SUMMARY
It is unclear how standing genetic variation affects the prognosis of prostate cancer patients. To provide one controlled answer to this problem, we crossed a dominant, penetrant mouse model of prostate cancer to Diversity Outbred mice, a collection of animals that carries over 40 million SNPs. Integration of disease phenotype and SNP variation data in 493 F1 males identified a metastasis modifier locus on Chromosome 8 (LOD=8.42); further analysis identified the genes Rwdd4, Cenpu, and Casp3 as functional effectors of this locus. Accordingly, analysis of over 5,300 prostate cancer patient samples revealed correlations between the presence of genetic variants at these loci, their expression levels, cancer aggressiveness, and patient survival. We also observed that ectopic overexpression of RWDD4 and CENPU increased the aggressiveness of two human prostate cancer cell lines. In aggregate, our approach demonstrates how well-characterized genetic variation in mice can be harnessed in conjunction with systems genetics approaches to identify and characterize germline modifiers of human disease processes.
Graphical Abstract
INTRODUCTION
It is estimated that 180,890 new prostate cancer cases will be diagnosed in the US in 2016 (Siegel et al. 2016). However, only approximately 14% of these men will succumb directly from prostate cancer. Serum measurement of prostate specific antigen (PSA) is commonly used to screen for prostate cancer, yet is limited in both its ability to identify patients at risk of disease progression, and to distinguish between aggressive and indolent disease at the time of diagnosis (Hayes et al. 2014; Romero et al. 2014). Consequently, these inaccuracies, as well as those of other prognostic tests, lead to overtreatment. The morbidity associated with definitive treatment of prostate cancer is high and the overall economic impact of over-treatment is substantial (Aizer et al. 2015; Wilt et al. 2015). Thus, advances in the molecular characterization of prostate cancer are needed to accurately identify men at risk of fatal prostate cancer.
Although non-hereditary, somatic mutations initiate prostate tumorigenesis and ultimately metastasis, we hypothesize that hereditary, germline variation has a modifying effect on each of these characteristics. Epidemiological studies support this hypothesis, with, for example, an earlier study of 1,304 father-son prostate cancer pairs demonstrating that prostate cancer-specific survival was significantly higher in sons with a father that had survived >60 months compared to those with a father who survived <24 months (HR = 0.62, 95% CI 0.41 – 0.94) (Hemminki et al. 2008). The findings of earlier family-based linkage studies, which employ ‘high-risk’ prostate cancer pedigrees where multiple individuals from the same family are affected, support a role for hereditary variation in aggressive prostate development. These studies have led to the identification of numerous genetic loci associated with aggressive disease susceptibility (reviewed in (Ostrander et al. 2006)). Since these regions of linkage typically encompass numerous genes, human genome-wide association studies (GWAS) have been employed to identify causal variants. However, GWAS have identified relatively few variants associated with aggressive prostate cancer, which likely reflects a host of confounding variables such as varied environmental exposures between subjects, small cohort sizes, case-control overlap, and the requirement for stringent correction for multiple testing (Bjorkegren et al. 2015).
A powerful complement to human GWAS are systems genetics approaches in mouse models, which allow for control of key confounding variables of human studies such as environmental variation. To study how germline variation modifies susceptibility to aggressive prostate cancer, we have crossed the C57BL/6-Tg(TRAMP)8247Ng/J (TRAMP) mouse model of prostate cancer (Gingrich et al. 1997) to other inbred mouse strains. Tumorigenesis in the TRAMP mouse is induced through prostate-specific expression of the small and large SV40-T antigens, which act to inactivate Rb and p53, thus resulting in the development of prostate cancer in 100% of male mice by 30 weeks. TRAMP mice develop neuroendocrine prostate tumors, which represents a form of prostate cancer that accounts for 25% of fatal cases (Beltran et al. 2012; Wang et al. 2014). Our earlier work demonstrated that introducing hereditary variation by breeding into the TRAMP mouse substantially modulates disease aggressiveness (Patel et al. 2013). In this ‘strain survey’ experiment, we crossed TRAMP to one of eight strains (five classical laboratory strains [C57BL/6J, A/J, 129S1/SvImJ, NOD/ShiLtJ, and NZO/HlLtJ] and three wild-derived lines [CAST/EiJ, PWK/PhJ, and WSB/EiJ]) and quantified tumor growth and metastasis in transgene positive F1 males. We observed a tremendous variation in disease aggressiveness, with, for example, a five-fold increase in tumor burden in (TRAMP × NOD/ShiLtJ) F1 males compared to wildtype TRAMP mice. Conversely, we observed profound suppression of tumorigenesis in (TRAMP × PWK/PhJ) F1 males, with only 1/37 animals developing macroscopic tumorigenesis at the 30 week experimental endpoint. Since tumorigenesis was initiated by the same somatic event (i.e., expression of the SV40-T transgene) in each mouse, and transgene expression was equal in each F1 strain we concluded that germline variation was impacting disease aggressiveness. Subsequent studies have since demonstrated that it is possible to identify modifiers of aggressive prostate cancer through quantitative trait locus (QTL) mapping (Lee et al. 2015; Williams et al. 2014). However, these earlier QTL studies were limited by the small amount of genetic variation encompassed in mouse mapping populations.
In our current study, we have overcome the limitations of low genetic diversity and poor mapping resolution by breeding TRAMP mice to ‘Diversity Outbred’ (J:DO) heterogeneous stock mice (Churchill et al. 2012), which are outbred stock derived from the same eight strains used in the ‘strain survey’ experiment described above. They are maintained by random mating and are superior to other mapping populations in that they carry over 40 million SNPs, have fine recombination block structure and a high average minor allele frequency (i.e., 1/8th). The striking degree of genetic variation seen in each genetically unique J:DO mouse substantially increases the number of segregating alleles and reduces linkage disequilibrium (LD) throughout the genome when compared to two-parent crosses (e.g., F2 intercrosses). This is of significance since the low levels of meiotic recombination in two-parent crosses results in low mapping resolutions and modifier loci that typically encompass hundreds of genes. Thus, identifying candidate modifiers within these broad loci is cumbersome and typically takes many years (Drinkwater et al. 2012). J:DO mice therefore represent a powerful means of identifying candidate genes when compared to two-parent crosses.
Here, we utilized the genetic variation present in J:DO mice and systems genetics methodologies to identify new germline modifiers of aggressive prostate cancer. Our approach, which is outlined in Fig. 1A, centers on high resolution modifier locus mapping in a population of (TRAMP × J:DO) F1 males. To the best of our knowledge, this study represents the first example of mapping modifiers of any complex trait in an F1 generation of Diversity Outbred mice. We identified three genes as aggressive prostate cancer modifiers: CENPU, which is also known as MLF1IP or PBIP1, and encodes a centromere component that is essential for mitosis; RWDD4, which is a poorly characterized gene that encodes the protein RWD Domain Containing 4, with an RWD domain being involved in protein-protein interactions; and CASP3, which is a cysteine-aspartic acid protease that mediates apoptosis. The relevance of these genes to aggressive human prostate cancer was validated in silico through comparison to human GWAS and primary tumor gene expression datasets, then in vitro and in vivo using human prostate cancer cell lines. This study demonstrates the utility of both systems and comparative genetics to investigate how hereditary variation influences complex traits such as susceptibility to aggressive prostate cancer, and defines new targets for improving outcomes in high risk prostate cancer patients.
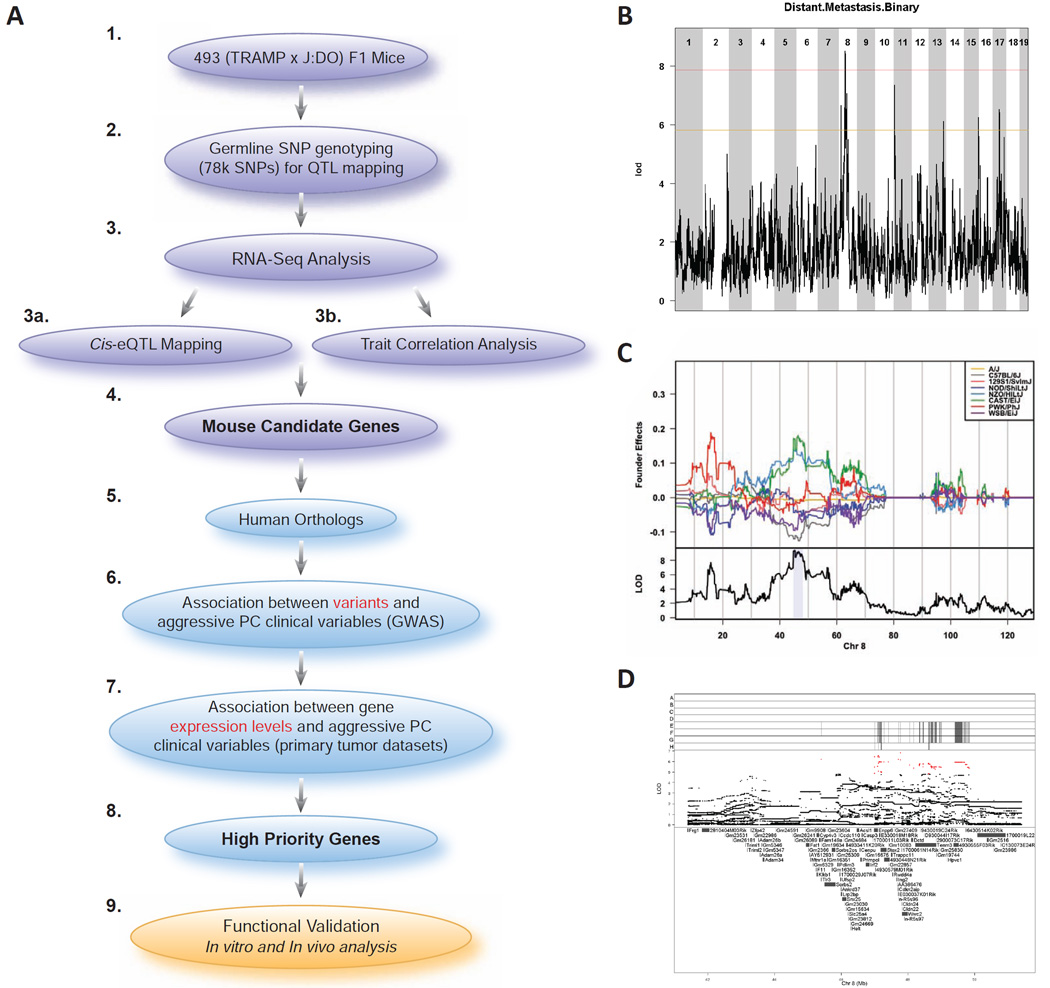
Figure 1.
Experimental strategy and aggressive prostate cancer modifier locus mapping in a cohort of 493 (TRAMP × J:DO) F1 mice. A) In order to identify candidate genes for susceptibility to aggressive prostate cancer, we crossed genetically diverse ‘Diversity Outbred’ (J:DO) male mice with female mice that were positive for the SV40 T antigen oncoprotein (TRAMP), resulting in the experimental (TRAMP × DO) F1 males (1). Over the course of 30 weeks, (TRAMP × J:DO) F1 mice were assessed for phenotypic characteristics associated with aggressive prostate cancer. At the conclusion of this experiment, genomic DNA extracted from (TRAMP × J:DO) F1 tails was used for germline SNP genotyping to map modifier loci associated with aggressive disease (2). Subsequently, RNA-seq analysis of 195 (TRAMP × J:DO) F1 tumors was performed (3), followed by tumor cis- eQTL mapping (3a) and analysis of correlations between patterns of primary tumor gene expression and aggressive disease traits (3b). The results of these analyses were integrated with the results of modifier locus mapping experiments to identify candidate aggressive PC modifier genes (4). The relevance of the human orthologs (5) of these candidate genes to aggressive human prostate cancer was assessed in a number of ways. First, we utilized human prostate cancer GWAS data to identify associations between aggressive disease clinical traits and the frequencies of candidate gene germline variants (6). Second, we identified associations between aggressive prostate cancer and changes in candidate gene mRNA expression levels using publically available human prostate tumor datasets (7). These analyses allowed us to prioritize candidate genes (8) for functional validation using in vitro and in vivo analysis of human prostate cancer cell lines expressing individual candidates (9). B) Genome scan of 493 Tg+ (TRAMP × J:DO) F1 males identified a locus on Chr. 8 associated with distant metastasis free survival. Red line indicates a significant association (genome-wide P < 0.05) and orange line represents a suggestive association (genome-wide P < 0.63). C) Upper panel: founder coefficient plot indicated that CAST/EiJ, NZO/HILtJ, and PWK/PhJ alleles are driving linkage across the Chr. 8 locus; lower panel: LOD score with Bayesian credible support interval shaded in blue. D) Association mapping was performed by additive regression on imputed SNP genotypes; red points denote scores above the P < 0.05 threshold. Upper panel shows founder strain SNP diplotypes (A – A/J; B – C57BL/6J; C – 129S1/SvlmJ; D – NOD/ShiLtJ; E – NZO/HILtJ; F – CAST/EiJ; G – PWK/PhJ; H – WSB/EiJ). Also see Fig. S1 and Fig. S2.
