, only a subset of BGIG genes is expected to be causal for BMD. We hypothesized that the causal subset could be identified based on their co-expression in bone. Thus, to pinpoint potentially causal genes, we mapped the mouse homologs of BGIG genes (N=148 of 167) onto a mouse cortical bone co-expression network. The network was constructed using cortical bone expression profiles from 96 inbred strains of the Hybrid Mouse Diversity Panel (HMDP) (Calabrese et al., 2012). The network is unique because samples profiled were marrow-free cortical bone fragments. As a result, the expression profiles primarily represented cells of the osteoblast lineage (osteocytes and mature bone-forming osteoblasts) (Bonewald, 2011). Osteocytes and osteoblasts along with bone-resorbing osteoclasts represent the three key cell-types responsible for bone modeling and remodeling (Seeman, 2009). The network consisted of 10,968 genes partitioned into 21 co-expression modules. Of the 148 BGIG homologs, 97 (66%) were members of at least one network module; a significant enrichment of genes mapping to the network over the number expected by chance (Odds Ratio (OR)=1.8; Fisher’s P= 8.12 × 10−6). Of the 21 network modules, two (modules 6 and 9) were significantly enriched for BGIG homologs (OR=3.2, Fisher’s P=3.0 × 10−4 and OR=4.7, Fisher’s P=4.0 × 10−4, respectively) (Figure 1B). A total of 22 (23%) BGIG homologs were members of modules 6 and 9 (N=14 and N=8, respectively) (Table 1).
TABLE 1.
OFM genes.
| GWAS SNP | Chr | SNP Position (Mbp) | Gene | Gene Position (Mbp) | Modulea | Groupb | Novelc | Referenced |
|---|---|---|---|---|---|---|---|---|
| rs7521902 | 1 | 22.164 | WNT4 | 22.143 | 9 | OFM_9 | NO | 25108526, 25108526 |
| rs6426749 | 1 | 22.385 | WNT4 | 22.143 | 9 | OFM_9 | NO | 25108526, 25108526 |
| rs17482952 | 1 | 68.174 | WLS | 68.233 | 9 | OFM_9 | NO | 22745162, 22745162 |
| rs12407028 | 1 | 68.182 | WLS | 68.233 | 9 | OFM_9 | NO | 22745162, 22745162 |
| rs479336 | 1 | 172.230 | DNM3OS | 172.145 | 6 | OFM_6 | NO | 18985749, NA |
| rs4233949 | 2 | 54.433 | SPTBN1 | 54.456 | 6 | OFM_6 | YES | NA |
| rs17040773 | 2 | 111.742 | MERTK | 111.899 | 0 | OFM_R | YES | NA |
| rs1346004 | 2 | 165.745 | GALNT3 | 165.794 | 0 | OFM_R | NO | 22912827, 25899975 |
| rs6532023 | 4 | 87.853 | MEPE | 87.821 | 6 | OFM_6 | NO | 12421822, 12421822 |
| rs1366594 | 5 | 89.080 | MEF2C | 88.904 | 6 | OFM_6 | NO | 17336904, 21652706 |
| rs13245690 | 7 | 121.145 | CPED1 | 120.989 | 6 | OFM_6 | YES | NA |
| rs13245690 | 7 | 121.145 | WNT16 | 121.325 | 6 | OFM_6 | NO | 25306234, 25306234 |
| rs3801387 | 7 | 121.335 | FAM3C | 121.396 | 6 | OFM_6 | YES | NA |
| rs3801387 | 7 | 121.335 | WNT16 | 121.325 | 6 | OFM_6 | NO | 25306234, 25306234 |
| rs2062377 | 8 | 118.995 | SAMD12 | 118.622 | 0 | OFM_R | YES | NA |
| rs2062377 | 8 | 118.995 | TNFRSF11B | 118.952 | 9 | OFM_9 | NO | 9108485, 9950424 |
| rs7851693 | 9 | 130.603 | FUBP3 | 130.580 | 0 | OFM_R | YES | NA |
| rs1373004 | 10 | 52.668 | DKK1 | 52.314 | 6 | OFM_6 | NO | 16753024, 16753024 |
| rs7071206 | 10 | 77.642 | KCNMA1 | 77.638 | 6 | OFM_6 | YES | NA |
| rs7071206 | 10 | 77.642 | DLG5 | 77.927 | 9 | OFM_9 | YES | NA |
| rs7108738 | 11 | 15.689 | INSC | 15.112 | 9 | OFM_9 | YES | NA |
| rs7932354 | 11 | 46.701 | LRP4 | 46.919 | 9 | OFM_9 | NO | 25733894, 25733894 |
| rs3736228 | 11 | 68.434 | LRP5 | 68.313 | 9 | OFM_9 | NO | 11956231, 11956231 |
| rs3736228 | 11 | 68.434 | PPP6R3 | 68.461 | 0 | OFM_R | YES | NA |
| rs2887571 | 12 | 1.529 | WNT5B | 1.617 | 6 | OFM_6 | NO | 26273529, NA |
| rs2016266 | 12 | 53.334 | SP7 | 53.345 | 9 | OFM_9 | NO | 11792318, 11792318 |
| rs736825 | 12 | 54.024 | HOXC6 | 54.017 | 6 | OFM_6 | NO | 1100234, NA |
| rs1053051 | 12 | 106.973 | TMEM263 | 106.956 | 3 | OFM_R | YES | NA |
| rs11623869 | 14 | 103.417 | MARK3 | 103.385 | 0 | OFM_R | YES | NA |
| rs1564981 | 16 | 50.952 | CYLD | 50.742 | 0 | OFM_R | NO | 18382763, NA |
| rs1566045 | 16 | 50.988 | CYLD | 50.742 | 0 | OFM_R | NO | 18382763, NA |
| rs4790881 | 17 | 2.166 | HIC1 | 2.055 | 0 | OFM_R | YES | NA |
| rs4790881 | 17 | 2.166 | SMG6 | 2.304 | 0 | OFM_R | YES | NA |
| rs4792909 | 17 | 43.721 | MEOX1 | 43.662 | 6 | OFM_6 | NO | 12538525, NA |
| rs4792909 | 17 | 43.721 | SOST | 43.759 | 6 | OFM_6 | NO | 11181578, 15024046 |
| rs227584 | 17 | 44.148 | HDAC5 | 44.124 | 1 | OFM_R | NO | 25271055, 19920351 |
| rs3790160 | 20 | 10.659 | BTBD3 | 11.891 | 6 | OFM_6 | YES | NA |
Coexpression network module from (Calabrese et al. Plos Genetics).
OFM gene group, OFM_6 = member of module 6, OFM_9 = member of module 9 and OFM_R = not a member of module 6 or 9, but expression was highly correlated with the module 6 or 9 eigengene.
Whether or not a gene is known to be involved in the regulation of BMD and/or osteoblast activity.
Reference PMID number demonstrating a known gene is involved in BMD (first #) and osteoblast activity (second #).
To determine if the enrichments for modules 6 and 9 were specific to the BMD GWAS, we identified 18 GWAS studies for a wide-range of diseases with a similar number (N>50 and <70) of genome-wide significant loci and defined gene lists and calculated enrichments using the same parameters as we did for the BMD GWAS. Modules 6 and 9 were not significantly (P≤2.7 × 10−3) enriched for genes implicated by any of the other 18 GWAS (Figure S1).
Modules 6 and 9 contain functionally-related genes
We previously observed that modules 6 and 9 shared a number of similarities (Calabrese et al., 2012). For instance, genes in both modules are preferentially expressed in bone-forming osteoblasts (Calabrese et al., 2012). To more formally evaluate the relationship between the two modules, we created a network based on the eigengene of each of the 21 modules (Figure 1C). In this network, the module 6 and 9 eigengenes clustered together, and as would be expected their eigengenes were highly correlated (r=0.63, P=3.4 × 10−12) (Figure 1D).
We next evaluated shared gene ontology (GO) enrichments to determine whether the similarity in the behavior meant that both modules contained functionally related genes. Modules 6 and 9 were nominally enriched (P-value<0.001) for 731 and 253 GO terms, respectively. Of these, 189 (74.7% of the total module 9 terms) terms were enriched (P<0.001) in both modules. Furthermore, we observed a positive correlation in fold enrichments and enrichment significance for shared terms (Figure 1E and 1F). Most of the ontologies shared between the modules described cellular components, biological processes and molecular functions pertinent to osteoblasts, such as “extracellular matrix” (module 6 P= 3.0 × 10−26 and module 9 P=1.5 × 10−22), “ossification” (module 6 P=2.9 × 10−14 and module 9 P=1.0 × 10−15), “bone development” (module 6 P=2.4 × 10−12 and module 9 P=2.5 × 10−6) and “osteoblast differentiation” (module 6 P=2.0 × 10−7 and module 9 P=3.5 × 10−7) (Figure 1E and 1F).
