Abstract
Zika virus is an emerging arthropod-borne flavivirus for which there are no vaccines or specific therapeutics. We screened a library of 2000 ‘bioactive’ compounds for their ability to block Zika virus infection in three distinct cell-types with two different strains of Zika virus. Using a microscopy-based assay, we validated 38 drugs that inhibited Zika virus infection, including FDA approved nucleoside analogs. Cells expressing high levels of the attachment factor AXL can be protected from infection with receptor tyrosine kinase inhibitors, while placental-derived cells that lack AXL expression are insensitive to this inhibition. Importantly, we identified nanchangmycin as a potent inhibitor of Zika virus entry across all cell types tested including physiologically relevant primary cells. Nanchanmycin was also active against other medically relevant viruses including West Nile, dengue, and chikungunya virus that use a similar route of entry. This study provides a resource of small molecules to study Zika virus pathogenesis.
Introduction
Zika virus (ZIKV) is a flavivirus transmitted by Aedes mosquitoes including A. aegypti which is globally widespread. The virus is rapidly spreading through South America, Central America, and the Caribbean. Epidemiologists predict that the virus will continue to spread widely in the coming months across the Western Hemisphere including the continental US, and locally-transmitted virus has now been detected in several areas in Florida. While ZIKV was discovered in Uganda in 1947 and there have been smaller outbreaks of ZIKV infection in the ensuing years, there has never been a widespread explosive pandemic like the one that is occurring presently. The World Health Organization has declared that the current ZIKV outbreak is a global emergency, due to the rapid spread of the virus and the association of viral infection with microcephaly (Oliveira Melo et al., 2016; Schuler-Faccini et al., 2016; Tetro, 2016). It is not understood why the 2015–2016 ZIKV pandemic has been so explosive, although the fact the virus is now endemic in immunologically naïve populations likely contributes. Given the public health crisis associated with the ongoing outbreak, new strategies must be identified rapidly to prevent or treat these infections. In most individuals, ZIKV-induced disease is mild or asymptomatic, however, during the most recent outbreak, the virus has been linked to dramatic increases in microcephaly and other congenital abnormalities as well as being linked to Guillain-Barre syndrome (Araujo et al., 2016; Coyne and Lazear, 2016).. Furthermore, recent data demonstrates sexual transmission and that the testes can harbor infectious virus for long periods (D’Ortenzio et al., 2016; Turmel et al., 2016).
There are no therapeutics or vaccines approved to treat ZIKV infection. The majority of successful antivirals have been developed against specific, well studied viral enzymes; however, this target-based approach does not interrogate other possible targets, including cellular factors essential for infection. Furthermore, the development of viral-enzyme targeted drugs takes years to develop. Targeting cellular factors may be advantageous because such treatments are less liable to be evaded by the high mutation rate of viral genomes. Indeed, there are a large number of drugs that target diverse human proteins that have been developed for use in humans, allowing them to be rapidly repurposed, bypassing the early stages of the development and safety testing in humans. Since all flaviviruses use similar cellular pathways for entry, translation and replication, host targeted therapies against ZIKV may also have utility against other flaviruses, such as dengue virus which is co-circulating in many areas (Blitvich and Firth, 2015; Lessler et al., 2016; Monaghan et al., 2016)
ZIKV displays broad tissue tropism which can result in diverse outcomes including fetal transmission as well as long term infection of the testes(Lazear and Diamond, 2016). Experimentally, it has been shown that many cell types are permissive to the virus, although it remains unclear how the virus is vertically transmitted. Since diverse cell types are targeted by ZIKV, it is important to identify inhibitors that are active in a wide range of cell types, particularly those that might impact vertical transmission or the establishment of a viral reservoir in the testes or other sites. To identify potent inhibitors of ZIKV infection we adapted a high-content 384 well-based assay for small molecule screening against ZIKV. We screened a library of existing FDA approved (~1000) and known bioactives (~1000) in three different cell types with two different strains of ZIKV to uncover inhibitors of viral infection. Downstream validation studies and dose-responses studies determined the efficacy and potency of the drugs for their ability to block ZIKV infection as well as their toxicity across cell types. Further study in physiologically relevant primary cells as well as mechanism-of-action studies reveal the dependencies of ZIKV in distinct cellular environments. Importantly, we discovered that AXL inhibitors block infection in cells that express high levels of AXL but not in cells devoid of the AXL expression. In addition, we found that nanchangmycin, a bacterially-derived natural product that has not been tested clinically, potently blocked ZIKV infection across diverse cell types. Studies with dengue virus (DENV) revealed the similarities in host factor dependencies of this closely related virus, which was also sensitive to nanchangmycin. Since we found that nanchangmycin blocks ZIKV entry, as well as DENV infection, and that these viruses use similar host pathways for entry, we tested whether West Nile virus (WNV), an additional flavivivirus, could be inhibited by nanchangmycin. We found that all three flaviviruses (ZIKV, DENV, WNV) were sensitive to nanchangmycin. Since alphaviruses use a similar pathway for entry to the flaviviviruses, we tested chikungunya virus (CHIKV) and sindbis virus (SINV) and found that these viruses could also be inhibited by nanchangmycin while parainfluenza virus 5, which enters by a completely different route, was not blocked by this drug. Altogether, these studies reveal insights into ZIKV infection and will ultimately inform strategies for antiviral interventions against this important emerging human pathogen.
