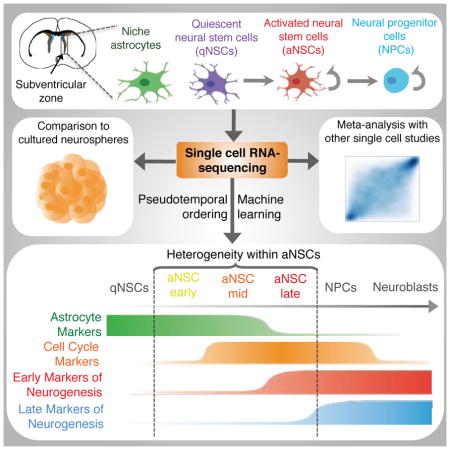
Summary
Neural stem cells (NSCs) in the adult mammalian brain serve as a reservoir for the generation of new neurons, oligodendrocytes, and astrocytes. Here we use single cell RNA-sequencing to characterize adult NSC populations and examine the molecular identities and heterogeneity of in vivo NSC populations. We find that cells in the NSC lineage exist on a continuum through the processes of activation and differentiation. Interestingly, rare intermediate states with distinct molecular profiles can be identified and experimentally validated, and our analysis identifies putative surface markers and key intracellular regulators for these subpopulations of NSCs. Finally, using the power of single cell profiling, we conduct a meta-analysis to compare in vivo NSCs and in vitro cultures, distinct fluorescent-activated cell sorting strategies, and different neurogenic niches. These data provide a resource for the field and contribute to an integrative understanding of the adult NSC lineage.
Keywords: Neural stem cells, adult neurogenesis, single cell RNA-seq, machine learning, pseudotemporal ordering
ETOC Blurb
Dulken et al. perform single cell transcriptomics on neural stem cells (NSCs) from adult mice. They use machine learning to identify rare intermediate cells in the continuum of the NSC lineage and perform a meta-analysis with other single cell transcriptomic data from in vitro or in vivo NSCs.
Introduction
Populations of neural stem cells (NSCs) in the adult brain represent a critical reservoir of regenerative cells with the potential to combat neuronal injury and neurodegeneration. The adult brain contains two NSC pools located in the sub-ventricular zone (SVZ) of the lateral ventricles and the dentate gyrus (DG) of the hippocampus (Zhao et al., 2008). Both NSC pools produce new neurons, which can integrate into functional circuits (Zhao et al., 2008). The NSCs of the SVZ have been identified as a subtype of sub-ependymal astrocyte (Doetsch et al., 1999; Garcia et al., 2004). The majority of NSCs are quiescent and express GFAP along with the marker CD133 (Prominin 1) (Codega et al., 2014; Fischer et al., 2011). These quiescent NSCs (qNSCs or type B1q cells) give rise to proliferative, activated neural stem cells (aNSCs or type B1a cells) that express epidermal growth factor receptor (EGFR) (Codega et al., 2014). Activated NSCs can in turn produce neural progenitor cells (NPCs or transient amplifying progenitor [TAPs] or type C cells), a proliferative cell population that expresses markers of early neuronal differentiation (Doetsch et al., 2002). Finally, the NPCs give rise to neuroblasts (type A cells), which migrate to the olfactory bulb where they become primarily interneurons (Garcia et al., 2004; Mirzadeh et al., 2008) (Figure 1A).
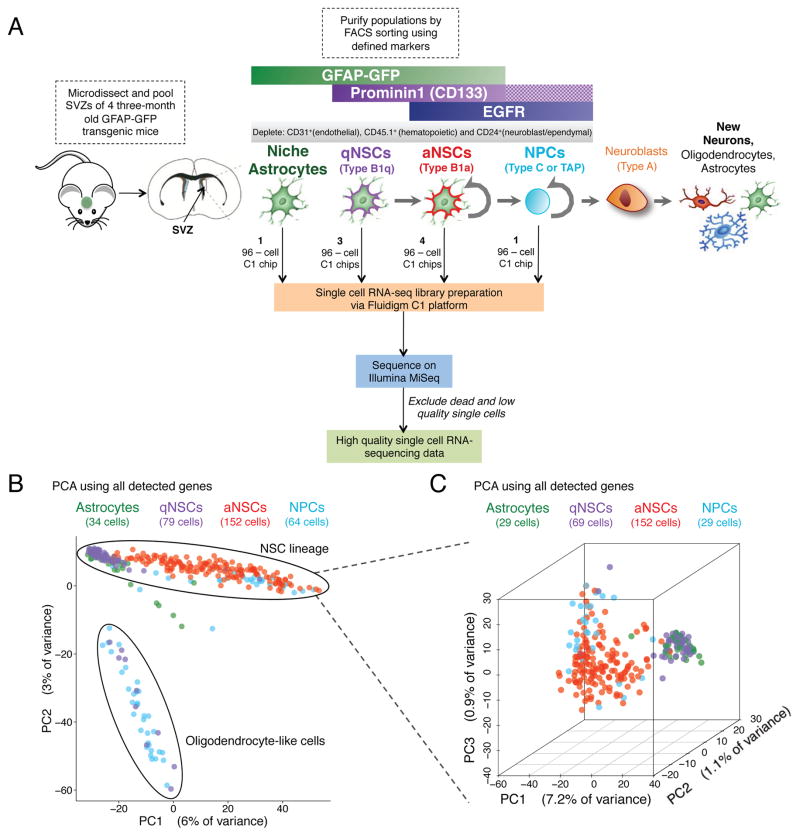
Figure 1. Single cell RNA-seq of 329 cells from four populations of FACS-purified cells from the SVZs of adult mice.
(A) FACS-sorting scheme for the enrichment of astrocytes, qNSCs, aNSCs, and NPCs from the SVZs of adult mice, and microfluidic-based single cell RNA-seq library generation and sequencing. Checkered bar in the FACS-sorting scheme indicates that the presence of Prominin 1 was not selected for. Note that while Prominin 1 enriches for NSCs, the astrocyte population could contain some qNSCs and the qNSC population could contain some astrocytes (Codega et al., 2014).
(B) Principal component analysis (PCA) on all 329 high-quality single cells.
(C) 3-dimensional PCA on all 288 cells excluding oligodendrocyte-like cells and seven outlying cells.
The purification of NSCs from their in vivo niche has been made possible by Fluorescence-Activated Cell Sorting (FACS) via the expression of transgenic markers and defined surface markers (Codega et al., 2014; Fischer et al., 2011; Garcia et al., 2004; Mich et al., 2014). Purification of cell populations, coupled to gene expression profiling, has begun to reveal the molecular identities of NSCs in the SVZ (Codega et al., 2014; Mich et al., 2014). However, population-based approaches have likely obscured underlying heterogeneity in the NSC lineage, thereby limiting the identification of new rare cell types or intermediates, and hindering the characterization of complex transcriptional dynamics. While recent single cell studies have started to reveal the complex composition of NSC populations in various neurogenic regions of the adult brain, the SVZ (Llorens-Bobadilla et al., 2015; Luo et al., 2015) and the DG (Shin et al., 2015), a comprehensive molecular understanding of the heterogeneity of the neural stem cell lineage still remains elusive.
Here we perform single cell RNA-sequencing on 329 high quality single cells from four different populations – niche astrocytes, qNSCs, aNSCs, and NPCs – freshly isolated from young adult mouse SVZs. Using machine learning and pseudotemporal ordering, we reveal subpopulations of NSCs along the spectrum of activation and differentiation, which we experimentally validate, and suggest putative markers for these subpopulations. Using the power of single cell transcriptomics, we compare our single cell dataset to other single cell datasets, including in vitro cultured NSCs and other in vivo NSC datasets. Our findings not only serve as a great resource for the field, but also provide an integrative understanding of the neural stem cell lineage, which is an essential step toward identifying new ways to reactivate dormant NSCs in the context of stroke and aging.
Results
Single cell RNA-seq from four populations of cells directly isolated from the SVZ regenerative region in the adult mouse brain
To define the molecular heterogeneity of the SVZ regenerative region in the adult mouse brain, we performed single cell RNA-sequencing from four cell populations – niche astrocytes, quiescent and activated NSCs, and more committed NPCs. We implemented a well-accepted FACS protocol to freshly isolate adult populations from the SVZ (Codega et al., 2014) using a transgenic line in which green fluorescent protein (GFP) is under the control of the human GFAP promoter (GFAP-GFP mice) (Zhuo et al., 1997). Single cells were dissociated from microdissected SVZs from young adult (3 months old) GFAP-GFP male mice and stained with markers of NSC identity and activation, including CD133/Prominin 1 [PROM1] and EGFR. This approach enabled us to isolate niche astrocytes (henceforth referred to as astrocytes) (GFAP-GFP+PROM1−EGFR−), qNSCs (GFAP-GFP+PROM1+EGFR−), aNSCs (GFAP-GFP+PROM1+EGFR+), and NPCs (GFAP-GFP−EGFR+), as described in (Codega et al., 2014) (Figure 1A, Figure S1A). Each of these enriched populations was used to prepare single cell RNA-sequencing libraries using the Fluidigm C1 Single-Cell Auto Prep microfluidic system (Wu et al., 2014). A total of 524 single cell libraries were sequenced on Illumina MiSeq, and a subset was also sequenced on Illumina HiSeq 2000 (Tables S1, S2, S3, S4). The majority of unique genes in each library were detected by MiSeq (Figure S1B) and there was good correlation between gene detection for libraries sequenced on MiSeq and HiSeq for all genes except those expressed at very low levels (Figure S1C), consistent with previous observations that high sequencing depth is not necessary to capture single cell library complexity (Pollen et al., 2014). We excluded low quality cells, based on a threshold for reads mapping to the transcriptome and number of genes detected (Figure S1D). On the remaining 329 cells, there was good correlation of gene expression between two representative single cells (Pearson correlation = 0.602) or pseudopopulations (Pearson correlation = 0.932) (Figure S1E). Furthermore, aggregated single cell pseudo-populations for each cell type cluster with population RNA-seq (Leeman et al.) for their associated cell type, and away from a cell type from an independent lineage (endothelial cells) (Figures S1F–G), underscoring the quality of the single cell RNA-seq libraries.