RESULTS
Modifier Locus Mapping in (TRAMP × J:DO) F1 Males Reveals a Region of Mouse Chromosome 8 Associated with Distant Metastasis Free Survival
Based on earlier observations (Patel et al. 2013), we defined ‘aggressive disease’ as a primary tumor burden over approximately 1 g, metastasis either locally to regional lymph nodes or to distant visceral organs (lung and liver); and/or early age of death. We hypothesized that high-resolution aggressive prostate cancer modifier locus mapping will be possible in the F1 male mapping cohort derived by crossing TRAMP females to J:DO males. Since prostate tumorigenesis in TRAMP mice is inherited in a dominant manner and is highly penetrant, every male mouse will develop prostate cancer. However, the aggressiveness will vary depending on the genetic background of each (TRAMP × J:DO) F1 mouse. Therefore, we aimed to map “modifier loci” by characterizing prostate cancer-associated traits in (TRAMP × J:DO) F1 mice. Aggressive disease traits were quantified in a cohort of 493 (TRAMP × J:DO) F1 males (Table S1 and Fig. S1), which were bred from 192 J:DO F0 males. Mice were maintained for 30 weeks, or until humane endpoints were reached, and tissues collected for analysis. Modifier locus mapping was performed by correlating patterns of single nucleotide polymorphism (SNP) variation with patterns of aggressive disease development in (TRAMP × J:DO) F1 males using an additive SNP model. A locus associated with distant metastasis free survival was identified on Chromosome (Chr.) 8 with a logarithm of the odds ratio (LOD) greater than the genome-wide P < 0.05 (peak region of linkage = ~47.8 Mb; LOD = 8.42; Fig. 1B). This locus spanned 3.29 Mb of Chr. 8 (support intervals 44.74 Mb to 48.03 Mb; Fig. 1C, lower panel) and encompassed 58 annotated transcripts. No associations of genome-wide significance were observed for other aggressive disease traits (Fig. S2A).
A coefficient plot was constructed to estimate J:DO founder effects across the Chr. 8 locus (Fig. 1C, upper panel). This revealed that (TRAMP × J:DO) F1 mice harboring CAST/EiJ, NZO/HILtJ, and PWK/PhJ variants across the Chr. 8 locus are more likely to develop distant metastasis. To identify causative variants, association mapping (Fig. 1D) using results from the additive SNP model was performed to define associations between the frequencies of individual variants (either genotyped or imputed) and distant metastasis free survival. This analysis identified 671 metastasis-associated variants with a LOD score greater than the genome-wide P < 0.05 (355 intragenic and 316 intergenic [Table S2]). Of the 355 intragenic SNPs, one non-synonymous coding polymorphism (Enpp6 R324K [rs37680350]) and three synonymous coding polymorphisms (Enpp6 H394H [rs36555850]; Stox2 S607S [rs38425327]; and Stox2 P307P [rs221539338]) were identified. The remaining 351 SNPs were either intronic, or within the 5’- or 3’-UTRs of 14 transcripts.
Candidate Gene Identification Through RNA-Seq Analysis of (TRAMP × J:DO) F1 Primary Tumors
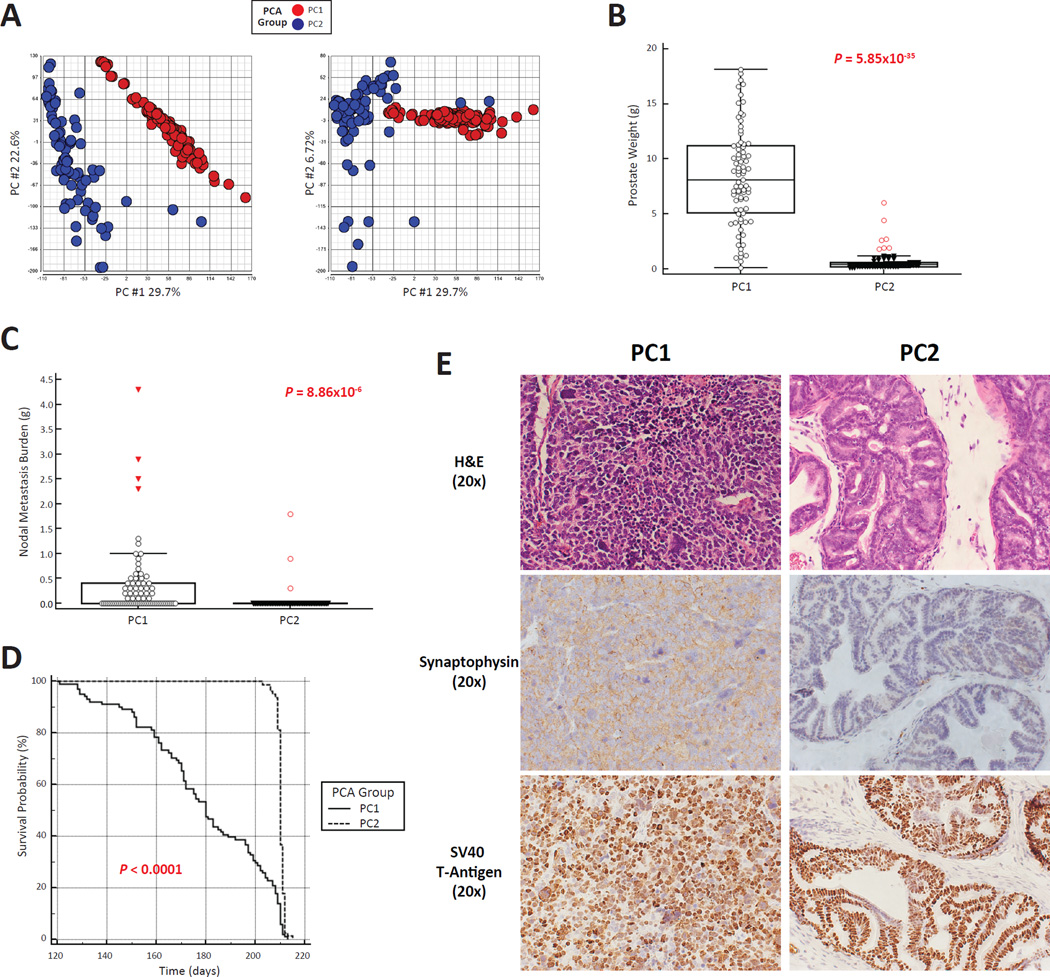
RNA-seq analysis of 195 randomly selected (TRAMP × J:DO) F1 prostates that had been harvested at experimental termination (210 days or humane endpoints) was performed to identify metastasis-associated transcriptomic changes. Principal component (PC) analysis revealed two distinct clusters of gene expression (PC1 [n = 107] and PC2 [n = 88]; Fig. 2A). Prostate cancer aggressiveness differed significantly between PC1 and PC2: primary tumor burden was significantly higher in PC1 (av. weight = 8.81 g ± 4.58 g vs. PC2 =0.69 g ± 0.90 g; P = 5.85×10−35; Fig. 2B); lymph node metastasis burden was significantly higher in PC1 (av. lymph node burden = 0.41g ± 0.78g vs. PC2 = 0.05 g ± 0.24 g; P = 8.86×10−6; Fig. 2C); distant metastasis frequency was higher in PC1 (26/107 [24.3%] vs. 9/88 [10.2%], respectively; Fisher’s exact P = 0.014); and finally, age at euthanasia (210 days or humane endpoints) was significantly earlier PC1 (average age = 179.8 days [174.9 days – 184.7 days] vs. PC2 = 210.2 days [209.9 days – 210.6 days]; χ2 = 91.22; P < 0.0001; Fig. 2D). H&E staining of representative PC1 prostates revealed an anaplastic tumor morphology, characteristic of aggressive NE tumorigenesis (Fig. 2E, upper left panel). Conversely, H&E staining of PC2 prostates revealed that these lesions are predominantly composed of ‘atypical hyperplasia of the T-antigen (Tag)’ (Chiaverotti et al. 2008), which is a benign, non-metastatic neoplastic lineage specific to the TRAMP mouse (Fig. 2E, upper right panel). Immunohistochemical (IHC) staining of prostates revealed strong expression for the NE marker Synaptophysin in PC1 but not PC2 prostates (Fig. 2E, middle left and middle right panels, respectively. Finally, IHC analysis for SV40-T antigen expression at the experimental endpoint (210 days of age) demonstrated that representative PC1 and PC2 prostates strongly express the SV40-T antigen transgene (Fig. 2E, lower left and lower right panels, respectively). These findings are consistent with our earlier work (Patel et al. 2013), which demonstrated that TRAMP tumors > ~ 1 g usually have NE characteristics. We thus conclude that PC1 tumors represent the NE lineage TRAMP tumors, and PC2 prostates represent the atypical hyperplasia of Tag lineage.
Figure 2.
RNA-Seq analysis of 195 (TRAMP × J:DO) F1 tumors reveals two distinct patterns of gene expression. A) Principal component analysis of global patterns of sub-divided tumor gene expression in two distinct groups (PC1 [red] & PC2 [blue]) when comparing PC1 vs. PC2 and PC1 vs. PC3. B) PC1 tumors were significantly larger than PC2 tumors (mean ± SD). C) Metastasis burden to regional lymph nodes was significantly more frequent in PC1 compared to PC2 mice (mean ± SD). D) Animals were aged to 210 days or humane end points. PC1 animals had a significantly earlier age at euthanasia compared to PC2 animals. E) H&E staining of representative PC1 and PC2 prostates at the experimental endpoint (210 days) revealed an anaplastic histological appearance indicative of NE tumorigenesis in PC1 tumors (upper right panel) and benign, atypical hyperplasia of Tag in PC2 prostates (upper left panel). IHC analysis revealed strong staining of endpoint PC1 tumors for the NE maker Synaptophysin (middle left panel) with low levels of this NE marker observed in endpoint PC2 prostates (middle right panel). Finally, IHC analysis of SV40-T antigen expression in endpoint PC1 and PC2 prostates (lower left and right panels, respectively) revealed equal levels of nuclear transgene expression (20X optical magnification).
To test whether the PC1/PC2 lineage dissociation was influenced by germline variation, we performed modifier locus mapping in (TRAMP × J:DO) F1 males by mapping primary tumor burden as a dichotomous trait. Specifically, the cohort of 493 mice was sub-divided into mice where prostate burden was ≥ 1 g and mice with prostates < 1 g. Although no loci achieved genome-wide significance, four loci associated with this trait surpassed the suggestive threshold for significance (α < 0.63; Fig. S2B–E) suggesting that the lineage dissociation trait is likely subject to a degree of genetic control.
Expression levels of the only gene harboring a non-synonymous coding polymorphism, Enpp6, were extremely low in PC1 and PC2 prostates (average transcripts per million [TPM] PC1 = 0.11 ± 0.10 and PC2 = 0.10 ± 0.15, not expressed in 12 of 195 prostates). Additionally, data from the Genotype-Tissue Expression (GTEx) project (http://www.gtexportal.org/) demonstrate that ENPP6 expression is limited to the brain, peripheral nerves, and the ovary in humans. Enpp6 therefore is unlikely to modulate metastasis owing to its very low primary and secondary site expression, and was excluded from further analyses. Thus, we hypothesized that Chr. 8 distant metastasis free survival modifiers act through expression-related mechanisms and possess both of the following: 1) a cis-expression quantitative locus (cis-eQTL); and 2) an expression level correlated with distant metastasis free survival. To investigate this hypothesis, cis-eQTL and trait correlation analyses were performed for the 33 transcripts within the Chr. 8 locus for PC1 tumors that were expressed above threshold in our RNA-seq dataset. PC2 tumors were excluded from further analysis since atypical hyperplasia of Tag lesions are both non-metastatic and of questionable relevance to human prostate cancer (Chiaverotti et al. 2008).