Results
Screen for inhibitors of ZIKV infection in human transformed osteosarcoma cells
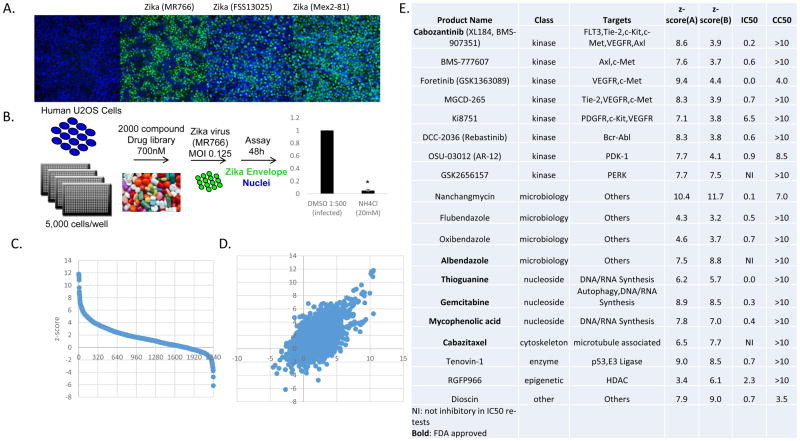
All viruses, including flaviviruses, must utilize a large number of cellular factors to complete their replication cycle in host cells. We and others have performed high-throughput genetic screens for host factors required for flavivirus infection (Hackett et al., 2015; Yasunaga et al., 2014; Zhang et al., 2016). Given that ZIKV is closely related to other flaviviruses we have screened (WNV, DENV), we adapted our previously published high-throughput image-based screening strategies to interrogate ZIKV dependencies in human cells. Since there are no approved vaccines or therapeutics to treat ZIKV we began by screening a library of ~2000 compounds of FDA approved drugs and known ‘bioactives’ for their ability to block ZIKV infection. We optimized this assay in human osteosarcoma cells (U2OS) since we have performed many screens in this cell type and these cells are permissive to all arthropod-borne viruses we have tested (Moy et al., 2014; Panda et al., 2013; Yasunaga et al., 2014) We first tested whether these cells were permissive to ZIKV infection using 3 different strains of ZIKV: the prototype Uganda/African strain MR766 from 1947, the FSS13025 Cambodian/Asian isolate from 2010, and an Americas-derived virus isolated in 2016, MEX 2-81. U2OS cells were susceptible to infection by all three strains of virus (Figure 1A).
Figure 1.
19 drugs robustly block ZIKV infection in human U2OS cells. (A) U2OS cells were infected with three different strains of ZIKA virus and the infection was monitored by microscopy (anti-flavivirus envelope, 4G2, green; nuclei, blue). (B) Schematic of the screening strategy with ZIKV (MR766) where ammonium chloride (NH4Cl) was used as a positive control. (C) Z scores of the percent infection of replicate 1. (D) Z scores of replicate 1 plotted against replicate 2. (E) The 19 drugs that inhibit infection with Z-scores>3, impacted infection >3-fold that are nontoxic. IC50 values for ZIKV (Mex2-81) (uM) and CC50 values (uM) shown.