To explore the molecular identities of individual single cells, we performed global principal component analysis (PCA) projection of all single cells profiled in this analysis. Most astrocytes, qNSCs, aNSCs, and NPCs clustered in a well-defined ‘band’, although a subpopulation of cells sorted as qNSCs and NPCs separated significantly from the majority of the single cells on the second principal component (PC) of the PCA (Figure 1B). Genes with the strongest contribution to this second PC were highly enriched for genes involved in myelination and oligodendrocyte function/identity (e.g. Mog, Plp1, and Mbp (Cahoy et al., 2008)) (Figure S1H). Thus, a minority of oligodendrocytes appears to be present in the population of cells sorted as qNSCs and NPCs, which was also observed in another single cell study (Llorens-Bobadilla et al., 2015).
To focus our analysis on the NSC lineage, we excluded all cells exhibiting an oligodendrocyte expression signature as well as a small number of outlying cells that clustered away from the NSC lineage (Figure 1B). PCA on the remaining cells revealed clustering of the more quiescent cell types (astrocytes and qNSCs) away from the active, proliferative cell types (aNSCs and NPCs) (Figure 1C). While there was no significant difference between astrocytes and qNSCs, consistent with previous studies (Codega et al., 2014), aNSCs separated from NPCs (Figure 1C). Interestingly, a range of aNSCs was observed between the quiescent and progenitor states (Figure 1C), raising the possibility that in vivo NSCs exist on a continuum of quiescence, activation, and differentiation.
Single cells from populations of qNSCs, aNSCs, and NPCs can be ordered through activation and differentiation, suggesting heterogeneity and intermediary states
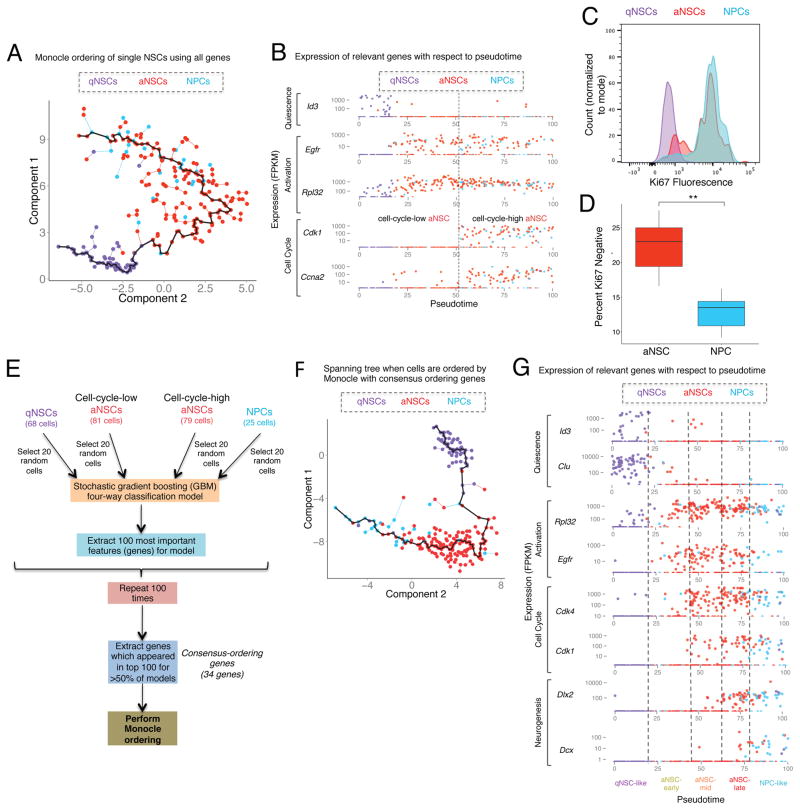
To explore the intermediary states in the continuum of NSCs and progeny, we performed pseudotemporal ordering of the single cells using Monocle (Trapnell et al., 2014). Because astrocytes and qNSCs could not be distinguished by PCA (Figure S2A) or differential expression (Table S5), we omitted astrocytes from the Monocle-ordering analysis. Monocle ordering on qNSCs, aNSCs, and NPCs using all detected genes revealed gene expression dynamics that recapitulate previous understanding of the activation of NSCs (Figure 2A–B). Indeed, qNSCs that highly express previously reported markers of this population such as Id3 (Bonaguidi et al., 2008; Mira et al., 2010) are ordered first and are followed by aNSCs that have upregulated Egfr (Figure 2B). As cells transition from qNSCs to aNSCs, they first upregulate genes important for ribosomal biogenesis (e.g. Rpl32), before expressing markers of the cell cycle (Figure 2B). This corroborates a recent study that described an early stage of biogenesis in aNSCs prior to cell cycle entry (Llorens-Bobadilla et al., 2015). To experimentally validate the existence of this population of “cell-cycle-low” aNSCs, we stained FACS-sorted populations of qNSCs, aNSCs, and NPCs with the cell cycle marker Ki67. Consistent with our single cell prediction, a fraction of aNSCs was negative for the Ki67 cell cycle marker (Figure 2C), and the proportion of Ki67-negative cells was significantly greater in the aNSC population than in NPCs (Figure 2D). These results indicate that a subpopulation of aNSCs is not cycling, but that these “cell-cycle-low” aNSCs are in fact already expressing the EGFR protein, based on the FACS approach we used, rather than merely expressing the Egfr transcript and preparing to enter an EGFR positive state.
Figure 2. Ordering of single cells from populations of qNSCs, aNSCs, and NPCs reveals transcriptional dynamics, and suggests intermediary states.
(A) Minimum spanning tree generated for all qNSCs, aNSCs, and NPCs ordered by Monocle using all detected genes.
(B) Expression of key genes associated with quiescence (Id3), activation (Egfr and Rpl32) and cell cycle (Cdk1 and Ccna2) (FPKM) in each cell plotted with respect to pseudotime produced by Monocle in Figure 2A. Cells are color-coded by their FACS-sorting identity.
(C) Histogram of Ki67 fluorescence values measured by intracellular FACS in purified populations of qNSCs, aNSCs, and NPCs. Histogram values normalized to mode.
(D) Percentage of Ki67-negative cells measured by intracellular FACS in purified populations of aNSCs and NPCs. Two-sided Wilcoxon signed-rank test. ** p≤0.005.
(E) Machine learning algorithm to obtain consensus-ordering genes. The list of consensus-ordering genes is in Table S7.
(F) Minimum spanning tree generated for all qNSCs, aNSCs, and NPCs ordered by Monocle using FPKM of the consensus-ordering genes (Table S7A).
(G) Expression (FPKM) of key genes related to quiescence (Id3), activation (Egfr and Rpl32), cell cycle (Cdk4 and Cdk1), and neuronal differentiation (Dlx2, Dcx) (FPKM) in each cell is plotted with respect to pseudotime produced by Monocle when all qNSCs, aNSCs, and NPCs are ordered using the consensus-ordering genes. Cells are color-coded by their FACS-sorting identity (indicated on top). Bottom: name of the intermediary states (qNSC-like, aNSC-early, aNSC-mid, aNSC-late, and NPC-like).
