Abstract
Studies have reported that interactions between keratins (KRTs) and other proteins initiate signaling cascades that regulate cell migration, invasion, and metastasis. In the current study, we found that expression of KRT19 was specifically high in breast cancers and significantly correlated with their invasiveness. Moreover, knockdown of KRT19 led to increased proliferation, migration, invasion, drug resistance, and sphere formation in breast cancer cells via an upregulated NOTCH signaling pathway. This was owing to reduced expression of NUMB, an inhibitory protein of the NOTCH signaling pathway. In addition, we found that KRT19 interacts with β-catenin/RAC1 complex and enhances the nuclear translocation of β-catenin. Concordantly, knockdown of KRT19 suppressed the nuclear translocation of β-catenin as well as β-catenin-mediated NUMB expression. Furthermore, modulation of KRT19-mediated regulation of NUMB and NOTCH1 expression led to the repression of the cancer stem cell properties of breast cancer patient-derived CD133high/CXCR4high/ALDH1high cancer stem-like cells (CSLCs), which showed very low KRT19 and high NOTCH1 expression. Taken together, our study suggests a novel function for KRT19 in the regulation of nuclear import of the β-catenin/RAC1 complex, thus modulating the NUMB-dependent NOTCH signaling pathway in breast cancers and CSLCs, which might bear potential clinical implications for cancer or CSLC treatment.
Introduction
Breast cancer is a multifactorial disease that can be initiated by genetic mutations, chronic inflammation, exposure to toxic compounds, and abundant stress factors.1 Despite of being a subject of concern across the world, the exact mechanism of breast cancer progression is not completely resolved yet.
Some genes of the keratin (KRT) family are important markers in reverse transcriptase polymerase chain reaction (RT–PCR)-mediated detection of tumors in the lymph nodes, peripheral blood, and bone marrows of breast cancer patients.2, 3 KRTs are intermediate filament proteins responsible for the structural integrity of epithelial cells and markers of the epithelial tissue.4 Further, they are subdivided into cytokeratins and hair KRTs and also into low molecular weight acidic type I and high molecular weight basic or neutral type II.5 KRTs are integrated in the cellular framework and interact with a range of cellular proteins including kinases, receptors, adaptors, and effectors. These interactions activate signaling networks that regulate cell migration, invasion, metastasis, cell cycle, and apoptosis.4, 6 For example, KRT8 and KRT18 bind with tumor necrosis factor receptor-2 in order to interact with other specific downstream proteins. Therefore, they affect tumor necrosis factor-dependent activation of downstream effectors such as c-JUN-NH2-kinase and NF-kB.7 In addition, KRT17 binds to the adaptor protein 14-3-3σ to upregulate protein synthesis and cell growth via the AKT/mTOR pathway.8
One of the smallest intermediate filament KRT proteins is a 40-kDa type I KRT, KRT19.9 It contains a highly conserved α-helical central domain, essential for filament formation, and does not have a C-terminal non-helical tail domain.10 In a previous study, KRT19-knocked out mice showed induced skeletal myopathy,11 whereby KRT19 was shown to regulate breast cancer properties by activating the AKT signaling pathway.12 However, recent studies have shown that modulation of KRT19 expression led to contrasting effects on cell proliferation, survival, invasion, migration, and apoptosis, depending on the cancer cell type.12, 13, 14 Therefore, extensive molecular studies on KRT19 are required to elucidate its role in cancer cells.
In this study, we demonstrate that knockdown of KRT19 leads to increased proliferation, migration, invasion, drug resistance, and sphere formation in breast cancer cells. We report for the first time, a novel function of KRT19 in the NOTCH signaling pathway. Our data show that KRT19 directly interacts with β-catenin/RAC1 complex to regulate the stability and translocation of β-catenin. β-Catenin, in turn, binds to the NUMB promoter and accelerates its expression in breast cancer cells. Modulation of NUMB expression by KRT19 is therefore involved in the NOTCH pathway-mediated regulation of breast cancer and cancer stem cell properties.
Results
Differential expression of the KRT family of genes in breast cancer cells
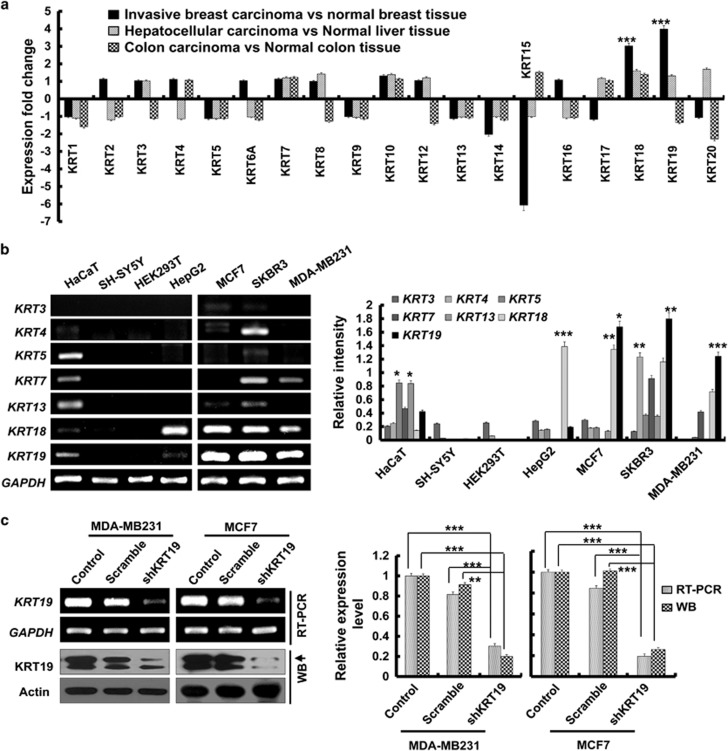
Using the Oncomine database (www.oncomine.org), we compared the expression patterns of the KRT family of genes (KRT1–20) between normal and cancer tissues. Several of these genes showed altered expression in carcinoma compared with that in normal tissues (Figure 1a and Supplementary Figure 1).15 Interestingly, invasive breast carcinoma tissues showed relatively high KRT18 and KRT19 expression. In particular, the fold change for KRT19 expression in invasive breast carcinoma versus normal breast tissue was significantly higher (P<0.00001) than that for the other KRT genes (Figure 1a and Supplementary Figure 1A). This suggested strong correlation of KRT19 expression with invasiveness of breast cancers. In order to confirm the specificity of our observation, we also examined the fold changes for the KRT genes in liver and colon cancer (Figure 1a and Supplementary Figures 1B and C).16, 17 The results concluded that indeed, KRT19 expression specifically correlates with the invasiveness of breast carcinoma (Figure 1a).
Figure 1.
Knockdown of KRT19 increases cell proliferation, migration, invasion, drug resistance, and sphere formation in breast cancer cell lines. Data were obtained from three independent experiments and presented as average values±s.d. (*P<0.05, **P<0.01, ***P<0.001). (a) Expression fold changes for KRT genes (KRT1–20) in invasive ductal breast carcinoma versus (vs) normal breast tissue, hepatocellular carcinoma vs normal liver tissue, and colon adenocarcinoma vs normal colon tissue, as obtained from the Oncomine database. (b) mRNA expression of KRT genes in breast cancer (MCF7, SKBR3, and MDA-MB231), hepatocellular carcinoma (HepG2), neuroblastoma (SH-SY5Y), immortalized human keratinocytes (HaCaT), and immortalized human embryonic kidney (HEK293T) cell lines. Bands for KRTs in b were quantified by scanning densitometry and normalized to that of GAPDH (right panel). (c) KRT19 expression analyzed by reverse transcription polymerase chain reaction (RT–PCR) and western blot analysis. Either GAPDH or actin expression was used as control. Both KRT19 mRNA and protein expression were quantified by scanning densitometry and normalized to that of GAPDH and actin, respectively (right panel). (d) Effect of KRT19 knockdown on cell proliferation analyzed by cell counting. Cells were counted up to 4 days. (e) Migration capacity of the indicated cells analyzed using wound-healing/migration assay. The number of cells in the enclosure was enumerated at the indicated time points. (f) Effect of KRT19 suppression on cell invasion assessed using CytoSelect 96-Wells Cell Invasion Assay Kit. Fluorescent intensities (RFUs) of the invading cells were plotted for control, scrambled shRNA (scramble), and shKRT19 MDA-MB231 and MCF7 cells. (g) Effect of KRT19 knockdown on drug resistance measured by cell counting after 24 h of doxorubicin treatment (0.5 μM). The mRNA expression level of drug-resistance marker genes was analyzed in the shKRT19 knockdown cells. (h) Cells were cultured in suspension in sphere-forming media (SFM) using non-coated plates. The number of spheres was counted on day 5. (i) mRNA expression levels of stemness marker genes were analyzed in the scramble and/or shKRT19 MDA-MB231 and MCF7 cells.
Using RT–PCR, we next evaluated the expression of KRT genes in several cell lines, including MCF7, SKBR3 and MDA-MB231 (breast cancer); HepG2 (hepatocellular carcinoma); SH-SY5Y (neuroblastoma); HaCaT (immortalized human keratinocytes); and HEK293T (immortalized human embryonic kidney cells). In this case, too, KRT19 was specifically overexpressed in the breast cancer cell lines MCF7, SKBR3, and MDA-MB231 (Figure 1b).
Knockdown of KRT19 increases cell proliferation, migration, invasion, drug resistance, and sphere formation
As the breast cancer cells showed significantly high expression of KRT19 compared with other KRT genes, we next assessed the effect of KRT19 knockdown in MDA-MB231 and MCF7 cells. Compared with KRT19 expression in the control (scrambled) short hairpin (sh)RNA-transduced cells, specific downregulation of KRT19 expression (~80%) was achieved in the KRT19-knocked down shKRT19 MDA-MB231 and shKRT19 MCF7 cells (Figure 1c).
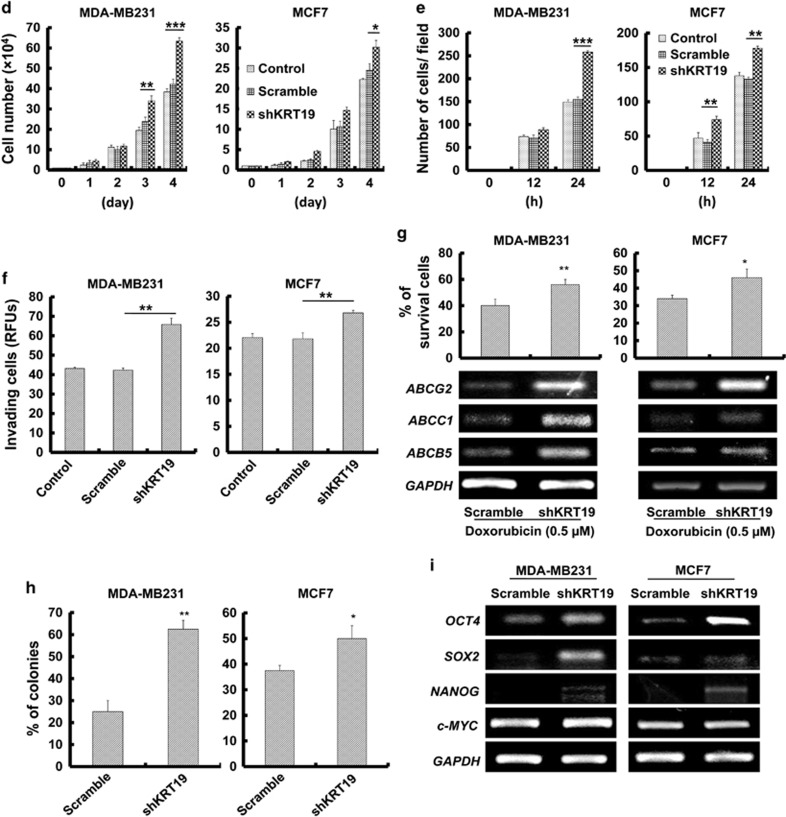
Although the analysis of Oncomine database revealed that the KRT19 expression positively correlates with the invasiveness of breast carcinoma (Figure 1a), knockdown of KRT19 expression resulted in the increased proliferation of the breast cancer cells (Figure 1d). Migration and invasiveness of MDA-MB231 and MCF7 cells were also significantly increased upon KRT19 knockdown (Figures 1e and f). Subsequent experiments in the presence of doxorubicin revealed significantly elevated drug resistance and its marker genes expression, including ABCG2, ABCC1, and ABCB5, in the KRT19-knocked down cells (Figure 1g). KRT19 knockdown also significantly increased colony formation and expression of stemness markers (OCT4, SOX2, NANOG, and c-MYC) in both the shKRT19 MDA-MB231 and shKRT19 MCF7 cells (Figures 1h and i).