We performed the screen using the prototypical ZIKV strain from Africa (MR766) optimizing assay conditions using the lysosomotropic inhibitor ammonium chloride (NH4Cl) which blocks acidification of the endo-lysosomal network thereby blocking fusion of flaviviruses schematized in Figure 1B (Randolph et al., 1990). Human U2OS cells were plated and subsequently treated with each drug in the library at 700nM as this dose minimizes off-target effects and toxicity observed when screening at higher concentrations. We also included a positive control (NH4Cl) and a vehicle control (0.1% DMSO). We pretreated cells for 1hr and infected with ZIKV (MOI 0.125) for 48h and subsequently fixed and processed for automated microscopy using an antibody against the viral glycoprotein (4G2). Automated imaging and automated image analysis were used to quantify cell number and the percentage of cells that were productively infected in four sites per well. This assay design identifies drugs that inhibit any stage in viral infection as we observe viral spread by 48h. We calculated z-scores for the percent infection shown in Figure 1C and performed the screen in duplicate (Figure 1D). The full datasets are in Table S1. Since we performed this assay by microscopy we also monitored the cell number in each well as a surrogate for cell viability.
Most drugs showed little to no effect on either ZIKV infection or cell density. In contrast, 19 drugs significantly inhibited ZIKV infection (z-score ≥3.00 and ≥3-fold decrease in infection) with modest to no toxicity (>60% of cells remaining) in both replicates of the screen (Figure 1E). We identified nucleosides which are known to block flavivirus infection including mycophenolic acid and gemcitabine (de Wispelaere et al., 2013a; Diamond et al., 2002). We identified 8 kinase inhibitors of which 5 are receptor tyrosine kinase inhibitors. This included the only two kinase inhibitors that are known to target AXL that are in the screening library we used. It has been established that AXL can function as an attachment factor for many flaviviruses including ZIKV (Hamel et al., 2016; Meertens et al., 2012) suggesting that these receptor tyrosine kinase inhibitors were blocking infection at the step of viral entry into these cells.
We re-purchased these drugs to validate their activity and performed dose-responses to determine their IC50s. Furthermore, we performed this validation analysis with a strain of ZIKV currently circulating in the Americas (Mex2-81) at 24h post infection, a time where there is no spread allowing us to focus only on inhibitors active during early steps in the viral lifecycle against two distinct strains of ZIKV. We validated 17 of these drugs and determined their potencies by IC50 analysis (Figure 1E), FDA approved drugs are shown in bold. In addition, we tested the toxicity of these drugs in the absence of infection using resazurin (Alamar blue) and report the CC50s (Figure 1E).
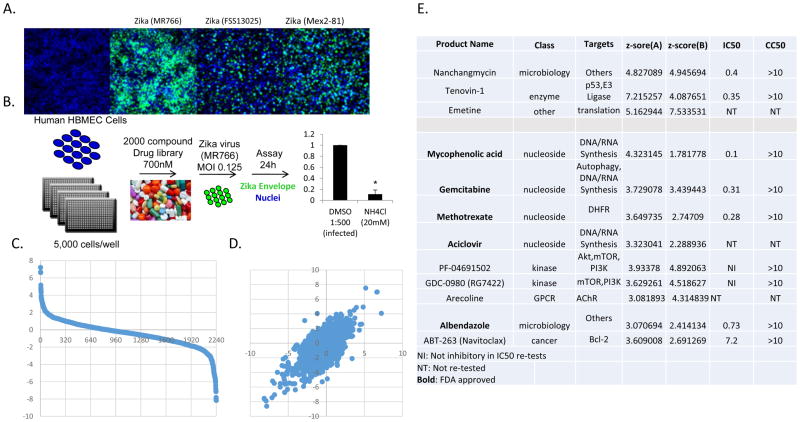
Screen for inhibitors of ZIKV infection in human brain microvascular endothelial cells
Since ZIKV can access the fetal brain during gestation, the virus may cross the blood-brain barrier, which is composed in part by microvascular endothelial cells. Therefore, we directly screened the inhibitor library in human microvascular endothelial cells (HBMEC), an immortalized model of the human blood-brain barrier microvasculature (Figure 2A). Again, NH4Cl was used as a positive control (Figure 2B) and the screen was performed using ZIKV (MR766, MOI 0.125) for 24h in duplicate (Figure 2B). This assay design identifies drugs that inhibit viral entry, translation, or RNA synthesis, but not late stages of infection including assembly and egress as we observe no spread at this time point. We calculated z-scores for the percent infection shown in Figure 2C and performed the screen in duplicate with good concordance (Figure 2D). The full datasets are in Table S2. 3 drugs inhibited infection by >3-fold with a z-score ≥3 and exhibited modest to no toxicity (>60% of cells remaining) in both replicates of the screen. Two of these compounds also inhibited ZIKV in U2OS cells. In addition, at a >2-fold decrease in infection, 9 additional drugs emerged, and included 4 nucleosides as well as albendazole which also inhibited in U2OS cells.