Monocle ordering could not place NPCs after aNSCs, perhaps because genes highly expressed in both cell types (e.g. cell cycle, metabolism genes) masks more subtle transcriptomic changes. Therefore, to increase the sensitivity of Monocle ordering to the process of lineage commitment/differentiation, we built machine learning models to identify the genes most important for defining the trajectory of cells through four states (Figure 2E): qNSCs, “cell-cycle-low” aNSC, “cell-cycle-high” aNSCs, and NPCs. We implemented a four-way stochastic gradient-boosting classification model (Friedman, 2002), using a subsampled set of 20 cells from each of these four groups (‘training set’) (Figure 2E, code available at https://github.com/bdulken/SVZ_NSC_Dulken_2). We bootstrapped this process by building 100 independent models using independently-sampled subsets of single cells (Figure 2E). In predicting the identity of cells that were not used to build the model (‘testing set’), the accuracy of the models was approximately 80% (Figure S2B), indicating that the models perform drastically better than random assignment in predicting cell state. Machine learning also identifies the genes that are most important for the construction of the models (Table S6). Of these, we selected the genes found in the top 100 most important features in at least half of the models, producing a list of 34 genes, several of which were previously known to be dynamically regulated during NSC activation and differentiation (e.g. Clu, Ccnd2, Dlx2, Dcx) (Table S7A). When Monocle-based cell ordering was conducted using this subset of 34 “consensus-ordering” genes, it resulted in a strikingly accurate recapitulation of the current understanding of activation and commitment/differentiation of NSCs and their progeny (Figure 2G, Figure S2C) (Codega et al., 2014; Doetsch et al., 2002; Llorens-Bobadilla et al., 2015). Monocle ordering with the consensus-ordering genes not only orders qNSCs first, followed by aNSCs negative for cell cycle markers but also captures the dynamics of differentiation (Figure 2G). Indeed, a subset of aNSCs expressing cell cycle markers, also exhibits expression of Dlx2, a pro-neural transcription factor known to promote neural differentiation (Doetsch et al., 2002; Petryniak et al., 2007; Suh et al., 2009). These cells are ordered later in pseudotime than other aNSCs, closely juxtaposed with NPCs (Figure 2G). Thus, a subpopulation of aNSCs may exhibit an early transcriptomic signature of neural differentiation. NPCs themselves are predominantly ordered last and express other important regulators and indicators of neurogenesis such as Dcx, Sp8, and Sp9 (Figure 2G, Figure S2C) (Hsieh, 2012; Long et al., 2009; Waclaw et al., 2006). Other important regulators of neurogenesis such as Ascl1 and Pax6 are expressed throughout the aNSC and NPC populations (Figure S2C), consistent with evidence that Ascl1 is both required for quiescent cells to enter the active state and for neuronal differentiation (Andersen et al., 2014). Together, the dynamic expression of key markers along this continuum of activation and differentiation suggest five distinct consecutive molecular states: qNSC-like (Egfr−), aNSC-early (Egfr+Cdk1−), aNSC-mid (Egfr+Cdk1+Dlx2low), aNSC-late (Egfr+Cdk1+Dlx2high), and NPC-like (Dlx2+Dcx+) (Figure 2G, Figure S2C, Figure S3B, Table S8).
Thus, machine learning identifies specific consensus-ordering genes that can order NSCs and progeny and suggests the existence of new intermediate states of activation and differentiation within the aNSC population.
Activated NSCs can be divided into specific subpopulations, defined by the expression of markers, along the spectrum of activation and differentiation
To independently corroborate the subpopulations identified by machine learning and Monocle ordering (qNSCs-like, aNSC-early, aNSC-mid, aNSC late, NPC-like), we used diffusion mapping, which has been recently developed to plot cells with respect to their molecular trajectories (Haghverdi et al., 2015). Diffusion mapping with the 2500 most variable genes (Figure 3A) or all detected genes (Figure S3A) clusters the cells in a similar manner as Monocle or PCA using the consensus-ordering genes (Figures 3B–D), confirming our machine learning approach.
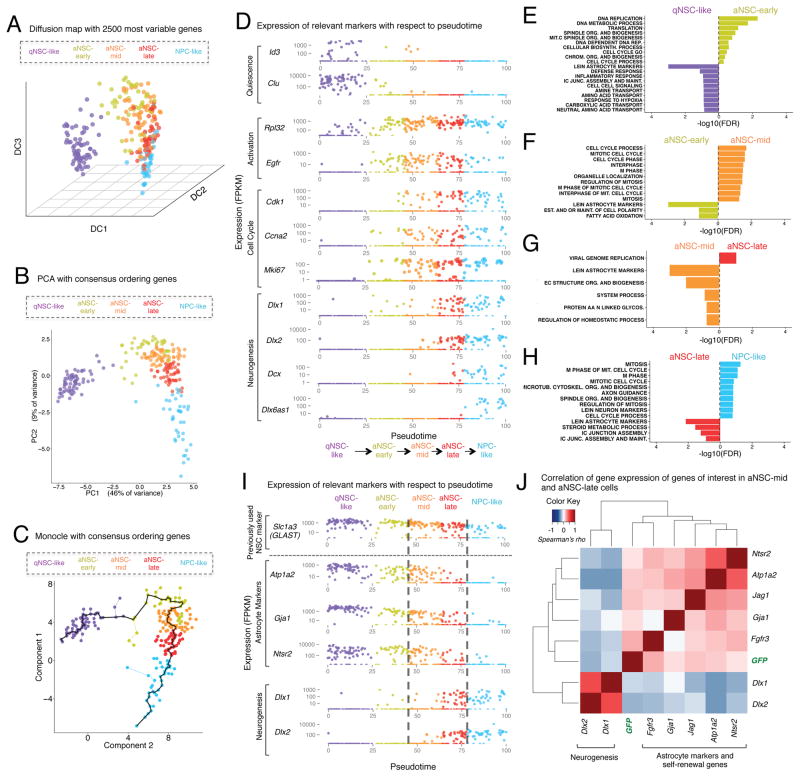
Figure 3. Activated NSCs can be divided into specific subpopulations, defined by the expression of defined genes, along the spectrum of activation and differentiation.
(A) Diffusion map using the 2500 most variable genes in the dataset for all qNSCs, aNSCs, and NPCs. Cells are colored by the identity of the intermediate states defined in Figure 2G.
(B) PCA using the consensus-ordering genes (Table S7A) for all qNSCs, aNSCs, and NPCs. Cells are colored as in Figure 3A.
(C) Spanning tree produced by Monocle when all qNSCs, aNSCs, and NPCs are ordered using the consensus-ordering genes (Table S7A). Black line represents pseudotime “track” through the single cell lineage. Cells are colored as in Figure 3A.
(D) Expression (FPKM) of genes relevant to the transition between the indicated stages in each cell, plotted with respect to pseudotime produced by Monocle when all qNSCs, aNSCs, and NPCs are ordered using the consensus-ordering genes (Table S7A). Cells are colored as in Figure 3A.
(E–H) Gene-set enrichments for genes ranked by Z-score for differential expression between cells in intermediate states defined in Figure 3A. Enrichments expressed as [−log10(FDR)] and directionality and color indicate the intermediate state in which the gene set is enriched. Gene sets presented are those for which FDR<0.2.
(I) Expression (FPKM) of markers of astrocytes (Atp1a2, Gja1, Ntsr2) and neurogenesis (Dlx1, Dlx2) in each cell plotted as a function of pseudotime. GLAST (Slc1a3), a marker of astrocytes that was previously used in FACS sorting studies is presented as a comparison on top. Cells are colored as in Figure 3A.
(J) Markers of astrocytes (Atp1a2, Ntsr2, Gja1) and mediators of self-renewal (Jag1, Fgfr3) are correlated with each other and are anticorrelated with early markers of neuronal differentiation (Dlx1, Dlx2) in aNSC-mid and aNSC-late cells. Carpet plot showing correlation (Spearman’s rho) between individual genes in all aNSC-mid and aNSC-late cells.
To define the gene expression changes occurring between all five states (qNSC-like, aNSC-early, aNSC-mid, aNSC-late, and NPC-like), we conducted differential expression analysis at each cell state transition using the single cell differential expression tool SCDE (Kharchenko et al., 2014) and assessed pathway enrichment using GSEA (Table S8). The transition from qNSC-like to aNSC-early is characterized by upregulation of genes belonging to ribosomal signatures (Figure 3D, E), confirming our earlier observations (Figure 2) and findings from another single cell study in the SVZ (Llorens-Bobadilla et al., 2015). As expected, the transition from aNSC-early to aNSC-mid is characterized by upregulation of genes belonging to cell cycle signatures (Figure 3D, F). The transition between the aNSC-mid and aNSC-late cell states is defined partly by the upregulation of Dlx1 and Dlx2, two genes normally associated with neuronal differentiation (Petryniak et al., 2007) (Figure 3D). However, aNSC-late cells did not express the other genes that are characteristic of the NPC-like population such as Dcx, Nrxn3, Dlx6as1, Sp8, and Sp9 (Figure 3D, Figure S3C), suggesting that aNSC late are distinct from NPCs. Interestingly, the transition from aNSC-mid to aNSC-late is characterized by downregulation of genes relating to astrocyte identity (Figure 3G), such as Atp1a2, Gja1, and Ntsr2 (Cahoy et al., 2008) (Figure 3I). Astrocytic markers are further downregulated as cells transition into the NPC-like state (Figures 3H–I). Thus, aNSCs that highly express cell cycle genes can be further sub-divided into two groups, a group still expressing astrocyte markers (characteristic of earlier cells in the lineage), and a group in which early neurogenesis markers begin to be expressed. These two states could represent the division between a self-renewing NSC and a lineage-committed NSC, primed for differentiation.
This analysis also enables us to identify putative markers or regulators that may be specific to these earlier, potentially self-renewing NSCs. Indeed, while GLAST (Slc1a3) has been previously used as a marker to detect NSCs (Llorens-Bobadilla et al., 2015; Mich et al., 2014), it is actually expressed in aNSC-mid, aNSC-late, and NPCs (Figure 3I). In contrast, other markers appear to be more specific to the aNSC-mid subtype, including the cell surface genes Atp1a2, Gja1, and Ntsr2 (Figure 3I). While these genes are also expressed in other cell types in the brain, including cortical astrocytes, they could serve to isolate the aNSC-mid group in combination with other markers of NSCs. Furthermore, Jagged1 and Fgfr3, which have been implicated in NSC self-renewal (Maric et al., 2007; Nyfeler et al., 2005), are among the genes elevated in the aNSC-mid cells (Figure S3D) and could also potentially serve as markers in combination with other NSC markers. Interestingly, genes that are enriched in the aNSC-mid population, including markers of astrocytes (Atp1a2, Ntsr2, Gja1) and mediators of self-renewal (Fgfr3, Jag1), are correlated with each other, and anti-correlated with genes associated with the aNSC-late population, Dlx1 and Dlx2, in the aNSC-mid and aNSC-late states (Figure 3J; Figure S3F). Collectively, these data support the notion that the division between the aNSC-mid and aNSC-late populations is associated with the loss of astrocytic gene signatures and the acquisition of a pro-neural gene expression signature.
