Abstract
Epigenetic inactivation of GPX3 has been identified in various cancers including leukemia. Moreover, aberrant DNA methylation was also found as a dominant mechanism of disease progression in myelodysplastic syndrome (MDS). This study intended to explore GPX3 promoter methylation and its clinical relevance in 110 patients with MDS. GPX3 methylation was examined by real‐time quantitative methylation‐specific PCR (RQ‐MSP) and bisulfite sequencing PCR (BSP). GPX3 methylation was identified in 15% (17/110) MDS patients, and significantly higher than controls, and lower than acute myeloid leukemia (AML) patients (P = 0.024 and 0.041). GPX3 methylated patients had older age and higher frequency of DNMT3A mutation (P = 0.015 and 0.066). Cases with GPX3 methylation showed significantly shorter overall survival (OS) time than those with GPX3 unmethylation analyzed with Kaplan–Meier analysis (P = 0.012). Moreover, Cox regression analysis revealed that GPX3 methylation might act as an independent prognostic indicator in MDS (HR = 1.847, P = 0.072). GPX3 methylation density was significantly increased during the progression from MDS to secondary acute myeloid leukemia (sAML) in three follow‐up paired patients. Our study concludes that GPX3 methylation in bone marrow is associated with adverse prognosis and leukemia transformation in MDS.
Keywords: Disease progression, GPX3, MDS, methylation, prognosis
Introduction
Myelodysplastic syndrome (MDS) is a heterogeneous hematopoietic stem cells (HSCs) disorder characterized by clonal hematopoiesis with quantitatively and qualitatively abnormal myeloid differentiation, and has a high risk of transforming to acute myeloid leukemia (AML) 1. The precise diagnosis of MDS is difficult, and its classification mainly depends on the cytopenias, blast percentage, morphology, and cytogenetics in bone marrow (BM) 2. Additionally, all these characteristics combined with red blood cell (RBC) transfusion dependence and patient performance status provides powerful information in the prognosis of MDS 2. Although the definite pathogenesis of MDS is not well defined, abnormal expression of proto‐oncogenes and tumor suppressor genes (TSGs) play crucial roles in the pathogenesis of MDS 3. Moreover, promoter hypermethylation of TSGs, the common mechanisms of silencing gene expression, are regarded as the drivers of MDS pathogenesis and progression to secondary AML (sAML) 4. Therefore, identifying promising biomarkers are useful for better diagnose and prognosis, to eventually plan precise therapy for MDS patients.
Glutathione peroxidase 3 (GPX3), a member from glutathione peroxidase (GPX) family (GPX1‐GPX8) located at the 5q23, accounts for almost all of the GPX activity in plasma, plays a crucial role in preventing oxidative damages through reducing redundant reactive oxygen species (ROS) 5. Tumor suppressor function of GPX3 in vivo and in vitro has been identified in a number of tumors 6, 7, 8. Additionally, reduced expression of GPX3 caused by its promoter hypermethylation has been observed in various solid tumors 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20. Simultaneously, our previous studies also have proved the epigenetic style of GPX3 in AML and chronic myeloid leukemia (CML), and further disclosed that hypermethylation of GPX3 promoter was a prognostically adverse indicator in AML 21, 22, 23. Here, we reported GPX3 promoter methylation and its clinical implication in patients with MDS.
Materials and Methods
Patients
This study was approved by the Institutional Review Board of the Affiliated People's Hospital of Jiangsu University. After written informed consent was obtained, BM was collected from 110 patients with a diagnosis of MDS according to the 2008 revised World Health Organization (WHO) criteria 1, 24. Risk groups were classified based on International Prognosis Scoring System (IPSS) 25. The treatment for MDS patients with lower IPSS scores (Low/Int‐1) was symptomatic and supportive treatment with/without thalidomide (100 mg/d), whereas patients with higher IPSS scores (Int‐2/High) received chemotherapy which included aclacinomycin (12 mg/m2/d, d1‐d4), cytarabine (10 mg/m2/12 h, d1‐d14), granulocyte colony‐stimulating factor (200 μg/m2/d, d0‐d14) together with symptomatic and supportive treatment. The control group was used as reported previously 22, 23.