Figure 2.
12 drugs robustly block ZIKV infection in human HBMEC cells. (A) HBMEC cells were infected with three different strains of ZIKV virus and the infection was monitored by microscopy (anti-flavivirus envelope, 4G2, green; nuclei, blue). (B) Schematic of the screening strategy where ammonium chloride (NH4Cl) was used as a positive control. (C) Z-scores of the percent infection of replicate 1. (D) Z-scores of replicate 1 plotted against replicate 2. (E) The 12 drugs that inhibit infection >3-fold that are nontoxic. IC50 values for ZIKV (Mex2-81) (uM) and CC50s are shown
We repurchased the drugs to validate the activity of these compounds and performed dose-responses for 9 drugs using the circulating strain of ZIKV (Mex2-81) at 24h post infection allowing us to identify inhibitors active during early steps in the viral lifecycle against two distinct strains of ZIKV. We found that 7 potently inhibited infection (Figure 2E), FDA approved compounds are shown in bold. The validated set includes the nucleoside analogs as well as albendazole, tenovin-1 and nanchangmycin which all also inhibited in U2OS cells. In addition, we tested the toxicity of these drugs in the absence of infection using resazurin (Alamar blue) and report the CC50s (Figure 2E).
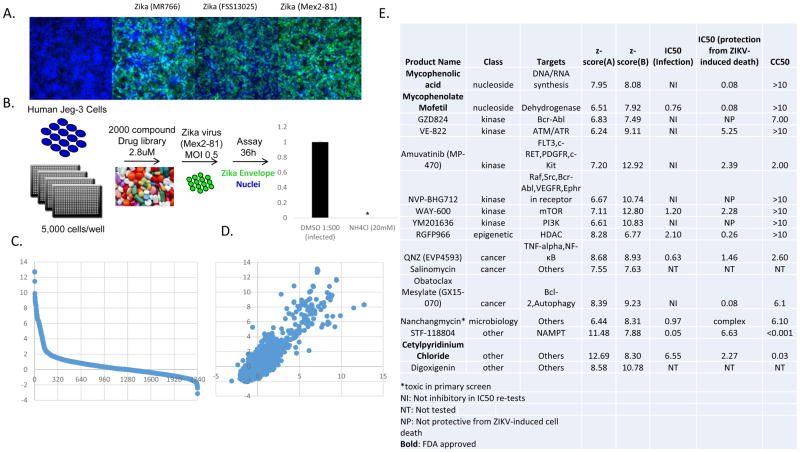
Screen for inhibitors of ZIKV infection in human placental trophoblast cells
The major complication associated with the ongoing ZIKV outbreak is severe congenital anomalies of the fetus during gestation. However, it remains unclear how the virus is vertically transmitted. Recent studies have shown that primary placental syncytiotrophoblasts isolated from full term placentas are refractory to infection due to the production of type III interferons and that these cells also resist infection in the first trimester(Bayer et al., 2016; Tabata et al., 2016). However, many commonly used trophoblast cell lines, most of which are derived from choriocarcinomas and serve as models of mononuclear cytotrophoblasts, are permissive to infection(Bayer et al., 2016; Tabata et al., 2016). Given that the syncytiotrophoblast layer could be breached by ZIKV by a number of mechanisms, such as breaks in the syncytium, and could thus reach the underlying cytotrophoblast layer that is more permissive to ZIKV, we also optimized a screen for inhibitors active in the human trophoblast cell line Jeg-3, which serve as a model of cytotrophoblasts (Figure 3A). Jeg-3 cells are highly permissive to ZIKV infection (all three strains tested) with prominent signs of virus-induced cell death not observed in the other cell lines tested (not shown). We performed the screen with ZIKV (Mex2-81, MOI=0.5) for 36h optimized with NH4Cl (Figure 3B). This assay design identifies drugs that inhibit any stage in viral infection as we observe viral spread by 36h. The screen was performed at 2.8uM, and we calculated z-scores for the percent infection shown in Figure 3C and performed the screen in duplicate (Figure 3D). The full datasets are in Table S3.