Experimental validation of single cell data prediction by purifying aNSCs subpopulations using level of GFAP-GFP expression
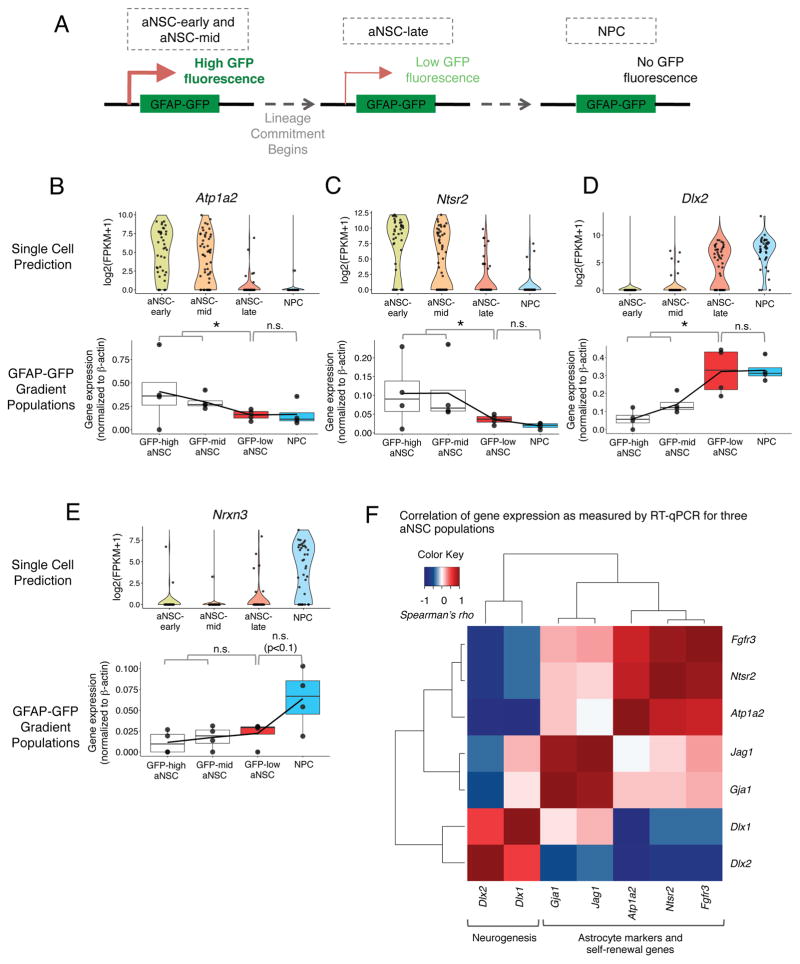
We next experimentally validated the existence of specific aNSC subpopulations. The GFAP-GFP transgene is known to be downregulated as NSCs commit to the NPC state (Doetsch et al., 2002; Pastrana et al., 2009) (Figure 4A). Indeed, GFP transcript levels from the GFAP-GFP transgene positively correlate with markers of astrocytes and negatively correlate with early markers of neurogenesis in aNSCs (Figure 3J). We therefore used FACS to sort different populations of aNSCs based on their level of GFP fluorescence from the GFAP-GFP transgene. Because we did not know the levels of GFP fluorescence to which aNSC transitions would correspond, we sorted three subpopulations of aNSCs: GFAP-high (GFAP-GFP(high)PROM1+EGFR+), GFAP-mid (GFAP-GFP(mid)PROM1+EGFR+), and GFAP-low (GFAP-GFP(low)PROM1+EGFR+), as well as NPCs (GFAP-GFP(neg)EGFR+). As predicted by the single cell data, FACS-sorted aNSCs with higher levels of GFP fluorescence expressed markers of astrocytes and self-renewal, such as Atp1a2 and Ntsr2 (Figures 4B–C). Consistent with single cell data, aNSCs with the lowest levels of GFP fluorescence had significantly higher expression of Dlx2 and Dlx1 (markers of early neurogenesis) (Figure 4D, Figure S4B) but did not yet express other later makers that were more exclusively expressed in NPCs such as Nrxn3 and Dcx (Figure 4E, Figure S4C). The populations expressed equal amounts of genes detected equally in all aNSCs subpopulations such as Egfr (Figure S4D). The subdivision of the aNSC population by GFP levels generally recapitulated the gene correlation module observed in Figure 3J, specifically the positive correlation between markers of astrocytes and mediators of self-renewal and anti-correlation between these genes and early mediators of neurogenesis (Dlx1, Dlx2) (Figure 4F, Figure S4I). In contrast, this sorting scheme could not distinguish the aNSC-early and aNSC-mid populations, which differed in their expression of cell cycle markers (Figure S4E–G), probably because these two populations express GFP at similar levels. Thus, the molecular states along the spectrum of activation and differentiation predicted by single cell analysis can be experimentally validated.
Figure 4. Experimental validation of the difference between aNSC-mid and aNSCs-late subpopulations by separating aNSCs based on level of GFAP-GFP expression by FACS.
(A) Predicted GFP fluorescence states of aNSCs from a GFP-high state in which the GFAP-GFP promoter is active to a GFP-low state in which the cells have committed to differentiation but retain some GFP, and finally to the NPC state which do not express GFP.
(B–E) (upper) Gene expression in single cells grouped by molecular subtype as defined in Figure 3. Gene expression expressed as log2(FPKM+1). (lower) Gene expression measured by RT-qPCR in subpopulations of aNSCs divided by their level of GFAP-GFP expression (GFAP-GFP high aNSC, GFAP-GFP mid aNSC, GFAP-GFP low aNSC) and NPCs. P-value from one-sided Wilcoxon signed-rank test. * p≤0.05.
(F) Correlation between expression of key markers of NSCs and neurogenesis in aNSC populations divided by GFAP-GFP. Carpet plot showing correlation (Spearman’s rho) between individual genes in all aNSC-subpopulations divided by level of GFAP-GFP. Color of box indicates correlation (Spearman’s rho) between a given gene pair (scale on upper left).
In the spectrum of NSC activation and differentiation, in vitro cultured NSCs resemble in vivo aNSCs but exhibit a signature of inflammation
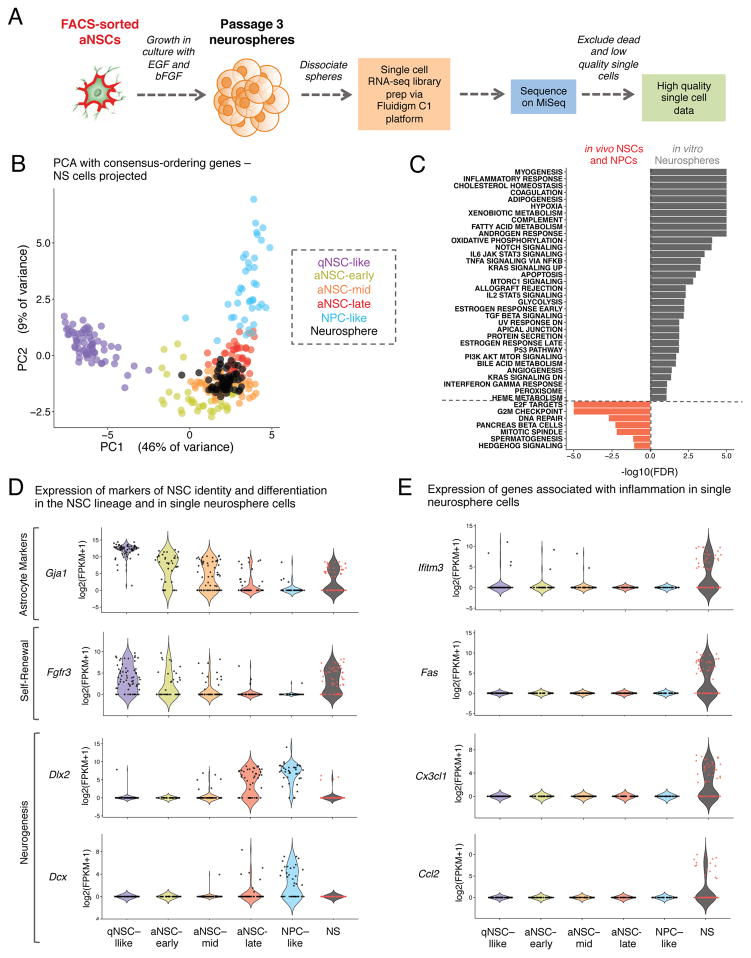
Cultures of primary NSCs as neurospheres have been used to study NSCs in vitro (Conti and Cattaneo, 2010; Hitoshi et al., 2002; Ma et al., 2014), though it is debated whether these cells are good models for in vivo NSCs (Conti and Cattaneo, 2010; Parker et al., 2005). To understand how cultured NSCs compare to their in vivo counterparts, we performed single cell RNA-sequencing of passage 3 neurospheres (NS) cultured from FACS-sorted aNSCs (Figure 5A). Single cells were filtered for quality in the same manner as for in vivo cells (Figure S5A), resulting in 62 high-quality single cell RNA-seq datasets. To determine where cultured NS single cells fall on the spectrum of activation and differentiation of in vivo neural progenitors, we performed PCA using the consensus-ordering genes (Table S7A) on all of our in vivo single qNSCs, aNSCs, and NPCs and projected the single NS cells onto this PCA space (Figure 5B). This analysis revealed that single NS cells most closely resemble the aNSC-mid population (proliferative aNSCs that have not yet begun to express neuronal differentiation markers) with respect to the expression of key genes that define the activation and differentiation of NSCs. However, when PCA was performed using all in vivo cells and in vitro neurosphere single cells, the neurospheres cluster separately from the in vivo lineage (Figure S5C), suggesting that there are also significant differences between the in vivo and in vitro states. Differential expression using SCDE between the cultured NS single cells and in vivo aNSCs or NPCs revealed that many of the genes significantly enriched in the in vivo populations are markers of neuronal differentiation such as Dlx2, Dcx, Nrxn3, and Dlx6as1 (Figure 5D, Figure S5B, Table S9). This is consistent with the notion that cultured neurospheres do not express markers of neuronal differentiation but express markers of astrocytes (Figure 5D, Figure S5B), likely representing an undifferentiated, self-renewing state.