RNA isolation, reverse transcription, and RQ‐PCR
BM mononuclear cells (BMMNCs) were focused in this study. Total RNA was isolated form BMMNCs and was reverse transcribed as reported in our previous report 21. GPX3 expression was evaluated by the transcript level of GPX3 determined by real‐time quantitative PCR (RQ‐PCR) as reported 21.
DNA isolation, chemical modification, and RQ‐MSP
Genomic DNA was isolated from BMMNCs and modified as reported previously 22, 23. The primers used for GPX3 methylation were reported previously 17. GPX3 methylation was detected by real‐time quantitative methyaltion‐specific PCR (RQ‐MSP) using AceQ qPCR SYBR Green Master Mix (Vazyme Biotech Co., Piscataway, NJ). RQ‐MSP was performed as our previous study 22, 23. Both positive [bisulfite‐modified DNA from K562 cells, cultured in RPMI 1640 medium containing 10% fetal calf serum (ExCell Bio, Shanghai, China)] and negative controls (ddH2O) were included in each assay. The normalized ratio (NM‐GPX3) was used to assess GPX3 methylation in each sample, and calculated using the following formula: 22, 23.
BSP
TaKaRa Taq™ Hot Start Version (Tokyo, Japan) kit was applied for bisulfite sequencing PCR (BSP) reaction with primers also as reported 22, 23. BSP conditions was performed as our previous study 22, 23. BSP products cloning sequencing was performed as described previously 22, 23. Six independent clones from each specimen were sequenced (BGI Tech Solutions Co., Shanghai, China).
HRMA and DNA sequencing
High‐resolution melting analysis (HRMA) and DNA sequencing was performed in the detection of IDH1/2, U2AF1, SF3B1, DNMT3A, and CEBPA mutations as reported previously 26, 27, 28, 29, 30, 31.
Statistical analyses
SPSS 20.0 software package (SPSS, Chicago, IL) was applied to the statistical analyses. Pearson Chi square analysis/Fisher's exact test was applied to compare the differences in categorical variables, whereas continuous variables were compared by Mann Whitney U test. Survival analyses were performed by both the Kaplan–Meier and the Cox regression analyses. For all analyses, a two‐tailed P value less than 0.05 was considered as statistically significant.
Results
GPX3 methylation in MDS
Our previous studies have detected GPX3 methylation in controls, AML, and CML patients, and the value of 0.184 was selected as the cutoff point to define GPX3 methylation and unmethylation 22, 23. According to the set point, GPX3 methylation was identified in 15% (17/110) MDS patients, and significantly higher than controls (P = 0.024). Notably, the percentage of GPX3 methylation in MDS was significantly lower than AML patients (P = 0.041). Furthermore, BSP was performed in one control at the cutoff point and one MDS patient with highest methylation level showed in Figure 1.
Figure 1.
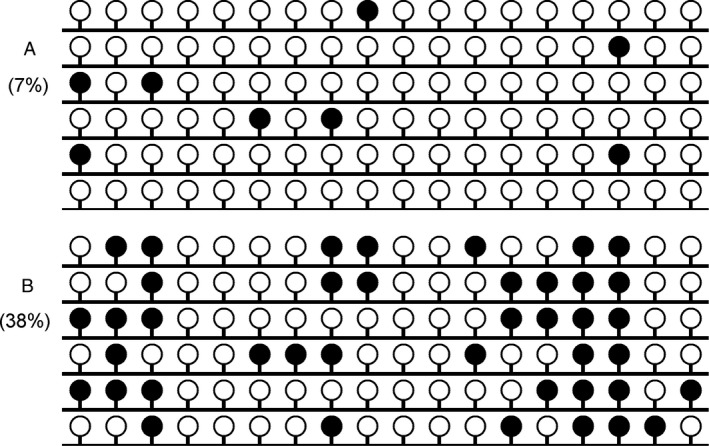
GPX3 methylation density in one control and one myelodysplastic syndrome(MDS) patient. (A) Control at the cutoff point. (B) MDS patient with highest methylation level. White cycle: unmethylated CpG dinucleotide; Black cycle: methylated CpG dinucleotide.