Figure 3.
16 drugs robustly block ZIKV infection in human Jeg-3 cells. (A) Jeg-3 cells were infected with three different strains of ZIKV and the infection was monitored by microscopy (anti-flavivirus envelope, 4G2, green; nuclei, blue). (B) Schematic of the screening strategy where ammonium chloride (NH4Cl) was used as a positive control. (C) Z-scores of the percent infection of replicate 1. (D) Z-scores of replicate 1 plotted against replicate 2. (E) The 15 drugs that inhibit infection >3-fold that are nontoxic. IC50 values for ZIKV (Mex2-81) (uM), IC50s for protection from ZIKV-induced death for ZIKV (Mex2-81) (uM), and CC50 values are shown.
Using this screening strategy we identified 14 drugs that led to a >3-fold decrease in infection (z-score >3) with low to modest toxicity (>50% of cells remaining) in both replicates of the screen (Figure 3E). We re-purchased 12 drugs and performed dose-responses with ZIKV (Mex2-81) at 24h post infection, a time where there is no spread allowing us to focus only on inhibitors active during early steps in the viral lifecycle against two distinct strains of ZIKV. We found that 7 are antiviral and the IC50s are shown. Since ZIKV infection in these cells leads to virus-induced cell death, we also analyzed the potency of the drugs for protection from infection-induced toxicity and found that 11 drugs were protective and their IC50s for protection is shown (Figure 3E). FDA approved compounds shown in bold. Nanchangmycin was modestly toxic in the primary screen but very efficacious in inhibiting viral infection. Further analysis revealed that nanchangmycin blocks ZIKV infection with low toxicity in the sub-micromolar range (Figure S1). In addition, we tested the toxicity of these drugs in the absence of infection using resazurin (Alamar blue) and report the CC50s (Figure 3E).
Receptor Tyrosine Kinase inhibitors are cell type specific
Kinases are the master regulators of signaling events in cells and play important roles in many viral infections (Ramage and Cherry, 2015), and kinase inhibitors have been shown to have activity against flaviviruses (Chu and Yang, 2007; de Wispelaere et al., 2013b; Zhang et al., 2012). Recent studies have shown that the receptor tyrosine kinase AXL can function as an attachment factor for many viruses including flaviviruses such as DENV and more recently, ZIKV (Hamel et al., 2015; Meertens et al., 2012; Savidis et al., 2016). Indeed, we identified and validated 5 receptor tyrosine kinase inhibitors in U2OS cells that included the only two inhibitors annotated as targeting AXL in our screening library. Many kinase inhibitors bind to the highly conserved ATP binding site. For this reason, many kinase inhibitors are not exclusively selective for their intended targets and it is likely that this group of inhibitors are acting on AXL.
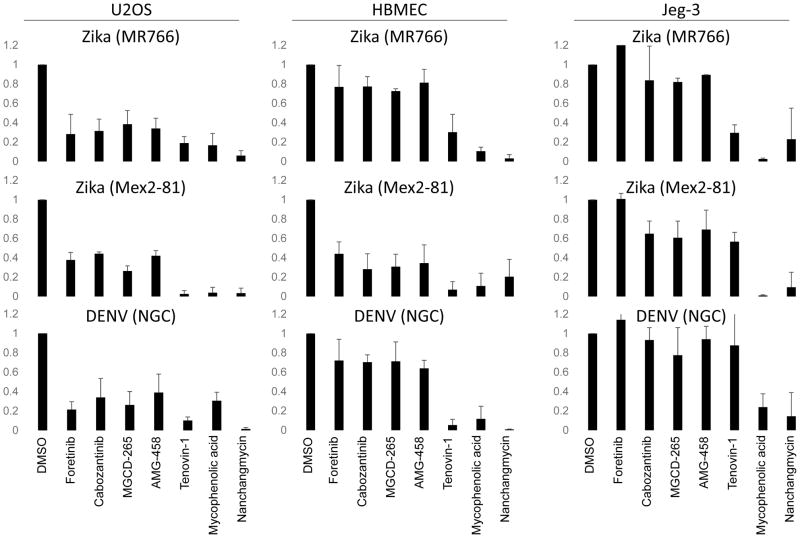
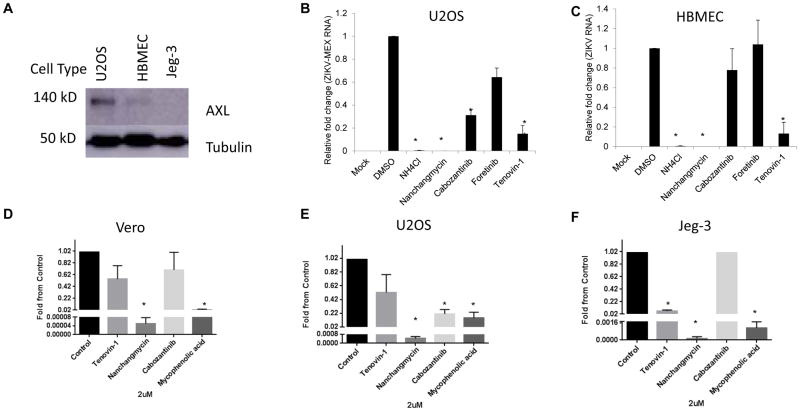