Cis-eQTLs were calculated for all expressed transcripts within the Chr. 8 locus using DOQTL, and were defined as a variant within 1 MB of either the transcription start site (TSS) or transcription end site (TES) of the cognate transcript, since 95% of enhancer elements fall within this range (Vavouri et al. 2006). Sixteen of 33 transcripts within the Chr. 8 locus were identified as harboring a cis-eQTL (Table S3). Associations between the expression levels of individual transcripts and distant metastasis free survival were assessed using a two-tailed Student’s t-test. Of these transcripts, 19 were observed to be associated with distant metastasis free survival (Table S4). Eleven of the 33 expressed transcripts were identified as candidate metastasis modifiers by intersecting cis-eQTL and distant metastasis free survival association data (Table 1). Correction for multiple testing was performed using Benjamini-Hochberg false discovery rates (FDR; (Benjamini et al. 2001)). An FDR of 10% defined significance owing to the loss of power resulting from the exclusion of PC2. Cis-eQTL founder effect coefficient plots for each of these 11 transcripts are shown in Fig. S3.
Table 1.
Candidate Gene Identification Using PC1 Tumor Gene Expression Data. Also see Fig. S3.
| Gene Symbol | Entrez Gene ID |
Transcript Start (Mb) |
Transcript End (Mb) |
cis-eQTL Analysis | Association with Distant Metastasis Free Suvival |
Human Ortholog |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| eQTL | eQTL Position |
LOD Score |
Percent Variance Explained |
P-Value | FDR | P-Value | FDR | |||||
| 1700029J07Rik | 69479 | 45.95 | 45.98 | rs6283436 | 44.99 | 6.69 | 25.01 | 6.79×10−5 | 0.002 | 3.58×10−4 | 0.009 | C4orf47 |
| Ufsp2 | 192169 | 45.98 | 46.00 | rs33292257 | 46.33 | 3.98 | 15.73 | 0.011 | 0.076 | 0.024 | 0.048 | UFSP2 |
| Cfap97 | 66756 | 46.15 | 46.20 | rs33528193 | 46.53 | 3.48 | 13.91 | 0.025 | 0.079 | 0.002 | 0.013 | CFAP97 |
| Acsl1 | 14081 | 46.47 | 46.54 | 46.53 | 3.50 | 14.00 | 0.024 | 0.079 | 0.013 | 0.029 | ACSL1 | |
| Cenpu | 71876 | 46.55 | 46.58 | 46.53 | 3.66 | 14.58 | 0.018 | 0.079 | 0.001 | 0.009 | CENPU | |
| Casp3 | 12367 | 46.62 | 46.64 | rs33292257 | 46.33 | 3.65 | 14.54 | 0.019 | 0.079 | 0.013 | 0.029 | CASP3 |
| Trappc11 | 320714 | 47.49 | 47.53 | rs33504741 | 47.60 | 3.27 | 13.12 | 0.035 | 0.089 | 0.001 | 0.009 | TRAPPC11 |
| Ing2 | 69260 | 47.67 | 47.68 | 47.60 | 3.81 | 15.14 | 0.014 | 0.079 | 0.001 | 0.009 | ING2 | |
| Rwdd4a | 192174 | 47.53 | 47.55 | rs30948998 | 48.33 | 5.68 | 21.68 | 4.73×10−4 | 0.007 | 0.005 | 0.017 | RWDD4 |
| Cdkn2aip | 70925 | 47.71 | 47.71 | UNC14692700 | 48.71 | 4.04 | 15.97 | 0.009 | 0.068 | 0.003 | 0.014 | CDKN2AIP |
| Tenm3 | 23965 | 48.23 | 48.84 | rs30967527 | 48.42 | 3.57 | 14.23 | 0.021 | 0.079 | 0.002 | 0.013 | TENM3 |
Analysis of Metastasis Modifier Candidate Genes in Human Prostate Cancer GWAS
We hypothesized that candidate gene orthologs with relevance to aggressive prostate cancer susceptibility in humans would: 1) harbor variants associated with aggressive prostate cancer; and 2) have primary tumor expression levels associated with aggressive prostate cancer. Accordingly, the relevance of the 11 candidate transcripts identified in (TRAMP × J:DO) F1 mice to aggressive prostate cancer in humans was approached using a three-stage in silico validation: first, we identified candidate gene germline variants associated with aggressive human prostate cancer in two human GWAS; second, we mapped eQTLs in normal human prostate tissue to determine whether candidate expression levels were associated with germline variation; and third, we characterized associations between candidate primary tumor gene expression levels and aggressive human prostate cancer in three datasets.
It has been suggested that biologically important modifiers acting in the latter stages of disease progression are less likely to exhibit genome-wide significance in GWAS analyses (Bjorkegren et al. 2015). Accordingly, the first stage of our validation involved performing case-only analyses of two publicly available human prostate cancer GWAS: 1) Cancer Genetic Markers of Susceptibility (CGEMS) GWAS that consists of 1,172 prostate cancer patients (Gohagan et al. 2000; Prorok et al. 2000); and 2) the International Consortium for Prostate Cancer Genetics (ICPCG) GWAS of familial prostate cancer that consists of 2,515 prostate cancer patients derived from high-risk prostate cancer pedigrees (Jin et al. 2012). In the CGEMS GWAS, prostate cancer cases are subdivided into non-aggressive (n = 484) and aggressive (n = 688) disease based on clinical stage (I/II vs. III/IV, respectively) and Gleason score (< 7 vs. ≥ 7, respectively). In the ICPCG GWAS, prostate cancer cases are subdivided into non-aggressive (n = 1,117) and aggressive (n = 1,398) disease based on clinical variables described elsewhere (Christensen et al. 2007; Schaid et al. 2006).
In the CGEMS cohort, 624 SNPs mapped to the human orthologs of the 11 candidate genes. Associations between aggressive disease occurrence and SNPs and/or haplotypes were examined using a generalized linear model (GLM). For single SNP analysis, 5 of 11 candidate genes were associated with aggressive disease development: SNPs in LD with ACSL1 and RWDD4 were associated with prostate cancer-specific mortality; and SNPs in LD with CDKN2AIP, ING2, and TENM3 were associated with nodal metastasis (Table 2). For haplotype analysis, 2 of 11 candidate genes that were not implicated in single SNP analysis were associated with aggressive disease: a haplotype in LD with CENPU was associated with pathological stage; and a haplotype in LD with CASP3 was associated with Gleason score (Table 2). Finally, haplotypes in LD with CDKN2AIP, ING2, and TENM3 were also associated with a variety of aggressive prostate cancer clinical variables (Table S5).
Table 2.
Candidate Gene Analysis in Human Prostate Cancer GWAS
| Gene | Cohort | Clinical Trait | SNP/ Haplotype |
Distance From Gene (bp) |
Frequency* | OR | 95% CI | P-Value | Permutation P-Value |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Aggressive | Non- Aggressive |
|||||||||
| CENPU | ICPCG | Aggressive vs. Non- Aggressive Prostate Cancer |
rs12644905 | 21,397 | 0.021 | 0.016 | 1.39 | 1.10–1.77 | 6.86×10−3 | 6.00×10−3 |
| CGEMS | Pathological Stage | rs3749232-T rs4862417-A rs3792311-C rs4069938-G rs4862419-G |
32,087 | 0.571 | 0.225 | 2.12 | 1.36–3.29 | 8.66×10−4 | 1.30×10−3 | |
| ING2 | ICPCG | Aggressive vs. Non- Aggressive Prostate Cancer |
rs8872-A rs4862213-A |
0 | 0.557 | 0.527 | 1.13 | 1.03–1.24 | 9.15×10−3 | 1.00×10−2 |
| CGEMS | Nodal Metastasis | rs13111484 | 69,645 | 0.001 | 0.388 | 1.05 | 1.01–1.08 | 8.76×10−3 | 9.60×10−3 | |
| RWDD4 | ICPCG | Aggressive vs. Non- Aggressive Prostate Cancer |
rs6830164-A rs7693110-A rs4862229-G rs4624700-G |
0 | 0.059 | 0.045 | 1.34 | 1.08–1.67 | 6.81×10−3 | 6.40×10−3 |
| CGEMS | Prostate Cancer Specific Mortality |
rs9312316 | 27,639 | 0.337 | 0.098 | 1.04 | 1.01–1.07 | 5.41×10−3 | 1.00×10−2 | |
| TENM3 | ICPCG | Aggressive vs. Non- Aggressive Prostate Cancer |
rs17073007 | 28,369 | 0.324 | 0.181 | 2.75 | 1.43–5.28 | 2.33×10−3 | 1.30×10−3 |
| CGEMS | Nodal Metastasis | rs13106786 | 0 | 0.000 | 0.373 | 1.05 | 1.02–1.09 | 2.79×10−3 | 3.70×10−3 | |
| ACSL1 | CGEMS | Prostate Cancer Specific Mortality |
rs4862451 | 42,780 | 0.095 | 0.034 | 1.07 | 1.03–1.12 | 1.31×10−3 | 4.60×10−3 |
| CASP3 | CGEMS | Gleason Score | rs13111483-G rs1918633-G rs956845-G rs766908-A |
79,964 | 0.317 | 0.487 | 1.20 | 1.05–1.36 | 5.80×10−3 | 6.00×10−3 |
| CDKN2AIP | CGEMS | Nodal Metastasis | rs955229 | 68,905 | 0.002 | 0.118 | 0.91 | 0.87–0.96 | 2.20×10−4 | 4.00×10−4 |
Four candidate genes (bold typeface) validated in both cohorts.
Minor allele frequencies are shown for associations with single alleles; overall frequency is shown for associations with haplotypes.
Abbreviations: OR = odds ratio, CI = confidence interval.
In the ICPCG GWAS, 1,749 SNPs mapped to the human orthologs of the 11 candidate genes. For single SNP analysis, one SNP in LD with CENPU, and one SNP in LD with TENM3 were associated with aggressive disease development (Table 2). For haplotype analysis, haplotypes in LD with ING2 and RWDD4 were associated with aggressive disease (Table 2). In addition, four haplotypes in LD with TENM3 were associated with aggressive disease (Table S5). In summary, four genes harbor variants associated with aggressive disease in both the CGEMS and ICPCG cohorts: CENPU, ING2, RWDD4, and TENM3. An additional three genes were associated with aggressive disease in the CGEMS cohort only: ASCL1, CASP3, and CDKN2AIP. Four candidate genes (C4orf47, CFAP97, TRAPCC11, and UFSP2) were not associated with aggressive prostate cancer in either cohort and were excluded from further analyses.
Candidate Gene Cis-eQTL Mapping in a Normal Prostate Tissue Dataset
It is plausible that the effects of hereditary variation of aggressive disease modifiers may exert themselves in the prostate prior to onset of tumorigenesis. Therefore, we hypothesized that a subset of the seven genes implicated in GWAS analysis will exhibit cis-eQTLs in normal prostate tissue. Thus, in the second element of our in silico validation we characterized cis-eQTLs in human normal prostate tissue derived from a cohort of 471 men (Thibodeau et al. 2015).
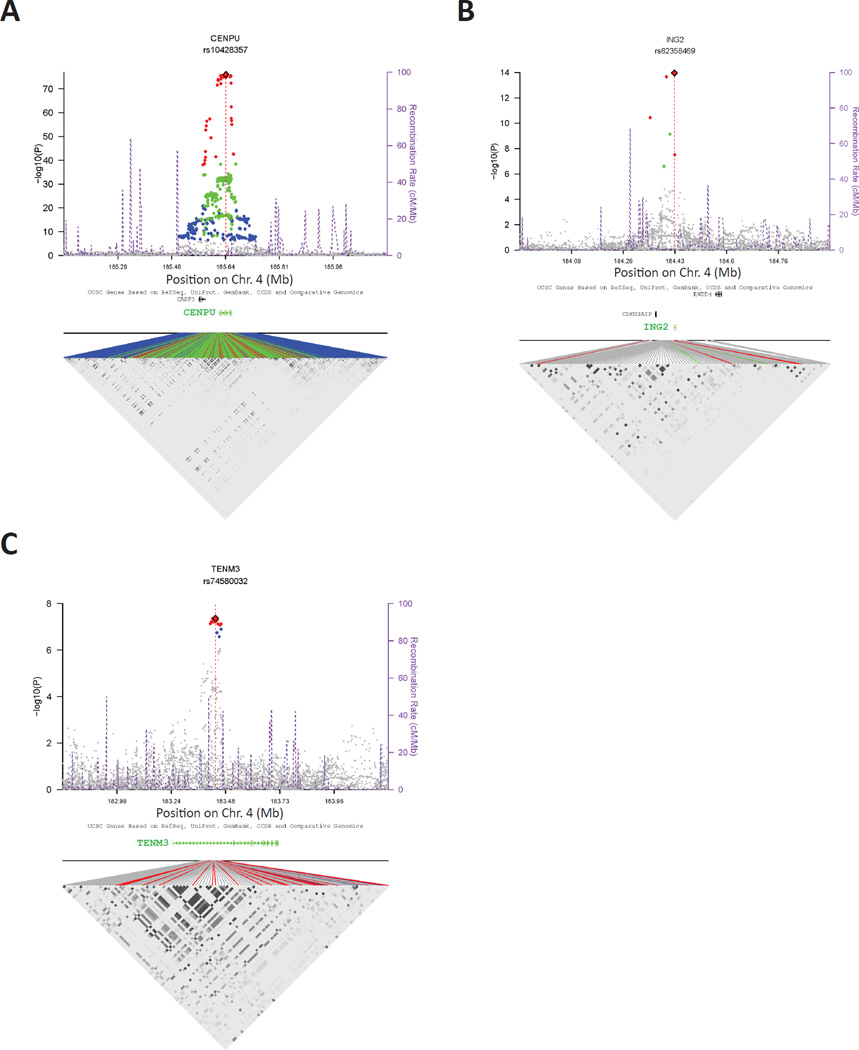
All SNPs within 1.1 Mb of either the TSS or TES were mapped to each of the 11 candidate genes, and cis-eQTLs were mapped by correlating RNA-seq transcript levels with SNP genotype. A total of 733 cis-eQTLs were observed after adjusting for covariates and meeting the Bonferroni threshold of 3.64×10−7 (Table S6). Three of the 7 candidate aggressive prostate cancer modifier genes exhibited statistically significant cis-eQTLs: CENPU (peak cis-eQTL SNP = rs10428357_A; P = 8.81×10−77; FDR = 4.49×10−73; percent variation of expression explained by the SNP, after adjusting for covariates = 53.24%; Fig. 3A); ING2 (peak cis-eQTL SNP = rs62358469_G; P = 1.12×10−14; FDR = 1.05×10−10; percent variation explained = 12.36%; Fig. 3B); and TENM3 (peak cis-eQTL SNP = rs74580032_T; P = 4.34×10−8; FDR = 4.34×10−5; percent variation explained = 6.41%; Fig. 3C). Statistically significant associations were not observed for the remaining four candidate genes, although ACSL1 and RWDD4 exhibited cis-eQTL signals that did not reach the Bonferroni threshold level of significance (Fig. S4).