KRT19 knockdown upregulates NOTCH signaling pathway by downregulating the expression of NUMB
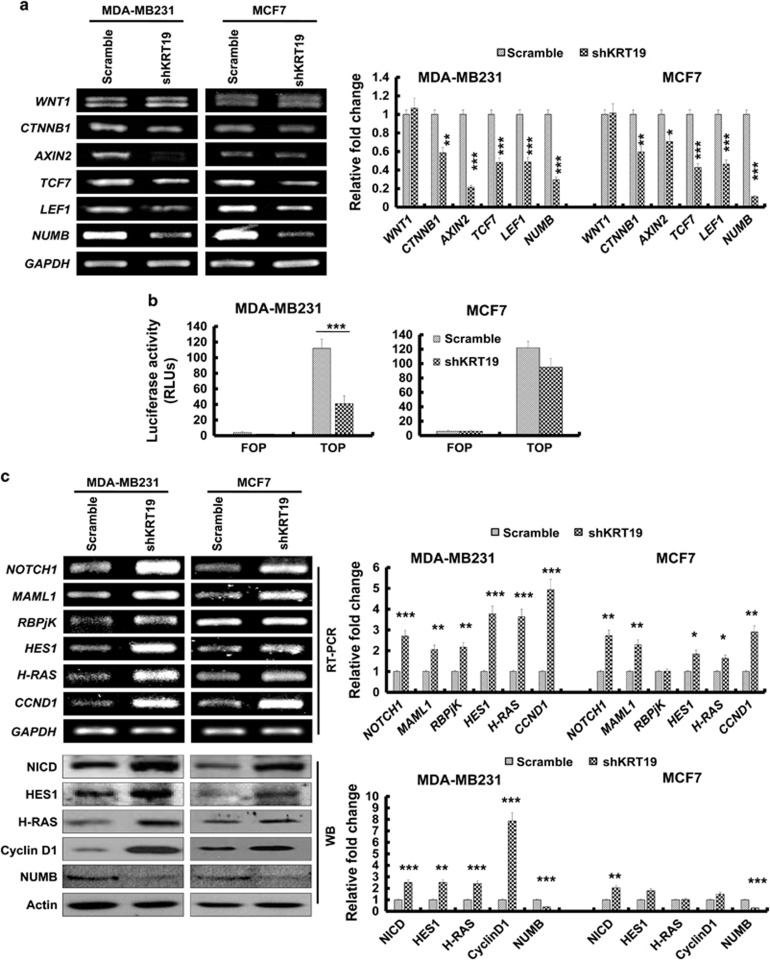
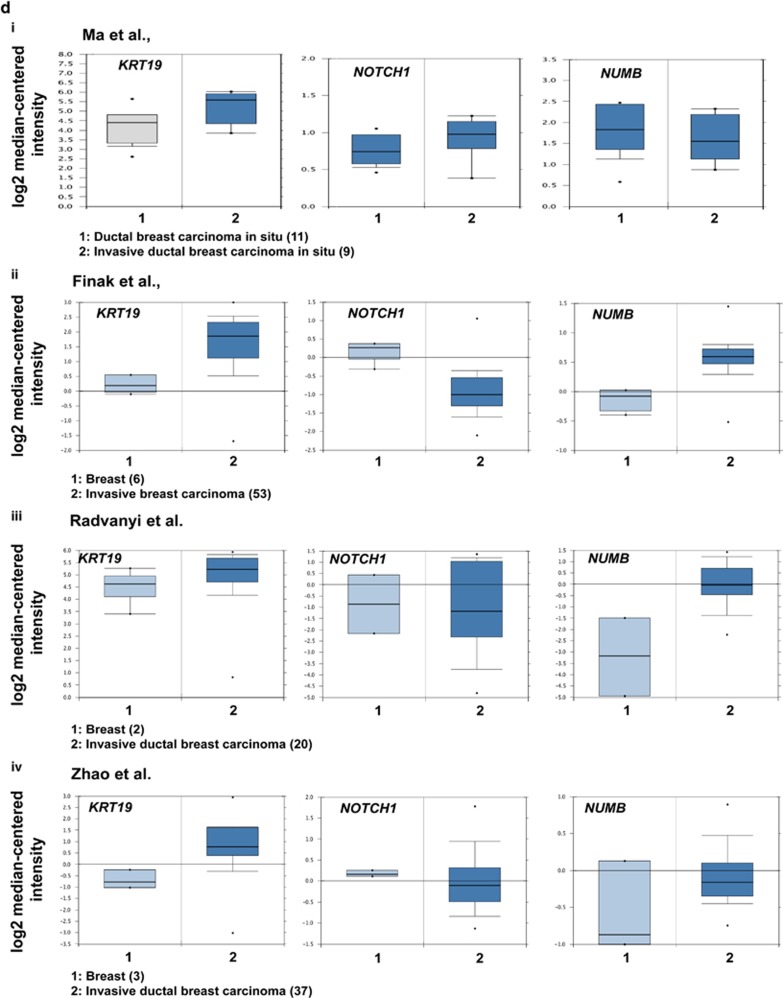
The unexpected findings that KRT19 knockdown led to enhancement of breast cancer properties led us to explore the molecular mechanism behind the phenomenon. Recent report demonstrated that KRT19 had a role in modulation of NOTCH signaling pathway during the invasion of hepatocellular carcinoma.13 More recently, a review article summarized on the crucial role of NOTCH and WNT signaling pathways and the cross-talk between NOTCH receptor and other oncogenic proteins such as Ras, c-Myc, and JAG1.18, 19 Actually, the WNT/β-catenin signaling pathway was reported to be positively or negatively cross-linked with the NOTCH pathway in cancer progression.20, 21, 22, 23, 24, 25 Therefore, we try to investigate whether the NOTCH and WNT signaling pathways are involved in molecular mechanism underlying the KRT19-silencing effect in breast cancer cells. First, in the KRT19-knocked down cells, we checked expression of the canonical WNT/β-catenin signaling target genes such as AXIN2, TCF7, LEF1, and NUMB. Although the canonical WNT signaling was generally suggested to be positively correlated with breast cancer cells,26, 27, 28 our results showed that expression of WNT/β-catenin signaling target genes was significantly downregulated (Figure 2a). Furthermore, the TCF/LEF-responsive TOP- or FOP-FLASH transcription assay confirmed the downregulation of WNT/β-catenin signaling pathway in KRT19-silenced cells (Figure 2b). Next, we analyzed the expression of NOTCH signaling pathway-related genes, such as NOTCH1, MAML1, RBPjK, HES1, H-RAS, and CCND1. KRT19 knockdown led to increased expression of NOTCH1 signaling pathway-interacting proteins, such as MAML1, RBPjK, and NOTCH1 itself (Figure 2c). In this context, several studies have shown that the NOTCH1 intracellular domain forms a complex with MAML1 and RBPjK that translocates to the nucleus to regulate target gene expression.29, 30, 31, 32 Concordantly, the expression of NOTCH1 target genes, such as HES1, CCND1, and H-RAS, were also increased in the KRT19-knocked down cells in this study (Figure 2c). Significantly, we observed that the expression of NUMB, a key inhibitory protein of the NOTCH signaling pathway,33 was significantly downregulated upon KRT19 knockdown (Figures 2a and c). As WNT/β-catenin pathway was reported to be interlinked with the NOTCH signaling pathway by NUMB,34, 35, 36, 37 we assumed that KRT19-mediated regulation of NUMB expression may be crucial for the cross-talk between the WNT and NOTCH signaling pathways. Furthermore, most Oncomine data analyses of normal breast tissue versus invasive ductal breast carcinoma supported the observation that KRT19 expression correlates positively and inversely with that of NUMB and NOTCH1, respectively (Figure 2d (ii–iv)),38, 39, 40 although a ductal breast carcinoma in situ versus invasive ductal breast carcinoma in situ data by Ma et al.15 reported an opposite result (Figure 2d (i)).
Figure 2.
Silencing of KRT19 expression upregulates the NOTCH signaling pathway by downregulating NUMB expression. Data were obtained from three independent experiments and presented as average values±s.d. (*P<0.05, **P<0.01, ***P<0.001). (a) Expression levels of WNT1, CTNNB1, AXIN2, TCF7, LEF1, and NUMB were analyzed using RT–PCR. Bands were quantified by scanning densitometry and normalized to that of GAPDH (right panel). (b) The TOP-/FOP-Flash assays were evaluated using luciferase assay system. The indicated cells were transfected with TOP- or FOP-Flash plasmid, and then luciferase activity was measured. (c) Expression levels of NOTCH1, MAML1, RBPjK, HES1, CCND1, and H-RAS were analyzed using RT–PCR; protein levels of NICD, HES1, cyclinD1, H-RAS, and NUMB were assessed using western blotting (lower panel). Bands were quantified by scanning densitometry and normalized to that of GAPDH or actin (right panel). (d) Relative levels of KRT19, NOTCH1, and NUMB expression were analyzed in normal and invasive breast carcinomas using different Oncomine databases. Box, whiskers and asterisks reflect the interquartile range, 10–90% range, and the minimum or maximum values, respectively. Data are normalized facilitating inter-study comparison. The image was downloaded from the Oncomine datasets provided by Ma et al.,15 Finak et al.,38 Radvanyi et al.39 or Zhao et al.40
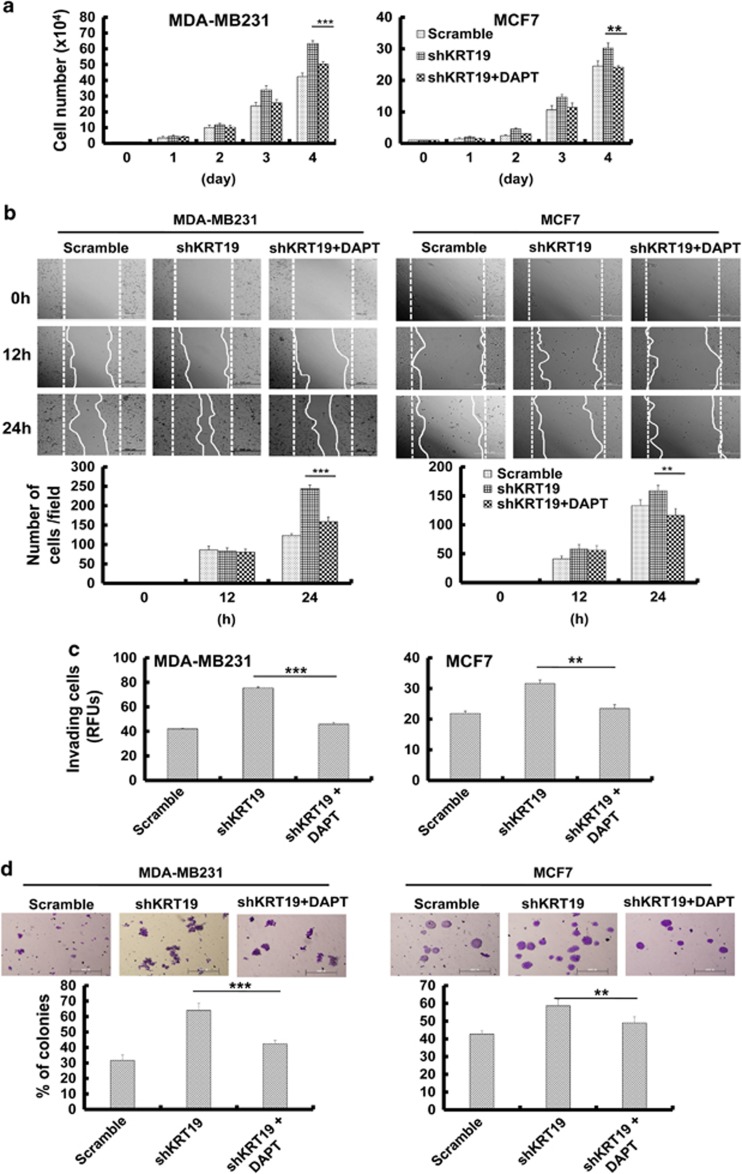
We then examined the effects of inhibiting the NOTCH signaling pathway by treating shKRT19 cells with N-((3,5-Difluorophenyl)acetyl)-l-alanyl-2-phenyl)glycine-1,1-dimethylethyl ester (DAPT), a γ-secretase inhibitor. The results revealed a suppression of shKRT19-induced expression of NOTCH1 target genes (Supplementary Figure 2A). Moreover, DAPT treatment could relieve the shKRT19-mediated increase of cell proliferation (Figure 3a), migration (Figure 3b), invasion (Figure 3c), and sphere formation (Figure 3d) in MCF7 cells, but more markedly in the highly invasive and metastatic MDA-MB231 cells. This demonstrated that the NOTCH signaling pathway is crucial for KRT19 knockdown to exert its cellular effects.