Association between GPX3 methylation and expression in MDS
GPX3 transcript was further examined in 20 MDS patients and 44 controls with available samples 21. GPX3 mRNA level was down‐regulated in MDS compared with 44 controls (median 0.042 vs. 0.348, P < 0.001). Although no significantly negative correlation was observed between GPX3 methylation and expression (R = −0.348, P = 0.133, Spearman test, n = 20), patients with GPX3 methylation (n = 5) showed markedly lower GPX3 mRNA level than those without GPX3 methylation (n = 15) (median 0.0002 vs. 0.0652, P = 0.002).
Association between GPX3 methylation and clinical characteristics in MDS
We further compared the clinical manifestations and laboratory features between GPX3 unmethylated and methylated MDS patients (Table 1). No significant differences were shown in sex, peripheral blood cells, WHO classifications, cytogenetics classifications, and IPSS scores classifications (P > 0.05). However, cases with GPX3 methylation had significantly older age than those with GPX3 unmethylation (P = 0.015). Gene mutations were detected in 106 patients, and GPX3 methylated patients tend to have higher frequency of DNMT3A mutation (P = 0.066).
Table 1.
Comparison of clinical manifestations and laboratory features between GPX3 unmethylated and methylated MDS patients
| Patients' parameter | Unmethylated (n = 93) | Methylated (n = 17) | Total (n = 110) | P value |
|---|---|---|---|---|
| Age (years)a | 57 (14–86) | 70 (38–84) | 60 (14–86) | 0.015 |
| Sex (male/female) | 54/39 | 10/7 | 64/46 | 1.000 |
| WBC (×109/L)a | 2.7 (0.6–82.4) | 3.6 (1.4–8.2) | 2.75 (0.6–82.4) | 0.140 |
| HB (g/L)a | 64 (26–128) | 65 (37–118) | 64 (26–128) | 0.634 |
| PLT (×109/L)a | 60 (0–1176) | 59 (1–754) | 60 (0–1176) | 0.853 |
| WHO | 0.149 | |||
| RA(RS) | 11 (12%) | 2 (12%) | 13 (12%) | |
| RCMD(RS) | 40 (43%) | 4 (24%) | 44 (40%) | |
| RAEB‐1 | 17 (18%) | 5 (29%) | 22 (20%) | |
| RAEB‐2 | 23 (24%) | 4 (24%) | 27 (25%) | |
| 5q‐ | 2 (2%) | 1 (6%) | 3 (3%) | |
| MDS‐U | 0 (0%) | 1 (6%) | 1 (1%) | |
| Cytogenetics | 0.724 | |||
| Good | 62 (67%) | 13 (76%) | 75 (68%) | |
| Intermediate | 16 (17%) | 1 (6%) | 17 (15%) | |
| Poor | 7 (8%) | 1 (6%) | 8 (7%) | |
| No data | 8 (9%) | 2 (12%) | 9 (8%) | |
| IPSS | 0.310 | |||
| Low | 8 (9%) | 3 (18%) | 11 (10%) | |
| Int‐1 | 55 (59%) | 6 (35%) | 61 (55%) | |
| Int‐2 | 14 (15%) | 4 (24%) | 18 (16%) | |
| High | 8 (9%) | 2 (12%) | 10 (9%) | |
| No data | 8 (9%) | 2 (12%) | 10 (9%) | |
| Gene mutations | ||||
| C/EBPA (±) | 2/87 | 1/16 | 3/103 | 0.411 |
| IDH1/2 (±) | 3/86 | 1/16 | 4/102 | 0.508 |
| DNMT3A (±) | 1/88 | 2/15 | 3/103 | 0.066 |
| U2AF1 (±) | 7/82 | 0/17 | 7/99 | 0.595 |
| SF3B1 (±) | 6/83 | 0/17 | 6/100 | 0.586 |
Median (range); WBC, white blood cells; HB, Hemoglobin; PLT, Platelet count; IPSS, International Prognostic Scoring System; WHO, World Health Organization; RA, refractory anemia; RARS, RA with ringed sideroblasts; RCMD, refractory cytopenia with multilineage dysplasia; RCMD‐RS, RCMD with ringed sideroblasts; RAEB, RA with excess of blasts. Int‐1, intermediate‐1; Int‐2: intermediate‐2.