Figure 3.
Regional association plots for aggressive prostate cancer candidate susceptibility genes with statistically significant cis-eQTLs. Cis-eQTLs in normal prostates derived from a cohort of 471 men are shown for A) CENPU, B) ING2, and C) TENM3. In each instance, the peak cis-eQTL SNP is marked as a red diamond. All SNPs reaching the Bonferroni are colored according to LD with the peak SNP (red: r2 > 0.5; green: [0.5, 0.2]; blue: < 0.2). The purple dotted lines represent recombination rates and positions. A LD heat map for significant regional SNPs is shown in the bottom part of each panel. The points are colored as above to signify LD with the peak cis-eQTL SNP. Also see Fig. S4.
Correlation of Candidate Gene Expression Levels with Aggressive Prostate Cancer Clinical Traits in Human Prostate Cancer Tumor Gene Expression Datasets
Since each of the candidate genes entered into our in silico validation pipeline had primary tumor transcript levels associated with distant metastasis free survival in (TRAMP × J:DO) F1 mice, we hypothesized that candidate gene expression levels within human primary prostate tumors would be associated with aggressive prostate cancer clinical variables. Accordingly, in the third element of in silico validation, we used logistic regression to correlate candidate gene expression levels with aggressive prostate cancer clinical variables in RNA-seq prostate cancer gene expression datasets (The Cancer Genome Atlas [TCGA] prostate adenocarcinoma [PRAD]; n = 497); and two microarray datasets (GSE21032 (Taylor et al. 2010) n = 150; and GSE49961 (Erho et al. 2013) n = 545). Logistic regression analysis demonstrated that expression levels of CASP3, CENPU, and RWDD4 were associated with various aggressive prostate cancer traits after adjusting the regression for recurrence, which was the one common clinical variable between the three cohorts. CASP3 was associated with nodal metastasis in TCGA (OR = 1.60, 95% CI = 1.19–2.15, P = 0.002, FDR = 0.028); CENPU was associated with pathological tumor stage in TCGA (OR = 2.44; 95% CI = 1.70–3.52; P = 1.00×10−4; FDR = 0.003); and RWDD4 was associated with Gleason Score in GSE21032 (OR = 2.16; 95% CI = 1.29–3.64; P = 0.004; FDR = 0.048). Results for the 7 candidate genes in each of the 3 cohorts are shown in Table S7. No associations were evident for ASCL1, CDKN2AIP, ING2, and TENM3, and we thus excluded these genes from further analysis.
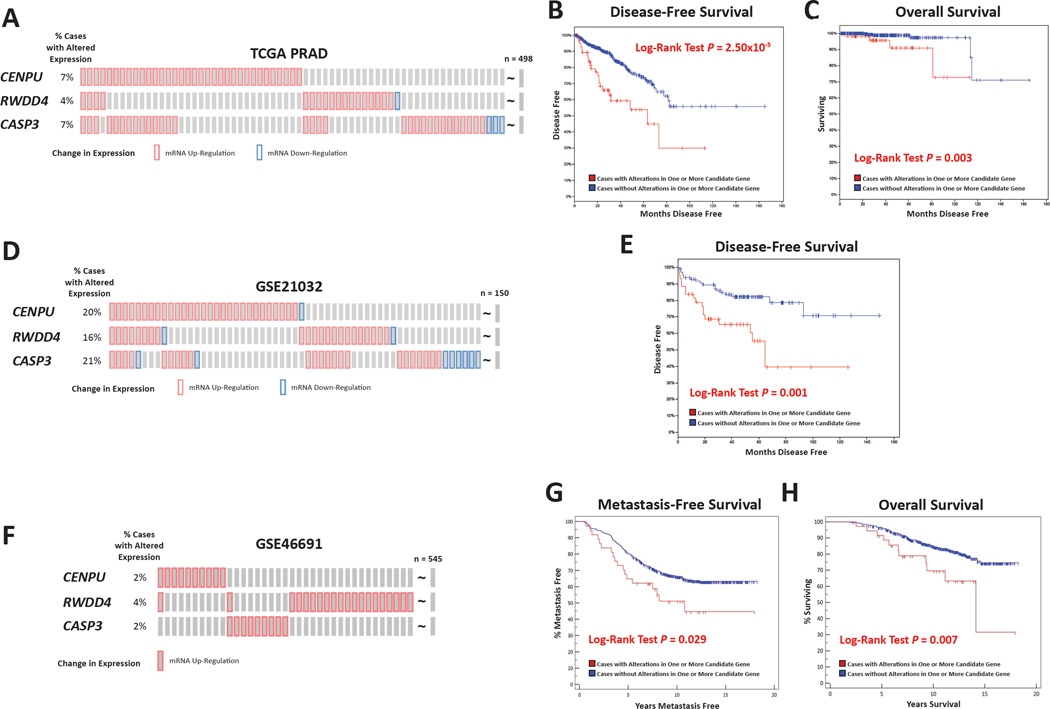
Kaplan-Meier survival analyses were performed to further evaluate the association of CASP3, CENPU, and RWDD4 with aggressive disease, by comparing survival in patients with and without significant expression level changes of one or more of these genes in primary tumor samples. Higher or lower levels of gene expression were defined by a z-score of > 2 or < −2, respectively. In TCGA cohort, 65 of 497 (13%) tumors had altered expression of one or more of the three candidates, with exclusively higher than average levels being observed in 61 of 65 cases (Fig. 4A). These predominantly higher than average levels of expression were associated with both a poorer disease free survival (log-rank P = 2.50×10−5; Fig. 4B) and poorer overall survival (log-rank P = 0.003; Fig. 4C). In the GSE20132 cohort, expression of one or more of the candidates was altered in 57 of 150 (38%) cases, with higher levels again prevailing (Fig. 4D). As with TCGA, disease free survival was poorer in cases with abnormal candidate levels (log-rank P = 0.026; Fig. 4E). Finally, in GSE46691, expression of one or more of the three candidates was significantly changed in 66 of 545 (12%) cases (Fig. S5A). However, the pattern of gene expression differed in this cohort, with 37 of 66 tumors exhibiting exclusively higher candidate gene expression levels, and 29 of 66 tumors exhibiting exclusively lower than average levels. No significant differences in either metastasis free survival (Fig. S5B) or overall survival (Fig. S5C) were observed when comparing these 66 cases to the rest of the cohort. However, since predominantly higher than average expression levels were observed in TCGA and GSE21032, survival was tested in similar cases in GSE46691 (Fig. 4F). Here, exclusively higher than average expression levels were associated with a poorer metastasis free survival (log-rank P = 0.029; Fig. 4G) and poorer overall survival (log-rank P = 0.007; Fig. 4H). Interestingly, a reciprocal effect was observed in 29 GSE46691 cases with exclusively lower than average levels of one or more of the three genes (Fig. S5D), with both metastasis free survival (Fig. S5E) and overall survival (Fig. S5F) being better in these cases (log-rank P = 0.014 and 0.037, respectively).
Figure 4.
Candidate gene expression levels were associated with patient outcome in multiple cohorts. The association of differential expression of CASP3, CENPU, and RWDD4 with patient outcome was tested using Kaplan-Meier survival analysis. In the TCGA PRAD gene expression dataset (n = 497), where predominantly higher than average levels of candidate gene expression were observed in 13% of patients (A). Here, the percentage of patients with dysregulation of individual genes is noted on the left of the diagram. Patients with higher than average level of these three genes had both a poorer disease-free survival (B) and overall survival (C) compared to patients with normal levels. A significantly different level of expression of the three candidates was seen in 38% of patients in the GSE21032 cohort (n = 150; D), with this differential expression being associated with poorer disease-free survival (E). Finally, a higher than average level of candidate expression was seen in 7% of patients in the GSE46619 cohort (n = 545); F). Patients with higher than average level of these three genes had both a poorer disease-free survival (G) and overall survival (H) compared to patients with normal levels. Also see Fig. S5 and Fig. S6.
Comparison of Candidate Gene Expression in Aggressive and Indolent Tumors, and Matched Normal Tissue in Mice and Humans
Having demonstrated that CENPU, CASP3, and RWDD4 dysregulation was associated with patient survival in multiple human gene expression cohorts, we hypothesized that similar associations will be observed in (TRAMP × J:DO) F1 mice. We calculated candidate gene z-scores in the combined PC1 and PC2 (TRAMP × J:DO) F1 RNA-seq tumor expression cohort (n = 195). Mice were then then sub-divided into one of two sub-groups (survival = 210 days vs. death or euthanasia < 210 days), and the association between changes in candidate gene expression and survival was calculated. In the combined PC1 and PC2 cohort, mice with changed candidate gene expression had a poorer disease-specific survival (log-rank P = 0.013; Fig. S6A). As was the case with the human studies, poorer survival was associated with a predominantly higher than average candidate gene expression. A similar effect was evident in PC1 subgroup, with predominantly higher than average expression levels being associated with a poorer disease-specific survival (log-rank P = 0.027; Fig. S6B).
Next, we analyzed patterns of gene expression in primary prostate tumors and normal prostate tissue in both mice and humans. In mice, we compared candidate gene expression in aggressive neuroendocrine PC1 tumors, atypical hyperplasia of Tag PC2 lesions, and a third group composed of RNA-seq data from normal prostates harvested from 85 (C56BL/6J × J:DO) F1 males aged 30 weeks. We found that Cenpu, Casp3, and Rwdd4 expression changed in a phenotype-dependent manner (PC1>PC2>normal; Fig. S6C). In humans, gene expression levels were analyzed in the GSE21032 and TCGA PRAD cohorts, where gene expression levels were compared between prostate cancer specimens and matched normal prostates. CENPU and RWDD4 were expressed at significantly higher levels in prostate cancer samples in both the GSE21032 dataset (CENPU P = 0.0001, RWDD4 P = 0.005; Figure S6D, upper panel) and in TCGA PRAD cohort (CENPU P = 2.85 × 10−9, RWDD4 P = 0.003; Fig. S6D, lower panel). Finally, CASP3 was more highly expressed only in the GSE21032 dataset (P = 0.027; Figure S6D, upper panel).
Analysis of Candidate Gene Dysregulation in Human Prostate Cancer Cell Lines
Since higher levels of both RWDD4 and CENPU were associated with a more aggressive phenotype in mice and humans, we over-expressed each candidate gene in the human prostate cancer cell lines LNCaP and PC-3 using lentiviral-mediated ectopic expression. We prioritized these two genes over CASP3 for two reasons: 1) CASP3 variants were associated with aggressive prostate cancer in only one of the two GWAS; and 2) its inclusion in Kaplan-Meier survival analyses was not essential in two of the three tumor gene expression datasets, with CENPU and RWDD4 expression levels alone predicting survival in TCGA and GSE20132 datasets (Fig. S5G-I and Fig. S5J-K, respectively).