Figure 3.
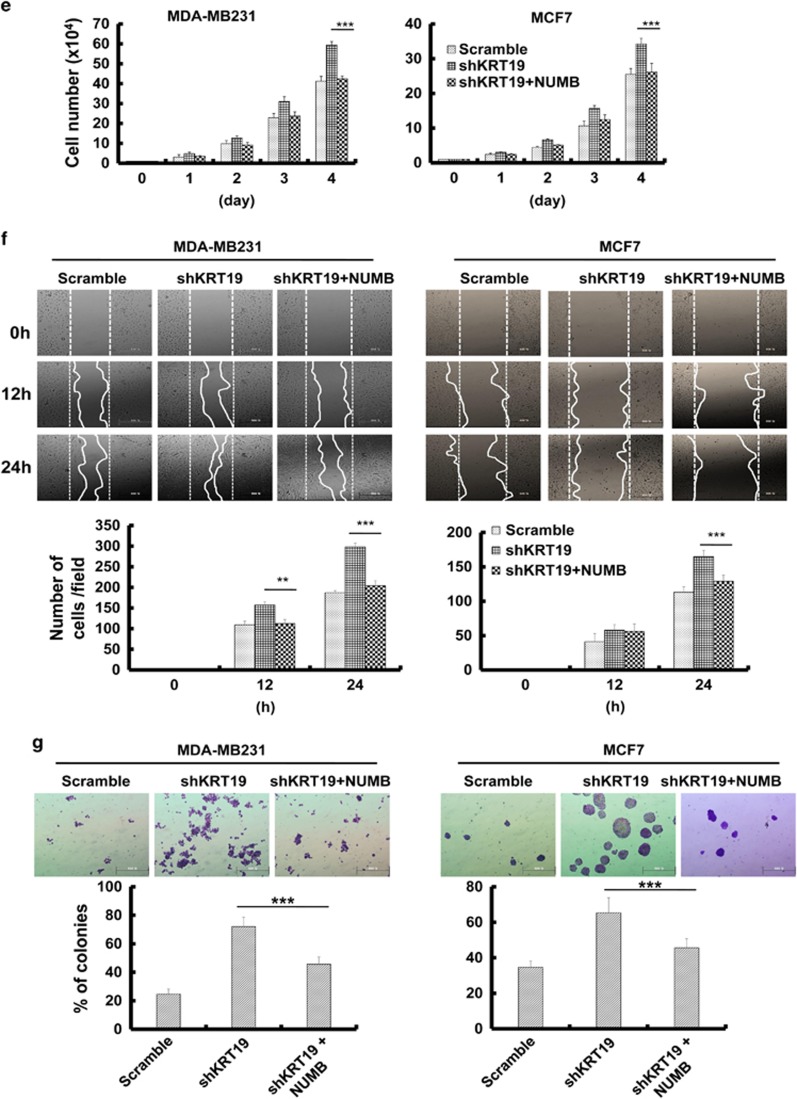
DAPT treatment and NUMB overexpression help to overcome the effects of KRT19 knockdown on proliferation, migration, invasion, and sphere formation in breast cancer cell lines. Data were obtained from three independent experiments and presented as average values±s.d. (*P<0.05, **P<0.01, ***P<0.001). (a) Cell proliferation upon DAPT treatment (20 μM) was analyzed by cell counting. Cells were counted up to 4 days. (b) Wound-healing/migration assay in DAPT-treated shKRT19 knockdown cells. Images were acquired at the indicated time points using bright-field microscopy (upper panel). The number of cells in the enclosure was counted (lower panel). (c) Invasiveness was analyzed in DAPT-treated shKRT19 knockdown cells. (d) The indicated cells were cultured in suspension in SFM using non-coated plates. The number of spheres was stained with crystal violet (upper panel), and counted (lower panel) on day 5. (e) Cell proliferation upon NUMB overexpression in shKRT19 cells was analyzed by cell counting. Cells were counted up to 4 days. (f) Wound-healing/migration assay was conducted in NUMB-overexpressed shKRT19 knockdown cells. Images were acquired at the indicated time points using bright-field microscopy (upper panel). The number of cells in the enclosure was counted (lower panel). (g) The indicated cells were cultured in suspension in SFM using non-coated plates. The number of spheres was stained with crystal violet (upper panel), and counted (lower panel) on day 5.
In addition, we tried to suppress the NOTCH signaling pathway by overexpressing NUMB in shKRT19 cells. We confirmed NUMB overexpression (Supplementary Figure 2B) and found that NOTCH intracellular domain and its target protein expression were downregulated in NUMB-overexpressing shKRT19 cells, compared with the shKRT19 cells (Supplementary Figure 2B). Notably, NUMB overexpression reversed the shKRT19-mediated increase of cell proliferation (Figure 3e), migration (Figure 3f), or sphere formation (Figure 3g), confirming that KRT19-mediated regulation of NOTCH signaling pathway and cancer properties is NUMB dependent.
KRT19 regulates nuclear localization of β-catenin
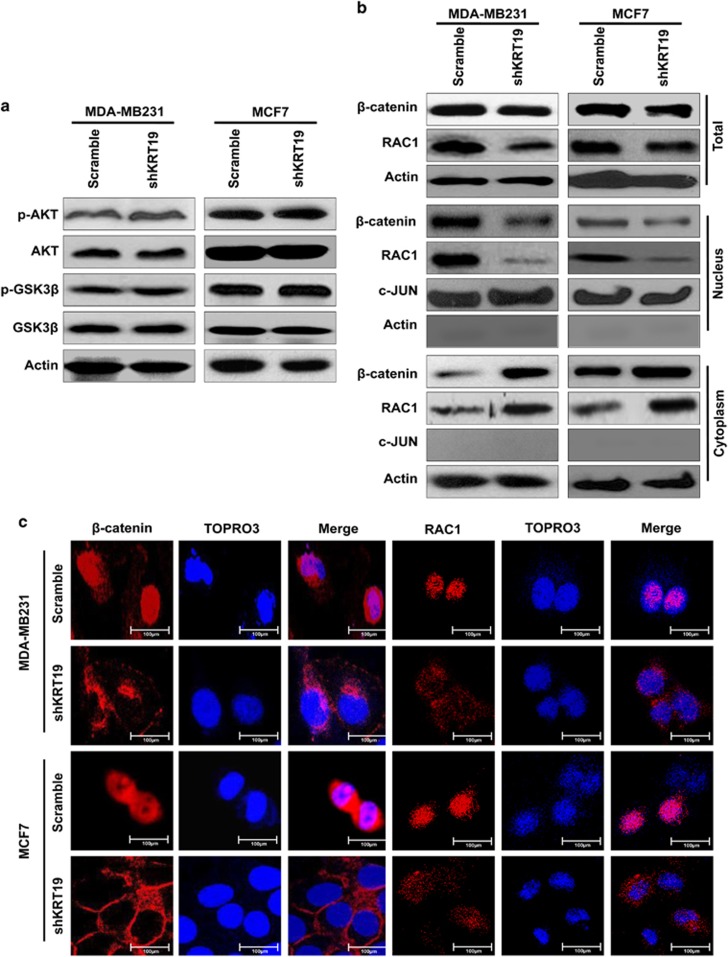
Next, we focused on the KRT19-dependent regulation of NUMB transcription. A previous study demonstrated that β-catenin translocates to the nucleus and binds the TCF4 region of the NUMB promoter, enhancing transcription of the latter.41 Based on this information, we investigated whether the β-catenin signaling pathway is involved in the KRT19-dependent regulation of NUMB transcription. The expression and phosphorylation (Ser 473 of AKT and Ser9 of GSK3β) of both AKT and GSK3β were not significantly altered in the KRT19-knocked down cells (Figure 4a), whereas the total amount of β-catenin and RAC1 proteins were slightly decreased (Figure 4b). However, the nuclear translocation of β-catenin was retarded upon KRT19 knockdown (Figure 4b). Previously, several studies depicted an intriguing but controversial role of Rac1 in regulation of β-catenin nuclear localization.42, 43, 44, 45 Our results revealed that KRT19 knockdown markedly suppressed nuclear translocation of RAC1, a binding partner of β-catenin that aids nuclear translocation of the latter.43, 46 Meanwhile, the cytoplasm presented higher amounts of β-catenin and RAC1 in the shKRT19 cells compared with that in control cells.
Figure 4.
KRT19 regulates nuclear translocation of the β-catenin-RAC1 complex. (a) Levels of phospho (p)-AKT, AKT, p-GSK3β, and GSK3β analyzed in the indicated cells using western blotting. (b) β-catenin and RAC1 levels in cytosolic and nuclear fractions as analyzed by western blotting. c-JUN and actin were used as nuclear and cytoplasmic markers, respectively. (c) Subcellular localization of β-catenin and RAC1 monitored by immunocytochemistry. Scale bar represents 100 μm.
Nuclear translocation of β-catenin and RAC1 was then monitored by immunocytochemistry. This confirmed that both overall and nuclear β-catenin and RAC1 levels were significantly downregulated upon KRT19 knockdown (Figure 4c), whereas that in the cytoplasm was increased. These results strongly suggested that regulation of β-catenin and RAC1 nuclear translocation is involved in the KRT19-dependent regulation of NUMB transcription.
KRT19 interacts with β-catenin and RAC1 to modulate nuclear translocation of the former
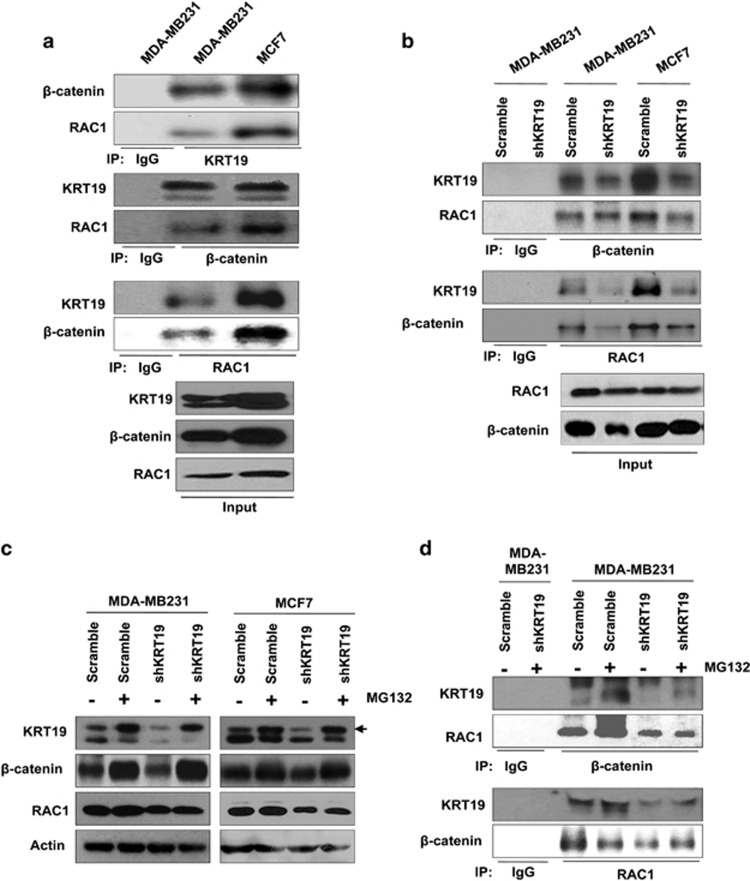
As RAC1 was reported to be a binding partner of β-catenin and may help nuclear translocation of β-catenin,43, 46 we performed coimmunoprecipitation experiments and revealed the interaction of KRT19 with β-catenin and RAC1 in the breast cancer cell lines (Figure 5a). However, KRT19 knockdown significantly inhibited these interactions (Figure 5b). Upon treatment with the proteasome inhibitor MG132, β-catenin levels were increased and KRT19 levels were slightly augmented, whereas that of RAC1 was not significantly altered (Figure 5c). The interactions among KRT19, β-catenin and RAC1 were only slightly enhanced upon treatment with MG132, but markedly decreased upon KRT19 knockdown even in presence of MG132 (Figure 5d). These results suggest the role of KRT19 in its interactions with β-catenin and RAC1.