Association between GPX3 methylation and prognosis in MDS
A total of 91 MDS patients with available survival data (range 1–113 months, median 25 months) were obtained. Kaplan–Meier analysis revealed that GPX3 methylated patients (median: 10 months, 95% CI: 3.277–16.723 months) had markedly shorter overall survival (OS) time than GPX3 unmethylated patients (median: 33 months, 95% CI: 15.777–50.223 months) (P = 0.012, Fig. 2). To investigate whether GPX3 methylation could act as an independent prognostic factor in MDS, Cox regression analyses (univariate and multivariate analyses) were further performed. The multivariate analysis included variables with P < 0.200 in univariate analysis, and showed that GPX3 methylation might serve as an independent risk predictor in MDS (HR = 1.847, P = 0.072, Table 2).
Figure 2.
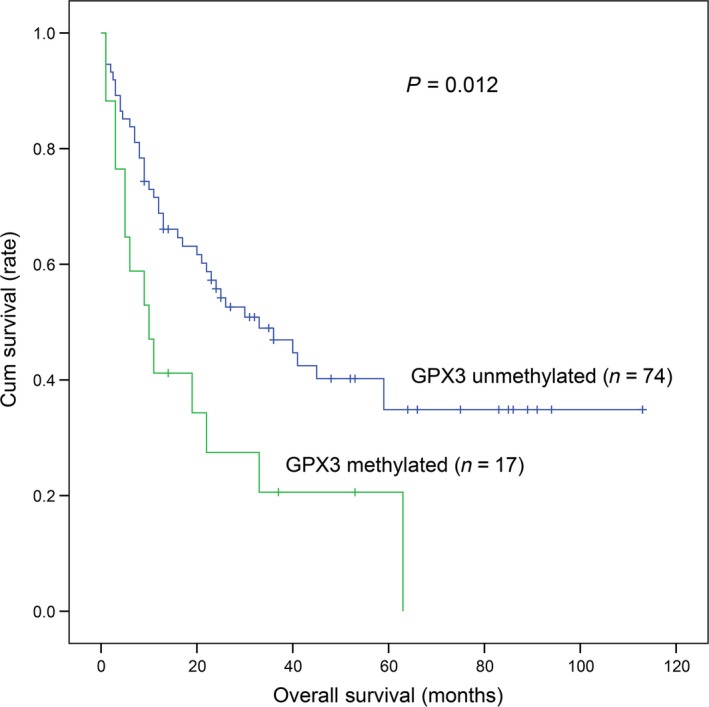
The impact of GPX3 methylation on overall survival in MDS patients.
Table 2.
Univariate and multivariate analyses on overall survival in MDS patients
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | |
| Age | 1.962 (1.128–3.412) | 0.017 | 2.052 (1.130–3.725) | 0.018 |
| IPSS | 1.547 (1.091–2.193) | 0.017 | 1.569 (1.108–2.223) | 0.011 |
| GPX3 methylation | 2.126 (1.155–3.913) | 0.015 | 1.847 (0.948–3.600) | 0.072 |
| DNMT3A mutation | 3.059 (0.936–9.998) | 0.064 | 1.838 (0.520–6.499) | 0.345 |
| CEBPA mutation | 0.414 (0.057–3.015) | 0.384 | – | – |
| IDH1/2 mutations | 0.951 (0.296–3.054) | 0.933 | – | – |
| U2AF1 mutation | 0.780 (0.280–2.170) | 0.634 | – | – |
| SF3B1 mutation | 1.570 (0.488–5.051) | 0.450 | – | – |
IPSS, International Prognostic Scoring System. Variables including age (≤60/>60 years old), IPSS risks (low/intermediate‐1/intermediate‐2/high), gene mutations (positive/negative), and GPX3 methylation (methylated/unmethylated).
Alterations in GPX3 methylation during the progression from MDS to sAML
In order to determine whether GPX3 methylation was involved in MDS progression to sAML, three patients with follow‐up data from MDS to sAML were further examined by BSP. As was shown in Figure 3, GPX3 methylation density was significantly increased during the progression from MDS to sAML.
Figure 3.