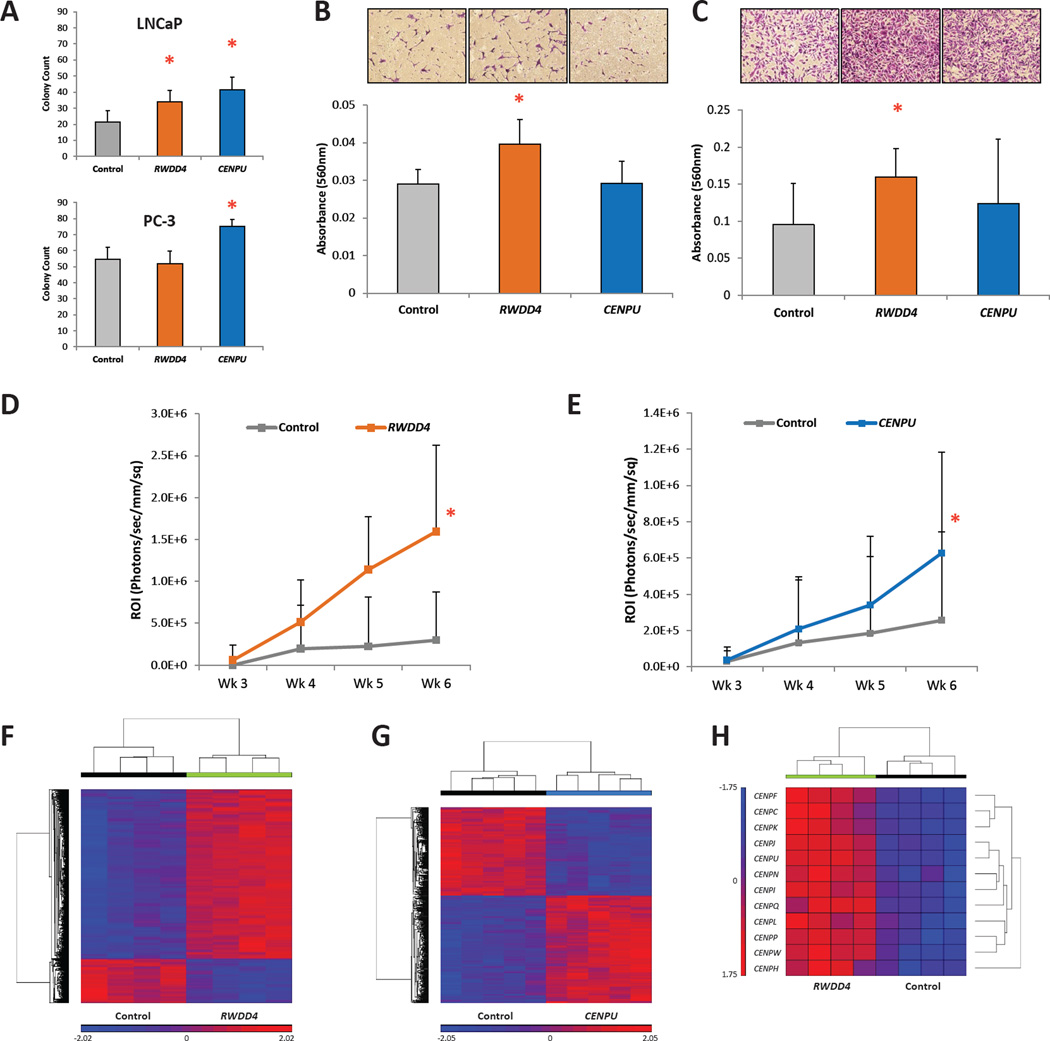
Following selection, stable over-expression of each gene was confirmed by quantitative real-time PCR (qPCR; Fig. S7A). Soft agar assays were performed to assess the effects of candidate gene dysregulation on anchorage-independent growth. Ectopic expression of both candidate genes in LNCaP cells significantly increased colony formation (RWDD4 av. colony count = 34.0 ± 7.1, P = 0.047; and CENPU av. count = 41.5 ± 7.7, P = 0.009, Fig. 5A upper panel) compared to control cells (av. count = 21.3 ± 7.4). In PC-3 cells, anchorage independent growth was increased only in cells ectopically expressing CENPU (av. count = 75.6 ± 4.3 vs. control av. count = 54.8 ± 7.2, P = 0.003, Fig. 5A lower panel). Trans- well assays were performed to assess the effects of candidate gene ectopic expression on invasion and migration. Over-expression of RWDD4 increased invasiveness in both LNCaP (av. absorbance 560 nm = 0.0396 ± 0.0066 vs. control av. absorbance 560 nm = 0.0290 ± 0.0039, P = 0.009; Fig. 5B) and PC-3 cells (av. absorbance 560 nm = 0.1596 ± 0.0385 vs. control av. absorbance 560 nm = 0.0950 ± 0.0562, P = 0.042; Fig. 5C). Ectopic expression of CENPU did not change the invasiveness of either cell line. No differences in migration were observed with either gene in both cell lines compared to controls (Fig. S7B). In LNCaP cells, analysis of in vitro cell growth rate revealed that over-expression of RWDD4 significantly increased proliferation (total cell count RWDD4 = 53.15 ± 11.29 vs. control = 29.075 ± 6.13; P < 0.001) while over-expression of CENPU had no effect on LNCaP cells (Fig. S7C). Neither gene had an effect on the in vitro proliferation rate of PC-3 cells (Fig. S7D). Finally, PC-3 cells co-expressing luciferase and either of the candidate genes or a control vector were injected into the left cardiac ventricle of NU/J mice to assess their ability to colonize distant sites. Dissemination of cells over-expressing either RWDD4 or CENPU was increased compared to controls (ANOVA P = 1.07×10−4 [Fig 5D] and ANOVA P = 0.015 [Fig 5E], respectively).
Figure 5.
In vitro and in vivo analysis of the effects of RWDD4 and CENPU dysregulation on LNCaP and PC-3 cells. (A) Soft agar anchorage-independent growth assays for LNCaP (upper panel) and PC-3 (lower panel) cells over-expressing either RWDD4 or CENPU (mean ± SD). (B) Trans-well invasion assay for LNCaP cells over-expressing either candidate gene (mean ± SD). (C) Trans-well invasion assay for PC-3 cells over-expressing either candidate gene (mean ± SD). (D) Intracardiac tumor dissemination assay for NU/J mice injected with either PC-3 cells over-expressing RWDD4 (n = 9) or a control cell line (n = 8) (mean ± SD). (E) Intracardiac tumor dissemination assay for NU/J mice injected with either PC-3 cells over-expressing CENPU (n = 20) or a control cell line (n = 16) (mean ± SD). (F) Microarray analysis of PC-3 cells over-expressing RWDD4 revealed dysregulation of 5,732 transcripts (fold change ± 1.5; FDR < 0.050). (G) Microarray analysis of PC-3 cells over-expressing CENPU revealed dysregulation of 682 transcripts (fold change ± 1.5; FDR < 0.050). (H) Multiple transcripts encoding centromere components are dysregulated in PC-3 cells over-expressing RWDD4. * P < 0.050. Also see Fig. S7.
Transcriptomic Analysis of Cells Over-Expressing Candidate Genes
Since germline-driven expression changes in both RWDD4 and CENPU were associated with aggressive prostate cancer, we hypothesized that human prostate cancer cells over-expressing these genes would exhibit characteristic transcriptomic changes. Accordingly, we performed microarray analysis of PC-3 clonal isolates over-expressing RWDD4 and CENPU. We identified 5,732 dysregulated transcripts in four clonal isolates of RWDD4 over-expressing cells (fold change ± 1.5; FDR < 0.050; Fig. 5F; Table S8), and 682 significantly dysregulated transcripts in CENPU over-expressing cells (Fig. 5G; Table S9). Among the dysregulated transcripts in RWDD4 cells were multiple genes encoding centromere components (Fig. 5H). These genes, all of which displayed strong up-regulation, included the other high priority candidate gene CENPU, as well as another centromere family member gene CENPF. CENPF is a master regulator of aggressive prostate cancer-associated gene expression, with higher levels of CENPF being associated with activation pathways associated with prostate cancer malignancy (Aytes et al. 2014). In our study, we confirmed by qPCR the significantly higher levels of both CENPF and CENPU in cells over-expressing RWDD4 (Fig. S7E). Finally, Ingenuity Pathway Analysis (IPA) was performed on the dysregulated transcripts in both the RWDD4- and CENPU-expressing cells to identify aberrant pathways (Fig. S7F and Fig. S7G, respectively), and as expected, many pathways critical to tumor progression were dysregulated. For example, in RWDD4 cells, multiple DNA damage response pathways were found to be affected (e.g., BRCA1-related DNA damage response and G2/M DNA damage checkpoint regulation), while in CENPU cells, affected pathways included those involved in cell cycle regulation, DNA replication, recombination and DNA repair.
DISCUSSION
In this study, we have defined common patterns of genetic and functional variation between mice and humans and identified biologically-relevant hereditary modifiers of a human disease that exhibits a complex inheritance pattern. Initially, our approach centered on a ‘discovery’ phase, which involved investigating how high levels of germline variation influenced disease patterns in a transgenic mouse model of aggressive prostate cancer. The relevance of the identified candidate transcripts to human prostate cancer was investigated through an in silico validation, which incorporated multiple human prostate cancer GWAS and tumor gene expression patient cohorts, and collectively encompassed over 5,300 prostate cancer patients. Finally, the biological relevance of these highest priority candidate genes was confirmed through functional and transcriptomic analysis of two prostate cancer cell lines. Accordingly, we have identified CENPU and RWDD4 as germline modifiers of aggressive prostate cancer.
The use of Diversity Outbred mice in combination with the TRAMP transgenic model of prostate cancer has proven central to this strategy, with a locus associated with distant metastasis free survival identified on Chr. 8 in a cohort of 493 (TRAMP × J:DO) F1 males. The comparatively small size of this locus, which spanned ~3.3 Mb of Chr. 8 and encompassed 58 transcripts, strongly illustrates the superiority of modifier locus mapping performed with J:DO mice compared to traditional two-parent crosses. For example, in an earlier study with aims similar to those presented here, we bred an F2 intercross population by crossing TRAMP females to NOD/ShiLtJ males (Williams et al. 2014) and identified eleven loci associated with aggressive prostate cancer traits. These loci spanned on average 34.5 Mb (range: 19.6 – 55.7 Mb), and candidate gene identification was consequently challenging, primarily because the number of candidate genes within each locus was approximately an order of magnitude higher compared to the (TRAMP × J:DO) F1 cross. Thus, our present study, as well as studies from other groups (e.g., (Church et al. 2015; French et al. 2015; Svenson et al. 2012)) demonstrates the power of using Diversity Outbred mice to map hereditary modifiers of a broad range of complex traits.
We assayed the full spectrum of tumorigenesis seen in (TRAMP × J:DO) F1 mice in our RNA-seq analysis, ranging from tumors representing the most benign to the most aggressive disease forms seen in this mouse model. These analyses demonstrated the existence of two very strong principal components, which was not an entirely unexpected finding given the seminal work of Chiaverotti et al. (Chiaverotti et al. 2008), which definitively described the disease process in the TRAMP mouse. Specifically, this study demonstrated that tumorigenesis dissociates into two lineages: a) a neuroendocrine lineage defined by aggressive prostatic tumorigenesis with metastasis; and b) an epithelial lineage termed ‘atypical hyperplasia of the T-antigen (Tag), which represents a relatively benign, non-metastatic form of disease. Our histological analysis of representative prostates (Fig. 2E) demonstrates that PC2 represent lesions falling predominantly into the atypical hyperplasia of Tag lineage. Thus, in the (TRAMP × J:DO) F1 cross, we appear to have transcriptomically captured the tumor lineage dissociation observed by Chiaverotti et al. PC2 lesions were therefore excluded since the majority of these neoplasms were non-metastatic and thus not informative for identifying transcriptomic modifiers of distant metastasis free survival. Accordingly, analysis of PC1 RNA-seq data, composed predominantly of NE tumors, demonstrated that Cenpu, Rwdd4, and Casp3 were three of 11 genes within the Chr. 8 metastasis modifier locus that harbored both a cis-eQTL and an expression level correlated with metastasis.