Identifying additional putatively causal BGIG genes
Membership within a co-expression module is based on a particular threshold in connection strength. However, in many cases a gene just missing this threshold of similarity for a particular module may still be functionally related to the genes in that module. Also, genes responsible for orchestrating module behavior (e.g. a transcription factor coordinating module co-expression) are not always members of the module due to having many weak or moderate correlations with module members as opposed to a few strong ones (Calabrese et al., 2012). Therefore, to more comprehensively identify putative causal genes in the BGIG, we identified all BGIG genes that were not members of module 6 and 9, but whose expression correlated (r≥∣0.40∣, P<5.0 × 10−5) with the module 6 and/or 9 eigengene. To increase specificity we also required the correlations with the eigengenes of modules 6 and/or 9 be the strongest of the 21 modules. There were 11 genes fitting these criteria (Table 1).
The Osteoblast Functional Module
Based on the data above, we merged the 22 BGIG genes mapping to modules 6 and 9 and the 11 additional BGIG genes correlated with the eigengene of either module into what we termed the Osteoblast Functional Module (OFM). We evaluated connectedness in the OFM to ensure it represented a cohesive set of co-expressed genes. In the OFM, the median correlation between genes was r=0.29. This represented a significant (P<0.001) enrichment over what would be expected by chance (Figure S2), indicating that as a group, OFM genes were co-expressed and highly interconnected (Figure 1G).
Of the 33 OFM genes, 18 (55%) have been shown to regulate BMD in humans or mice and 13 of the 18 (72%) have been directly implicated in the function of osteoblast-lineage cells (Table 1). This includes genes such as LDL receptor related protein 5 (LRP5), Sp7 transcription factor (SP7; aka osterix), tumor necrosis factor receptor superfamily member 11b (TNFRSF11B; aka osteoprotegerin) and sclerostin (SOST), which are among the best-known and most well characterized genes involved in the regulation of osteoblast-lineage cells and BMD (Balemans et al., 2001; Brunkow et al., 2001; Gong et al., 2001; Little et al., 2002; Nakashima et al., 2002; Simonet et al., 1997). To compare the recovery of known BMD genes to what would be expected by chance, we searched whole-body BMD data on 1928 mouse mutants (selected at random with respect to known involvement in BMD) from the International Mouse Phenotyping Consortium (IMPC) (Brown and Moore, 2012a; 2012b). In the IMPC, 6.3% (122 of 1928) of mouse knockouts exhibited a BMD phenotype compared to 55% recovery of known regulators of BMD in the OFM (OR=8.6, Fisher’s P=9.6 × 10−10), confirming that with respect to the regulation of BMD, the OFM is not a random gene set.
OFM genes have characteristics of a causal gene set
Proximity to lead BMD GWAS SNPs
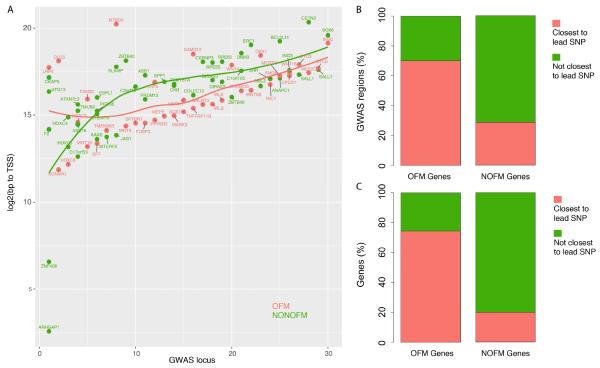
We next set out to generate additional evidence that OFM genes are likely to be causal. It has recently been shown that for a given GWAS locus the gene nearest to the lead SNP is not always causal (Claussnitzer et al., 2015; Smemo et al., 2014). However, we would expect across multiple GWAS loci that causal genes would tend to be located closer to lead SNPs than non-causal genes, as shown in studies mapping local expression quantitative trait locus (eQTL) (Veyrieras et al., 2008). Therefore, for the 30 GWAS that contain at least one OFM gene (referred to as OFM loci), we calculated the distance between the lead GWAS SNP and the distance to the transcription start site (TSS) for each gene in the locus. When plotted starting from the locus with the shortest distance from lead SNP to nearest TSS to the locus with the longest distance between lead SNP and the nearest TSS there was a clear trend for OFM genes to be closer to lead SNPs than non-OFM genes (Figure 2A). In fact, OFM genes were the closest to the lead SNP at 21 (70.0%) of the 30 OFM loci (OR=2.3, Fisher’s P=0.07) (Figure 2B). Additionally, of the 33 OFM genes in OFM loci, 26 (76%) were the closest gene in at least one association, whereas only 8 (20%) of the 40 non-OFM genes in OFM loci were the closest to at least one lead GWAS SNP (OR=3.9, Fisher’s P=3.2 × 10−3) (Figure 2C).
Figure 2.
OFM genes are in close proximity to lead BMD GWAS SNPs. A). OFM (pink) and non-OFM genes (green) were plotted based on their distance from the lead GWAS SNP in each of 30 OFM loci. Loess lines indicate that OFM genes tend to be closer to lead SNPs across most associations. B). The percentage of OFM GWAS loci in which OFM or non-OFM (NOFM) genes are the closest to the lead SNP. C) The percentage of OFM or NOFM genes closest to the lead SNP at any association.
OFM SNPs overlap regulatory elements in osteoblasts
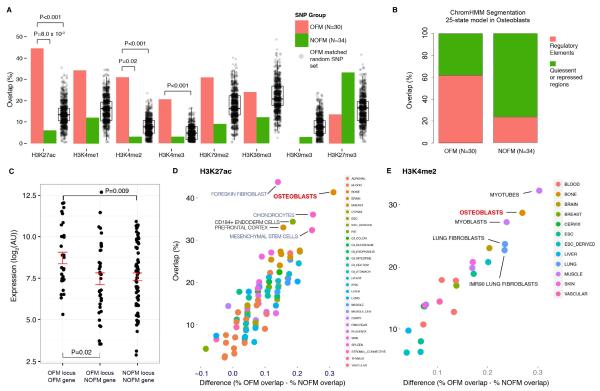
Of the 30 OFM loci, only three harbored lead SNP proxies (r2>0.7) that were nonsynonymous variants, suggesting that most of the OFM loci influence BMD by altering gene regulation. Thus, if OFM genes are truly causal and their mechanism of action is to influence osteoblast activity, we would expect that the associations would influence BMD by altering gene expression in osteoblasts. To determine if this is the case, we assessed the overlap between lead SNPs in OFM loci (N=30) and all non-OFM loci (N=34) and histone modifications marking regulatory elements in primary human osteoblasts (ENCODE Project Consortium et al., 2012; Roadmap Epigenomics Consortium et al., 2015). We observed a higher level of overlap with six histone modifications that are marks of active transcription for OFM lead SNPs as compared to non-OFM lead SNPs (Figure 3A). The opposite was seen for two repressive histone modifications (Figure 3A). This was most prominent for H3K27ac and H3K4me2, which showed significant increases in overlap with OFM SNPs as compared to non-OFM SNPs (Fisher’s P=0.02). H3K27ac and H3K4me2 binding is associated with enhancer elements and regions of transcription factor binding, respectively (ENCODE Project Consortium et al., 2012). For both modifications, as well as H3K4me3 (associated with promoter elements), we observed more overlap with OFM lead SNPs than would be expected by chance (P<0.001) by comparison with 1000 sets of 30 randomly selected SNPs, matched with OFM SNPs on minor allele frequency and distance from the nearest TSS (Figure 3A). OFM SNPs were also more likely to lie in locations annotated as regulatory elements using ChromHMM genome segmentations in osteoblasts (Ernst and Kellis, 2012) (Figure 3B). We also observed that OFM genes were on average 2-fold more highly expressed in either osteoblasts or bone than non-OFM genes in either OFM (P=0.02) or non-OFM loci (P=9 × 10−3) (Figure 3C). Similar overlap was observed using lead SNPs and proxies at both r2=1.0 and r2=0.8 (data not shown).
Figure 3.
Lead SNPs in OFM loci overlap active regulatory elements specifically in primary human osteoblasts. A). Overlap between lead GWAS SNPs and histone modifications at OFM and non-OFM (NOFM) loci in primary human osteoblasts and 1000 randomly selected SNP sets. B). ChromHMM state segmentations in osteoblasts overlaps with lead GWAS SNPs in OFM and NOFM loci. C). Expression of OFM genes and NOFM genes in OFM loci and NOFM genes in NOFM loci in bone and primary human osteoblast samples. D) Difference in % OFM overlap versus % NOFM overlap (from A) plotted against % OFM overlap in the 97 cell lines and tissues for the H3K27ac modification. E) Same as in D for H3K4me2.