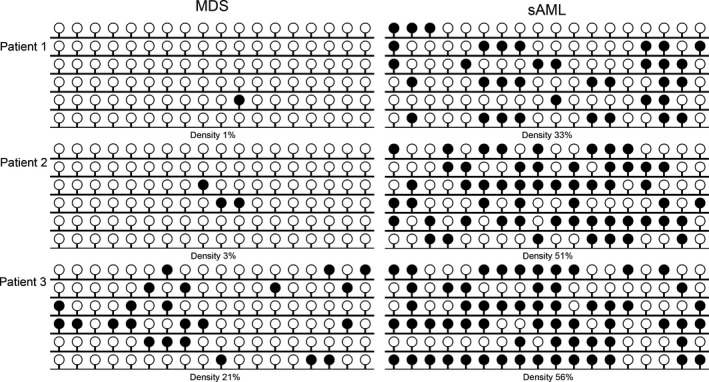
Alterations in GPX3 methylation density during the progression from MDS to secondary AML (sAML) in three follow‐up paired patients. White cycle: unmethylated CpG dinucleotide; Black cycle: methylated CpG dinucleotide.
Discussion
Tumor suppressor function of GPX3 through the prevention of cancer initiation, promotion, and metastasis has been gradually identified in solid tumors 32. A recent study revealed that overexpression of GPX3 suppressed tumor invasiveness, both in vivo and in vitro 8. In addition, the study also described the therapeutic value of the engineered hiPSC‐MSCs delivering GPX3 in hepatocellular carcinoma 8. GPX3 inhibits prostate tumorigenesis in transgenic adenocarcinoma of the mouse prostate (TRAMP) mice 33. Moreover, function role of GPX3 has also been investigated in leukemia. Herault et al. have revealed that GPX3 plays a vital role in the competitiveness of leukemic stem cells (LSCs) and self‐renewal activity of HSCs 34. Recently, increasing studies have demonstrated the reduced expression of GPX3 and hypermethylation of GPX3 promoter in various types of malignancies 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19. Furthermore, epigenetic mechanism regulating GPX3 expression was also identified in human cancer cells including esophageal squamous cell carcinoma (ESCC), cervical cancer (CC), gastric carcinoma (GC), and multiple myeloma (MM) 9, 13, 17, 19. With regard to leukemia, our previous studies also confirmed the epigenetic style in myeloid leukemia patients and leukemic cell lines including K562 and THP1 22, 23. These results together suggested that epigenetically silenced GPX3 may play a vital role in both solid tumors and hematological malignancies.
In this study, GPX3 methylation was found in 15% MDS and higher than controls but lower than AML, which indicated that GPX3 methylation was involved in MDS pathogenesis, and might be more significant in AML. In addition, GPX3 expression down‐regulation probably caused by promoter hypermethylation was also observed in MDS. Interestingly, Herault et al. reported that leukemia with a high frequency of LSCs expressed high levels of GPX3 by promoter hypomethylation and low levels of ROS, which enhanced the competitiveness of LSCs 34. However, GPX3 showed down‐regulation in clinical AML cases compared with normal CD34+ cells, especially in patients with favorable/intermediate karyotypes 20, 34. Taken together, these may probably be due to GPX3 playing different roles in initial development of cancer, and GPX3 expressed heterogeneously among different cells types like in CD34+ cells and mesenchymal stem cells.
DNA methylation, the most common epigenetic alteration in regulating gene expression, has been used as promising biological markers for early diagnosis, prognosis, disease progression, therapeutic stratification, and post‐therapeutic monitoring in human cancers 35. Clinical significance of GPX3 hypermethylation has been demonstrated in a number of human cancers. The early diagnostic value of GPX3 methylation has been revealed in ESCC 36. In addition to the diagnostic value, an increasing number of studies have also disclosed the prognostic impact of GPX3 methylation in different types of malignancies. Peng et al. disclosed that GPX3 methylation in GC was associated with lymph node metastasis 13. Chen et al. revealed that GPX3 hypermethylation in head and neck cancer (HNC) correlated with chemoresistance (cisplatin‐based chemotherapy), and could serve as a potential marker predicting the prognosis of HNC patients 15. Kaiser et al. also demonstrated the negative effect of GPX3 hypermethylation on prognosis in multiple myeloma 19. Moreover, the prognostic value of GPX3 hypermethylation was also identified in non‐M3 AML patients 22. In this study, we also demonstrated that GPX3 methylation might be a potential biomarker for assessing treatment outcome in MDS patients. Obviously, prospective studies are required before GPX3 methylation can be used routinely as a novel marker for risk stratification in MDS.