Figure 5. In the spectrum of NSC activation and differentiation in vivo, in vitro cultured NSCs resemble aNSCs but exhibit a signature of inflammation.
(A) Preparation of single cell RNA-sequencing libraries from passage 3 neurospheres (NS) derived from FACS-sorted aNSCs.
(B) PCA with qNSCs, aNSCs, and NPCs using expression [log2(FPKM+1)] of the consensus-ordering genes from machine learning models (Table S7A). NS single cells are projected onto the resulting principal component space. Cells are colored by identity defined in Figure 2G and NS single cells are black.
(C) Gene-set enrichments for genes ranked by Z-score for differential expression between single NS cells and in vivo aNSCs and NPCs. Enrichments expressed as [−log10(FDR)] and directionality and color indicate the intermediate state in which the gene set is enriched (FDR<0.2).
(D) Expression of genes associated with astrocyte identity, self-renewal, and neurogenesis in in vitro NS single cells and in vivo NSCs. Violin plots showing gene expression in the cellular states defined in Figure 2G as well as in NS single cells.
(E) Expression of genes associated with inflammatory signatures in single NS cells and in vivo NSCs. Data presented as in Figure 5D.
To identify global pathways that are different between cultured NS cells and in vivo NSCs, we performed GSEA on genes differentially expressed between the in vivo and in vitro states (Table S9). Strikingly, pathways associated with inflammation and cytokine signaling were among those upregulated in the cultured NS cells (Figure 5C). Furthermore, genes associated with inflammatory signaling, such as Fas and Ifitm3, were highly expressed in many in vitro single cells but were not consistently detected in vivo (Figure 5E, Figure S5B). Thus, while cultured NSCs resemble aNSC-mid cells on the spectrum of NSC activation and differentiation, there are important differences between cultured neurospheres and in vivo NSCs, such as the expression of markers of inflammation. Understanding these differences could help better model NSCs in vitro.
Meta-analysis of single cells isolated by different FACS methods using the power of single cell transcriptomics
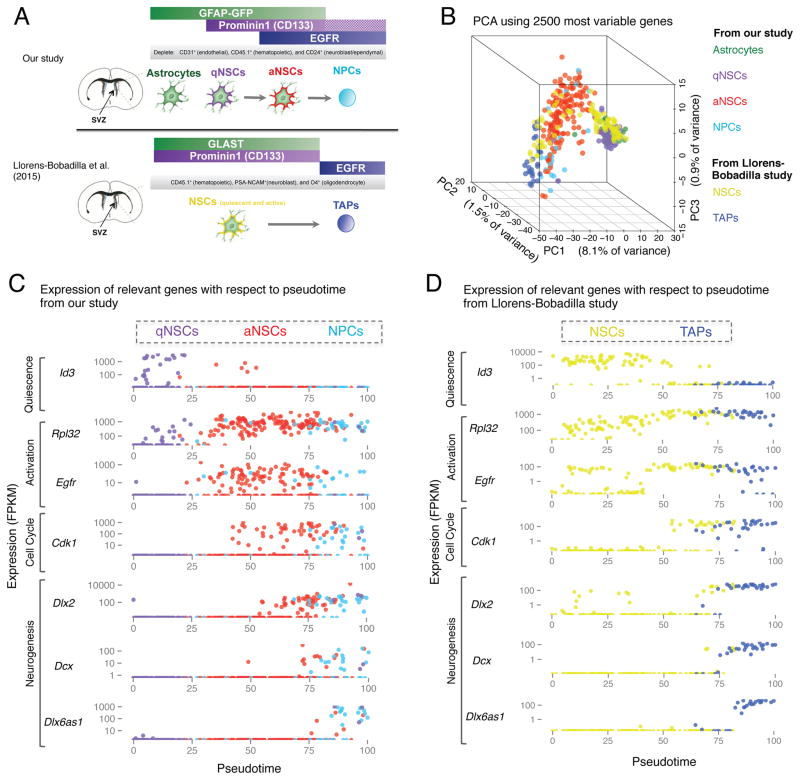
A single cell characterization of NSCs in the SVZ was recently published (Llorens-Bobadilla et al., 2015) using a different dissociation method (trypsin instead of papain) and a distinct FACS strategy (Llorens-Bobadilla et al., 2015) (Figure 6A). This provides a unique opportunity to address questions regarding the identity of cells isolated by different approaches. The Llorens-Bobadilla study isolated two populations by FACS from wild-type mice: GLAST+PROM1+ (NSCs) and GLAST−PROM1−EGFR+ (transient amplifying progenitors [TAPs]) (Figure 6A) whereas we isolated 4 populations by FACS from GFAP-GFP transgenic mice: GFAP-GFP+PROM1−EGFR− (niche astrocytes), GFAP-GFP+PROM1+EGFR− (qNSCs), GFAP-GFP+PROM1+EGFR+ (aNSCs), and GFAP-GFP−EGFR+ (NPCs/TAPs). One main difference is that the Llorens-Bobadilla study used the surface protein GLAST to purify NSCs from wild-type mice (Llorens-Bobadilla et al., 2015), while we isolated them using GFP from GFAP-GFP transgenic mice. Another main difference is that the Llorens-Bobadilla study did not differentiate between qNSCs and aNSCs (Llorens-Bobadilla et al., 2015), whereas we used the marker EGFR to distinguish aNSCs from qNSCs (Figure 6A). The method of cell dissociation and marker choices for FACS have been areas of active debate in the field of NSC biology (Codega et al., 2014; Luo et al., 2015; Mich et al., 2014). To compare these single cell datasets, we independently mapped the raw sequencing data from the Llorens-Bobadilla study using our pipeline. When we conducted global PCA using all cells from both studies, the primary axis of variation is defined by the study, likely due to differences in library preparation and sequencing depth (Figure S6A). However, when we projected cells from the Llorens-Bobadilla study onto a PCA with either the consensus-ordering genes (Table S7A) or the most variable genes from our study, we observed an alignment of the cell types profiled in each study (Figure 6B, Figure S6B). Furthermore, Monocle ordering with the consensus-ordering genes on the NSCs and TAPs from (Llorens-Bobadilla et al., 2015) revealed that the dynamic expression of key genes with respect to pseudotime is very similar between the two datasets (Figures 6C–D; Figures S6C–D). In both datasets, quiescent NSCs high in Id3 and Clu are ordered earliest, and activation is accompanied by an upregulation of genes important for ribosome biogenesis followed by the upregulation of cell cycle genes (Figures 6C–D). Interestingly, a subset of aNSCs from the Llorens-Bobadilla study expresses high levels of cell cycle markers (Cdk1) as well as Dlx2 transcript (Figure 6D). This state is reminiscent of the aNSC-late cells described in Figure 3. Moreover, the transition from aNSCs to NPCs (TAPs), characterized by expression of neuronal associated genes such as Dcx and Dlx6as1, is also highly conserved in both datasets (Figures 6C–D; Figures S6C–D). Importantly, though NPCs (TAPs) express some markers usually associated with type A neuroblasts (e.g. Dcx), they also express cell cycle markers (Figure S6E, F) unlike neuroblasts which do not express cell cycle markers (Figure 6E) (Llorens-Bobadilla et al., 2015). Thus, the transcriptional dynamics of NSC regulators captured in these divergent FACS-sorting approaches are very similar with respect to the expression of key genes dynamically regulated along the processes of activation and differentiation.
Figure 6. Meta-analysis to compare single cell identities of SVZ NSCs isolated using divergent FACS-sorting strategies.
(A) Comparison of FACS sorting schemes implemented in our study and in the Llorens-Bobadilla study.
(B) PCA on all qNSCs, aNSCs, and NPCs from our study, using the expression [log2(FPKM+1)] of the 2500 most variable genes in these cells. All NSCs and TAPs from the Llorens-Bobadilla study are projected on to the resulting principal component space. Cells colored by FACS sorting identity indicated on the right.
(C–D) Regulators of activation and differentiation exhibit similar dynamics in NSCs and progeny isolated by divergent FACS sorting schemes. Expression (FPKM) of key markers of activation and differentiation in each cell plotted as a function of pseudotime generated by Monocle ordering using the consensus-ordering genes identified by machine learning (Table S7A) for (C) all qNSCs, aNSCs, and NPCs from our study and (D) NSCs and TAPs analyzed in Llorens-Bobadilla, et al. 2015. Cells colored by FACS-sorting identity, indicated on top.