To determine if the increase in overlap with histone modifications was specific to osteoblasts, we focused on data for H3K27ac and H3K4me2 in a large number of cell-types from the NIH Epigenomics Roadmap project (Roadmap Epigenomics Consortium et al., 2015). We focused on H3K27ac and H3K4me2 because they demonstrated the most significant enrichments with lead OFM SNPs in primary human osteoblasts (Figure 3A) and, as mentioned above, they are known to mark enhancers (ENCODE Project Consortium et al., 2012). For both modifications, we observed that osteoblasts were among the cell-types with the largest difference in overlap between the OFM and non-OFM SNP groups as well as having the highest level of OFM overlap (upper right quadrant) (Figure 3D). Moreover, the cell types that grouped with osteoblasts were highly related cell-types such as chondrocytes, fibroblasts and mesenchymal stem cells (MSCs; osteoblasts are derived from MSCs). Similarly for H3K4me2, myotubes and myoblasts grouped with osteoblasts in the upper right quadrant, both of which are derived from MSCs (Figure 3E). Together, these data are consistent with OFM loci influencing BMD by altering gene expression in osteoblasts, which strengthens our prediction that OFM genes in these loci are more likely to be causal than non-OFM genes.
Using the OFM to inform GWAS
Based on the data above, OFM genes are strong candidates to underlie their respective associations. To use the OFM to inform GWAS, we first grouped the 30 OFM loci into three categories based on whether the OFM gene(s) in the locus was known, previously demonstrated to regulate BMD in humans or mice, or novel, not previously shown to regulate BMD. The three groups included loci containing: 1) only known OFM genes, 2) known and novel OFM genes and 3) only novel OFM genes (Table 1). A total of 21 of the 30 (70%) OFM loci fell into one of the first two groups. We expect that many of the known OFM genes to be causal at their respective association. The remaining nine associations harbored only novel OFM genes. These nine loci are high priority for functional follow-up given their potential to provide new insight on the regulation of BMD. As a proof-of-principle, we selected two of the nine (Chrs. 2p16.2 and 14q32.32) loci for experimental interrogation.
SPTBN1 is a candidate causal gene for the BMD GWAS locus on Chr. 2p16.2
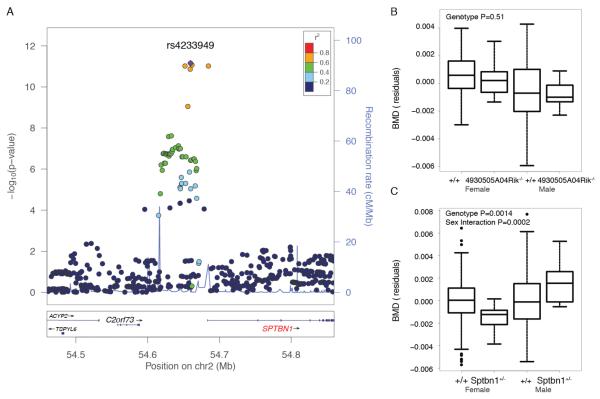
Chr. 2p16.2 contained SNPs associated with LSBMD (discovery P=5.0 × 10−12 and discovery + replication P=2.25 × 10−18) (Figure 4A). This locus was also one of six in the GEFOSII meta-analysis that were associated with osteoporotic fracture (P=2.6 × 10−8) (Estrada et al., 2012). The lead SNP, rs4233949, was located within an intergenic region ~23 Kbp from the spectrin, beta, nonerythrocytic 1 (SPTBN1) TSS and ~100 Kbp from the TSS for chromosome 2 open reading frame 73 (C2orf73; 4930505A04Rik is its homolog in the mouse). C2orf73 encodes for a protein of unknown function and SPTBN1 encodes for beta spectrin, a molecular scaffolding protein that links the actin cytoskeleton to the plasma membrane (Viel and Branton, 1996). C2orf73 and SPTBN1 represented the locus in the BGIG list and SPTBN1 was a member of the OFM (4930505A04Rik was not part of the bone network) (Table 1). Consistent with its known function, the 50 genes most strongly connected to Sptbn1 in the bone network included genes encoding extracellular matrix (P=2.2 × 10−7) proteins and proteins involved in alpha-actinin binding (P=1.6 × 10−5), focal adhesion (P=0.001) and cell adhesion (P=0.001).
Figure 4.
SPTBN1 is a strong candidate causal gene for a GWAS association on Chr. 2p16.2. A). GEFOSII GWAS discovery phase P-values for LSBMD in the GWAS locus on Chr. 2p16.2. Of the two genes, C2orf73 and SPTBN1, in the locus Sptbn1 was a member of the OFM. B). Residuals of whole body BMD adjusted for batch and body weight by genotype and sex for mice homozygous for a gene-trap allele of 4930505A04Rik, the mouse homolog of C2orf73. C). Residuals of whole body BMD adjusted for batch and body weight by genotype and sex for mice heterozygous for a Sptbn1 gene-trap allele.
Based on its membership in the OFM, we predicted that SPTBN1 was causal. To test this prediction, we first evaluated the broad tissue expression profiles for both genes in mice. The expression of 4930505A04Rik was restricted to testis (Figure S3). In contrast, Sptbn1 was broadly expressed in many tissues and cell-types, including high expression in osteoblasts (Figure S3). The IMPC (Brown and Moore, 2012a; 2012b) had collected BMD data on knockouts for both genes. At 14 weeks of age, mice of both sexes homozygous for a gene-trap allele of 4930505A04Rik (4930505A04Riktm1b(KOMP)Wtsi; hereafter referred to as 4930505A04Rik−/−) displayed no difference in BMD relative to controls (Figure 4B). However, male mice heterozygous for a gene-trap allele of Sptbn1 (Sptbn1tm1a(EUCOMM)Wtsi; hereafter referred to as Sptbn1+/−) had increased whole body BMD and female Sptbn1+/− mice had decreased whole body BMD relative to controls (genotype P=0.0014 and sex interaction P=0.002) (Figure 4C). These data are consistent with the prediction that SPTBN1 is involved in the regulation of BMD and is potentially responsible for the 2p16.2 BMD GWAS locus.
Mark3 is a causal gene for the BMD GWAS locus on Chr. 14q32.32
As further proof-of-principle, we performed a more detailed investigation of the BMD GWAS locus on Chr. 14q32.32. We selected this locus because it contained multiple genes, one of which was a non-OFM gene known to regulate BMD (brain-type creatine kinase (CKB)); thus, we reasoned it would provide a rigorous validation of our approach. The locus was associated with both FNBMD (discovery P=1.3 × 10−6 and discovery + replication P=5.2 × 10−16) and LSBMD (discovery P=7.5 × 10−4 and discovery + replication P=5.1 × 10−11) in GEFOSII (Estrada et al., 2012) (Figure 5A). The lead SNP, rs11623869, was located in the second intron of the MAP/microtubule affinity-regulating kinase 3 (MARK3) gene; however, rs11623869 proxy SNPs (r2>0.7) spanned a 157 Kbp interval also containing CKB and tRNA methyltransferase 61 homolog A (TRMT61A). Of the three, CKB has been demonstrated to play a role in the activity of bone-resorbing osteoclasts and Ckb deficient mice are protected against bone loss induced by ovariectomy and inflammatory challenges (Chang et al., 2008). TRMT61A is a tRNA methyltransferase (Ozanick et al., 2005) and MARK3 is a member of the AMPK family of kinases (Bright et al., 2009). TRMT61A and MARK3 have not been implicated in the regulation of BMD.
Figure 5.
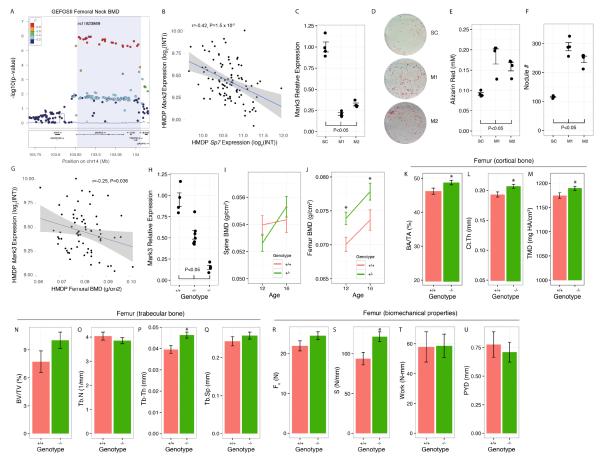
SPTBN1 is a strong candidate causal gene for a GWAS association on Chr. 2p16.2. A). GEFOSII GWAS discovery phase P-values for FNBMD in the locus on Chr. 14q32.32. Of the three genes, MARK3, CKB and TRMT61A, located in the association Mark3 was a member of the OFM. B). In the HMDP, Mark3 expression is negatively correlated with Sp7 (osterix), a key transcription factor required for osteoblast differentiation. C). Knockdown of Mark3 in primary calvarial osteoblasts using two siRNAs (M1 and M2) compared to a scrambled control (SC). D). Images of cultures stained with Alizarin Red 10 days after transfection with SC, M1 or M2. Quantification of E) Alizarin Red staining and F) nodule counts 10 days after transfection with SC, M1 or M2. G). In the HMDP, Mark3 expression is negatively correlated with femoral BMD. H). Mark3 transcript levels in embryos are reduced in mice harboring the tm1a gene-trap allele. I) Spine and J) femur BMD in male heterozygous gene-trap mice (+/−) and wild-type littermates (+/+) at 12 and 16 weeks of age. K) Cortical bone fraction (BA/TA), L) cortical thickness (Ct.Th) and M) tissue mineral density (TMD) in gene-trap mice (−/−) and wild-type littermates (+/+). N) Trabecular bone fraction (BV/TV), O) trabecular number (Tb.N), P) trabecular thickness (Tb.Th) and Q) trabecular separation (Tb.Sp) in male homozygous gene-trap (−/−) and wild-type littermates (+/+) at 12 weeks of age. R) Biomechanical strength (F), S) stiffness (S), T) work and U) post-yield deflection in gene-trap mice (−/−) and wild-type littermates (+/+). Plotted values in I-U are lsmeans ± s.e.m. *P<0.05.