To date, the underlying mechanism of leukemia transformation in MDS remains poorly defined. Genetic alterations including chromosomal abnormalities and gene mutations are considered as progression‐related drivers 37. However, these changes could not be observed among the overall process. Notably, Jiang et al. reported that epigenetic modifications of aberrant DNA methylation were a dominant mechanism in MDS progression to sAML 4. Following this study, we further determined GPX3 methylation in follow‐up paired patients with disease progression, and demonstrated that GPX3 methylation played a vital role in progression of MDS to sAML. However, the limited followed up paired patients calls for further studies to be able to confirm and expand our results.
Previous evidences showed that several methylation‐related gene mutations including ASXL1, EZH2, IDH1/2, TET2, and DNMT3A have been identified contributing to epigenetic alterations in hematological malignancies 38, 39. DNMT3A mutations played a crucial role in leukemogenesis and led to poor prognosis in AML and MDS 26, 40. Our study further found that GPX3 methylation might be associated with DNMT3A mutation in MDS patients. Overall, these results suggested that GPX3 methylation might be involved in MDS pathogenesis caused by DNMT3A mutation. Further investigations are needed to determine the specific role in MDS.
In summary, GPX3 methylation in bone marrow is associated with adverse prognosis and leukemia transformation in MDS.
Conflict of Interest
The authors stated that there are no conflicts of interest regarding the publication of this article.
Acknowledgments
This study was supported by National Natural Science foundation of China (81270630), Science and Technology Special Project in Clinical Medicine of Jiangsu Province (BL2012056), 333 Project of Jiangsu Province (BRA2013136), Six talent peaks project in Jiangsu Province (2015‐WSN115), Science and Technology Infrastructure Program of Zhenjiang (SS2012003), Social Development Foundation of Zhenjiang (SH2014086, SH2014044, SH2015058), Clinical Medical Science, Development Foundation of Jiangsu University (JLY20140018), Key Medical Talent Program of Zhenjiang City.
Cancer Medicine 2017, 6(1):267–274
Contributor Information
Zhao‐Qun Deng, Email: zqdeng2002@gmail.com.
Jun Qian, Email: qianjun0007@hotmail.com.
References
- 1. Swerdlow, S. H. , Campo E., Harris N. L., Jaffe E. S., Pileri S. A., Stein H.. et al. 2008. WHO classification of tumours of haematopoietic and lymphoid tissues. IARC, Lyon. [Google Scholar]
- 2. Malcovati, L. , and Nimer S. D.. 2008. Myelodysplastic syndromes: diagnosis and staging. Cancer Control 15(Suppl):4–13. [DOI] [PubMed] [Google Scholar]
- 3. Harada, H. , and Harada Y.. 2015. Recent advances in myelodysplastic syndromes: molecular pathogenesis and its implications for targeted therapies. Cancer Sci. 106:329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jiang, Y. , Dunbar A., Gondek L. P., Mohan S., Rataul M., O'Keefe C., et al. 2009. Aberrant DNA methylation is a dominant mechanism in MDS progression to AML. Blood 113:1315–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brigelius‐Flohé, R. , and Maiorino M.. 1830. Glutathione peroxidases. Biochim. Biophys. Acta 2013:3289–3303. [DOI] [PubMed] [Google Scholar]
- 6. Yu, Y. P. , Yu G., Tseng G., Cieply K., Nelson J., Defrances M., et al. 2007. Glutathione peroxidase 3, deleted or methylated in prostate cancer, suppresses prostate cancer growth and metastasis. Cancer Res. 67:8043–8050. [DOI] [PubMed] [Google Scholar]