We set out to determine the breadth of the antiviral activity of these kinase inhibitors by testing their activity in the three cell types against two strains of ZIKV (MR766 and Mex2-81) outside of the screening platform. While these drugs block the three ZIKV strains tested in U2OS cells (Figure 4, not shown), they have a mild phenotype in HBMEC cells and no activity in Jeg-3 cells (Figure 4). This reflects the expression levels of AXL: immunblot analysis reveals highest levels in U2OS cells with intermediate expression in HBMEC and no detectable expression in Jeg-3 cells as has been recently reported (Tabata et al., 2016) (Figure 5A). Since Jeg-3 cells are the highly permissive to ZIKV these data suggest that AXL expression does not define tropism for this virus.
Figure 4.
Anti-ZIKV inhibitors show activity against DENV. The indicated cell lines were treated with the indicated drugs at 2uM one hour prior to infection with the indicated flaviviruses and the percent infection was quantified using automated microscopy from 3 independent experiments with mean±SD of the fold change in infection shown.
Figure 5.
Receptor tyrosine kinase inhibitors block infection only in cells that express high levels of the entry factor AXL. (A) The indicated cell lines were processed for immunoblot for AXL and the control tubulin. A representative experiment is shown. U2OS (B) or HBMEC (C) cells were treated with the indicated drugs and infected with ZIKV (Mex2-81, MOI=2.5) and processed for RT-qPCR analysis to monitor the level of infection from 3 independent experiments with mean±SD shown. (D–F) The indicated cell type was treated with the indicated drugs and infected with ZIKV (Mex2-81) and TCID50s were calculated for 3 independent experiments with mean±SD shown. *p,0.05.
Furthermore, studies have shown that DENV can also use AXL as an attachment factor and thus we tested DENV (serotype 2; New Guinea C) and found that this trend was similar, with receptor tyrosine kinase inhibitors having the strongest effect in U2OS. These data also suggest that in cells that express AXL, inhibitors of AXL activity significantly reduce infection.
Nanchangmycin potently blocks ZIKV infection across cell types
While the receptor tyrosine kinase inhibitors targeting AXL showed cell type specificity, the strongest inhibitor we identified was nanchangmycin. This drug potently reduces infection of all three strains of ZIKV across all three cell types (Figures 1–4). The IC50s for infection are between 0.1 and 0.4 uM while nanchangmycin has low toxicity in these ranges (Figure S1). In addition, we found that DENV is inhibited by nanchangmycin across cell types (Figure 4).
In addition to the microscopy-based assay we also tested for inhibition using a RT-qPCR assay, which validated that nanchangmycin blocked infection (Fig 5B–C) while the receptor tyrosine kinase inhibitors were active in U2OS but not HBMEC cells. Furthermore, we determined the impact of these drugs on infectious virus production in diverse cell lines. In Vero cells we observed a 4-log reduction in viral titers upon treatment with nanchangmycin (Figure 5D, Figure S2). This was greater than the inhibition observed with mycophenolic acid. We observed a similar trend in U2OS and Jeg-3 cells (Figure 5E–F, Figure S2). By this assay, cabozantinib modestly impacted infection in U2OS cells but had no activity in the other cell types tested.