Figure 5.
KRT19 interacts with β-catenin and RAC1. (a) Lysates from the indicated cells were used for immunoprecipitation using Protein A/G Sepharose, antibodies specific for KRT19, β-catenin, and RAC1, and normal IgG. The immunoprecipitates were analyzed by western blotting with the antibodies indicated. For inputs, lysates were analyzed by western blotting with the indicated antibodies. (b) Immunoprecipitation conducted with the indicated antibodies using lysates from the indicated cells. For inputs, lysates were analyzed by western blotting with the indicated antibodies. (c) Cells were treated with MG132 and the protein levels of KRT19, β-catenin, and RAC1 were assessed by western blotting. (d) Immunoprecipitation assay was performed after MG132 treatment, using the antibodies indicated.
Patient-derived CD133high/CXCR4high/ALDH1high cancer stem-like cells (CSLCs) have decreased KRT19 expression and highly enhanced CSLC properties
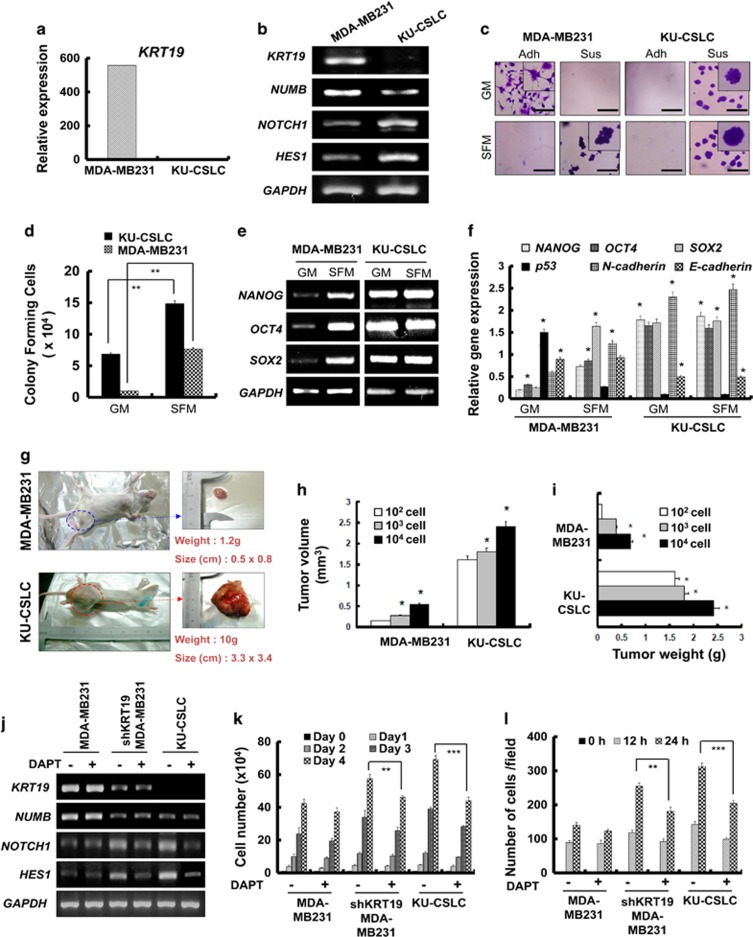
Interestingly, we observed a marked suppression of KRT19 expression in the CD133high/CXCR4high/ALDH1high cancer stem-like cells (KU-CSLCs), which were prepared from chemo-treated breast cancer patients at the Konkuk University hospital (Figure 6a). Compared with the MDA-MB231 cells, KU-CSLCs showed lower NUMB, but higher NOTCH1 and HES1 (target of NOTCH1) expression (Figure 6b). Our data also demonstrated that KU-CSLCs have remarkably high sphere-forming capacity not only in sphere-forming media, but also in normal growth media (Figures 6c and d). KU-CSLCs also showed significantly enhanced expression of stemness and epithelial-to-mesenchymal transition markers, such as NANOG, OCT4, SOX2, and N-cadherin, with reduction in p53 and E-cadherin levels (Figures 6e and f). Examination of KU-CSLCs using a xenograft mouse model revealed that these cells possess highly enhanced tumorigenic capacity in vivo (Figures 6g, h, and i).
Figure 6.
Patient-derived CD133high/CXCR4high/ALDH1high cancer stem-like cells (KU-CSLCs) show significantly decreased expression of KRT19 and highly enhanced CSLC properties. Data were obtained from three independent experiments and presented as average values±s.d. *P<0.05, **P<0.01, ***P<0.001). (a) mRNA expression of KRT19 analyzed by RNA sequencing in MDA-MB231 cells and KU-CSLCs. (b) Expression levels of KRT19, NUMB, NOTCH1, and HES1 analyzed using reverse transcription polymerase chain reaction (RT–PCR) and western blotting. (c and d) Sphere-formation analysis in either growth (GM) or SFM medium using non-coated plates. Spheres from either MDA-MB-231 cells or KU-CSLCs were collected after 5 days of culture, stained with crystal violet (c) and counted (d). (e and f) mRNA expression levels of stemness (OCT4, SOX2, and NANOG) and epithelial-to-mesenchymal (EMT; N- and E-cadherin) markers analyzed using RT–PCR (e) and quantitative (qPCR) (f) in MDA-MB231 cells and KU-CSLCs cultured in either GM or SFM. (g) Representative images of the MDA-MB231- or KU-CSLC-injected SCID mice (left), and the excized tumors (right) 4 weeks post tumor formation (n=5). Tumor volume (mm3) (h) and weight (g) (i) assessed after biopsy. (j) Expression of KRT19, NUMB, NOTCH1, and HES1 analyzed in the indicated cells after treatment with DAPT (20 μM). (k) Proliferation of the indicated cells was assessed for 4 days after DAPT treatment. (l) Wound-healing/migration assay in control and DAPT-treated cells. The number of cells in the enclosure was enumerated at the time points indicated.
As KU-CSLCs showed either very low or no KRT19 expression together with high NOTCH1 expression, similar to the shKRT19 MDA-MB231 cells, we next investigated whether KRT19 can regulate cancer stem cell properties by modulating the NOTCH signaling pathway. First, we treated the cells with DAPT (a NOTCH signaling inhibitor), which led to significant decrease in the expression of NOTCH1 and HES1. However, DAPT did not affect NUMB expression in both the shKRT19 MDA-MB231 and KU-CSLCs (Figure 6j). Importantly, DAPT treatment suppressed proliferation and migration of KU-CSLCs, similar to that in the shKRT19 MDA-MB231 cells (Figures 6k and l; Supplementary Figure 3A). The cancer stem cell properties, including expression of stemness/epithelial-to-mesenchymal transition markers, and sphere-formation capacity of KU-CSLCs were also suppressed by DAPT treatment (Supplementary Figures 3B and C). KU-CSLCs showed KRT19 and NOTCH1 expression lower and higher, respectively, than the shKRT19 MDA-MB231 cells did (Figure 6j). The DAPT-mediated effect was also more marked in KU-CSLCs than in the shKRT19 MDA-MB231 cells (Figures 6–l). These results supported KRT19-dependent regulation of the NOTCH signaling pathway.
Either overexpression of KRT19 or knockdown of NOTCH1 led to decreased proliferation, migration, drug resistance, and sphere formation in the patient-derived KU-CSLCs
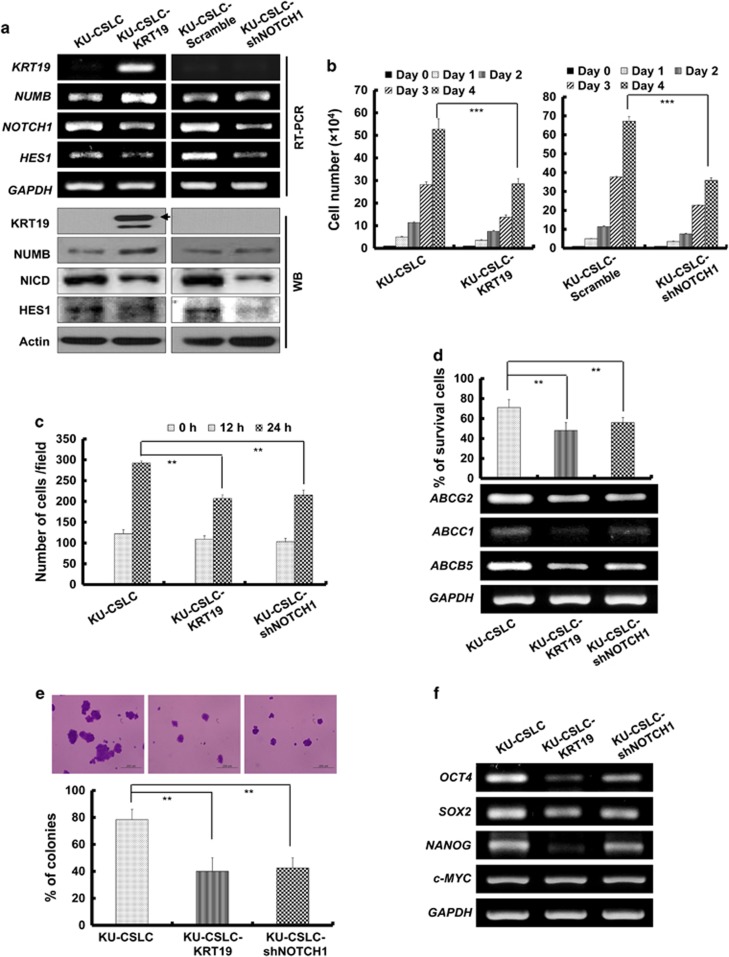
Finally, to confirm the function of KRT19 in regulating the NOTCH signaling pathway, we either overexpressed KRT19 or knocked down NOTCH1 in the KU-CSLCs, both of which were successfully achieved (Figure 7a). Overexpression of KRT19 increased NUMB and decreased NOTCH1 and HES1 expression. Meanwhile, NOTCH1 knockdown led to decreased HES1 expression, without affecting either NUMB or KRT19 levels (Figure 7a). Importantly, both KRT19 overexpression and NOTCH1 knockdown significantly suppressed cell proliferation (Figure 7b) and migration (Figure 7c and Supplementary Figure 4). Drug resistance, expression of drug-resistance markers (Figure 7d), sphere formation (Figure 7e), and expression of stemness markers (Figure 7f) were also suppressed upon KRT19 overexpression and NOTCH1 knockdown in KU-CSLCs. These results strongly supported that KRT19 modulates the NUMB-dependent NOTCH signaling pathway in breast cancers and patient-derived CD133high/CXCR4high/ALDH1high/KRT19low CSLCs, thus harboring potential clinical implications.
Figure 7.