CKB was a strong candidate for this locus based on its known role in the regulation of BMD; however, based on its membership in the OFM we predicted that MARK3 was at least partially responsible for the effects of the locus. To determine how MARK3 may influence BMD, we went back to the OFM and evaluated the relationship between Mark3 and other OFM genes. Mark3 expression was negatively correlated with all other OFM genes. It was most strongly negatively correlated with the expression of Sp7, a key transcription factor required for osteoblast differentiation (Figure 5B), suggesting that Mark3 was a negative regulator of osteoblast differentiation.
To test this hypothesis, we first confirmed that Mark3 was expressed in osteoblasts. Across 96 different mouse tissues and cell-types, we observed that Mark3 expression was ubiquitously expressed, with strong expression in osteoblasts (Figure S4). In primary calvarial osteoblasts, Mark3 expression started high and decreased as a function of differentiation (Figure S4). We also observed its expression in 3T3-E1 cells, a mouse osteoblast cell line, by immunofluorescence and its expression in osteoblast-lineage cells in bone was confirmed by immunohistochemistry (Figure S4).
We then used two siRNAs (M1 and M2) to knockdown the expression of Mark3 in mouse primary calvarial osteoblasts. At 48 hours post-transfection in undifferentiated cells, Mark3 transcript levels were reduced to 21% and 32% of the scrambled control in M1 and M2 transfected cells, respectively (P<0.05) (Figure 5C). Primary osteoblasts can be induced to mineralize in vitro, which is a marker of the rate of differentiation/maturation. At 10 days post-differentiation, Mark3 knockdown resulted in dose dependent increases in the formation of mineralized nodules (Figure 5D) as measured by alizarin red staining (which stains mineralized nodules, P<0.05, Figure 5E) and the number of nodules formed (P<0.05, Figure 5F).
We next measured the correlation between Mark3, Ckb and Trmt61a expression and femoral BMD in the HMDP (population used to generate the bone network). Of the three, Mark3 was the only gene correlated with femoral BMD (r=−0.25, P=0.036) (Figure 5G). These data, together with the in vitro experiments, suggested that Mark3 was a negative regulator of BMD.
To test this prediction, we generated mice carrying a Mark3 gene trap allele (Mark3tm1a(KOMP)Mbp; hereafter referred to as Mark3−/−). The gene trap allele decreased Mark3 levels by 47% and 85% in Mark3+/− and Mark3−/− male mice, respectively (Figure 5H). We generated two cohorts of male mice. In the first, we measured BMD in Mark3+/− and Mark3+/+ male mice at 12 and 16 weeks of age. A decrease in Mark3 levels had no effect on BMD at the spine at either age (Figure 5I). However, at both time points we observed significantly (P<0.05) increased femoral BMD in Mark3+/− mice compared to Mark3+/+ littermates (Figure 5J). This is consistent with the stronger association between rs11623869 and FNBMD, as compared to LSBMD, in GEFOSII (Estrada et al., 2012).
We used μCT in the second cohort to evaluate the effect of a decrease in Mark3 on cortical and trabecular microarchitecture of the femur in 12-week old male Mark3−/− and Mark3+/+ mice. These data revealed that the decrease in Mark3 levels primarily increased cortical bone, with significant (P<0.05) increases in cortical bone area fraction (BA/TA), cortical thickness (Ct.Th) and tissue mineral density (TMD) in male Mark3−/− compared to Mark3+/+ littermates (Figures 5K-M). Trabecular bone volume fraction (BV/TV), trabecular number (Tb.N) and trabecular separation (Tb.Sp) were not altered in the distal femur of Mark3−/− mice (P>0.05), but we did observe a significant (P<0.05) increase in trabecular thickness (Tb.Th) (Figures 5N-Q). The increase in cortical bone mass was associated with a suggestive increase in bone strength (P=0.10) (Figure 5R) and a significant (P<0.05) increase in femoral stiffness in male Mark3−/− mice (Figure 5S). No differences were observed in Work (a measure of toughness) (Figure 5T) and post-yield deflection (a measure of ductility) (Figure 5U).
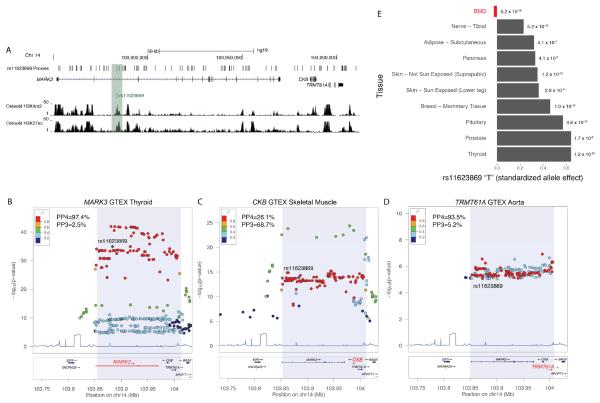
During the course of our study, eQTL results from the Gene Tissue Expression (GTEx) project became available, so we went back and further investigated the associated region. Of the 117 SNP proxies (r2>0.7) for rs11623869, only two were coding and they were synonymous variants in CKB, suggesting that the variant(s) underlying the association with BMD is regulatory. Many of these proxies were located in putative regulatory regions (marked by H3K4me2, H3H27ac, etc.) in human osteoblasts (Figure 6A) and other cell-types (data not shown). We queried GTEx to determine if any of three genes in the region were regulated by the same variants associated with BMD. SNPs in the region were significantly (P<1.0 × 10−5) associated with the expression of MARK3 in nine different tissues, CKB in eight different tissues and TRMT61A in seven different tissues (Figures 6B-D). We used a Bayesian colocalization test to evaluate whether the genetic data best fit a model in which the eQTLs (most significant for each gene) and BMD associations were due to a single shared pleiotropic variant (model 4) or two independent variants (model 3). For MARK3 the strongest eQTL was in thyroid tissue and the posterior probability for the model of a single shared variant (PP4) underlying the eQTL and BMD association was 97.4% compared to the model of two independent variants (PP3=2.5%) (Figure 6B). The two independent variant model was favored for the CKB eQTL in skin tissue (PP3=68.7% versus PP4=26.1%) (Figure 6C) and the single shared variant model was favored for the TRMT61A eQTL in aorta tissue (PP4=93.5% versus PP3=5.2%) (Figure 6D). Importantly, as seen in mice, the “T” allele of rs11623869 was associated with increased (P<1.0 × 10−7) MARK3 levels, across many tissues, and decreased FNBMD (Figure 6E). These data indicate that lower levels of MARK3 in both humans and mice are associated with increased BMD. Though they do not exclude the involvement of other genes in the region, these data are consistent with the hypothesis that MARK3 is a causal BMD GWAS gene and at least partially responsible for the effects of the Chr. 14q32.32 association.
Figure 6.
MARK3 expression in multiple tissues is regulated by the same variants associated with BMD. A). H3K4me2 and H3K27ac modifications in primary human osteoblasts across the Chr. 14q32.32 association. Local eQTL influencing B) MARK3 expression in thyroid tissue, C) CKB expression skeletal muscle and D) TRMT61A expression in aorta. In B-D, PP4 is the posterior probability that the eQTL and BMD associations share a single common variant and PP3 is the posterior probability for the region harboring two independent variants, each affecting either gene expression or BMD. E) Effect of the “T” allele of rs11623869 (lead GWAS SNP at the Chr. 14q32.32 locus) on BMD (in red) and the expression of MARK3 in multiple tissues.
DISCUSSION
Our goal was to use a network-driven approach to predict putative causal genes for 64 genomic regions robustly associated with BMD. Using a disease-relevant co-expression network to inform GWAS, we were able to identify putative causal genes for 30 of 64 BMD GWAS loci. Based on the function of known genes within the OFM, and modules 6 and 9 as a whole, we were also able to infer that OFM genes impacted BMD by altering the activity of bone-forming osteoblasts. Our predictions represent a key first step in systematically interrogating each locus and generating a much deeper understanding of the genes and cellular processes that regulate BMD in humans.