Nanchangmycin blocks viral entry
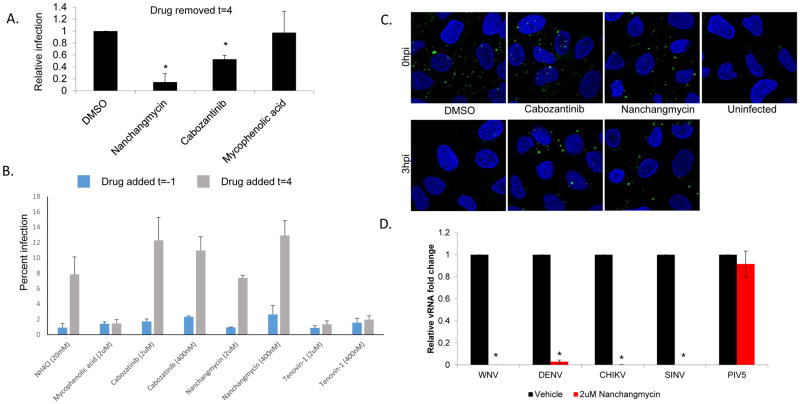
Nanchangmycin is a natural product of Streptomyces nanchangensis that was shown to have insecticidal activity against silkworms and anti-bacterial activity in vitro (Ouyang L, 1993; Sun et al., 2002). However, there is little known about this drug and its potential mechanism of action against ZIKV. We set out to determine if the drug was blocking a pre- or post-entry step in the viral life cycle. We treated cells with nanchangmycin, the AXL inhibitor cabozantinib or nucleoside mycophenolic acid at the time of challenge and removed the drug at 4 hpi, a time after entry has occurred. The level of infection was monitored 24h later by automated microscopy. Both the AXL inhibitor and nanchangmycin inhibited infection when removed 4hpi while mycophenolic acid, which is known to block RNA replication downstream of entry, was not inhibitory when removed at 4hpi (Figure 6A). These data suggest that cabozantinib, as expected, and nanchangmycin, block an early step in the viral lifecycle.
Figure 6.
Nanchangmycin potently blocks infection at the level of entry. (A) U2OS cells were treated with the indicated drugs and infected with ZIKV (Mex2-81, MOI 25), 4 hpi the drugs were washed out and the cells were processed for immunofluorescence 24hpi from 3 independent experiments with mean±SD shown. (B) U2OS cells were treated with the indicated drugs either one hour prior to infection (blue) or 4hpi (grey) with ZIKV (Mex2-81, MOI=1). NH4Cl was added at 4hpi to block further entry. The infection was processed at 24hpi for microscopy and the percent infection shown for a representative experiment of three. (C) U2OS cells were treated with the indicated drugs and then infected with ZIKV (Mex2-81, MOI=100) at 4C for one hour for binding or released at 37C for 3h to allow uptake. The cells were fixed without permeablization to monitor extracellular virions (anti-envelope, green; nuclei, blue). Images were taken by confocal microscopy and are representative of 3 independent experiments. (D) U2OS cells were treated with vehicle or nanchangmycin (2uM) and infected with the indicated viruses for 24h and processed for RT-qPCR with mean±SD shown. * p<0.05.
If nanchangmycin was only inhibiting during entry we reasoned that if we added the drug after entry, at 4hpi, we would observe no inhibition. Indeed, addition of NH4Cl before infection was inhibitory while addition at 4hpi had no impact on infection. In contrast, mycophenolic acid, which blocks viral replication downstream of entry, remained inhibitory even when added 4hpi (Figure 6B). Under these conditions, we found that both cabozantinib, as expected, and nanchangmycin were only active when added during the entry stages of infection (Figure 6B).
These data suggest that nanchangmycin is an inhibitor of viral entry. Entry is dependent on viral binding to cells as well as viral uptake. We adapted a fluorescence-based uptake assay which we previously validated for WNV entry (Hackett et al., 2015). For these studies we treated U2OS cells with the indicated drugs and then prebound ZIKV to cells at 4C (t=0) or released the cells to allow viral uptake at 37C for 3h. We then fixed the cells without permeabilization to exclusively monitor extracellular virions. Under these conditions we observed punctate staining at t=0 which was lost at 3hpi in the vehicle control treated cells (Figure 6C). While cabozantinib and nanchangmycin had no apparent impact on ZIKV binding, both drugs blocked uptake as virions remained on the cell surface (Figure 6C).