In summary, we have utilized both comparative and systems genetics to identify multiple novel susceptibility genes for aggressive prostate cancer, which is a disease that kills over 26,000 men annually in the US. To the best of our knowledge, this study represents the first published example of an F1 cross involving Diversity Outbred mice. Accordingly, we have presented a new conceptual framework for extending high resolution modifier locus mapping to other transgenic mouse models, which will facilitate identification of new susceptibility genes for a wide variety of human diseases. Identifying this type of germline susceptibility gene has proven challenging using conventional approaches such as GWAS, with most complex diseases exhibiting varying degrees of ‘missing heritability’. There are many plausible explanations for this, including over-estimation of the degree of complex disease heritability, under-estimation of allelic effect sizes, and yet-to-be identified rare variants with large effect sizes. Therefore, it is likely that multi-faceted approaches such as the study presented here are required in order to more fully understand how the germline influences different disease processes. It has been suggested that biologically relevant modifiers, which achieve nominal but not genome-wide significance and likely act in a more ‘context dependent’ fashion, are being overlooked (Bjorkegren et al. 2015; Farber 2013). In aggregate, our work demonstrates that GWAS data can be mined using systems genetics approaches to produce biological meaningful observations based on associations that reach nominal but not genome-wide significance.
STAR METHODS
KEY RESOURCES TABLE
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author, Dr. Nigel Crawford (crawforn@mail.nih.gov).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse Studies
All animals were handled, housed, and used in the experiments humanely in accordance with the guidelines approved by the NHGRI Animal Care and Use Committee. Six week old TRAMP females, which were hemizygous for PB-TAg transgene (Tg), and J:DO males (generations 7–14) were obtained from The Jackson Laboratory (Bar Harbor, ME). A total of 198 F0 J:DO males were used to breed 493 (TRAMP × J:DO) F1 males and 85 (C57BL/6J × J:DO) F1 males. (C57BL/6J × J:DO) F1 males were the transgenenegative littermates of transgene-positive (TRAMP × J:DO) F1 mice. Transgene status was assessed by genotyping mouse tail genomic DNA, which was extracted from F1 progeny using the HotSHOT method (Alsarraj et al. 2011). To identify hemizygous PB-TAg transgene positive F1 mice, PCR screening was performed as previously described (Hurwitz et al. 2001).
Human Studies
For GWAS analyses, PCa cohort data were obtained from dbGAP. The clinical characteristics of both GWAS cohorts have been described extensively within dbGAP (CGEMS cohort – dbGaP Study Accession: phs000207.v1.p1; (Yeager et al. 2007); ICPCG cohort - dbGaP Study Accession: phs000733.v1.p1 (Teerlink et al. 2014)). Detailed clinical characteristics of the Mayo Clinic cohort used for cis-eQTL mapping have been described previously (Thibodeau et al. 2015). Briefly, normal prostate tissue was obtained from an archival collection derived from 471 patients undergoing either radical prostatectomy or cystoprostatectomy, where the Gleason score was < 7 for the presenting tumor. Normal regions of the prostate were defined by histological re-examination (Thibodeau et al. 2015). Finally, for human tumor gene expression analyses, three datasets were available for analysis (TCGA PRAD [N=497 PCa cases]; GSE46691 [N =545 PCa cases]; and GSE21032 [N =150 PCa cases]). Comparison of clinical features revealed that disease recurrence was the only clinical variable between the three datasets (see STAR Methods Table 1). Accordingly, we adjusted each regression analysis for this trait. Tumor gene expression levels for TCGA and GSE21032 were obtained from cBioPortal for Cancer Genomics (Cerami et al. 2012; Gao et al. 2013). Expression data for GSE46691 were obtained from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE46691). Clinical characteristics for these cohorts can be found at the respective websites.
Cell Culture
PC-3 and LNCaP cells were purchased from ATCC (Manassas, VA). Cells were maintained in either DMEM (PC-3) or RPMI (LNCaP) supplemented with 10% FBS and 1% penicillin-streptomycin (Gibco) and incubated in 5% CO2 at 37 °C.
METHODS DETAIL
F1 Mouse Tissue Collection, Phenotypic Quantification, and High Density SNP Genotyping
TRAMP × J:DO) and (C57BL/6J × J:DO) F1 male mice were sacrificed by pentobarbital injection at 210 days of age or humane endpoint, whichever was achieved first. For (TRAMP × J:DO) F1 mice, prostate and seminal vesicle tumors, lungs, liver, and lymph nodes were collected at the time of euthanasia. Primary tumor burden was quantified by weighing the prostate and seminal vesicles. Visibly enlarged para-aortic lymph nodes were enumerated and weighed to quantify lymph node metastasis burden. For distant metastases, lung and liver lesions were quantified for macroscopic count at the time of necropsy and lung histology slides were examined for microscopic count. For histology, collected tissues were fixed in neutral buffered formalin (10% w/v phosphate buffered formaldehyde, Fisher Scientific, Waltham, MA) overnight and then transferred to 70% ethanol. Fixed tissues were embedded in paraffin, sectioned to a thickness of 4 µm and stained with hematoxylin and eosin (H&E). Histology slides were scanned with Scanscope Digital microscope (Aperio, Vista, CA). To calculate distant metastasis free survival, the presence or absence of either pulmonary or hepatic metastasis was converted into a binary trait. For (C57BL/6J × J:DO) F1 mice, prostates were collected and snap frozen in liquid nitrogen.
For high density SNP genotyping, tail biopsies were obtained from (TRAMP × J:DO) and (C57BL/6J × J:DO) F1 mice at the time of necropsy. Genomic DNA was extracted from tail tissue using the Qiagen DNeasy Blood and Tissue Kit per the manufacturer's instructions (Qiagen, Valencia, CA). Each (TRAMP × J:DO) F1 mouse was genotyped at 77,808 markers using the MegaMUGA genotyping array, and each (C57BL/6J mouse was genotyped at 143,259 markers using the GigaMUGA genotyping array (Morgan et al. 2015). Genotyping was performed at Neogen/GeneSeek (Lincoln, NE). Samples were either sent directly or sent through the UNC Systems Genetics Core Facility to Neogen/Geneseek (Lincoln, NE).
Mouse Tumor and Human PCa Cell Line RNA Extraction
As described previously (Williams et al. 2014), total RNA was extracted from snap frozen (TRAMP × J:DO) F1 bulk tumors and (C57BL/6J × J:DO) normal prostates using TRIzol Reagent (Life Technologies, Inc.) according to the manufacturer’s protocol, and purified using RNeasy mini kit (QIAGEN) according to the manufacturer’s protocol. In instances where tumors were macroscopically small or absent at experimental endpoints, RNA was extracted from the ventral prostate since this is the primary site for aggressive NEPC in the TRAMP model (Chiaverotti et al. 2008). For cell lines, RNA was extracted using RNeasy mini kit (QIAGEN) according to the manufacturer’s protocol. The quality and quantity of both primary tumor and cell line RNA was assessed using the Bioanalyzer (Agilent, Inc., Santa Clara, CA) and NanoDrop (Thermo Scientific, Inc., Waltham, MA), respectively.
RNA-Seq Library Preparation
RNA-Seq libraries were constructed from 1 µg total RNA after rRNA depletion using Ribo-Zero GOLD (Illumina). The Illumina TruSeq RNA Sample Prep V2 Kit was used according to the manufacturer’s instructions. The cDNAs were fragmented to ~275 bp using a Covaris E210. Amplification was performed using 10 cycles, which was optimized for the input amount and to minimize the chance of overamplification. Unique barcode adapters were applied to each library for cataloging. Libraries were pooled together for sequencing. The pooled libraries were sequenced on multiple lanes of a HiSeq 2500 using version 4 chemistry to achieve a minimum of 43 million 126 base read pairs. The data was processed using RTA version 1.18.64 and CASAVA 1.8.2.
RNA-Seq Analysis of Mayo Clinic Normal Prostates and Germline SNP Genotyping
Following histological re-examination, normal prostate tissue was isolated (Thibodeau et al. 2015). RNA and DNA were extracted from these samples to perform transcriptomic analysis and genome-wide SNP genotyping, respectively. RNA-seq reads (51 bp) were generated on an Illumina HiSeq 2000, and analyzed using the MAP-R-Seq pipeline (Kalari et al. 2014). For germline SNP genotyping, Illumina Infinium 2.5M bead arrays were used, per the manufacturer’s protocol (Illumina, San Diego, CA).
Immunohistochemical Staining of (TRAMP × J:DO) F1 Prostates
To determine if prostates from either the PC1 or PC2 group express the SV40 T antigen or the NE marker Synaptophysin, five prostates weighing approximately 0.1 g were randomly chosen at necropsy from mice aged 30 weeks for formalin fixation and paraffin embedding. For comparison, a prostate weighing over 4.5 grams from the PC1 group was also collected for immunohistological analysis. Tissue blocks were dissected at 5µm, adhered to a frosted glass slide and immersed in Clear-Rite™ (Thermo Fischer Scientific) for 2 changes of 10 minutes each. Tissue sections were rehydrated in gradient alcohols and endogenous enzyme activity was quenched using 0.3% H2O2 at RT and rinsed in TBST, followed by 1 hour of protein blocking using Superblock T20 Blocking Buffer (Thermo Scientific, cat#37356) at room temperature (RT). Primary antibodies mouse anti-SV40 LargeT-antigen (TAg) (BD Pharmingen, cat#61095) and mouse anti-synaptophysin (Thermo Fischer cat# MA5-16402), were diluted at 1:400 and 1:1500, respectively, in Superblock T20 blocking buffer for incubation overnight at 4°C. The next day, slides were washed in TBST for 3 × 2 minutes each followed by 1 hour incubation using EnVision™ + Dual link labelled HRP polymer (Dako, cat#K4065) at RT. Staining was visualized using DAB chromagen in DAB substrate buffer (Dako, cat#K4065) for 1 minute and counterstained with Haematoxylin for 2 minutes.
Generation of Stable Cell Lines
Lentiviral vectors for CENPU and RWDD4 were purchased from GE Dharmacon (Lafayette, CO). Lentiviral particles were generated as described previously (Lee et al. 2014b) and cells over-expressing candidate genes were selected using 20 µg/mL blasticidin for PC-3 cells and 3 µg/mL blasticidin for LNCaP cells. Control cells were generated by transducing with virus generated from an empty lentiviral vector.
Microarray and qPCR Expression Analyses
Microarray gene expression analysis was performed to analyze patterns of gene expression in PC-3 and LNCaP cell lines overexpressing candidate genes, as described (Lee et al. 2015). Briefly, 200 ng of total RNA was used for labelling in conjunction with the recommended protocol for the Affymetrix Human GeneChip 2.0 ST chips (Santa Clara, CA). Hybridization and subsequent processing of the chips were performed as previously described (Williams et al. 2014). Quantitative real-time PCR was performed as previously described (Lee et al. 2015). Reverse transcription was performed using cDNA Synthesis Kit (Bio-Rad, Hercules, CA). The obtained cDNA was diluted 10-fold, and 1 µL was used for each 5 µL realtime PCR reaction. Expression data are presented as mean fold change over control cells ± SD.
In Vitro Migration and Invasion, Soft Agar, and Cell Proliferation Assays
Cell proliferation assays were performed as previously described (Lee et al. 2014a; Lee et al. 2015). Soft agar assays were performed as previously described (Lee et al. 2014a; Lee et al. 2015), using 2 × 103 cells per 24-well in 0.33% bacto-agar and incubated for 14 days. For in vitro migration and invasion assays, PC-3 or LNCaP cells were starved in serum-free media for 12 hours. A total of 5 × 105 cells were seeded into the upper chamber of a 24-well plate containing an 8.0 µM cell insert membrane (Thermo Scientific, Inc.). Insert wells were placed in 24-well tissue culture plates containing cell culture media supplemented with 10% FBS, which serves as an attractant to the starved cells. For cell migration assays, membranes were pre-coated with collagen I, and for invasion assays, insert membranes were pre-coated with Matrigel (BD Biosciences, San Jose, CA). Forty-eight hours later, cells from the upper chamber were removed using a cotton swab and cells that invaded/migrated and attached to the lower surface were fixed with 4% paraformaldehyde, and stained with crystal violet (0.05% in ethanol). Snapshots of migratory cells were taken and stained cells were de-stained in 2% SDS. Absorbance was read at 560nm using a microplate reader (Molecular Devices, Sunnyvale, CA). For cell growth assays, cells were plated at 2.5 × 104 per 12-well plate and counted at the same time each day in duplicate for 5 days using a cellometer slide and automated counter. Statistical analyses were performed using Student’s t-test, and data are presented as mean + SD where P < 0.05 was considered significant.
In Vivo Tumor Dissemination Assay
The ability of PC-3 cells overexpressing either CENPU or RWDD4 to disseminate to distant sites was analyzed by intracardiac injections, as described (Campbell et al. 2012). Briefly, six week old male NU/J mice (The Jackson Laboratory, Bar Harbor, ME; Stock 002019) were anaesthetized under isoflurane and marked slightly left of the midway point between the top of the rib cage and the xyphoid process. A total of 1 × 105 cells in 100ul of sterile PBS were injected into the left cardiac ventricle, followed by recovery under a heat lamp. Mice were monitored for 7 days post-injection before being transferred to the mouse imaging facility on day 8 to measure bioluminescence on day 10.