Overexpression of KRT19 or knockdown of NOTCH1 leads to decreased CSLC properties in KU-CSLCs. Data were obtained from three independent experiments and presented as average values±s.d. (*P<0.05, **P<0.01, ***P<0.001). (a) Expression of KRT19, NUMB, NOTCH1, and HES1 analyzed in the KRT19-overexpressing or NOTCH1-knocked down KU-CSLCs using reverse transcription polymerase chain reaction (RT–PCR; upper panel). Protein levels of KRT19, NUMB, NICD, and HES1 were assessed by western blotting. (b) Proliferation of the indicated cells was assessed for 4 days. (c) Wound-healing/migration assay for the indicated cells. The number of cells in the enclosure was counted at the time points indicated. (d) Cell survival measured by cell counting after 24 h of treatment with doxorubicin (0.5 μM) (upper panel) and expression of drug-resistance markers analyzed by RT–PCR (lower panel). (e) Sphere formation in SFM using non-coated plates. Spheres from the indicated cells were collected after 5 days of culture, stained with crystal violet (upper panel) and counted (lower panel). (f) Expression of stemness markers in the indicated cells.
Discussion
A prominent type I filament protein, KRT19 is abundantly expressed in epithelial tumor cells and is a marker for metastatic tumors.47, 48, 49 Its overexpression in circulating tumor cells (CTCs) governs its crucial role in regulating their cancerous properties. However, a previous gene profiling microarray study showed that KRT19 is differentially expressed in different breast cancer cell lines. Moreover, its expression in these cell lines showed reciprocal correlation with their cancer properties (that is, high KRT19 with low CD44 and vice versa).50 In fact, several reports have demonstrated the contradictory effects of KRT19 on cancer properties, depending on its concentration and the cell type involved.12, 13, 14 For instance, cancer properties were positively regulated through the KRT19/HER2 signaling pathway by very high KRT19 levels in HER2-overexpressed breast cancer.51 Meanwhile, negative regulation was observed in the BT474 and SKBR3 breast cancer cells, through the KRT19/Egr-1 signaling pathway, at either very low or no KRT19 concentration.12 Another study demonstrated that overexpression of KRT19 in the BT549 breast cancer cell line (expresses low levels of endogenous KRT19) inhibits cell proliferation and induces cell arrest by regulating endoplasmic reticulum stress and its related signaling pathway. Meanwhile, KRT19 knockdown yielded the opposite results in MDA-MB231 cells (which express high endogenous KRT19).52 Furthermore, KRT19 also has a critical role in the proliferation and invasiveness of some subtypes of hepatocellular carcinomas,13, 53 functioning as a cancer stem cell marker in the same.14 Overall, therefore, the role of KRT19 in cancer (that is, progression or suppression) remains to be resolved through extensive molecular studies.
In this study, we compared the expression patterns of KRT genes (KRT1–20) in normal and cancer tissues, and found specific high expression of KRT19 in invasive breast cancer. Although the analysis of Oncomine database suggested strong correlation of KRT19 expression with breast cancer invasiveness, knockdown of KRT19 expression yielded increased cell proliferation, migration, invasion, drug resistance and sphere formation in breast cancer cells, which showed specifically enhanced KRT19 expression. Our results demonstrate that high KRT19 levels may negatively affect proliferation and invasiveness of breast cancers, though several KRT genes (including KRT19) are reported as important markers for RT–PCR-mediated detection of cancers.2, 3 Importantly, KRT19 expression was markedly suppressed in CD133high/CXCR4high /ALDH1high KU-CSLCs, which were prepared from chemo-treated breast cancer patients and showed abnormally elevated cell proliferation, migration, invasion, drug resistance, and sphere formation. In addition, KRT19 overexpression in KU-CSLCs significantly suppressed their proliferation and migration (Figure 7), strongly supporting the anticancer/-CSLC function of KRT19.
The underlying molecular mechanism of KRT19 must be studied in order to understand better its functions in cancer cells. We report here, for the first time, a novel function of KRT19 in regulation of the NUMB-dependent NOTCH signaling pathway in breast cancer cells and CSLCs. NOTCH possesses oncogenic functions in several cancers, including leukemia and breast cancer,54, 55 and also regulates differentiation, proliferation, and growth of either cancer or stem cells.56, 57 Several studies also reported that NUMB-mediated regulation of the NOTCH signaling pathway modulates migration and invasion in different tumor cell types, including pancreatic,58 ovarian,59 and prostate.60 NUMB also suppresses the NOTCH signaling pathway through proteolytic degradation of the NOTCH intracellular domain.33 Furthermore, it regulates asymmetric cell division in mammalian development, cell adhesion and migration, as well as a number of signaling pathways.35, 61, 62 Recent studies confirmed that the NOTCH signaling pathway is suppressed by overexpressed NUMB, suggesting that the latter acts as a tumor suppressor.63, 64 In the current study, KRT19 knockdown-mediated suppression of NUMB expression led to increased NOTCH activity as well as NOTCH-dependent proliferation of breast cancer cells. Importantly, we demonstrated the crucial function of KRT19 in modulating the NOTCH signaling pathway and related cancer and CSLC properties, not only in breast cancer cells (MCF7 and MDA-MB231), but also in patient-derived CD133high/CXCR4high/ALDH1high/KRT19low CSLCs.
At the molecular level, we demonstrated that KRT19 functionally interacts with the β-catenin-RAC1 complex, and promotes stability and nuclear translocation of the same. This regulation is crucial to the expression of NUMB, which suppresses the NOTCH signaling pathway. NUMB, a direct transcriptional target of the WNT signaling pathway, could ubiquitously degrade NOTCH intracellular domain and negatively regulate NOTCH signaling pathway.35, 36 Furthermore, previous studies demonstrated that inhibition of NUMB resulted in activation of NOTCH signaling pathway.65, 66 KRT19-mediated regulation of NUMB expression may be crucial for cross-linking and balancing of the canonical WNT and NOTCH signaling pathways, which will be characterized in detail in the further study. Essentially, our results demonstrated that silencing of KRT19 expression inhibits formation of the β-catenin-RAC1 complex, and positively regulates NOTCH signaling by inhibiting β-catenin-mediated NUMB transcription. A recent study reported that the canonical WNT signaling pathway upregulates NUMB expression in myoblasts. This occurs when β-catenin binds to regulatory regions of the NUMB promoter at TCF sites 545 and 640 bp upstream of the transcriptional start site.41 Moreover, RAC1 genetically interacts with β-catenin and controls its nuclear localization to the TCF3/4 sites of the target gene promoter.43, 44 Furthermore, active/inactive Rac1 could shift Rac1–β-catenin complexes toward the nucleus.67, 68, 69 In addition, a protein complex composed of RAC1B, β-catenin and disheveled functions to enhance nuclear translocation of transcription factors required for increased interaction with the target promoters.46 These studies corroborate our proposed mechanism that KRT19 directly interacts with the β-catenin-RAC1 complex to promote nuclear translocation of β-catenin and activate NUMB transcription.
Our data also showed that the effect of KRT19 knockdown is more marked in the highly invasive and metastatic MDA-MB231 breast cancer cells (‘basal' type, triple negative (ER−PR−HER2−)), compared with that in the MCF7 cells (‘luminal' type, ER+PR+).70 A previous report demonstrated that KRT19+ CTCs differentially affect breast cancer patients with ER-negative and -positive (basal- and luminal-like CTCs, respectively) tumors. Despite of similar KRT19 expression in CTCs of both tumor types, the clinical outcomes of prognosis differed depending on the ER expression, especially at the early stages.2 Specifically, breast cancer patients with ER− CTCs showed poor prognosis than those with ER+ CTCs.2, 71, 72, 73, 74, 75, 76 Concomitantly, in this study, KRT19-dependent regulation of the NOTCH signaling pathway significantly modulated the cancer/CSLC properties of KU-CSLCs, which are ER−. ERα is known to increase the expression of KRT19, (ref. 77, 78, 79, 80) and NOTCH signaling activity is higher in ERα− cells than in ERα+ ones.81, 82 Furthermore, one previous study has reported an important and differential role of NOTCH signaling pathway in regulation of cancer/CSLC properties either in ERα+ or ERα− and triple negative breast cancer.83 Further studies are required to elucidate the effects of ER, PR, and HER2 expression on KRT19-mediated regulation of the NOTCH signaling pathway as well as cancer/CSLC properties.
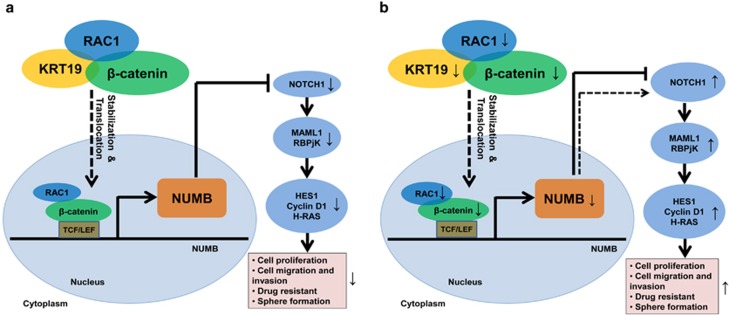
In conclusion, our study shows that KRT19 directly interacts with the β-catenin-RAC1 complex, and stabilizes ubiquitination and proteasomal degradation of β-catenin to increase its nuclear translocation. NUMB expression is enhanced in the process, which then downregulates NOTCH signaling; its target genes; and consequently, cell proliferation, migration, invasion, drug resistance, and sphere formation (Figure 8). Our data might be crucial to understand the molecular basis of KRT19- and NOTCH-mediated regulation of cancer/CSLC properties, and might hold potential clinical implications as well.
Figure 8.
Schematic diagram depicting the KRT19-mediated regulation of NUMB expression, NOTCH signaling pathway and cancer/cancer stem-like cells (CSLC) properties in KRT19-overexpressing breast cancer cells. (a) KRT19 directly interacts with the β-catenin-RAC1 complex and stabilizes the ubiquitination and proteasomal degradation of β-catenin. This increases the nuclear translocation of β-catenin, enhancing NUMB expression in the process, which downregulates NOTCH signaling, its target genes, and consequently, cell proliferation, migration, invasion, drug resistance, and sphere formation. Patient-derived CD133high/CXCR4high/ALDH1high KU-CSLCs, which showed significantly reduced KRT19 expression, (b) displayed highly enhanced CSLC properties.
Materials and methods
Cell culture
Human cell lines for breast cancer (MDA-MB231, SKBR3, and MCF7), hepatocellular carcinoma (HepG2), neuroblastoma (SH-SY5Y), immortalized human keratinocytes (HaCaT), and embryonic kidney cells (HEK293T) are obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). All cell lines and breast cancer patient-derived CD133high/CXCR4high/ALDH1high high sphere-forming KU-CSLCs were cultured in either high glucose-Dulbecco's Modified Eagle Medium or Roswell Park Memorial Institute medium (DMEM or RPMI) 1640 (Sigma-Aldrich, Saint Louis, MO, USA) supplemented with 10% fetal bovine serum. Cells were maintained in a humidified incubator at 5% CO2 and 37 °C. All cell lines were authenticated by short tandem repeat profiling and were tested for mycoplasma contamination using BioMycoX Mycoplasma PCR Detection Kit (Cat. No. CS-D-25) (Cellsafe, Yeongtong-gu, Suwon, Republic of Korea).
CD133high/CXCR4high/ALDH1high KU-CSLCs were prepared from breast tumor tissue specimens, which were collected from chemo-treated patients who underwent mammotomy for breast tumors at the Breast Cancer Center, Konkuk University Hospital, Seoul. The study was performed with approval from the Institutional Review Board (IRB, KUH 1020003).
Knockdown and overexpression of KRT19 and NOTCH1
Sense and antisense oligonucleotides for control (scrambled), KRT19- (shKRT19),12 and NOTCH1-specific (shNOTCH1) shRNA,84 containing BamHI and EcoRI restriction sites, were annealed by using annealing buffer (Tris-EDTA buffer (pH 8), 100 mM NaCl (Sigma-Aldrich)). Next, the annealed oligonucleotides were inserted into the pGreenPuro lentiviral expression vector (System Biosciences, Mountain View, CA, USA) to construct KRT19-, NOTCH1-knocked down, or control vectors. The oligonucleotide sequences of the shRNAs are listed in Table 1.