We identified two network modules (6 and 9) that contained more genes implicated by GWAS than would be expected by chance. From prior work, we know that each of the 21 network modules are enriched for genes involved in particular biological processes. Modules 6 and 9 were enriched for processes specific to osteoblasts, such as “osteoblast differentiation” and “osteoblast proliferation” (Calabrese et al., 2012). The bone co-expression network was generated using transcriptomic profiles of marrow-free cortical bone fragments. Bone fragments primarily (>90%) consist of cells of the osteoblast-lineage (mainly osteocytes) (Bonewald, 2011). This likely underlies the reason that a similar “osteoclast” functional module was not found, even though bone mass is the net product of formation of bone by the osteoblast and resorption by the osteoclast (Boyle et al., 2003), and several genes implicated by GWAS (particularly at the 34 non-OFM loci), such as TNFSF11 (Yasuda et al., 1998) and PLEKHM1 (Van Wesenbeeck et al., 2007), play key roles in osteoclast biology. In fact, in our previous characterization of the co-expression network, none of the 21 modules demonstrated a strong “osteoclast” signature, likely due to the low numbers of osteoclasts that would have been included in the expression profiles used to construct the network (Calabrese et al., 2012). One might expect that an osteoclast-specific network would yield a functional module potentially informative for the non-OFM GWAS loci.
For our analysis we used a mouse bone co-expression network. Using a mouse network had its limitations, such as missing genes due to the lack of homologs and we could not consider non-coding RNAs as potentially causal genes. However, there were advantages to using this network. For example, the network had been well characterized (Calabrese et al., 2012; Farber et al., 2011; Mesner et al., 2014) and it was generated from cortical bone samples free of marrow. This last point is important since the profiles primarily represented bone cells, instead of marrow cells, and similar data has not been generated from a large number of human samples. We do expect, however, that networks derived from human bone transcriptomic data would lead to more informative networks for the purpose of informing BMD GWAS.
We predicted that the OFM was highly enriched for truly causal genes based on the following observations: 1) OFM genes were located in BMD GWAS loci, 2) OFM genes were co-expressed in bone, 3) 55% of OFM genes were known regulators of BMD, 4) OFM genes were closer to lead GWAS SNPs than non-OFM genes and 5) lead SNPs in OFM loci were located within osteoblast-specific regulatory elements. The overlap of lead OFM SNPs and osteoblast regulatory elements suggest that OFM loci impact BMD by altering the target genes in osteoblasts and the OFM provides a list of potential targets. These data strengthen the assertion that OFM genes are responsible for the effects of OFM loci. Our results also suggest that a significant fraction of the genetic signal for BMD identified to date influences BMD by altering the activity of osteoblastlineage cells.
We observed a number of BMD GWAS loci that harbored multiple OFM genes. There are at least two possible explanations for this observation. First, it is highly likely that not all OFM genes are responsible for BMD GWAS signals. Though such genes may be involved in osteoblast activity due to their membership in the OFM, the localization of some OFM genes in GWAS regions is expected to be coincidental. A second explanation is that at a subset of BMD GWAS loci there are multiple functional genes. An example is the Chr. 7q31.31 region, which contained two independent associations in GEFOSII and three OFM genes; WNT16, FAM3C and CPED1. This region appears to be quite genetically complex and there is evidence from other GWASs that it contains more than just the two associations identified by GEFOSII (Chesi et al., 2015; Cho et al., 2009; Estrada et al., 2012; Kemp et al., 2014; Medina-Gómez et al., 2012; Moayyeri et al., 2014; Zhang et al., 2013a; Zheng et al., 2012). As a result, it is possible that all three OFM genes located in the 7q31.31 region are responsible for independent genetic associations with BMD.
The BMD GWAS locus on Chr. 2p16.2 harbors variants spanning an ~10 Kbp intergenic region flanked by two genes, C2orf73 and SPTBN1. Sptbn1 was a member of the OFM, and of the two genes, BMD was altered in mice with perturbed Sptbn1 expression. SPTBN1 is a molecular scaffolding protein that links the actin cytoskeleton to the plasma membrane (Zhang et al., 2013b). Through this role, it has been implicated in the regulation of cell shape, adhesion and TGF-β signaling (Zhang et al., 2013b). It was most strongly connected to genes involved in alpha-actinin binding and cell adhesion in the bone co-expression network, suggesting that it plays the same role in bone. In mice the effect of Sptbn1 perturbation was sexually dimorphic, however, the Chr. 2p16.2 locus affected BMD equally in both sexes. In mice sexually dimporphic effects of genetic perturbations is common (as an example (Mesner et al., 2014)). In humans, however, there is little evidence that genetic effects on BMD differ between sexes (Liu et al., 2012a). There are many potential explanations as to why global knockouts in mice show sexually dimorphic effects on bone. Most importantly though, the data presented link Sptbn1 to the regulation of BMD. Although further work is needed to determine how the Chr. 2p16.2 locus influences BMD, our results suggest that SPTBN1 plays a role in the 2p16.2 association.
MARK3 is one of four mammalian homologs of the C. elegans partitioning defective 1 (par1) kinase that regulates cell polarity (Drewes et al., 1998). MARK3 is a member of the AMP-kinase (AMPK) family, which has been found to participate in diverse cellular functions (Bright et al., 2009). Mice deficient in Mark3 have been shown to be resistant to high fat diet induced obesity, hepatic steatosis and diabetes (Lennerz et al., 2010). We demonstrated that in both humans and mice, MARK3 transcript levels are negatively correlated with BMD. Using GTEx data, we observed that MARK3 was regulated by the same eQTL in nearly every tissue. Even though bone tissue and bone cells are not included in GTEx, it is likely that the pan-tissue local eQTL we identified is also operative in bone cells. In the mouse, Mark3 expression started out high in primary osteoblasts and significantly decreased as differentiation progressed. Furthermore, its expression was inversely correlated with key genes involved in osteoblast differentiation. These data suggest that Mark3 may act as a “brake” for osteoblast differentiation and its decrease early in differentiation may remove this brake enabling differentiation to progress. All together, our results provide strong evidence that MARK3 is responsible for the effects of the BMD GWAS locus on Chr. 14q32.32.
In summary, through the integration of GWAS data and a bone co-expression network, we provide predictions of potential causal genes at 30 BMD GWAS loci and use this information to identify causal genes for two GWAS loci. Based on our results, we believe that significant insight into complex disease etiology can be gained using networks to inform GWAS.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Charles R. Farber
Center for Public Health Genomics
P.O. Box 800717
University of Virginia
Charlottesville, VA 22908
Tel: 434-243-8584
Fax: 434-982-1815
crf2s@virginia.edu
EXPERIMETNAL MODEL AND SUBJECT DETAILS
Generation and characterization of Mark3 gene trap mice
The study was carried out in strict accordance with NIH’s Guide for the Care and Use of Laboratory Animals. Additionally, the University of Virginia Institutional Animal Care and Use Committee approved all animal procedures. Mark3 gene trap mice were generated using targeted embryonic stem cell (JM8A1.N3) clones heterozygous for the Mark3tm1a(KOMP)Mbp gene trap allele obtained from the International Knockout Mouse Project (KOMP; https://www.komp.org). Two (H02 and C06) KOMP ES clones were karyotyped and injected using a XYClone Laser (Hamilton Thorne, Beverly, MA) into B6N-Tyrc-Brd/BrdCrCrl (Charles River, Wilmington, MA) 8-cell stage embryos to create chimeric mice. Resultant chimeras were bred to B6N-Tyrc-Brd/BrdCrCrl mice to obtain germline transmission of the Mark3 gene trap allele. Confirmation of transmission in resultant black or brown pups was determined by PCR using the following two sets of primer pairs: Common-lox-P-F/Mark3-3’R and Mark3-5′ F/Common-en2-3’R (Table S4). The first and second primer sets resulted in amplicons of 156 bp and 373 bp, respectively, both denoting the presence of the gene-trap allele. Albino littermates served as negative controls.
Mice heterozygous for the Mark3tm1a(KOMP)Mbp gene-trap allele were intercrossed to generate two cohorts of experimental mice. PCR was used to genotype experimental mice using primers Mark 3-172-5' and Mark3-3’R (Table S4), which resulted in a 238 bp amplicon from the gene-trap allele and 204 bp amplicon from the wildtype allele. BMD was measured in the first experimental cohort using DEXA (described below). At weaning, F2 mice were randomly allocated to a cage irrespective of genotype. Mice were housed in Thoren (Thoren Caging Systems, Hazelton, PA) ventilated racks and provided irradiated Teklad diet (Envigo, Huntingdon, UK). Mice from the second cohort were euthanized and dissected femurs were subjected to μCT and biomechanical testing (described below). For all procedures the experimenter was blinded to genotype.
Osteoblast cell cultures
MC3T3-E1 clone 4 (MC4) cells were obtained from the American Type Culture Collection (Manassas, VA). These cells were maintained in αMEM media (Cellgro, Herndo, VA), supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), penicillin (50 IU/ml) and streptomycin (50μg/ml).