Bioluminescence imaging of mice was performed once weekly for 6 weeks on the In-Vivo Xtreme Imager (Bruker, Billerica, MA). Mice were injected intraperitoneally with luciferin (150mg/kg), anaesthetized under isoflurane and imaged five minutes post-injection with 1 minute exposure time (luminescence) and 1 second exposure time (reflectance). After the final week, mice were humanely sacrificed and tissues collected in neutral buffered formalin. The minimum and maximum ROI (photons/second/mm/sq) for each luminescence image were adjusted and kept constant to eliminate signal background and saturation (min: 6.4 × 105; max: 1.1 × 107). Luminescence images were analyzed for mean ROI (photons/second/mm/sq) per mouse using the Bruker Molecular Imaging Software (Bruker MI SE, version 7.13, Billerica, MA) and overlaid with the reflectance image. Mice with luminescence signal detected in the chest cavity were excluded for analyses due to cell spillage at time of injection. ANOVA was performed to test the differences among groups with data presented as mean ± SD.
QUANTIFICATION AND STATISTICAL ANALYSIS
F1 Mouse Haplotype Reconstruction and Modifier Locus Mapping
MegaMUGA array genotypes were used to reconstruct the diplotypes of each (TRAMP × J:DO) and (C57BL/6J × J:DO) F1 mouse using a hidden Markov model (HMM), which produced a probabilistic reconstruction of the mouse’s genome in terms of the eight founder strain haplotypes. At each marker, we estimated the posterior probability that each mouse was in one of the eight possible genotype states. Linkage mapping was performed by fitting a mixed effects linear model at each marker, where we regressed the aggressive disease traits in Table S1 on the diplotype probabilities and the DO outbreeding generation. A kinship matrix was included as a random effects term to adjust for the relatedness between animals. Significance thresholds were determined via permutation of the phenotype values while holding the diplotype probabilities fixed (Gatti et al. 2014), and the support interval for significant peaks was established using the Bayesian Credible Interval (Sen et al. 2001). Within the support interval of significant peaks, we imputed the SNPs from the eight founders onto the individualized haplotype blocks of the J:DO chromosomes carried by each F1 male. Association mapping was performed by regressing the disease traits on each imputed SNP and the J:DO F0 outbreeding generation. In addition, a kinship matrix was included in this analysis. SNPs with the highest LOD score were selected, and candidate genes were nominated by identifying transcripts that fell within a 1 Mb interval of either side of the SNPs. All of these procedures were implemented in the DOQTL software package.
Generation of Individualized Genome Sequences and Gene Annotations in F1 Mice
Seqnature software was used to produce individualized genome sequences and gene models specific to each (TRAMP × J:DO) or (C57BL/6J × J:DO) F1 mouse, since highly polymorphic RNA-seq reads derived from these mice may not map effectively to a single reference sequence (Munger et al. 2014). The posterior probabilities calculated during haplotype reconstruction were used to impute allelic contribution from each of the eight J:DO founder strains at a given locus. Mouse genome reference sequence (GRCm38) and gene models (release 81, no ab initio predictions) were downloaded from Ensembl (ftp://ftp.ensembl.org/pub). Known sequence variation (SNPs and indels) for the J:DO founder strains was obtained from the Sanger Mouse Genomes Project website (ftp://ftpmouse.sanger.ac.uk/REL-1303-SNPs_Indels-GRCm38). According to the reconstructed haplotypes for each (TRAMP X J:DO) F1 mouse, founder variation was incorporated into the mouse reference genome sequence and gene annotations, thus generating mouse-specific diploid genome sequence along with corresponding gene annotations.
RNA-Seq Mapping and Quantification
Trimmomatic (v. 0.30; (Bolger et al. 2014)) was used to trim the 3’-most base and the first 17 bases from the 5’ end of each raw RNA-seq read. RSEM (v. 1.2.20) was used to obtain normalized read counts for downstream RNA-seq analyses (Li et al. 2011). An initial iteration of rsem-prepare-reference was used to generate allele-specific transcriptome sequences from the diploid genome sequences and corresponding gene annotations. These transcriptome sequences were used as input to a second iteration of rsem-prepare-reference, which was run to create a Bowtie2 index of allele-specific transcripts for each (TRAMP X J:DO) or (C57BL/6J × J:DO) F1 mouse using the --allele-to-gene-map option and supplying a table to link allelic variants to an annotated transcript. Mapping of RNA-seq reads to the allele-specific transcriptome and subsequent quantification was performed using rsem-calculateexpression, and expected and normalized (transcripts per million, TPM) gene-level counts were used for subsequent RNA-seq analyses.
Cis-eQTL Mapping
Expression QTL mapping was performed only for (TRAMP × J:DO) F1 tumors. eQTL mapping was performed using the DOQTL software package (Gatti et al. 2014). Only those genes with a non-zero expected read count in ≥ 85% of (TRAMP X J:DO) F1 samples, and at least one sample having ≥ 100 reads, were included for eQTL mapping. The normalized read counts (TPMs) of these genes were transformed into normal scores using the rankZ function in DOQTL. eQTLs were mapped using a linear mixed model regressing at each marker of the transformed expression scores on the diplotype probabilities obtained during haplotype reconstruction. A kinship matrix was included to account for relatedness between mice. For identifying cis-eQTLs, LOD scores and P-values were extracted for markers within 1Mb upstream and downstream of annotated genes. A Benjamini-Hochberg FDR was calculated using these P -values and those loci with a FDR ≤ 0.1 were considered significant. The percent variance explained by each eQTL was calculated using the ‘bayesint’ function of DOQTL.
Statistical Analysis of Human PCa GWAS
All SNPs analyzed either resided inside or within 100 kb of either the transcription start site (TSS) or transcription end site (TES) for each aggressive PCa candidate gene. Hardy-Weinberg equilibrium P-values were estimated using PLINK (Chang et al. 2015), and SNPs were omitted if P < 0.001. All SNPs and genes were mapped to GRCh37/hg19. For the CGEMS cohort, associations between SNP and/or haplotype frequency and aggressive PCa were defined using the following comparisons of clinical variables: for pathological stage, stage I+II vs. stage III+IV; for tumor stage, T1+T2 vs. T3+T4; for nodal metastasis, N0 vs. N1+N2; for distant metastasis, M0 vs. M1A+M1B+M1C; and for Gleason score, <7 vs. > 7. For the ICPCG GWAS, cases are pre-coded in dbGAP, and we compared variant frequencies between aggressive (cases coded ‘aggressive’) and ‘non-aggressive’ (coded ‘moderate’, ‘insignificant’ or ‘unknown’) (Christensen et al. 2007; Schaid et al. 2006). Individual clinical data points are not publicly available for this cohort. A GLM was used to define associations between aggressive PCa phenotype and SNP and/or haplotype. Age and PC1, PC2 and PC3 were included as covariates in the GLM. Correction for compounding of type I error was performing a permutation test (Churchill et al. 1994) using the GLM on NIH Biowulf super cluster computer system (http://biowulf.nih.gov). Specifically, permutation testing (n = 10,000 permutations) was performed by rearranging phenotype labels for SNPs in the same LD block for each subject. Permutation tests were performed only when the nominal P < 0.010. Genome-wide LD blocks were estimated by using the Solid Spine algorithm of Haploview (Barrett et al. 2005) with the default parameters. For haplotype analysis, fastPHASE (Stephens et al. 2001) was performed to generate haplotypes for each individual based on the LD blocks on NIH Biowulf super cluster computer system (http://biowulf.nih.gov). FDR P-values were calculated by the MULTITEST package of R. All analyses were performed using R.
Cis-eQTL Mapping in Mayo Clinic Normal Human Prostate Tissue Dataset
Untyped germline SNPs and missing genotypes for typed SNPs were imputed using SHAPEIT (Delaneau et al. 2013) and IMPUTE2 (Delaneau et al. 2013; Howie et al. 2012), and imputation quality assessed using BEAGLE (Browning et al. 2009). Cis-eQTLs were mapped by correlating patterns of gene expression with the genotypes of SNPs located within 1.1 Mb of the candidate gene TSS or TES, using Matrix eQTL (Shabalin 2012). Covariates used for eQTL mapping included histological variates (percent lymphocytic population and percent epithelium present) and 14 principal components defined by analysis of the normalized gene expression matrix. A Bonferroni adjustment was used to determine statistical significance (P < 3.64×10−7). Regional association plots were generated using a combination of LocusZoom (Pruim et al. 2010) and locally written R functions with LD estimates obtained from PLINK v1.9 (Chang et al. 2015). To determine the percent variation of expression explained after adjusting for covariates, we first regressed gene expression on covariates to compute an adjusted expression, i.e., residuals from this regression model: (y-y^), and regressed the SNP dosage on covariates to compute the adjusted SNP effect. Then, we regressed adjusted expression on the adjusted SNP to obtain the model R2 after adjusting for covariates.
Candidate Gene Analysis in Human and Mouse Tumor Gene Expression Datasets
Logistic regression analysis was performed to determine associations between the expression levels of the seven transcripts identified in GWAS analysis with dichotomized aggressive PCa clinical variables performed using the software package MedCalc (Ostend, Belgium). Clinical traits were dichotomized into ‘aggressive’ and ‘non-aggressive’ based on the following distinctions: for pathological stage, stage I+II vs. stage III+IV; for tumor stage, T1+T2 vs. T3+T4; for nodal metastasis, N0 vs. N1+N2; for distant metastasis, M0 vs. M1A+M1B+M1C; for Gleason score, < 7 vs. > 7; and for biochemical recurrence, recurrent vs. non-recurrent. For each dataset, candidate gene expression levels were presented as z-scores. For TCGA, z-scores were generated from RNA-seq read counts by calculating the standard deviation (SD) of transcript expression levels in each case compared to the mean transcript expression in diploid tumors. For GSE46691, z-scores were calculated using microarray gene expression data, by calculating the SD of the levels of transcript in each case compared to the mean transcript expression in all tumors. Finally, z-scores in GSE21032 were calculated by generating SDs for the comparison of mean transcript expression in cases compared to the average transcript expression level in matched normal prostates (n=149). In (TRAMP × J:DO) F1 data, z-scores were calculated by generating SDs for the comparison of mean transcript expression in individual mice compared to the average transcript expression level in either the combined PC1/PC2 cohort (n = 195) or PC1 cohort (n = 108). In human datasets, correction for multiple testing was performed by calculating a Benjamini-Hochberg FDR for the univariate logistic regression P -values with the threshold for significance being an FDR of 5%. Kaplan–Meier survival analysis was performed by using Medcalc by comparing the survival time in all cohorts with higher or lower levels of tumor candidate gene expression versus all other cases. Higher or lower levels of gene expression were defined by a z-score of > 2 or < −2, respectively. Significance of survival analyses was performed by using the Cox F test. For comparison of gene expression between prostate tumor and matched normal prostates, we utilized normalized expression values for each candidate (mouse tissue: RNA-seq TPM counts; GSE21032: normalized Affymetrix microarray gene expression values; TCGA PRAD: RNA-seq FPKM counts). Expression levels were compared between groups using a two-tailed Student’s t-test.
DATA AND SOFTWARE AVAILABILITY
F1 mouse RNA-seq and human PCa cell line microarray data have been deposited in Gene Expression Omnibus under accession number GSE87491.
ADDITIONAL RESOURCES
STAR Methods Table 1.
Comparison of clinical characteristics of human gene expression cohorts
| Clinical Characteristic | Cohort | ||
|---|---|---|---|
| TCGA | GSE21032 | GSE46691 | |
| Age at Diagnosis | X | X | |
| Death from Prostate Cancer | X | X | |
| Distant Metastasis | X | X | |
| Extra Capsular Extension | X | ||
| Gleason Score | X | X | |
| Nodal Metastasis | X | X | |
| PSA at Diagnosis | X | ||
| Recurrence | X | X | X |
| Seminal Vesicle Invasion | X | ||
| Tumor Stage | X | X | |
Common characteristics are shown in bold typeface
Supplementary Material
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health [HG200366-05]. DMG and GAC were funded by NIGMS grant R01GM070683. SKMcD, ZCF, MCL, AJF, DJS, and SNT were funded by grants from the DOD (W81XWH-11-1-0261), from the U.S. Public Health Service, National Institutes of Health (CA151254, CA157881 and CA15083), and U01 CA 89600. We would like to thank Darla Miller and the University of North Carolina Systems Genetics Core Facility for the assistance with MegaMUGA genotyping, and Steven Munger for his advice regarding Seqnature software. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization – NPSC, DJS, SNT, GAC; data analysis – JMW, DEG, DMG, YH, SZ, JCM, TGW, SKMcD, ZCF, MCL, AJF, NPSC; experimental procedures – JMW, JPA, KAW, ML; manuscript first draft – NPSC, DEG, DMG, JMW; manuscript edits and revisions – JMW, KAW, ML, TGW, DJS, SNT, NPSC.