- 7. Barrett, C. W. , Ning W., Chen X., Smith J. J., Washington M. K., Hill K. E., et al. 2013. Tumor suppressor function of the plasma glutathione peroxidase GPX3 in colitis‐associated carcinoma. Cancer Res. 73:1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qi, X. , Ng K. T., Lian Q. Z., Liu X. B., Li C. X., Geng W., et al. 2014. Clinical significance and therapeutic value of glutathione peroxidase 3 (GPx3) in hepatocellular carcinoma. Oncotarget 5:11103–11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang, X. , Zheng Z., Yingji S., Kim H., Jin R., Renshu L., et al. 2014. Downregulation of glutathione peroxidase 3 is associated with lymph node metastasis and prognosis in cervical cancer. Oncol. Rep. 31:2587–2592. [DOI] [PubMed] [Google Scholar]
- 10. Yang, Z. L. , Yang L., Zou Q., Yuan Y., Li J., Liang L., et al. 2013. Positive ALDH1A3 and negative GPX3 expressions are biomarkers for poor prognosis of gallbladder cancer. Dis. Markers 35:163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Agnani, D. , Camacho‐Vanegas O., Camacho C., Lele S., Odunsi K., Cohen S., et al. 2011. Decreased levels of serum glutathione peroxidase 3 are associated with papillary serous ovarian cancer and disease progression. J. Ovarian Res. 4:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mohamed, M. M. , Sabet S., Peng D. F., Nouh M. A., El‐Shinawi M., and El‐Rifai W.. 2014. Promoter hypermethylation and suppression of glutathione peroxidase 3 are associated with inflammatory breast carcinogenesis. Oxid. Med. Cell Longev. 2014:787195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peng, D. F. , Hu T. L., Schneider B. G., Chen Z., Xu Z. K., and El‐Rifai W.. 2012. Silencing of glutathione peroxidase 3 through DNA hypermethylation is associated with lymph node metastasis in gastric carcinomas. PLoS ONE 7:e46214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Min, S. Y. , Kim H. S., Jung E. J., Jung E. J., Jee C. D., and Kim W. H.. 2012. Prognostic significance of glutathione peroxidase 1 (GPX1) down‐regulation and correlation with aberrant promoter methylation in human gastric cancer. Anticancer Res. 32:3169–3175. [PubMed] [Google Scholar]
- 15. Chen, B. , Rao X., House M. G., Nephew K. P., Cullen K. J., and Guo Z.. 2011. GPX3 promoter hypermethylation is a frequent event in human cancer and is associated with tumorigenesis and chemotherapy response. Cancer Lett. 309:37–45. [DOI] [PubMed] [Google Scholar]
- 16. Falck, E. , Karlsson S., Carlsson J., Helenius G., Karlsson M., and Klinga‐Levan K.. 2010. Loss of glutathione peroxidase 3 expression is correlated with epigenetic mechanisms in endometrial adenocarcinoma. Cancer Cell Int. 10:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He, Y. , Wang Y., Li P., Zhu S., Wang J., and Zhang S.. 2011. Identification of GPX3 epigenetically silenced by CpG methylation in human esophageal squamous cell carcinoma. Dig. Dis. Sci. 56:681–688. [DOI] [PubMed] [Google Scholar]
- 18. Lee, O. J. , Schneider‐Stock R., McChesney P. A., Kuester D., Roessner A., Vieth M., et al. 2005. Hypermethylation and loss of expression of glutathione peroxidase‐3 in Barrett's tumorigenesis. Neoplasia 7:854–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kaiser, M. F. , Johnson D. C., Wu P., Walker B. A., Brioli A., Mirabella F., et al. 2013. Global methylation analysis identifies prognostically important epigenetically inactivated tumor suppressor genes in multiple myeloma. Blood 122:219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Melo, C. F. , Gigek C. O., da Silva J. N., Cardoso Smith Mde A., de Araújo R. M., Burbano R. R., et al. 2014. Association of COX2 gene hypomethylation with intestinal type gastric cancer in samples of patients from northern Brazil. Tumour Biol. 35:1107–1111. [DOI] [PubMed] [Google Scholar]
- 21. Zhou, J. D. , Wen X. M., Zhang Y. Y., Yang L., Ma Y. J., Ma J. C., et al. 2015. Down‐regulation of GPX3 is associated with favorable/intermediate karyotypes in de novo acute myeloid leukemia. Int. J. Clin. Exp. Pathol. 8:2384–2391. [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou, J. D. , Yao D. M., Zhang Y. Y., Ma J. C., Wen X. M., Yang J., et al. 2015. GPX3 hypermethylation serves as an independent prognostic biomarker in non‐M3 acute myeloid leukemia. Am. J. Cancer Res. 5:1786–1794. [PMC free article] [PubMed] [Google Scholar]