Meta-analysis of global gene expression in different single cell studies, including SVZ and DG
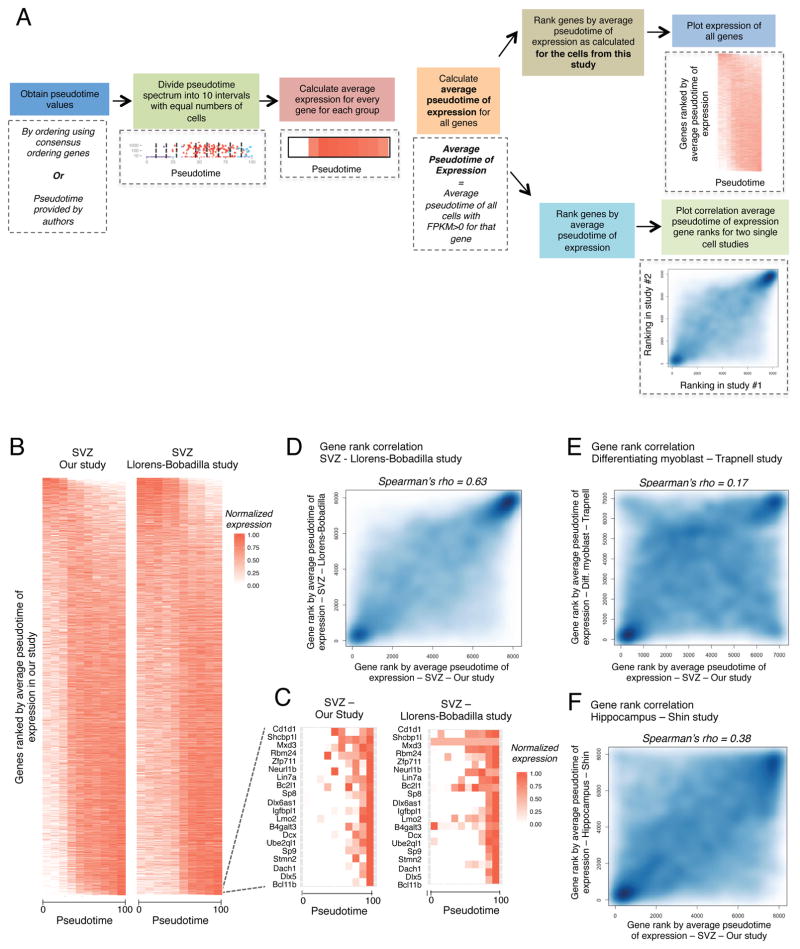
We next performed a global assessment of the similarities between NSC lineages in our study and the Llorens-Bobadilla study using all genes. We first ranked all detected genes in our dataset by their Average Pseudotime of Expression (APE) (Figure 7A). APE represents the average pseudotime of all cells expressing a given gene for all qNSCs, aNSCs, and NPCs ordered by Monocle using the consensus-ordering genes (Table S7A). Pseudotime expression heatmaps (see supplemental experimental procedures) for the qNSCs, aNSCs, and NPCs in our study and for the NSCs and TAPs from (Llorens-Bobadilla et al., 2015) revealed that most detected genes show high similarity in their expression profile (Figure 7B). Furthermore, the genes exclusively expressed in NPCs (or TAPs) are highly conserved between the two datasets (Figure 7C). The correlation between the APE rankings, when genes are independently ranked by APE using the two datasets, was excellent between our dataset and the Llorens-Bobadilla dataset (Figure 7D, Spearman’s rho = 0.63). The agreement between the global expression profiles of these cells is striking, considering the different FACS isolation protocols and the different depths to which the cells were sequenced. The correlation between the independent APE gene rankings for the cells from our study and the differentiating myoblasts from (Trapnell et al., 2014) was still positive but much lower (Figure 7E, Spearman’s rho = 0.17). Thus, the correlation between the SVZ NSC datasets cannot be solely attributed to cell cycle entry. Similar results were obtained when we performed Monocle ordering using the consensus-ordering genes with the normalized expression values provided in (Llorens-Bobadilla et al., 2015) and (Trapnell et al., 2014) (Figures S7B–C, S7F–G). The concordance between our study and that of Llorens-Bobadilla suggests global similarities between the lineages isolated in these two studies. Because our RNA-seq libraries were sequenced at much lower depth than those from the Llorens-Bobadilla study, these results also suggest that low-throughput sequencing is sufficient to capture complex transcriptional dynamics in single cells.
Figure 7. Meta-analysis to compare global pseudotime-dependent gene expression in various single cell datasets.
(A) Schematic outlining the approaches for generating pseudotime expression heatmaps and for correlating gene rankings by average pseudotime of expression (APE, see Supplemental Experimental Methods) for different single cell datasets.
(B) Heatmap representing the expression of all detected genes ranked by APE defined in our study. Expression plotted as a function of pseudotime. Left panel: expression from our study (qNSCs, aNSCs, and NPCs), pseudotime defined by Monocle ordering using consensus-ordering genes identified by machine learning (Table S7A). Right panel: expression from the Llorens-Bobadilla study (NSCs and TAPs), pseudotime defined by Monocle ordering using consensus-ordering genes (Table S7A).
(C) Heatmap representing expression of the 20 genes with highest values of APE (expressed most exclusively in NPCs) in our dataset. Left panel: expression from our study (qNSCs, aNSCs, and NPCs), pseudotime defined as in Figure 7B. Right panel: expression from the Llorens-Bobadilla study (NSCs and TAPs), pseudotime defined as in Figure 7B.
(D) Smooth scatter plot representing gene ranks by average pseudotime of expression (APE) in (x-axis) qNSCs, aNSCs, and NPCs from our study ordered by Monocle using the consensus-ordering genes identified by machine learning (Table S7A) and (y-axis) NSCs and TAPs from the Llorens-Bobadilla study ordered by Monocle using the consensus-ordering genes identified by machine learning (Table S7A) (Spearman’s rho = 0.63, p < 2.2e−16).
(E) Smooth scatter plot representing gene rankings by average pseudotime of expression (APE) in (x-axis) qNSCs, aNSCs, and NPCs from the current study ordered as in Figure 7D and (y-axis) differentiating myoblasts ordered by Monocle from (Trapnell et al., 2014) (Spearman’s rho = 0.17, p < 2.2e−16).
(F) Smooth scatter plot representing gene rankings by average pseudotime of expression (APE) in (x-axis) qNSCs, aNSCs, and NPCs from our study ordered as in Figure 7D and (y-axis) hippocampal NSCs ordered by Waterfall in the study by Shin and colleagues (Shin et al., 2015) (Spearman’s rho = 0.38, p < 2.2e−16).
We next extended this type of analysis to other NSC single cell datasets. Shin and colleagues generated 142 single cell RNA-seq datasets from the hippocampal NSCs (Shin et al., 2015). The overall gene expression pattern in single NSCs from the hippocampus was similar to that of the SVZ (Figure S7D), and there was positive correlation in independent gene rankings by APE for our study and hippocampal NSCs profiled in (Shin et al., 2015) (Figure 7F, Spearman’s rho = 0.38). This correlation was higher than the gene ranking correlation between SVZ NSCs and differentiating myoblasts, suggesting similarities between neurogenic niches beyond general processes of cell proliferation. Similar results were observed using the consensus-ordering genes (Figures S7E, S7H). Thus, the primary gene signatures of quiescence and activation may be conserved in the neurogenic niches in the adult brain.
This meta-analysis indicates that the NSC lineages identified by divergent FACS sorting schemes resulted in the isolation of very similar cells and suggests similarities between the gene signatures of quiescence and activation in the two different adult neurogenic niches.
Discussion
Our single cell RNA-sequencing on cells from four purified populations from the adult mouse SVZ – niche astrocytes, qNSCs, aNSCs, and NPCs – reveals heterogeneity and transcriptional dynamics in the adult neural stem cell lineage. Our data revealed that FACS-sorted aNSCs can be divided into three groups along the process of activation and differentiation ‘aNSC-early’, ‘aNSC-mid’, and ‘aNSC-late’. The ‘aNSC-late’ subpopulation can be enriched by sorting aNSCs with low levels of GFAP-GFP. In the future, excluding the population of cells expressing GFAP-GFP at low levels may allow for the enrichment of the earliest, putatively self-renewing stem cells using FACS.
The power of single cell profiling also allowed us to perform global comparison with other single cell studies. The high correlation between the identities of the single cells profiled in the Llorens-Bobadilla study and our study is highly instructive for FACS-sorting protocols for in vivo NSCs. In our protocol (based on (Codega et al., 2014)), we used the enzyme papain to digest the SVZ for FACS sorting, and we have found that papain cleaves GLAST (B.D., D.E.L., and A.B., unpublished data), the marker that was used in the Llorens-Bobadilla study (Llorens-Bobadilla et al., 2015). Thus, an enzyme other than papain should be used when sorting by GLAST (and indeed Llorens-Bobadilla and colleagues used trypsin (Llorens-Bobadilla et al., 2015)). Our meta-analysis of single cell data is encouraging for the field of NSC biology as it suggests that divergent methods for isolating or identifying the SVZ NSCs that use either GFAP-GFP (Beckervordersandforth et al., 2010; Codega et al., 2014; Fischer et al., 2011) or GLAST (Calzolari et al., 2015; Llorens-Bobadilla et al., 2015; Mich et al., 2014) isolate very similar cell types from the SVZ. Furthermore, the similarities of the pseudotime-related expression profiles of quiescent and active NSCs from the hippocampus (Shin, et al., 2015) and SVZ (Llorens-Bobadilla, et al., 2015 and our study) suggests that the molecular phenotypes of quiescence and activation in these cell types are at least partially conserved.