Table 1. List of primers used for knockdown and overexpression of KRT19 and NOTCH1, respectively.
| Accession no. | shRNA | Oligomers (5′–3′) | Target site |
|---|---|---|---|
| NM_002276.4 | KRT19 | AACCATGAGGAGGAAATCA | 791–809 |
| NM_017617.3 | NOTCH1 | GTCCAGGAAACAACTGCAA | 782–800 |
| Scrambled | CCTAAGGTTAAGTCGCCCTCGCTC | Non-specific | |
| Plasmid name | Gene | Forward (5′–3′) | Reverse (5′–3′) |
| pGEMR-T easy vector | KRT19 | CCTCGCCATGACTTCCTACA | TCCCATCCCTCTACCCAGAA |
| pCDH-EF1-MCS-T2A-copGFP | KRT19 | CGCtctagaCCTCGCCATGACTTCCTACA XbaI | CGCgaattcGGAGGACCTTGGAGGCAGACA EcoRI |
Underlined sequence is for restriction sites.
For overexpression of KRT19, its complete coding sequence was cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA) using the primers listed in Table 1. The complete coding sequence was further subcloned into the pCDH-EF1-MCS-T2A-copGFP lentiviral vector (System Biosciences) using XbaI and EcoRI restriction enzymes and the primers listed in Table 1.
HEK293T cells were transfected with the specific lentiviral vector and viral packaging plasmids, RRE and REV, using calcium phosphate transfection protocol. After 72 h of incubation at 5% CO2 and 37 °C, viruses were collected and concentrated using an ultracentrifuge (Optima L-90K, Beckman Coulter Inc., CA, USA). MDA-MB231 and MCF7 cells were then infected with either scrambled shRNA or shKRT19 lentiviruses, whereas KU-CSLCs were infected with either scrambled shRNA, shNOTCH1 or KRT19-expressing lentiviruses and incubated overnight at 5% CO2 and 37 °C. For virus infection, ~8.0 × 108 IU/ml viral particles were used to get stably expressing cells. To measure virus titer, we use the following equation, titer in IU/ml=(((no. of cells at starting time) × (dilution factor) × (percent infection))/(volume virus solution added expressed in ml)) as described previously.85 Next, the media were substituted with normal growth media (high glucose DMEM/RPMI 1640 with 10% fetal bovine serum). The virus infection efficiency was calculated to be <95%, and knockdown or overexpression was assessed by RT–PCR and western blot analyses.
For NUMB overexpression, MDA-MB231 and MCF7 KRT19-knocked down cells were incubated overnight at a density of 2 × 105 cells per wells in 24-well culture plate and transfected with the expression vectors (pEF-BOS-myc-NUMB and control vector, kind gifts from professor Hee-Sae Park (Chonnam National University) using Lipofectamine in 1:3 ratio (Invitrogen, Carlsbad, CA, USA). After 48 h of post-transfection, half of cells were harvested for western blotting and the remained cells were used for cell proliferation, migration and sphere-formation analyses.
Total RNA extraction and RT–PCR analyses
Total RNA was extracted from the cells using Easy-Blue total RNA extraction kit (iNtRON Biotechnology, Republic of Korea) according to the manufacturer's instructions. The concentration of total RNA was measured using Nanodrop (ND1000) spectrophotometer (Nanodrop Technologies Inc., Wilmington DE, USA). Subsequently, 2 μg of total RNA was used for cDNA synthesis with M-MLV reverse transcriptase (Promega) according to the manufacturer's instructions. Sequences of the primers used are given in Table 2. After the PCR reaction, the products were analyzed on either 1 or 1.5% agarose gels. Quantitative real-time RT–PCR was done using a PTC-200 thermal cycler with Chromo4 optical detector (MJ Research/Bio-Rad, CA, USA) using Fast SYBR Green Master Mix (Applied Biosystems, Stockholm, Sweden). mRNA expression was normalized to that of the control housekeeping gene, GAPDH as described previously.86
Table 2. List of primers used for quantification of specific gene expression.
| Accession no. | Gene | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|---|
| NM_057088.2 | KRT3 | GAGCGGGAACAGATCAAGAC | TCCACTTGGTCTCCAGGACT |
| NM_002272.3 | KRT4 | AGATACCTTGGGCAATGACA | CTTGTTCAGGTAGGCAGCAT |
| NM_000424.3 | KRT5 | CACCAAGACTGTGAGGCAGA | CCTTGTTCATGTAGGCAGCA |
| NM_005556.3 | KRT7 | CTCCGGAATACCCGGAATGAG | ATCACAGAGATATTCACGGCTCC |
| NM_153490.2 | KRT13 | ACGCCAAGATGATTGGTTTC | CGACCAGAGGCATTAGAGGT |
| NM_000224.2 | KRT18 | AGATCGAGGCTCTCAAGGAGG | CACGGTCAACCCAGAGCTG |
| NM_002276.4 | KRT19 | GCGAGCTAGAGGTGAAGATC | CGGAAGTCATCTGCAGCCA |
| NM_001005743.1 | NUMB | TCCCACCTCTCCTACTTCTG | TGCCTCCCCTTCTACTTCTG |
| NM_005430.3 | WNT1 | CTTCGGCCGGGAGTTCGTGG | CACCGTGCATGAGCCGGACA |
| NM_001904.3 | CTNNB1 | AAAATGGCAGTGCGTTTAG | TTTGAAGGCAGTCTGTCGTA |
| NM_004655.3 | AXIN2 | CTCCTTGGAGGCAAGAGC | GGCCACGCAGCACCGCTG |
| NM_003202.3 | TCF7 | GACATCAGCCAGAAGCAAG | CACCAGAACCTAGCATCAAG |
| NM_016269.4 | LEF1 | CCTGGTCCCCACACAACTG | GGCTCCTGCTCCTTTCTCTG |
| NM_017617.3 | NOTCH1 | GGGTACAAGTGCGACTGTGA | CGGCAACGTCGTCAATACAC |
| NM_014757.4 | MAML1 | CACCAGCCACCGAGTAACTT | AACAGGGAGTTCTGCTCGTG |
| NM_005349.3 | RBPjK | GAACAAATGGAACGCGATGG | GATGACTTTTATCCGCTTGCTG |
| NM_005524.3 | HES1 | GGCTAAGGTGTTTGGAGGCT | GGTGGGTTGGGGAGTTTAGG |
| NM_001130442.1 | H-RAS | TTCTACACGTTGGTGCGTGA | CACAAGGGAGGCTGCTGAC |
| NM_053056.2 | CCND1 | CACACGGACTACAGGGGAGT | ATGGTTTCCACTTCGCAGCA |
| NM_002046.5 | GAPDH | AATCCCATCACCATCTTCCAG | CACGATACCAAAGTTGTCATGG |
| Drug-resistance marker | |||
| NM_004827.2 | ABCG2 | TTATCCGTGGTGTGTCTGGAG | TCCTGCTTGGAAGGCTCTATG |
| NM_004996.3 | ABCC1 | GCCGGTGAAGGTTGTGTACT | CTGACGAAGCAGATGTGGAA |
| NM_001163993.2 | ABCB5 | GAGAGACAGTCGCCTTGGTC | CCACGATTGTAGTCCGACCT |
| Stemness marker | |||
| NM_001285986.1 | OCT4 | GTCCCAGGACATCAAAGCTC | CTCCAGGTTGCCTCTCACTC |
| NM_003106.3 | SOX2 | ACACCAATCCCATCCACACT | GCAAGAAGCCTCTCCTTGAA |
| NM_024865.3 | NANOG | ATACCTCAGCCTCCAGCAGA | GCAGGACTGCAGAGATTCCT |
| NM_002467.4 | c-MYC | CTCGGATTCTCTGCTCTC | TCGCCTCTTGACATTCTC |
Western blot analysis
Cells were lysed using lysis buffer (1% Triton X-100 (Sigma-Aldrich), 100 mM Tris-HCl (pH 7.5), 10 mM NaCl, 10% glycerol (Amresco, Solon OH, USA), 50 mM sodium fluoride (Sigma-Aldrich), 1 mM phenylmethylsulfonyl fluoride (PMSF; Sigma-Aldrich), 1 mM p-nitrophenyl phosphate (Sigma-Aldrich), and 1 mM sodiumorthovanadate (Sigma-Aldrich)) and the lysates were centrifuged at 13 000 rpm for 15 min at 4 °C. The protein supernatant was quantified using Bradford protein assay reagent (BioRad), and the proteins were resolved by either 10 or 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis. The separated proteins were then transferred onto nitrocellulose membranes (Bio-Rad). After blocking with 5% skimmed milk in Tris-buffered saline for 1 h, the membranes were incubated with appropriate primary antibodies against KRT19 (SC-53258, 1:500), cyclinD1 (SC-717, 1:1000), H-RAS (SC-35, 1:1000), NUMB (SC-15590, 1:200), β-catenin (SC-7199, 1:2000), RAC1 (SC-217, 1:500), AKT (SC-1619, 1:1000), phospho (p)-AKT (SC-16646-R, 1:1000), GSK3β (SC-9166, 1:1000), p-GSK3β (SC-11757, 1:1000), HES1 (SC-13844, 1:500), actin (SC-1616, 1:1000) (Santa Cruz Biotechnology, Dallas, TX, USA), and intracellular domain (ab83232, 1:1000) (Abcam, Cambridge, UK) overnight at 4 °C. This was followed by a 2 h incubation with anti-mouse (SC-2005), -goat (SC-2020), -rabbit (SC-2004) or –rat (SC-2006) IgGs (1:1000) tagged with horse radish peroxidase (Santa Cruz Biotechnology). Protein signals were detected using an enhanced chemiluminescence kit (Amersham Bioscience, Piscataway, NJ, USA) as described previously.86
Cell proliferation, invasion, wound-healing/migration and drug-resistance assays
Cell proliferation was performed as described.87 For analysis of cell proliferation, 2 × 104 cells/well were seeded in 12-well dishes. For 4 days, the cells (in triplicates) were trypsinized and enumerated every 24 h using a hemocytometer.
Invasion assay was conducted using CytoSelect 96-Wells Cell Invasion Assay Kit (Cell Biolabs Inc., CA, USA) as described.88 Cells (1 × 105) were seeded in 96-well culture plates, and the assay was conducted according to the manufacturer's protocol.
Wound-healing/migration assay was performed as described.89 In brief, 1 × 106 cells were seeded in 60-mm plates and grown until 90% confluent. Prior to the experiment, the cells were incubated with mitomycin C (10 μg/ml), which inhibits cell division. After 2–3 h of incubation at 5% CO2 and 37 °C, the cell monolayer was wounded with a 200 μl pipette tip. Cells were then washed thrice with phosphate-buffered saline (PBS), and images of this wound were acquired by Nikon eclipse TE2000-U microscopy (Nikon Instruments Inc., Melville, NY, USA) at various time points (0, 12 and 24 h).
Drug-resistance analysis was performed as described previously.90 In brief, 1 × 105 cells were seeded in 12-well dishes. Following overnight incubation at 5% CO2 and 37 °C, they were treated with 0.5 μM doxorubicin and incubated for another 48 h. They were then enumerated and represented as percent (%) of surviving cells.
Sphere-formation assay
Cells (1 × 104) were cultured and suspended in either growth medium (DMEM/RPMI 1640 (Sigma-Aldrich) with 10% fetal bovine serum) or SM (sphere-formation medium: serum-free DMEM/F12 supplemented with B27-supplement (1:50; Invitrogen)) supplemented with 20 ng/ml epidermal growth factor (Sigma-Aldrich), 10 μg/ml insulin (Invitrogen), and 0.4% bovine serum albumin (Sigma-Aldrich), using non-coated plates. To propagate the spheres in vitro, they were collected by gentle centrifugation, dissociated into single cells as described previously,91, 92 and then cultured to obtain next-generation spheres. Spheres from MDA-MB-231, MCF7 cells, and KU-CSLCs were collected after 5 days of culture, stained with crystal violet (Sigma-Aldrich) and photographed by Nikon eclipse TE2000-U microscopy (Nikon Instruments Inc.) prior to colony counting.
Luciferase reporter assay
For luciferase reporter assays, 1 × 105 MDA-MB-231 or MCF7 cells were seeded in 12-well plate and transiently transfected with either 200 ng of TOP- or FOP-FLASH (kind gifts from professor Keun Il Kim (Sookmyung Women's University) by using Lipofectamine 2000 reagents (1:3 ratio) (Invitrogen).93 After 48 h post transfection, the luciferase activity using a luciferase assay system (Promega) was analyzed by a luminometer (Veritas microplate luminometer, Turner Biosystems, CA, USA) and transfection efficiency was normalized by β-galactosidase expression.