Primary calvarial osteoblasts were isolated from 3-9 day old C57BL6/J (Jackson Laboratory, Stock #000664) neonates using sequential Collagenase P digestions. Cells were plated into 6 well plates at 300,000 cells in 2ml sterile plating media (DMEM, 10% heat-inactivated FBS, 100 U/ml penicillin, 100 μg/ml streptomycin) per well. After 24 hours, confluent cells were washed 1x with DPBS (Gibco) and placed in sterile differentiation media (MEM alpha, 10% heat inactivated FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 50 ug/ml ascorbic acid, 4 mM B-glycerophosphate). Every 48 hours thereafter cells were washed one time with DPBS (Gibco) and differentiation media was replaced until cells were collected for analysis at day 10.
METHODS DETAILS
Mark3 immunohistochemistry
Femurs were fixed in 10% neutral buffered formalin (Fisher Scientific) and decalcified with 4% EDTA for 15 days, dehydrated and cleared on a tissue processor and embedded in paraffin. Five-micron sections were deparaffinized in xylene followed by increasing grades of ethanol and rehydrated on slides. Antigen retrieval was accomplished by placing the slides in Coplin jars in 60°C citrate buffer (pH 6) and maintained at 60°C in a water bath for 20 minutes. Endogenous peroxidase was quenched using 3% hydrogen peroxide and non-specific staining was blocked using 0.1%BSA and 5% normal goat serum in Tris buffered saline. Sections were treated with rabbit anti-MARK3 polyclonal antibody (1:100 dilution, GeneTex, Irvine, CA) overnight at 4°C in a sealed moist chamber. The slides were washed and incubated with a goat anti-rabbit peroxidase labeled secondary antibody (1:100 dilution) for 1 hr. at room temperature in the dark. The slides were than washed and treated with DAB (DAB Development Kit, Vector) for 2-5 minutes. The slides were washed, counterstained with hematoxylin, dehydrated and cover-slipped.
Mark3 immunofluorescence
Mark3 immunofluorescence was performed in MC3T3 cells as previously described (Hebert and Stains, 2013) using a rabbit anti-MARK3 polyclonal antibody (GeneTex). Actin was labeled by incubation of the fixed cells with 1μM TRITC-labeled phalloidin for 30 minutes prior to mounting. Once collected images were processed (contrast, brightness, and merged) in Adobe Photoshop CS4 (Adobe Systems, Inc, San Jose, CA).
Mark3 knockdown and quantification of osteoblast activity
For siRNA transfection experiments, primary calvarial osteoblasts were plated at 150,000 cells/well followed by transfection ~24 hours post-plating with Lipofectamine 2000 Reagent (Invitrogen) according to manufacturer’s directions with differentiation commencing 72 hours post-plating. Stealth Select RNAi siRNAs (Invitrogen) targeting Mark3 were used to knockdown its expression in calvarial osteoblasts (Table S4). The Stealth RNAi Negative Control Duplex (Invitrogen) was used as a scrambled control.
qPCR was used to measure Mark3 gene expression. The following primer sets were used Mark3-F/Mark3-R and 36B4-F/36B4-R (Table S4). Relative quantification was determined by the 2(-Delta Delta CT)) method using 36B4 as the reference gene (Livak and Schmittgen, 2001).
Mineralized nodule formation was measured by staining cultures at 10 days post-differentiation with Alizarin Red (40 mM) (pH 5.6). The stained cells were imaged and nodule number was measured using ImageJ (NIH) (Schneider et al., 2012). Alizarin Red was quantified by destaining cultures with 5% Perchloric acid and determining the optical density (405 nM) of the resulting solution against a standard curve. All results were obtained from three independent experiments.
DEXA and μCT
At 12 and 16 weeks of age BMD was measured in male Mark3tm1a(KOMP)Mbp wild-type (Mark3+/+; N=12) and Mark3tm1a(KOMP)Mbp heterozygous (Mark3+/−; N=18) gene trap mice using a Lunar PIXImus II Mouse Densitometer (GE Medical Systems Model 51045; Madison, WI, USA) as described in (Mesner et al., 2014). Distal femurs from 12 week old male Mark3+/+ (N=8) and Mark3tm1a(KOMP)Mbp homozygous gene trap mice (Mark3−/−; N=13) were scanned using a vivaCT 40 imaging system (Scanco Medical) to measure trabecular bone volume fraction and microarchitecture of the distal femur and cortical microarchitecture of the femoral midshaft. The samples were scanned completely submerged in 95% ethanol. Scans were performed at energy level of 55 kVp and intensity of 145 μA and an isotropic voxel size of 12.5 μm. Trabecular microarchitecture were evaluated from approximately 200 consecutive slices of the secondary spongiosa. Cortical bone scans were performed at the midpoint of each femur. A total of 50 consecutive slices were scanned.
Biomechanical testing
The right femurs from 12-week old male Mark3 wild-type (+/+; N=8) and homozygous (−/−; N=13) gene trap mice were loaded to failure in four-point bending. All whole bone tests were conducted by loading the femur in the anterior to posterior direction, such that the posterior quadrant was subjected to tensile loads. The widths of the lower and upper supports of the four-point bending apparatus were 7 mm and 3 mm, respectively. Tests were conducted with a deflection rate of 0.05 mm/s using a servohydraulic materials test system (Instron Corp., Norwood, MA). The load and mid-span deflection were acquired directly at a sampling frequency of 200 Hz. Load-deflection curves were analyzed for strength (maximum load), stiffness (the slope of the initial portion of the curve), post-yield deflection, and total work. Femora were tested at room temperature and kept moist with phosphate buffered saline during all tests.
QUANTIFICATION AND STATISTICAL ANALYSIS
Defining the BGIG
All analyses were performed in the R language and environment for statistical computing (Team). For each of the 64 lead BMD GWAS SNPs identified by (Estrada et al., 2012) we used Plink v1.9 (Chang et al., 2015) and data from the 1000 Genome Project Phase 3 ((1000 Genomes Project Consortium, 2010) to identify proxy SNPs (r2<0.7). Each GWAS locus was physically defined by the positions of the left and right-most proxy SNPs. We then downloaded a catalog of all RefSeq transcripts from the USCS Genome Browser (https://genome.ucsc.edu). All SNP and gene locations were relative to the hg38 genome assembly. All RefSeq transcripts without a clear mouse homolog were removed. We then identified all remaining RefSeq genes located in or overlapping with one of the 64 defined GWAS loci. In addition, we included the closest gene up- and downstream (based on distance to TSS). The topGO R package (Alexa and Rahnenfuhrer, 2010) was used to measure gene ontology enrichments for the BGIG.
Identifying and characterizing enriched modules
The bone co-expression network used in this study was extensively described in (Calabrese et al., 2012). To identify enriched modules, we identified mouse homologs for all BGIG genes. The module membership for each mouse homolog was determined. Some homologs mapped to more than one module due to different microarray probes for the same gene belonging to different modules. A Fisher’s exact test was used to identify modules with significantly (Bonferroni P<2.3 × 10−3) enriched in BGIG genes. The topGO R package (Alexa and Rahnenfuhrer, 2010) was used to measure gene ontology enrichments for modules 6 and 9.
Characterizing OFM genes and loci
Osteoblast gene expression profiles for OFM genes were assessed using data on purified primary calvarial osteoblasts using data from GEO (GSE54461) and BioGPS (http://biogps.org). We used epigenomics data generated by the ENCODE and NIH Epigenomics Roadmap Consortiums to quantify the overlap between lead BMD GWAS SNPs and epigenetic marks (ENCODE Project Consortium et al., 2012; Roadmap Epigenomics Consortium et al., 2015). All data were downloaded from the Epigenomics Roadmap Consortium webportal (http://www.roadmapepigenomics.org/data/). The data for histone modifications were downloaded as BED files in GappedPeak format. The primary analysis used data on primary human osteoblasts. A secondary analysis comparing H3K27ac and H3K4me2 was performing comparing all Epigenomics Roadmap samples were those modifications had been measured. The analysis of the ChromHMM generated core 15-state epigenome model used the data from primary human osteoblasts (Ernst and Kellis, 2012). We downloaded dbSNP142 from the UCSC genome browser (https://genome.ucsc.edu) and 1000 sets of 30 random SNPs selected to match the lead OFM SNPs on minor allele frequence and distance to the nearest TSS. The random SNP sets were used to evaluate overlap at histone modification for lead OFM SNPs. To compare the expression of genes in OFM loci and non-OFM loci we used microarray expression data from human bone biopsies described in (Reppe et al., 2010).
Analysis of BMD data from the IMPC
The International Mouse Knockout Consortium (Skarnes et al., 2011) and the IMPC have generated and phenotyped mice harboring null alleles for 4930505A04Rik (4930505A04Riktm1b(KOMP)Wtsi) (C2orf73 homolog) (N=8 females and N=8 males) and Sptbn1 (Sptbn1tm1a(EUCOMM)Wtsi) mutant mice (N=7 females and N=10 males). Phenotypes for the appropriate controls (C57BL/6) were also collected (controls for 4930505A04Rik, N=252 females and N=257 males and controls for Sptbn1, N=777 females and N=814 males). A description of the battery of phenotypes collected on mutants can be found at (http://www.mousephenotype.org/impress/procedures/4). The mice were 14 weeks of age at DEXA scanning and both sexes for both mutants were included. We downloaded raw BMD, body weight and metadata for both mutants from the IMPC webportal (http://www.mousephenotype.org). These data were analyzed using the PhenStat R package (Kurbatova et al., 2015). PhenStat was developed to analyze data generated by the IMPC in which a large number of wild-type controls are phenotyped across a wide-time range in batches and experimental mutant animals are tested in small groups interspersed among wild-type batches. We used the Mixed Model framework in PhenStat to analyze BMD data on the mutants. The mixed model framework starts with a full model (with fixed effects of genotype, sex, genotype × sex and weight and batch as a random effect) and ends with final reduced model and genotype effect evaluation procedures (Kurbatova et al., 2015; West et al., 2014).