As a technology for sequencing hundreds and even thousands of single cells emerge (Cadwell et al., 2016; Fan et al., 2015; Habib et al., 2016; Klein et al., 2015; Macosko et al., 2015), it is likely that the single cell characterization of the adult NSC lineage will continue to improve. These developments will complement other methods for characterizing in vivo cell heterogeneity, such as lineage tracing, to provide more complete definitions of adult stem cell lineages (Goodell et al., 2015; Merkle et al., 2014). The knowledge of transcriptional dynamics and cell fate decisions as NSCs activate and commit to differentiation should provide key targets for recruiting NSCs or directing their differentiation. The improved definition of the NSC lineage at the single cell level should also facilitate the study of NSCs in the context of aging and disease.
Summary of Experimental Procedures
NSC isolation from adult mouse brains
For single cell RNA-seq library generation of in vivo cells, four 3-month old male GFAP-GFP mice (Jax cat #003257) were euthanized, and brains were immediately harvested. As described in (Codega et al., 2014), the SVZ from each hemisphere was micro-dissected. The SVZ was dissociated with enzymatic digestion with papain for 10 min at a concentration of 14U/mL. The dissociated SVZ was then titrated in a solution containing 0.7mg/mL ovomucoid, and 0.5 mg/mL DNAseI in DMEM/F12. The dissociated SVZ was then centrifuged through 22% Percoll in PBS to remove myelin debris. Following centrifugation through Percoll solution, cells were washed with FACS buffer (HBSS, 1% BSA, 1% Glucose). Antibody staining was carried out in FACS buffer at the following dilutions: Prom1-Biotin (eBioscience Cat.#13-1331-80 [1:300]), EGF-AlexaFluor 647 (Life Technologies Cat. #E35351 [1:300]), CD24-PacBlue (eBioscience Cat.#48-0242-80 [1:400]), CD31-PE (eBioscience Cat.#12-0311-81 [1:50]), CD45-BV605 (Biolegend Cat.#110737 [1:50]), Strep-PECy7 (eBioscience Cat.#25-4517-82 [1:500]). FACS sorting was performed on a BD FACS Aria II sorter, using a 100μm nozzle at 13.1 PSI. Cell gates were defined as follows (Codega et al., 2014):
Astrocytes: (GFAP-GFP)+ PROM1−CD31−CD24−CD45−
qNSCs: (GFAP-GFP)+PROM1+EGFR−CD31−CD24−CD45−
aNSCs: (GFAP-GFP)+PROM1+EGFR+CD31−CD24−CD45−
NPCs: (GFAP-GFP)−EGFR+CD31−CD24−CD45−
Endothelial Cells: (GFAP-GFP)−CD31−
Cells were sorted into catching media: DMEM/F12 with B27 (1:50), B27 supplement (ThermoFisher, no Vitamin A, 1:50), N2 supplement (ThermoFisher, 1:100), 15mM HEPES buffer, 0.6% glucose, Penicillin-Streptomycin-Glutamine (Life Technologies, 1:100), and Insulin-Transferrin-Selenium (Life Technologies, 1:1000). Cells were then spun down at 300×g at 4°C and resuspended in catching media at a concentration of 300 cells/μL.
Single cell RNA-seq library preparation
A 300 cell/μL cell solution was mixed at a 7:3 ratio with the Fluidigm C1 Suspension reagent and this solution was loaded onto a small size (5–10μm) Fluidigm C1 Single-Cell Auto Prep chip for all in vivo single cells studied and medium size (10–17μm) Fluidigm C1 Single-Cell Auto Prep chip for in vitro cultured neurosphere derived single cells. Live/dead staining was performed using the Fluidigm Live/Dead Cell Staining Solution as described in the Fluidigm C1 mRNA seq protocol and imaged using a Leica DMI4000B microscope. Reverse transcription was performed directly on the chip using the SMARTer chemistry from Clontech, and PCR was also performed on the chip using the Advantage PCR kit (SMARTer Ultra Low RNA Kit for the Fluidigm C1, Clontech #634832). Resulting cDNA was transferred to a 96 well-plate and a subset of representative samples were analyzed by bioanalyzer. A quarter of the cDNA for each library was quantified using the Quant-iT PicoGreen dsDNA Assay Kit (ThermoFisher Cat.# P11496) and verified to be within a range of 0.1–0.5ng/μL (or diluted when necessary with the C1 DNA dilution buffer). Sequencing libraries were prepared directly in a 96-well plate using the Nextera XT Library Preparation Kit (Illumina Cat. # FC-131-1024). Each library was individually barcoded using the Nextera XT 96-Sample Index Kit (Illumina Cat. # FC-131-1002), and all 96 bar-coded libraries from each chip were pooled into single multiplexed libraries. The DNA concentration of multiplexed libraries was measured using BioAnalyzer. These multiplexed libraries were sequenced using either the Illumina MiSeq (Illumina) or HiSeq2000 (Illumina) at a concentration of 2 pM. Details can be found in Table S1.
Construction of machine learning model and determination of consensus-ordering genes
We carried out a four-way classification between the following groups that correspond to key states/subpopulations: qNSCs, ‘cell-cycle low’ aNSCs, ‘cell-cycle high’ aNSCs, and NPCs. Classification was carried out by implementing a stochastic gradient boosted classification model using the R CRAN package GBM v2.1.1. Briefly, 20 single cells from each group (training set) were randomly selected and subjected to GBM modeling as implemented by the Caret package v.6.0–58 in R. Accuracy of the model was tested on cells that were not used for the training set. The GBM classification was bootstrapped by repeatedly sampling 20 cells from each group and building an independent model. In total, 100 GBM models were built. Following construction of the models, the top 100 features from each of the 100 models were obtained. A consensus set of ordering genes was built using genes that were in the top 100 most important features of at least half of the classification models, or in the top 100 most important features of at least 25% of the models (Table S7).
Ordering cells with Monocle using consensus-ordering genes
Monocle ordering was conducted for all qNSC, aNSC, and NPC cells using the set of consensus-ordering genes (Table S7A) identified by machine learning. The expression of genes of interest was plotted with respect to pseudotime. The resulting pseudotime expression spectrum was divided according to the expression of genes of interest. The approach used to divide the pseudotime expression spectrum is enumerated below:
qNSC-like to aNSC-early – Earliest pseudotime at which Rpl4, Rpl32, and Egfr are predominantly expressed.
aNSC-early to aNSC-mid – Earliest pseudotime at which Ccna2, Cdk1, and Ccnb2 are predominantly expressed.
aNSC-mid to aNSC-late– Earliest pseudotime at which Dlx1 and Dlx2 are predominantly expressed.
aNSC-late to NPC-like – Earliest pseudotime at which Nrxn3, Dlx6as1, and Dcx are predominantly expressed.
Differential expression between the putative groups was conducted using the R package SCDE v1.2.1 (Kharchenko et al., 2014) and genes were ranked by Z-score for differential expression between groups. Pathway enrichment was performed on ranked lists using GSEA, using GO Biological Process and lists related neuroepithelial cell identity (Lein et al., 2007) lists.
Supplementary Material
Highlights.
Single cell RNA-seq to characterize adult neural stem cell populations
Machine learning and pseudotemporal ordering shows a continuum in the lineage
Validation of an intermediate state in the neural stem cell population
Meta-analysis with other in vitro and in vivo single cell datasets
Acknowledgments
We thank Aaron Daugherty and Bérénice Benayoun for guidance on machine learning and statistical analysis. We thank Fluidigm Corporation for their help with single cell RNA-sequencing libraries. We thank the Stanford Shared FACS Facility and Cathy Carswell-Crumpton for technical support. We thank Theo Palmer, Tom Rando, Anshul Kundaje, and Vittorio Sebastiano for guidance. We thank Bérénice Benayoun, Xiaoai Zhou, Dana Pe’er, and Philip Brennecke for critical reading of the manuscript, and Bérénice Benayoun, Salah Mahmoudi, and Andrew McKay for their help double-checking code. Supported by P01 AG036695 (A.B), an NIH training grant (T32 GM7365) (B.W.D.), and the Stanford MSTP program (B.W.D.).
Footnotes
Author Contributions
B.D. designed, performed the experiments, and analyzed the data with guidance from A.B.. D.L. optimized the FACS-sorting of NSCs and provided the RNA-seq population datasets. S.B. helped performed single-cell library preparation. K.H. assisted with statistical analysis and code. B.D. wrote the paper with the help of A.B., and all the authors read the paper and provided comments.