Immunocytochemistry
Immunocytochemistry was performed as described previously.86 In brief, Cells (1 × 104) were seeded and incubated for 24 h at 5% CO2 and 37 °C. They were then washed with PBS, fixed with 4% paraformaldehyde, and permeabilized with 0.2% Triton X-100 for 10 min. The fixed cells were blocked with 10% normal goat serum (Vector Lab, Burlingame CA, USA) in PBS for 1 h, followed by incubation with primary antibody, diluted in PBS, overnight at 4 °C. After washing thrice with PBS, the cells were incubated with secondary antibody for 1 h and washed with PBS again. Nuclei were stained with TOPRO3 (10 μg/ml; Invitrogen) for an additional 10–15 min. Next, the cells were washed thrice with PBS. Immunofluorescence was monitored with a Leica TCS SP5 II laser scanning confocal microscope (Leica Microsystems, Wetzlar, Germany).
Cell fractionation
Cell fraction was performed as described previously.12 In brief, cells were washed with ice-cold PBS and harvested in cytoplasmic extraction buffer (10 mM HEPES (pH 7.9; Sigma-Aldrich) 10 mM KCl (Sigma-Aldrich), 0.1 mM EDTA (Sigma-Aldrich), 0.1 mM EGTA (Sigma-Aldrich), 1 mM dithiothreitol (DTT; Invitrogen), and 0.5 mM PMSF (Sigma-Aldrich)). The cell suspension was agitated for 10 min at 4 °C, followed by further agitation for another 10 min at 4 °C with NP-40 (final 0.5% (Sigma-Aldrich)). They were then centrifuged at 13 000 rpm at 4 °C for 5 min. The supernatant was collected as the cytoplasmic fraction. Next, the nuclear pellets were washed 2–3 times with cold PBS and resuspended in nuclear extraction buffer (20 mM HEPES (pH 7.9), 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and 1 mM PMSF). The suspended sample was agitated for 10 min at 4 °C, followed by centrifugation at 13 000 rpm at 4 °C for 10 min. The supernatants were then collected as the nuclear fraction.
Coimmunoprecipitation analysis
To investigate the protein–protein interactions, cell lysates (400 μg) were precleaned with 50 μl Protein A/G sepharose (Santa Cruz Biotechnology) for 2 h with agitation at 4 °C, and then centrifuged at 3000 rpm for 1 min. The supernatant was collected and incubated with either primary antibodies or normal rabbit/mouse IgG overnight at 4 °C. Next, another 50 μl of protein A/G sepharose was added and incubated for another 4 h at 4 °C. The pellet was then washed thrice with cell lysis buffer, and the immunoprecipitated proteins were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis and subsequent western blotting as described previously.12
In vivo tumorigenicity analysis
Either MDA-MB231 cells or KU-CSLCs (100, 1000 or 10 000) were injected subcutaneously into the lower flanks of 5-month-old female severe combined immunodeficient mice,94 purchased from KRIBB (Korean Research Institute of Bioscience and Biotechnology, Republic of Korea). The mice were monitored for tumor formation and growth. The mice were killed 4 weeks post tumor formation, and the tumor weights and volume determined as described previously.95 For statistical analysis, at least five mice were used in each group. During the study, the mice were excluded if they died or did not form tumor. According to body weight of mice, they were evenly divided into two groups but not randomly. During the experiment, investigator was blinded for allocation of group. The mice were cared for and treated in compliance with the institutional guidelines (KU11039).
Statistical analyses
All experiments were conducted independently at least thrice. Data were analyzed using GraphPad InStat version 3 program (Graphpad, Sandiego, CA, USA) and presented as average values±s.d. Statistical analyses were done using two-tailed Student's t-test for two groups and, for multiple comparison test, analysis of variance was performed with adjusting Tukey–Kramer to compare treated versus control. A P-value<0.05 was considered statistically significant.
Acknowledgments
This work was supported by grants from the National Research Foundation (NRF) funded by the Korean government (2013M3A9D3045880 and 2015R1A5A1009701).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Lopez de Silanes I, Quesada MP, Esteller M. Aberrant regulation of messenger RNA 3'-untranslated region in human cancer. Cell Oncol 2007; 29: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatiadis M, Xenidis N, Perraki M, Apostolaki S, Politaki E, Kafousi M et al. Different prognostic value of cytokeratin-19 mrna–positive circulating tumor cells according to estrogen receptor and HER2 status in early-stage breast cancer. J Clin Oncol 2007; 25: 5194–5202. [DOI] [PubMed] [Google Scholar]
- Bozionellou V, Mavroudis D, Perraki M, Papadopoulos S, Apostolaki S, Stathopoulos E et al. Trastuzumab administration can effectively target chemotherapy-resistant cytokeratin-19 messenger RNA–positive tumor cells in the peripheral blood and bone marrow of patients with breast cancer. Clin Cancer Res 2004; 10: 8185–8194. [DOI] [PubMed] [Google Scholar]
- Coulombe PA, Wong P. Cytoplasmic intermediate filaments revealed as dynamic and multipurpose scaffolds. Nat Cell Biol 2004; 6: 699–706. [DOI] [PubMed] [Google Scholar]
- Omary MB, Ku N-O, Tao G-Z, Toivola DM, Liao J. ‘Heads and tails' of intermediate filament phosphorylation: multiple sites and functional insights. Trends Biochem Sci 2006; 31: 383–394. [DOI] [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Chu Y-W, Trevor KT, Seftor RE. Role of intermediate filaments in migration, invasion and metastasis. Cancer Metastasis Rev 1996; 15: 507–525. [DOI] [PubMed] [Google Scholar]
- Caulin C, Ware CF, Magin TM, Oshima RG. Keratin-dependent, epithelial resistance to tumor necrosis factor-induced apoptosis. J Cell Biol 2000; 149: 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Wong P, Coulombe PA. A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature 2006; 441: 362–365. [DOI] [PubMed] [Google Scholar]
- Wu Y-J, Rheinwald JG. A new small (40 kd) keratin filament protein made by some cultured human squamous cell carcinomas. Cell 1981; 25: 627–635. [DOI] [PubMed] [Google Scholar]
- Fradette J, Germain L, Seshaiah P, Coulombe PA. The type I keratin 19 possesses distinct and context-dependent assembly properties. J Biol Chem 1998; 273: 35176–35184. [DOI] [PubMed] [Google Scholar]
- Stone MR, O'Neill A, Lovering RM, Strong J, Resneck WG, Reed PW et al. Absence of keratin 19 in mice causes skeletal myopathy with mitochondrial and sarcolemmal reorganization. J Cell Sci 2007; 120: 3999–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju J-h, Yang W, Lee K-m, Oh S, Nam K, Shim S et al. Regulation of cell proliferation and migration by keratin19-induced nuclear import of early growth response-1 in breast cancer cells. Clin Cancer Res 2013; 19: 4335–4346. [DOI] [PubMed] [Google Scholar]
- Govaere O, Komuta M, Berkers J, Spee B, Janssen C, de Luca F et al. Keratin 19: a key role player in the invasion of human hepatocellular carcinomas. Gut 2014; 63: 674–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Yasuchika K, Ishii T, Katayama H, Yoshitoshi EY, Ogiso S et al. Keratin 19, a cancer stem cell marker in human hepatocellular crcinoma. Clin Cancer Res 2015; 21: 3081–3091. [DOI] [PubMed] [Google Scholar]
- Ma XJ, Dahiya S, Richardson E, Erlander M, Sgroi DC. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res 2009; 11: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler S, Jia H-L, Budhu A, Forgues M, Ye Q-H, Lee J-S et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res 2010; 70: 10202–10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, Jarosz D et al. Correction: modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PloS One 2010; 5: 10.1371/annotation/1378c585739-a585354-585734fc585739-a585737d585730-d585735ae585726fa585706ca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y-F, Yong X, Tang B, Qin Y, Zhang J-W, Zhang D et al. Notch and Wnt signaling pathway in cancer: crucial role and potential therapeutic targets (Review). Int J Oncol 2016; 48: 437–449. [DOI] [PubMed] [Google Scholar]
- Bernemann C, Hülsewig C, Ruckert C, Schäfer S, Blümel L, Hempel G et al. Influence of secreted frizzled receptor protein 1 (SFRP1) on neoadjuvant chemotherapy in triple negative breast cancer does not rely on WNT signaling. Mol Cancer 2014; 13: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyanan A, Civenni G, Ciarloni L, Morel C, Mueller N, Lefort K et al. Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism. Proc Natl Acad Sci USA 2006; 103: 3799–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps J, Pitt JJ, Emons G, Hummon AB, Case CM, Grade M et al. Genetic amplification of the NOTCH modulator LNX2 upregulates the WNT/β-catenin pathway in colorectal cancer. Cancer Res 2013; 73: 2003–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H-A, Koo B-K, Cho J-H, Kim Y-Y, Seong J, Chang HJ et al. Notch1 counteracts WNT/β-catenin signaling through chromatin modification in colorectal cancer. J Clin Invest 2012; 122: 3248–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, Cheng P, King IN, Andersen P, Shenje L, Nigam V et al. Notch post-translationally regulates β-catenin protein in stem and progenitor cells. Nat Cell Biol 2011; 13: 1244–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peignon G, Durand A, Cacheux W, Ayrault O, Terris B, Laurent-Puig P et al. Complex interplay between β-catenin signalling and Notch effectors in intestinal tumorigenesis. Gut 2011; 60: 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodilla V, Villanueva A, Obrador-Hevia A, Robert-Moreno À, Fernández-Majada V, Grilli A et al. Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proc Natl Acad Sci USA 2009; 106: 6315–6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R, Ablett MP, Spence K, Landberg G, Sims AH, Clarke RB. Wnt pathway activity in breast cancer sub-types and stem-like cells. PloS One 2013; 8: e67811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkenburg KC, Graveel CR, Zylstra-Diegel CR, Zhong Z, Williams BO. Wnt/β-catenin signaling in normal and cancer stem cells. Cancers 2011; 3: 2050–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang G-B, Kim J-Y, Cho S-D, Park K-S, Jung J-Y, Lee H-Y et al. Blockade of Wnt/β-catenin signaling suppresses breast cancer metastasis by inhibiting CSC-like phenotype. Sci Rep 2015; 5: 12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci 2009; 66: 1631–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Täuber B, Dobner T, Bourteele S, Kostezka U, Adler G et al. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol Cell Biol 2001; 21: 7761–7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallberg AE, Pedersen K, Lendahl U, Roeder RG. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol Cell Biol 2002; 22: 7812–7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint Just RM, Hansson M, Wallberg A. A proline repeat domain in the Notch co-activator MAML1 is important for the p300-mediated acetylation of MAML1. Biochem J 2007; 404: 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulino A, Di Marcotullio L, Screpanti I. The multiple functions of Numb. Exp Cell Res 2010; 316: 900–906. [DOI] [PubMed] [Google Scholar]
- Boulter L, Govaere O, Bird TG, Radulescu S, Ramachandran P, Pellicoro A et al. Macrophage derived Wnt signalling opposes Notch signalling in a Numb mediated manner to specify HPC fate in chronic liver disease in human and mouse. Nat Med 2012; 18: 572–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M, Katoh M. NUMB is a break of WNT-Notch signaling cycle. Int J Mol Med 2006; 18: 517–521. [PubMed] [Google Scholar]
- Cheng X, Huber TL, Chen VC, Gadue P, Keller GM. Numb mediates the interaction between Wnt and Notch to modulate primitive erythropoietic specification from the hemangioblast. Development 2008; 135: 3447–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strazzabosco M, Fabris L. The balance between Notch/Wnt signaling regulates progenitor cells' commitment during liver repair: mystery solved? J Hepatol 2013; 58: 181–183. [DOI] [PubMed] [Google Scholar]
- Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med 2008; 14: 518–527. [DOI] [PubMed] [Google Scholar]