REFERENCES
- 1.Aizer AA, Gu X, Chen MH, Choueiri TK, Martin NE, Efstathiou JA, Hyatt AS, Graham PL, Trinh QD, Hu JC, et al. Cost implications and complications of overtreatment of low-risk prostate cancer in the United States. J.Natl.Compr.Canc.Netw. 2015;13:61–68. doi: 10.6004/jnccn.2015.0009. [DOI] [PubMed] [Google Scholar]
- 2.Alsarraj J, Walker RC, Webster JD, Geiger TR, Crawford NP, Simpson RM, Ozato K, Hunter KW. Deletion of the proline-rich region of the murine metastasis susceptibility gene Brd4 promotes epithelial-to-mesenchymal transition- and stem cell-like conversion. Cancer Res. 2011;71:3121–3131. doi: 10.1158/0008-5472.CAN-10-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aytes A, Mitrofanova A, Lefebvre C, Alvarez MJ, Castillo-Martin M, Zheng T, Eastham JA, Gopalan A, Pienta KJ, Shen MM, et al. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell. 2014;25:638–651. doi: 10.1016/j.ccr.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 5.Beltran H, Tagawa ST, Park K, MacDonald T, Milowsky MI, Mosquera JM, Rubin MA, Nanus DM. Challenges in recognizing treatment-related neuroendocrine prostate cancer. J.Clin.Oncol. 2012;30:e386–e389. doi: 10.1200/JCO.2011.41.5166. [DOI] [PubMed] [Google Scholar]
- 6.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav.Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 7.Bjorkegren JL, Kovacic JC, Dudley JT, Schadt EE. Genome-wide significant loci: how important are they? Systems genetics to understand heritability of coronary artery disease and other common complex disorders. J.Am.Coll.Cardiol. 2015;65:830–845. doi: 10.1016/j.jacc.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am.J.Hum.Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell JP, Merkel AR, Masood-Campbell SK, Elefteriou F, Sterling JA. Models of bone metastasis. J.Vis.Exp. 2012:e4260. doi: 10.3791/4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiaverotti T, Couto SS, Donjacour A, Mao JH, Nagase H, Cardiff RD, Cunha GR, Balmain A. Dissociation of epithelial and neuroendocrine carcinoma lineages in the transgenic adenocarcinoma of mouse prostate model of prostate cancer. Am.J.Pathol. 2008;172:236–246. doi: 10.2353/ajpath.2008.070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen GB, Camp NJ, Farnham JM, Cannon-Albright LA. Genome-wide linkage analysis for aggressive prostate cancer in Utah high-risk pedigrees. Prostate. 2007;67:605–613. doi: 10.1002/pros.20554. [DOI] [PubMed] [Google Scholar]
- 15.Church RJ, Gatti DM, Urban TJ, Long N, Yang X, Shi Q, Eaddy JS, Mosedale M, Ballard S, Churchill GA, et al. Sensitivity to hepatotoxicity due to epigallocatechin gallate is affected by genetic background in diversity outbred mice. Food Chem.Toxicol. 2015;76:19–26. doi: 10.1016/j.fct.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Churchill GA, Gatti DM, Munger SC, Svenson KL. The Diversity Outbred mouse population. Mamm.Genome. 2012;23:713–718. doi: 10.1007/s00335-012-9414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat.Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 19.Drinkwater NR, Gould MN. The long path from QTL to gene. PLoS.Genet. 2012;8:e1002975. doi: 10.1371/journal.pgen.1002975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erho N, Crisan A, Vergara IA, Mitra AP, Ghadessi M, Buerki C, Bergstralh EJ, Kollmeyer T, Fink S, Haddad Z, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS.One. 2013;8:e66855. doi: 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farber CR. Systems-level analysis of genome-wide association data. G3.(Bethesda.) 2013;3:119–129. doi: 10.1534/g3.112.004788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.French JE, Gatti DM, Morgan DL, Kissling GE, Shockley KR, Knudsen GA, Shepard KG, Price HC, King D, Witt KL, et al. Diversity Outbred Mice Identify Population-Based Exposure Thresholds and Genetic Factors that Influence Benzene-Induced Genotoxicity. Environ.Health Perspect. 2015;123:237–245. doi: 10.1289/ehp.1408202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci.Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gatti DM, Svenson KL, Shabalin A, Wu LY, Valdar W, Simecek P, Goodwin N, Cheng R, Pomp D, Palmer A, et al. Quantitative trait locus mapping methods for diversity outbred mice. G3.(Bethesda.) 2014;4:1623–1633. doi: 10.1534/g3.114.013748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gingrich JR, Barrios RJ, Kattan MW, Nahm HS, Finegold MJ, Greenberg NM. Androgen-independent prostate cancer progression in the TRAMP model. Cancer Res. 1997;57:4687–4691. [PubMed] [Google Scholar]
- 26.Gohagan JK, Prorok PC, Hayes RB, Kramer BS. The Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial of the National Cancer Institute: history, organization, and status. Control Clin.Trials. 2000;21:251S–272S. doi: 10.1016/s0197-2456(00)00097-0. [DOI] [PubMed] [Google Scholar]
- 27.Hayes JH, Barry MJ. Screening for prostate cancer with the prostate-specific antigen test: a review of current evidence. JAMA. 2014;311:1143–1149. doi: 10.1001/jama.2014.2085. [DOI] [PubMed] [Google Scholar]
- 28.Hemminki K, Ji J, Forsti A, Sundquist J, Lenner P. Concordance of survival in family members with prostate cancer. J.Clin.Oncol. 2008;26:1705–1709. doi: 10.1200/JCO.2007.13.3355. [DOI] [PubMed] [Google Scholar]
- 29.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat.Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hurwitz AA, Foster BA, Allison JP, Greenberg NM, Kwon ED. The TRAMP mouse as a model for prostate cancer. Curr.Protoc.Immunol. 2001 doi: 10.1002/0471142735.im2005s45. Chapter 20:Unit. [DOI] [PubMed] [Google Scholar]
- 31.Jin G, Lu L, Cooney KA, Ray AM, Zuhlke KA, Lange EM, Cannon-Albright LA, Camp NJ, Teerlink CC, FitzGerald LM, et al. Validation of prostate cancer risk-related loci identified from genome-wide association studies using family-based association analysis: evidence from the International Consortium for Prostate Cancer Genetics (ICPCG) Hum.Genet. 2012;131:1095–1103. doi: 10.1007/s00439-011-1136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalari KR, Nair AA, Bhavsar JD, O'Brien DR, Davila JI, Bockol MA, Nie J, Tang X, Baheti S, Doughty JB, et al. MAP-RSeq: Mayo Analysis Pipeline for RNA sequencing. BMC.Bioinformatics. 2014;15:224. doi: 10.1186/1471-2105-15-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee M, Dworkin AM, Gildea D, Trivedi NS, Moorhead GB, Crawford NP. RRP1B is a metastasis modifier that regulates the expression of alternative mRNA isoforms through interactions with SRSF1. Oncogene. 2014a;33:1818–1827. doi: 10.1038/onc.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee M, Dworkin AM, Lichtenberg J, Patel SJ, Trivedi NS, Gildea D, Bodine DM, Crawford NP. Metastasis-associated Protein Ribosomal RNA Processing 1 Homolog B (RRP1B) Modulates Metastasis Through Regulation of Histone Methylation. Mol.Cancer Res. 2014b doi: 10.1158/1541-7786.MCR-14-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee M, Williams KA, Hu Y, Andreas J, Patel SJ, Zhang S, Crawford NP. GNL3 and SKA3 are novel prostate cancer metastasis susceptibility genes. Clin.Exp.Metastasis. 2015;32:769–782. doi: 10.1007/s10585-015-9745-y. [DOI] [PubMed] [Google Scholar]
- 36.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC.Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morgan AP, Fu CP, Kao CY, Welsh CE, Didion JP, Yadgary L, Hyacinth L, Ferris MT, Bell TA, Miller DR, et al. The Mouse Universal Genotyping Array: From Substrains to Subspecies. G3.(Bethesda.) 2015;6:263–279. doi: 10.1534/g3.115.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munger SC, Raghupathy N, Choi K, Simons AK, Gatti DM, Hinerfeld DA, Svenson KL, Keller MP, Attie AD, Hibbs MA, et al. RNA-Seq alignment to individualized genomes improves transcript abundance estimates in multiparent populations. Genetics. 2014;198:59–73. doi: 10.1534/genetics.114.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ostrander EA, Kwon EM, Stanford JL. Genetic susceptibility to aggressive prostate cancer. Cancer Epidemiol.Biomarkers Prev. 2006;15:1761–1764. doi: 10.1158/1055-9965.EPI-06-0730. [DOI] [PubMed] [Google Scholar]
- 40.Patel SJ, Molinolo AA, Gutkind S, Crawford NP. Germline genetic variation modulates tumor progression and metastasis in a mouse model of neuroendocrine prostate carcinoma. PLoS.One. 2013;8:e61848. doi: 10.1371/journal.pone.0061848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prorok PC, Andriole GL, Bresalier RS, Buys SS, Chia D, Crawford ED, Fogel R, Gelmann EP, Gilbert F, Hasson MA, et al. Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin.Trials. 2000;21:273S–309S. doi: 10.1016/s0197-2456(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 42.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romero OJ, Garcia GB, Campos JF, Touijer KA. Prostate cancer biomarkers: an update. Urol.Oncol. 2014;32:252–260. doi: 10.1016/j.urolonc.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 44.Schaid DJ, McDonnell SK, Zarfas KE, Cunningham JM, Hebbring S, Thibodeau SN, Eeles RA, Easton DF, Foulkes WD, Simard J, et al. Pooled genome linkage scan of aggressive prostate cancer: results from the International Consortium for Prostate Cancer Genetics. Hum.Genet. 2006;120:471–485. doi: 10.1007/s00439-006-0219-9. [DOI] [PubMed] [Google Scholar]
- 45.Sen S, Churchill GA. A statistical framework for quantitative trait mapping. Genetics. 2001;159:371–387. doi: 10.1093/genetics/159.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J.Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 48.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am.J.Hum.Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Svenson KL, Gatti DM, Valdar W, Welsh CE, Cheng R, Chesler EJ, Palmer AA, McMillan L, Churchill GA. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics. 2012;190:437–447. doi: 10.1534/genetics.111.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teerlink CC, Thibodeau SN, McDonnell SK, Schaid DJ, Rinckleb A, Maier C, Vogel W, Cancel-Tassin G, Egrot C, Cussenot O, et al. Association analysis of 9,560 prostate cancer cases from the International Consortium of Prostate Cancer Genetics confirms the role of reported prostate cancer associated SNPs for familial disease. Hum.Genet. 2014;133:347–356. doi: 10.1007/s00439-013-1384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thibodeau SN, French AJ, McDonnell SK, Cheville J, Middha S, Tillmans L, Riska S, Baheti S, Larson MC, Fogarty Z, et al. Identification of candidate genes for prostate cancer-risk SNPs utilizing a normal prostate tissue eQTL data set. Nat.Commun. 2015;6:8653. doi: 10.1038/ncomms9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vavouri T, McEwen GK, Woolfe A, Gilks WR, Elgar G. Defining a genomic radius for long-range enhancer action: duplicated conserved non-coding elements hold the key. Trends Genet. 2006;22:5–10. doi: 10.1016/j.tig.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 54.Wang HT, Yao YH, Li BG, Tang Y, Chang JW, Zhang J. Neuroendocrine Prostate Cancer (NEPC) Progressing From Conventional Prostatic Adenocarcinoma: Factors Associated With Time to Development of NEPC and Survival From NEPC Diagnosis-A Systematic Review and Pooled Analysis. J.Clin.Oncol. 2014;32:3383–3390. doi: 10.1200/JCO.2013.54.3553. [DOI] [PubMed] [Google Scholar]
- 55.Williams KA, Lee M, Hu Y, Andreas J, Patel SJ, Zhang S, Chines P, Elkahloun A, Chandrasekharappa S, Gutkind JS, et al. A systems genetics approach identifies CXCL14, ITGAX, and LPCAT2 as novel aggressive prostate cancer susceptibility genes. PLoS.Genet. 2014;10:e1004809. doi: 10.1371/journal.pgen.1004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilt TJ, Dahm P. PSA Screening for Prostate Cancer: Why Saying No is a High-Value Health Care Choice. J.Natl.Compr.Canc.Netw. 2015;13:1566–1574. doi: 10.6004/jnccn.2015.0182. [DOI] [PubMed] [Google Scholar]
- 57.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat.Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.