Analysis of GTEx data
We queried GTEx for eQTL results for all genes in the Chr. 2q16.2 and Chr. 14q32.32 BMD GWAS regions. A description of the GTEx eQTL analysis methods can be found at (http://www.gtexportal.org/home/). Results from the version 6 analyses for all significant SNP expression pairs were downloaded from the GTEx web portal (http://www.gtexportal.org/home/) and used for downstream analyses. The “coloc” R package was used for the Bayesian localization analysis using default parameters (Giambartolomei et al., 2014).
Additional statistical analysis
The results of the siRNA analysis between groups were compared using the Student’s t-test. Phenotypic data in Mark3 mutant and wild-type mice were analyzed using the lsmeans R package (Lenth, 2016). Input for the lsmeans function was a linear model including terms for genotype and weight at sacrifice. Weight at sacrifice was a significant covariate for all evaluated bone phenotypes. Values reported were means ± s.e.m. for siRNA experiments and lsmeans ± s.e.m. for bone phenotypes. Comparisons at P<0.05 were deemed significant.
Supplementary Material
ACKNOWLEDGEMENTS
Research reported in this publication was supported in part by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number R01AR057759 to CRF. The authors acknowledge Wenhao Xu (University of Virginia) and the Genetically Engineered Mouse Models (GEMM) core for their technical assistance in generating the Mark3 gene trap mouse. Mark3 targeted ES cells were generated by the trans-NIH KOMP project and obtained from the KOMP Repository. NIH grants to Velocigene at Regeneron Inc (U01HG004085) and the CSD Consortium (U01HG004080) funded the generation of gene-targeted ES cells for 8500 genes in the KOMP Program and archived and distributed by the KOMP Repository at UC Davis and CHORI (U42RR024244). We thank the IMPC for accessibility to BMD data on Sptbn1 and 4930505A04Rik knockout mice. The data used for the analyses described in this manuscript were obtained from the IMPC Portal on 10/15/15. The GTEx Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health. Additional funds were provided by the NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. Donors were enrolled at Biospecimen Source Sites funded by NCI\SAIC-Frederick, Inc. (SAIC-F) subcontracts to the National Disease Research Interchange (10XS170), Roswell Park Cancer Institute (10XS171), and Science Care, Inc. (X10S172). The Laboratory, Data Analysis, and Coordinating Center (LDACC) was funded through a contract (HHSN268201000029C) to The Broad Institute, Inc. Biorepository operations were funded through an SAIC-F subcontract to Van Andel Institute (10ST1035). Additional data repository and project management were provided by SAIC-F (HHSN261200800001E). The Brain Bank was supported by a supplements to University of Miami grants DA006227 & DA033684 and to contract N01MH000028. Statistical Methods development grants were made to the University of Geneva (MH090941 & MH101814), the University of Chicago (MH090951, MH090937, MH101820, MH101825), the University of North Carolina - Chapel Hill (MH090936 & MH101819), Harvard University (MH090948), Stanford University (MH101782), Washington University St Louis (MH101810), and the University of Pennsylvania (MH101822). The data described in this manuscript were obtained from the GTEx Portal on 01/15/16.
Footnotes
DATA AND SOFTWARE AVAILABILITY
The R script used for generating the BGIG and network mapping is available from https://github.com/charlesfarber/GWAS_networks
AUTHOR CONTRIBUTIONS
CRF conceived the experiments. GMC, LDM, JPS, SMT, MCH, CJM and CRF performed the experiments. CRF analyzed the data. CRF wrote the paper and received feedback and edits from all other authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A, Rahnenfuhrer J. topGO: enrichment analysis for gene ontology (R package version) 2010.
- Balemans W, Ebeling M, Patel N, Van Hul E, Olson P, Dioszegi M, Lacza C, Wuyts W, Van Den Ende J, Willems P, et al. Increased bone density in sclerosteosis is due to the deficiency of a novel secreted protein (SOST) Hum Mol Genet. 2001;10:537–543. doi: 10.1093/hmg/10.5.537. [DOI] [PubMed] [Google Scholar]
- Black DM, Rosen CJ. Clinical Practice. Postmenopausal Osteoporosis. N Engl J Med. 2016;374:254–262. doi: 10.1056/NEJMcp1513724. [DOI] [PubMed] [Google Scholar]
- Bonewald LF. The amazing osteocyte. J. Bone Miner. Res. 2011;26:229–238. doi: 10.1002/jbmr.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- Bright NJ, Thornton C, Carling D. The regulation and function of mammalian AMPK-related kinases. Acta Physiol (Oxf) 2009;196:15–26. doi: 10.1111/j.1748-1716.2009.01971.x. [DOI] [PubMed] [Google Scholar]
- Brown SDM, Moore MW. The International Mouse Phenotyping Consortium: past and future perspectives on mouse phenotyping. Mamm Genome. 2012a;23:632–640. doi: 10.1007/s00335-012-9427-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SDM, Moore MW. Towards an encyclopaedia of mammalian gene function: the International Mouse Phenotyping Consortium. Dis Model Mech. 2012b;5:289–292. doi: 10.1242/dmm.009878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkow ME, Gardner JC, Van Ness J, Paeper BW, Kovacevich BR, Proll S, Skonier JE, Zhao L, Sabo PJ, Fu Y, et al. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. Am. J. Hum. Genet. 2001;68:577–589. doi: 10.1086/318811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese G, Bennett BJ, Orozco L, Kang HM, Eskin E, Dombret C, De Backer O, Lusis AJ, Farber CR. Systems genetic analysis of osteoblast-lineage cells. PLoS Genet. 2012;8:e1003150. doi: 10.1371/journal.pgen.1003150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Califano A, Butte AJ, Friend S, Ideker T, Schadt E. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nat. Genet. 2012;44:841–847. doi: 10.1038/ng.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 2015. 2015;4:1. doi: 10.1186/s13742-015-0047-8. 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang E-J, Ha J, Oerlemans F, Lee YJ, Lee SW, Ryu J, Kim HJ, Lee Y, Kim H-M, Choi JY, et al. Brain-type creatine kinase has a crucial role in osteoclast-mediated bone resorption. Nat Med. 2008;14:966–972. doi: 10.1038/nm.1860. [DOI] [PubMed] [Google Scholar]
- Chesi A, Mitchell JA, Kalkwarf HJ, Bradfield JP, Lappe JM, McCormack SE, Gilsanz V, Oberfield SE, Hakonarson H, Shepherd JA, et al. A trans-ethnic genome-wide association study identifies gender-specific loci influencing pediatric aBMD and BMC at the distal radius. Hum Mol Genet. 2015;24:5053–5059. doi: 10.1093/hmg/ddv210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban H-J, Yoon D, Lee MH, Kim D-J, Park M, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. 2009;41:527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- Claussnitzer M, Dankel SN, Kim K-H, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V, et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N Engl J Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes G, Ebneth A, Mandelkow EM. MAPs, MARKs and microtubule dynamics. Trends Biochem. Sci. 1998;23:307–311. doi: 10.1016/s0968-0004(98)01245-6. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nature Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada K, Styrkarsdottir U, Evangelou E, Hsu Y-H, Duncan EL, Ntzani EE, Oei L, Albagha OME, Amin N, Kemp JP, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. 2012;44:491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber CR. Identification of a gene module associated with BMD through the integration of network analysis and genome-wide association data. J. Bone Miner. Res. 2010;25:2359–2367. doi: 10.1002/jbmr.138. [DOI] [PubMed] [Google Scholar]
- Farber CR. Systems-level analysis of genome-wide association data. G3 (Bethesda) 2013;3:119–129. doi: 10.1534/g3.112.004788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber CR, Bennett BJ, Orozco L, Zou W, Lira A, Kostem E, Kang HM, Furlotte N, Berberyan A, Ghazalpour A, et al. Mouse genome-wide association and systems genetics identify Asxl2 as a regulator of bone mineral density and osteoclastogenesis. PLoS Genet. 2011;7:e1002038. doi: 10.1371/journal.pgen.1002038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- Gass M, Dawson-Hughes B. Preventing osteoporosis-related fractures: an overview. Am. J. Med. 2006;119:S3–S11. doi: 10.1016/j.amjmed.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh K-I, Cusick ME, Valle D, Childs B, Vidal M, Barabasi A-L. The human disease network. Proc Natl Acad Sci USA. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- Gustafsson M, Gawel DR, Alfredsson L, Baranzini S, Björkander J, Blomgran R, Hellberg S, Eklund D, Ernerudh J, Kockum I, et al. A validated gene regulatory network and GWAS identifies early regulators of T cell-associated diseases. Sci Transl Med. 2015;7:313ra178–313ra178. doi: 10.1126/scitranslmed.aad2722. [DOI] [PubMed] [Google Scholar]