- Radvanyi L, Singh-Sandhu D, Gallichan S, Lovitt C, Pedyczak A, Mallo G et al. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci USA 2005; 102: 11005–11010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Langerød A, Ji Y, Nowels KW, Nesland JM, Tibshirani R et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell 2004; 15: 2523–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X-H, Wu Y, Yao S, Levine AC, Kirschenbaum A, Collier L et al. Androgens up-regulate transcription of the notch inhibitor numb in c2c12 myoblasts via wnt/β-catenin signaling to t cell factor elements in the numb promoter. J Biol Chem 2013; 288: 17990–17998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esufali S, Bapat B. Cross-talk between Rac1 GTPase and dysregulated Wnt signaling pathway leads to cellular redistribution of β-catenin and TCF/LEF-mediated transcriptional activation. Oncogene 2004; 23: 8260–8271. [DOI] [PubMed] [Google Scholar]
- Wu X, Tu X, Joeng KS, Hilton MJ, Williams DA, Long F. Rac1 Activation Controls Nuclear Localization of β-catenin during Canonical Wnt Signaling. Cell 2008; 133: 340–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps RA, Chidester S, Dehghanizadeh S, Phelps J, Sandoval IT, Rai K et al. A two-step model for colon adenoma initiation and progression caused by APC loss. Cell 2009; 137: 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myant KB, Cammareri P, McGhee EJ, Ridgway RA, Huels DJ, Cordero JB et al. ROS production and NF-κB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell stem cell 2013; 12: 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pethe VV, Charames GS, Bapat B. Rac1b recruits Dishevelled and β-catenin to Wnt target gene promoters independent of Wnt3A stimulation. Int J Oncol 2011; 39: 805–810. [DOI] [PubMed] [Google Scholar]
- Stathopoulos E, Sanidas E, Kafousi M, Mavroudis D, Askoxylakis J, Bozionelou V et al. Detection of CK-19 mRNA-positive cells in the peripheral blood of breast cancer patients with histologically and immunohistochemically negative axillary lymph nodes. Ann Oncol 2005; 16: 240–246. [DOI] [PubMed] [Google Scholar]
- Yang X-R, Xu Y, Shi G-M, Fan J, Zhou J, Ji Y et al. Cytokeratin 10 and cytokeratin 19: predictive markers for poor prognosis in hepatocellular carcinoma patients after curative resection. Clin Cancer Res 2008; 14: 3850–3859. [DOI] [PubMed] [Google Scholar]
- Chen T-F, Jiang G-L, Fu X-L, Wang L-J, Qian H, Wu K-L et al. CK19 mRNA expression measured by reverse-transcription polymerase chain reaction (RT-PCR) in the peripheral blood of patients with non-small cell lung cancer treated by chemo-radiation: an independent prognostic factor. Lung Cancer 2007; 56: 105–114. [DOI] [PubMed] [Google Scholar]
- Charafe-Jauffret E, Ginestier C, Monville F, Finetti P, Adelaide J, Cervera N et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene 2006; 25: 2273–2284. [DOI] [PubMed] [Google Scholar]
- Ju J, Oh S, Lee K, Yang W, Nam K, Moon H et al. Cytokeratin19 induced by HER2/ERK binds and stabilizes HER2 on cell membranes. Cell Death Differ 2014; 22: 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambang IF, Lu D, Li H, Chiu L-L, Lau QC, Koay E et al. Cytokeratin 19 regulates endoplasmic reticulum stress and inhibits ERp29 expression via p38 MAPK/XBP-1 signaling in breast cancer cells. Exp Cell Res 2009; 315: 1964–1974. [DOI] [PubMed] [Google Scholar]
- Kim H, Choi GH, Na DC, Ahn EY, Kim GI, Lee JE et al. Human hepatocellular carcinomas with ‘Stemness'-related marker expression: keratin 19 expression and a poor prognosis. Hepatology 2011; 54: 1707–1717. [DOI] [PubMed] [Google Scholar]
- Pui C-H. T cell acute lymphoblastic leukemia: NOTCHing the way toward a better treatment outcome. Cancer Cell 2009; 15: 85–87. [DOI] [PubMed] [Google Scholar]
- Zardawi SJ, Zardawi I, McNeil CM, Millar EK, McLeod D, Morey AL et al. High Notch1 protein expression is an early event in breast cancer development and is associated with the HER-2 molecular subtype. Histopathology 2010; 56: 286–296. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science 1999; 284: 770–776. [DOI] [PubMed] [Google Scholar]
- Bolos V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocr Rev 2007; 28: 339–363. [DOI] [PubMed] [Google Scholar]
- Wang Z, Banerjee S, Li Y, Rahman KW, Zhang Y, Sarkar FH. Down-regulation of Notch-1 inhibits invasion by inactivation of nuclear factor-κB, vascular endothelial growth factor, and matrix metalloproteinase-9 in pancreatic cancer cells. Cancer Res 2006; 66: 2778–2784. [DOI] [PubMed] [Google Scholar]
- Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc Natl Acad Sci USA 2008; 105: 6392–6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li Y, Banerjee S, Kong D, Ahmad A, Nogueira V et al. Down-regulation of Notch-1 and Jagged-1 inhibits prostate cancer cell growth, migration and invasion, and induces apoptosis via inactivation of Akt, mTOR, and NF-kappaB signaling pathways. J Cell Biochem 2010; 109: 726–736. [DOI] [PubMed] [Google Scholar]
- Carter S, Vousden KH. A role for Numb in p53 stabilization. Genome Biol 2008; 9: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Kaibuchi K. Numb controls integrin endocytosis for directional cell migration with aPKC and PAR-3. Dev Cell 2007; 13: 15–28. [DOI] [PubMed] [Google Scholar]
- Westhoff B, Colaluca IN, D'Ario G, Donzelli M, Tosoni D, Volorio S et al. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci USA 2009; 106: 22293–22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaczyn A, Bani-Yaghoub M, Tremblay R, Kubu C, Cowling R, Adams TL et al. Two novel human NUMB isoforms provide a potential link between development and cancer. Neural Dev 2010; 5: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem 2003; 278: 23196–23203. [DOI] [PubMed] [Google Scholar]
- Frise E, Knoblich JA, Younger-Shepherd S, Jan LY, Jan YN. The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc Natl Acad Sci USA 1996; 93: 11925–11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esufali S, Charames GS, Bapat B. Suppression of nuclear Wnt signaling leads to stabilization of Rac1 isoforms. FEBS Lett 2007; 581: 4850–4856. [DOI] [PubMed] [Google Scholar]
- Buongiorno P, Pethe VV, Charames GS, Esufali S, Bapat B. Rac1 GTPase and the Rac1 exchange factor Tiam1 associate with Wnt-responsive promoters to enhance beta-catenin/TCF-dependent transcription in colorectal cancer cells. Mol Cancer 2008; 7: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson C, Lui C, Brocardo MG, Martino-Echarri E, Henderson BR. Rac1 augments Wnt signaling by stimulating β-catenin–lymphoid enhancer factor-1 complex assembly independent of β-catenin nuclear import. J Cell Sci 2015; 128: 3933–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday DL, Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res 2011; 13: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien H, Hsiao Y, Lin Y, Yao Y, Juan H, Kuo W et al. Molecular signatures of metaplastic carcinoma of the breast by large-scale transcriptional profiling: identification of genes potentially related to epithelial–mesenchymal transition. Oncogene 2007; 26: 7859–7871. [DOI] [PubMed] [Google Scholar]
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 2005; 365: 671–679. [DOI] [PubMed] [Google Scholar]
- Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst 2006; 98: 262–272. [DOI] [PubMed] [Google Scholar]
- Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med 2007; 356: 217–226. [DOI] [PubMed] [Google Scholar]
- Buyse M, Loi S, Van't Veer L, Viale G, Delorenzi M, Glas AM et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst 2006; 98: 1183–1192. [DOI] [PubMed] [Google Scholar]
- Patsialou A, Wang Y, Pignatelli J, Chen X, Entenberg D, Oktay M et al. Autocrine CSF1R signaling mediates switching between invasion and proliferation downstream of TGFβ in claudin-low breast tumor cells. Oncogene 2015; 34: 2721–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir NN, Rönnstrand L, Kazi JU. Keratin 19 expression correlates with poor prognosis in breast cancer. Mol Biol Rep 2014; 41: 7729–7735. [DOI] [PubMed] [Google Scholar]
- Tee MK, Rogatsky I, Tzagarakis-Foster C, Cvoro A, An J, Christy RJ et al. Estradiol and selective estrogen receptor modulators differentially regulate target genes with estrogen receptors α and β. Mol Biol Cell 2004; 15: 1262–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moggs JG, Tinwell H, Spurway T, Chang H-S, Pate I, Lim FL et al. Phenotypic anchoring of gene expression changes during estrogen-induced uterine growth. Environ Health Perspect 2004; 112: 1589–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moggs JG, Ashby J, Tinwell H, Lim FL, Moore DJ, Kimber I et al. The need to decide if all estrogens are intrinsically similar. Environ Health Perspect 2004; 112: 1137–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo P, Miao H, D'Souza G, Osipo C, Yun J, Zhao H et al. Cross-talk between notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches. Cancer Res 2008; 68: 5226–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CW, Simin K, Liu Q, Plescia J, Guha M, Khan A et al. A functional Notch-survivin gene signature in basal breast cancer. Breast Cancer Res 2008; 10: R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-Q, Russo J. ERα-negative and triple negative breast cancer: molecular features and potential therapeutic approaches. Biochim Biophys Acta Rev Cancer 2009; 1796: 162–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Zhao X, Zhang X, Luo M, Zuo X, Huang S et al. Notch1 signaling regulates the epithelial–mesenchymal transition and invasion of breast cancer in a Slug-dependent manner. Mol Cancer 2015; 14: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon P, Trono D. Production and titration of lentiviral vectors. Curr Protoc Hum Genet 2007; 12.10: 1–24. [DOI] [PubMed] [Google Scholar]
- Dayem AA, Kim B, Gurunathan S, Choi HY, Yang G, Saha SK et al. Biologically synthesized silver nanoparticles induce neuronal differentiation of SH-SY5Y cells via modulation of reactive oxygen species, phosphatases, and kinase signaling pathways. Biotechnol J 2014; 9: 934–943. [DOI] [PubMed] [Google Scholar]
- Moon S-H, Kim D-K, Cha Y, Jeon I, Song J, Park K-S. PI3K/Akt and Stat3 signaling regulated by PTEN control of the cancer stem cell population, proliferation and senescence in a glioblastoma cell line. Int J Oncol 2013; 42: 921–928. [DOI] [PubMed] [Google Scholar]
- Leung W-H, Vong QP, Lin W, Janke L, Chen T, Leung W. Modulation of NKG2D ligand expression and metastasis in tumors by spironolactone via RXRγ activation. J Exp Med 2013; 210: 2675–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujawski M, Kortylewski M, Lee H, Herrmann A, Kay H, Yu H. Stat3 mediates myeloid cell–dependent tumor angiogenesis in mice. J Clin Invest 2008; 118: 3367–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua G, Liu Y, Li X, Xu P, Luo Y. Targeting glucose metabolism in chondrosarcoma cells enhances the sensitivity to doxorubicin through the inhibition of lactate dehydrogenase-A. Oncol Rep 2014; 31: 2727–2734. [DOI] [PubMed] [Google Scholar]
- Shaw FL, Harrison H, Spence K, Ablett MP, Simões BM, Farnie G et al. A detailed mammosphere assay protocol for the quantification of breast stem cell activity. J Mammary Gland Biol Neoplasia 2012; 17: 111–117. [DOI] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007; 131: 1109–1123. [DOI] [PubMed] [Google Scholar]
- Lee JM, Kim IS, Kim H, Lee JS, Kim K, Yim HY et al. RORα attenuates Wnt/β-catenin signaling by PKCα-dependent phosphorylation in colon cancer. Mol Cell 2010; 37: 183–195. [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Konno M, Hamabe A, Hasegawa S, Kano Y, Fukusumi T et al. Surgically resected human tumors reveal the biological significance of the gastric cancer stem cell markers CD44 and CD26. Oncol Lett 2015; 9: 2361–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JP, Goldwasser R, Mark S, Schwartz J, Orion I. A mathematical model for tumor volume evaluation using two-dimensions. J Appl Quant Methods 2009; 4: 455–462. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.