- 23. Yao, D. M. , Zhou J. D., Zhang Y. Y., Yang L., Wen X. M., Yang J., et al. 2015. GPX3 promoter is methylated in chronic myeloid leukemia. Int. J. Clin. Exp. Pathol. 8:6450–6457. [PMC free article] [PubMed] [Google Scholar]
- 24. Bennett, J. M. , Catovsky D., Daniel M. T., Flandrin G., Galton D. A., Gralnick H. R., et al. 1982. Proposals for the classification of the myelodysplastic syndromes. Br. J. Haematol. 51:189–199. [PubMed] [Google Scholar]
- 25. Greenberg, P. , Cox C., LeBeau M. M., Fenaux P., Morel P., Sanz G., et al. 1997. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood 89:2079–2088 [Erratum in: Blood 1998; 91: p. 1100]. [PubMed] [Google Scholar]
- 26. Lin, J. , Yao D. M., Qian J., Chen Q., Qian W., Li Y., et al. 2011. Recurrent DNMT3A R882 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS ONE 6:e26906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin, J. , Yao D. M., Qian J., Chen Q., Qian W., Li Y., et al. 2012. IDH1 and IDH2 mutation analysis in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. Ann. Hematol. 91:519–525. [DOI] [PubMed] [Google Scholar]
- 28. Qian, J. , Yao D. M., Lin J., Qian W., Wang C. Z., Chai H. Y., et al. 2012. U2AF1 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS ONE 7:e45760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang, J. , Qian J., Lin J., Yang X. F., Qian W., Chen Q., et al. 2013. Development of a high‐resolution melting analysis for the detection of the SF3B1 mutations. Genet. Test Mol. Biomarkers 17:342–347. [DOI] [PubMed] [Google Scholar]
- 30. Wen, X. M. , Hu J. B., Yang J., Qian W., Yao D. M., Deng Z. Q., et al. 2015. CEBPA methylation and mutation in myelodysplastic syndrome. Med. Oncol. 32:605. [DOI] [PubMed] [Google Scholar]
- 31. Wen, X. M. , Lin J., Yang J., Yao D. M., Deng Z. Q., Tang C. Y., et al. 2014. Double CEBPA mutations are prognostically favorable in non‐M3 acute myeloid leukemia patients with wild‐type NPM1 and FLT3‐ITD. Int. J. Clin. Exp. Pathol. 7:6832–6840. [PMC free article] [PubMed] [Google Scholar]
- 32. Brigelius‐Flohé, R. , and Kipp A.. 2009. Glutathione peroxidases in different stages of carcinogenesis. Biochim. Biophys. Acta 1790:1555–1568. [DOI] [PubMed] [Google Scholar]
- 33. Chang, S. N. , Lee J. M., Oh H., and Park J. H.. 2016. Glutathione peroxidase 3 inhibits prostate tumorigenesis in TRAMP mice. Prostate 76:1387–1398. [DOI] [PubMed] [Google Scholar]
- 34. Herault, O. , Hope K. J., Deneault E., Mayotte N., Chagraoui J., Wilhelm B. T., et al. 2012. A role for GPX3 in activity of normal and leukemia stem cells. J. Exp. Med. 209:895–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taby, R. , and Issa J. P.. 2010. Cancer epigenetics. CA Cancer J. Clin. 60:376–392. [DOI] [PubMed] [Google Scholar]
- 36. Li, X. , Zhou F., Jiang C., Wang Y., Lu Y., Yang F., et al. 2014. Identification of a DNA methylome profile of esophageal squamous cell carcinoma and potential plasma epigenetic biomarkers for early diagnosis. PLoS ONE 9:e103162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dan, C. , Chi J., and Wang L.. 2015. Molecular mechanisms of the progression of myelodysplastic syndrome to secondary acute myeloid leukaemia and implication for therapy. Ann. Med. 47:209–217. [DOI] [PubMed] [Google Scholar]
- 38. Woods, B. A. , and Levine R. L.. 2015. The role of mutations in epigenetic regulators in myeloid malignancies. Immunol. Rev. 263:22–35. [DOI] [PubMed] [Google Scholar]
- 39. Shih, A. H. , Abdel‐Wahab O., Patel J. P., and Levine R. L.. 2012. The role of mutations in epigenetic regulators in myeloid malignancies. Nat. Rev. Cancer 12:599–612. [DOI] [PubMed] [Google Scholar]
- 40. Ley, T. J. , Ding L., Walter M. J., McLellan M. D., Lamprecht T., Larson D. E., et al. 2010. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 363:2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]