- Hebert C, Stains JP. An intact connexin43 is required to enhance signaling and gene expression in osteoblast-like cells. J Cell Biochem. 2013;114:2542–2550. doi: 10.1002/jcb.24603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan T, Meng Q, Saleh MA, Norlander AE, Joehanes R, Zhu J, Chen BH, Zhang B, Johnson AD, Ying S, et al. Integrative network analysis reveals molecular mechanisms of blood pressure regulation. Mol Syst Biol. 2015;11:799. doi: 10.15252/msb.20145399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P, Zhao Z. Network.assisted analysis to prioritize GWAS results: principles, methods and perspectives. Hum Genet. 2014;133:125–138. doi: 10.1007/s00439-013-1377-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp JP, Medina-Gómez C, Estrada K, St Pourcain B, Heppe DHM, Warrington NM, Oei L, Ring SM, Kruithof CJ, Timpson NJ, et al. Phenotypic dissection of bone mineral density reveals skeletal site specificity and facilitates the identification of novel loci in the genetic regulation of bone mass attainment. PLoS Genet. 2014;10:e1004423. doi: 10.1371/journal.pgen.1004423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurbatova N, Mason JC, Morgan H, Meehan TF, Karp NA. PhenStat: A Tool Kit for Standardized Analysis of High Throughput Phenotypic Data. PLoS ONE. 2015;10:e0131274. doi: 10.1371/journal.pone.0131274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MDM, Eldridge JV, Ramachandran S, Raphael BJ. Network analysis of GWAS data. Curr. Opin. Genet. Dev. 2013;23:602–610. doi: 10.1016/j.gde.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennerz JK, Hurov JB, White LS, Lewandowski KT, Prior JL, Planer GJ, Gereau RW, Piwnica-Worms D, Schmidt RE, Piwnica-Worms H. Loss of Par-1a/MARK3/C-TAK1 kinase leads to reduced adiposity, resistance to hepatic steatosis and defective gluconeogenesis. Mol Cell Biol. 2010 doi: 10.1128/MCB.01472-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenth RV. Least-squares means: the R Package lsmeans. J Stat Softw. 2016 [Google Scholar]
- Little RD, Carulli JP, Del Mastro RG, Dupuis J, Osborne M, Folz C, Manning SP, Swain PM, Zhao S-C, Eustace B, et al. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am. J. Hum. Genet. 2002;70:11–19. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C-T, Estrada K, Yerges-Armstrong LM, Amin N, Evangelou E, Li G, Minster RL, Carless MA, Kammerer CM, Oei L, et al. Assessment of gene-by-sex interaction effect on bone mineral density. J. Bone Miner. Res. 2012a;27:2051–2064. doi: 10.1002/jbmr.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C-T, Karasik D, Zhou Y, Hsu Y-H, Genant HK, Broe KE, Lang TF, Samelson EJ, Demissie S, Bouxsein ML, et al. Heritability of prevalent vertebral fracture and volumetric bone mineral density and geometry at the lumbar spine in three generations of the Framingham study. 2012b;27:954–958. doi: 10.1002/jbmr.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mäkinen V-P, Civelek M, Meng Q, Zhang B, Zhu J, Levian C, Huan T, Segrè AV, Ghosh S, Vivar J, et al. Integrative genomics reveals novel molecular pathways and gene networks for coronary artery disease. PLoS Genet. 2014;10:e1004502. doi: 10.1371/journal.pgen.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Gómez C, Kemp JP, Estrada K, Eriksson J, Liu J, Reppe S, Evans DM, Heppe DHM, Vandenput L, Herrera L, et al. Meta-analysis of genome-wide scans for total body BMD in children and adults reveals allelic heterogeneity and age-specific effects at the WNT16 locus. PLoS Genet. 2012;8:e1002718. doi: 10.1371/journal.pgen.1002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesner LD, Ray B, Hsu Y-H, Manichaikul A, Lum E, Bryda EC, Rich SS, Rosen CJ, Criqui MH, Allison M, et al. Bicc1 is a genetic determinant of osteoblastogenesis and bone mineral density. J Clin Invest. 2014;124:2736–2749. doi: 10.1172/JCI73072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayyeri A, Hsu Y-H, Karasik D, Estrada K, Xiao S-M, Nielson C, Srikanth P, Giroux S, Wilson SG, Zheng H-F, et al. Genetic determinants of heel bone properties: genome-wide association meta-analysis and replication in the GEFOS/GENOMOS consortium. Hum Mol Genet. 2014;23:3054–3068. doi: 10.1093/hmg/ddt675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Nayak RR, Kearns M, Spielman RS, Cheung VG. Coexpression network based on natural variation in human gene expression reveals gene interactions and functions. 2009;19:1953–1962. doi: 10.1101/gr.097600.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, Floratos A, Sham P-C, Li MJ, Wang J, et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- Ozanick S, Krecic A, Andersland J, Anderson JT. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. Rna. 2005;11:1281–1290. doi: 10.1261/rna.5040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Development Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; [Google Scholar]
- Ralston SH, de Crombrugghe B. Genetic regulation of bone mass and susceptibility to osteoporosis. Genes Dev. 2006;20:2492–2506. doi: 10.1101/gad.1449506. [DOI] [PubMed] [Google Scholar]
- Ralston SH, Uitterlinden AG. Genetics of osteoporosis. Endocrine Reviews. 2010;31:629–662. doi: 10.1210/er.2009-0044. [DOI] [PubMed] [Google Scholar]
- Reppe S, Refvem H, Gautvik VT, Olstad OK, Høvring PI, Reinholt FP, Holden M, Frigessi A, Jemtland R, Gautvik KM. Eight genes are highly associated with BMD variation in postmenopausal Caucasian women. Bone. 2010;46:604–612. doi: 10.1016/j.bone.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman E. Bone modeling and remodeling. Crit. Rev. Eukaryot. Gene Expr. 2009;19:219–233. doi: 10.1615/critreveukargeneexpr.v19.i3.40. [DOI] [PubMed] [Google Scholar]
- Simonet WS, Lacey DL, Dunstan CR, Kelley M, Chang MS, Lüthy R, Nguyen HQ, Wooden S, Bennett L, Boone T, et al. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell. 1997;89:309–319. doi: 10.1016/s0092-8674(00)80209-3. [DOI] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smemo S, Tena JJ, Kim K-H, Gamazon ER, Sakabe NJ, Gómez-Marín C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wesenbeeck L, Odgren PR, Coxon FP, Frattini A, Moens P, Perdu B, MacKay CA, Van Hul E, Timmermans J-P, Vanhoenacker F, et al. Involvement of PLEKHM1 in osteoclastic vesicular transport and osteopetrosis in incisors absent rats and humans. J Clin Invest. 2007;117:919–930. doi: 10.1172/JCI30328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrieras J-B, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, Pritchard JK. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viel A, Branton D. Spectrin: on the path from structure to function. Current Opinion in Cell Biology. 1996;8:49–55. doi: 10.1016/s0955-0674(96)80048-2. [DOI] [PubMed] [Google Scholar]
- West BT, Welch KB, Galecki AT. Linear Mixed Models: A Practical Guide Using Statistical Software. CRC Press; 2014. [Google Scholar]
- Yasuda H, Shima N, Nakagawa N, Yamaguchi K, Kinosaki M, Mochizuki S, Tomoyasu A, Yano K, Goto M, Murakami A, et al. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc Natl Acad Sci USA. 1998;95:3597–3602. doi: 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- Zhang L, Choi HJ, Estrada K, Leo PJ, Li J, Pei Y-F, Zhang Y, Lin Y, Shen H, Liu Y-Z, et al. Multistage genome-wide association meta-analyses identified two new loci for bone mineral density. Hum Mol Genet. 2013a doi: 10.1093/hmg/ddt575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Zhang C, Zhao Q, Li D. Spectrin: structure, function and disease. Sci China Life Sci. 2013b;56:1076–1085. doi: 10.1007/s11427-013-4575-0. [DOI] [PubMed] [Google Scholar]
- Zheng H-F, Spector TD, Richards JB. Insights into the genetics of osteoporosis from recent genome-wide association studies. Expert Rev Mol Med. 2011;13:e28. doi: 10.1017/S1462399411001980. [DOI] [PubMed] [Google Scholar]
- Zheng H-F, Tobias JH, Duncan E, Evans DM, Eriksson J, Paternoster L, Yerges-Armstrong LM, Lehtimäki T, Bergstrom U, Kähönen M, et al. WNT16 influences bone mineral density, cortical bone thickness, bone strength, and osteoporotic fracture risk. PLoS Genet. 2012;8:e1002745. doi: 10.1371/journal.pgen.1002745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.