Accession Numbers
All data is available online at BioProject Accession # PRJNA324289 Code is available at https://github.com/bdulken/SVZ_NSC_Dulken_2
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen J, Urbán N, Achimastou A, Ito A, Simic M, Ullom K, Martynoga B, Lebel M, Göritz C, Frisén J, et al. A Transcriptional Mechanism Integrating Inputs from Extracellular Signals to Activate Hippocampal Stem Cells. Neuron. 2014;83:1085–1097. doi: 10.1016/j.neuron.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckervordersandforth R, Tripathi P, Ninkovic J, Bayam E, Lepier A, Stempfhuber B, Kirchhoff F, Hirrlinger J, Haslinger A, Lie DC, et al. In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell. 2010;7:744–758. doi: 10.1016/j.stem.2010.11.017. [DOI] [PubMed] [Google Scholar]
- Bonaguidi MA, Peng CY, McGuire T, Falciglia G, Gobeske KT, Czeisler C, Kessler JA. Noggin expands neural stem cells in the adult hippocampus. J Neurosci. 2008;28:9194–9204. doi: 10.1523/JNEUROSCI.3314-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell CR, Palasantza A, Jiang X, Berens P, Deng Q, Yilmaz M, Reimer J, Shen S, Bethge M, Tolias KF, et al. Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat Biotech. 2016;34:199–203. doi: 10.1038/nbt.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzolari F, Michel J, Baumgart EV, Theis F, Gotz M, Ninkovic J. Fast clonal expansion and limited neural stem cell self-renewal in the adult subependymal zone. Nat Neurosci. 2015;18:490–492. doi: 10.1038/nn.3963. [DOI] [PubMed] [Google Scholar]
- Codega P, Silva-Vargas V, Paul A, Maldonado-Soto AR, Deleo AM, Pastrana E, Doetsch F. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron. 2014;82:545–559. doi: 10.1016/j.neuron.2014.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L, Cattaneo E. Neural stem cell systems: physiological players or in vitro entities? Nat Rev Neurosci. 2010;11:176–187. doi: 10.1038/nrn2761. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Petreanu L, Caille I, Garcia-Verdugo JM, Alvarez-Buylla A. EGF converts transit-amplifying neurogenic precursors in the adult brain into multipotent stem cells. Neuron. 2002;36:1021–1034. doi: 10.1016/s0896-6273(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Fan HC, Fu GK, Fodor SPA. Combinatorial labeling of single cells for gene expression cytometry. Science. 2015:347. doi: 10.1126/science.1258367. [DOI] [PubMed] [Google Scholar]
- Fischer J, Beckervordersandforth R, Tripathi P, Steiner-Mezzadri A, Ninkovic J, Gotz M. Prospective isolation of adult neural stem cells from the mouse subependymal zone. Nat Protoc. 2011;6:1981–1989. doi: 10.1038/nprot.2011.412. [DOI] [PubMed] [Google Scholar]
- Friedman JH. Stochastic gradient boosting. Comput Stat Data Anal. 2002;38:367–378. [Google Scholar]
- Garcia AD, Doan NB, Imura T, Bush TG, Sofroniew MV. GFAP-expressing progenitors are the principal source of constitutive neurogenesis in adult mouse forebrain. Nat Neurosci. 2004;7:1233–1241. doi: 10.1038/nn1340. [DOI] [PubMed] [Google Scholar]
- Goodell MA, Nguyen H, Shroyer N. Somatic stem cell heterogeneity: diversity in the blood, skin and intestinal stem cell compartments. Nat Rev Mol Cell Biol. 2015;16:299–309. doi: 10.1038/nrm3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N, Li Y, Heidenreich M, Swiech L, Trombetta JJ, Zhang F, Regev A. Div-Seq: A single nucleus RNA-Seq method reveals dynamics of rare adult newborn neurons in the CNS. Science. 2016 doi: 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Buettner F, Theis FJ. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics. 2015;31:2989–2998. doi: 10.1093/bioinformatics/btv325. [DOI] [PubMed] [Google Scholar]
- Hitoshi S, Alexson T, Tropepe V, Donoviel D, Elia AJ, Nye JS, Conlon RA, Mak TW, Bernstein A, van der Kooy D. Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 2002;16:846–858. doi: 10.1101/gad.975202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J. Orchestrating transcriptional control of adult neurogenesis. Genes Dev. 2012;26:1010–1021. doi: 10.1101/gad.187336.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein Allon M, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz David A, Kirschner Marc W. Droplet Barcoding for Single-Cell Transcriptomics Applied to Embryonic Stem Cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeman D, Hebestreit K, Webb A, Pollina E, Dulken B, Devarajan K, Brunet A. Proteostatic differences between neural stem cell populations are linked to age-associated transcriptional change. Manuscript in preparation. [Google Scholar]
- Llorens-Bobadilla E, Zhao S, Baser A, Saiz-Castro G, Zwadlo K, Martin-Villalba A. Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells that Become Activated upon Brain Injury. Cell Stem Cell. 2015;17:329–340. doi: 10.1016/j.stem.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Long JE, Swan C, Liang WS, Cobos I, Potter GB, Rubenstein JLR. Dlx1&2 and Mash1 transcription factors control striatal patterning and differentiation through parallel and overlapping pathways. J Comp Neurol. 2009;512:556–572. doi: 10.1002/cne.21854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Coskun V, Liang A, Yu J, Cheng L, Ge W, Shi Z, Zhang K, Li C, Cui Y, et al. Single-cell transcriptome analyses reveal signals to activate dormant neural stem cells. Cell. 2015;161:1175–1186. doi: 10.1016/j.cell.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CY, Yao MJ, Zhai QW, Jiao JW, Yuan XB, Poo MM. SIRT1 suppresses self-renewal of adult hippocampal neural stem cells. Development. 2014;141:4697–4709. doi: 10.1242/dev.117937. [DOI] [PubMed] [Google Scholar]
- Macosko Evan Z, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas Allison R, Kamitaki N, Martersteck Emily M, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maric D, Fiorio Pla A, Chang YH, Barker JL. Self-renewing and differentiating properties of cortical neural stem cells are selectively regulated by basic fibroblast growth factor (FGF) signaling via specific FGF receptors. J Neurosci. 2007;27:1836–1852. doi: 10.1523/JNEUROSCI.5141-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Fuentealba LC, Sanders TA, Magno L, Kessaris N, Alvarez-Buylla A. Adult neural stem cells in distinct microdomains generate previously unknown interneuron types. Nat Neurosci. 2014;17:207–214. doi: 10.1038/nn.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mich JK, Signer RA, Nakada D, Pineda A, Burgess RJ, Vue TY, Johnson JE, Morrison SJ. Prospective identification of functionally distinct stem cells and neurosphere-initiating cells in adult mouse forebrain. eLife. 2014;3:e02669. doi: 10.7554/eLife.02669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira H, Andreu Z, Suh H, Lie DC, Jessberger S, Consiglio A, San Emeterio J, Hortiguela R, Marques-Torrejon MA, Nakashima K, et al. Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell. 2010;7:78–89. doi: 10.1016/j.stem.2010.04.016. [DOI] [PubMed] [Google Scholar]
- Mirzadeh Z, Merkle FT, Soriano-Navarro M, Garcia-Verdugo JM, Alvarez-Buylla A. Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell. 2008;3:265–278. doi: 10.1016/j.stem.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyfeler Y, Kirch RD, Mantei N, Leone DP, Radtke F, Suter U, Taylor V. Jagged1 signals in the postnatal subventricular zone are required for neural stem cell self-renewal. EMBO J. 2005;24:3504–3515. doi: 10.1038/sj.emboj.7600816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MA, Anderson JK, Corliss DA, Abraria VE, Sidman RL, Park KI, Teng YD, Cotanche DA, Snyder EY. Expression profile of an operationally-defined neural stem cell clone. Exp Neurol. 2005;194:320–332. doi: 10.1016/j.expneurol.2005.04.018. [DOI] [PubMed] [Google Scholar]
- Pastrana E, Cheng LC, Doetsch F. Simultaneous prospective purification of adult subventricular zone neural stem cells and their progeny. Proc Natl Acad Sci U S A. 2009;106:6387–6392. doi: 10.1073/pnas.0810407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryniak MA, Potter GB, Rowitch DH, Rubenstein JLR. Dlx1 and Dlx2 Control Neuronal versus Oligodendroglial Cell Fate Acquisition in the Developing Forebrain. Neuron. 2007;55:417–433. doi: 10.1016/j.neuron.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, Li N, Szpankowski L, Fowler B, Chen P, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotech. 2014;32:1053–1058. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming GL, et al. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell. 2015;17:360–372. doi: 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y, Obernier K, Holzl-Wenig G, Mandl C, Herrmann A, Worner K, Eckstein V, Ciccolini F. Interaction between DLX2 and EGFR regulates proliferation and neurogenesis of SVZ precursors. Mol Cell Neurosci. 2009;42:308–314. doi: 10.1016/j.mcn.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotech. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waclaw RR, Allen ZJ, II, Bell SM, Erdélyi F, Szabó G, Potter SS, Campbell K. The Zinc Finger Transcription Factor Sp8 Regulates the Generation and Diversity of Olfactory Bulb Interneurons. Neuron. 2006;49:503–516. doi: 10.1016/j.neuron.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Wu AR, Neff NF, Kalisky T, Dalerba P, Treutlein B, Rothenberg ME, Mburu FM, Mantalas GL, Sim S, Clarke MF, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014;11:41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Zhuo L, Sun B, Zhang CL, Fine A, Chiu SY, Messing A. Live astrocytes visualized by green fluorescent protein in transgenic mice. Dev Biol. 1997;187:36–42. doi: 10.1006/dbio.1997.8601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.