Since nanchangmycin blocked ZIKV uptake as well as infection by ZIKV and DENV we reasoned that it may be blocking the internalization pathway used by these viruses which is clathrin-mediated endocytosis (Pierson and Kielian, 2013). Therefore, we tested whether nanchangmycin could block infection by an additional flavivirus (WNV, Kunjin) or alphaviruses (Chikungunya (CHIKV, 181/25), Sindbis (SINV), HRsp) which all use clathrin-mediated endocytosis for entry. We found that each virus was sensitive to nanchangmycin (Figure 6D). In addition, we tested parainfluenza 5 (PIV5) which does not depend on clathrin-mediated endocytosis for entry and found that nanchangmycin does not block infection of this virus (Figure 6D).
These data suggest that nanchangmyin blocks clathrin-mediated endocytosis. Therefore, we tested whether uptake of transferrin, which is a classic endogenous cargo dependent on clathrin-mediated endocytosis for internalization, was impacted by nanchangmycin. While we found that treatment with dynasore, a known inhibitor of this pathway, completely abrogated uptake, nanchangmycin had no impact on transferrin internalization (Figure S3). This suggests that nanchangmycin is not broadly blocking clathrin-mediated endocytosis but rather blocking a virus-specific aspect of uptake.
Nanchangamycin blocks ZIKV infection of primary cells
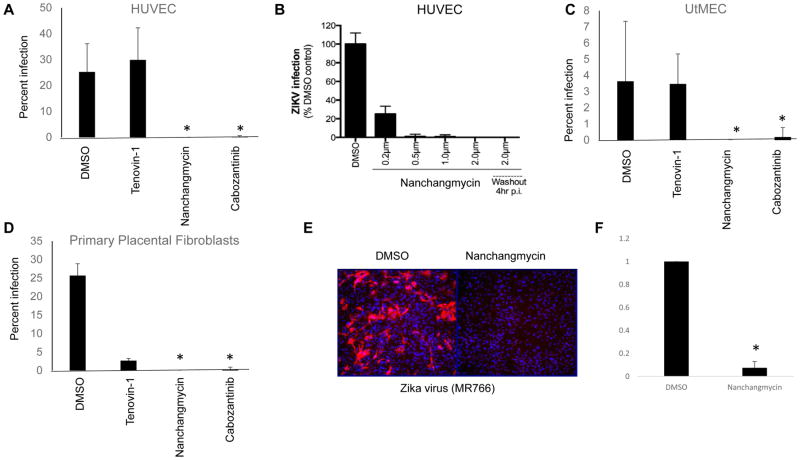
Given how efficacious nanchangmycin was observed to be in Vero, U2OS, HBMEC and Jeg-3 cells, we set out to test its efficacy in other primary cells thought to be relevant to ZIKV infection. Previous studies have shown that ZIKV might access the fetal compartment via infection of extravillous trophoblasts (EVTs), which are buried in the maternal decidua and anchor the placenta to the uterine wall (Tabata 2016). In order to access these cells, ZIKV might directly replicate in the maternal microvasculature, which are in close proximity to EVTs. Therefore, we utilized human primary uterine microvascular endothelial cells (UtMEC). Once ZIKV has breaches the placental barrier, it then targets cells types located in either the core of the villous trees of the human placenta such as placental fibroblasts (Jurado et al., 2016) and also likely targets fetal microvasculare cells to reach the fetal systemic circulation. Therefore, we also tested the effects of compounds in primary placental fibroblasts and in human umbilical vein endothelial cells (HUVEC). Similar to our results in HBMEC, cabozantinib and nanchangmycin effectively blocked ZIKV infection in HUVEC cells (Figure 7A). Moreover, serial dilutions of nanchangmycin revealed high potency in these cells (Figure 7B). Furthermore, when nanchangmycin was removed at 4hpi, it remained potently antiviral in HUVEC, further suggesting a role for entry in this cell type. Similar to HUVECs, we found that cabozantinib and nanchangmycin effectively blocked ZIKV infection in UtMECs (Figure 7C). Finally, we found that tenovin-1, nanchangmycin and cabozantinib all potently inhibited ZIKV infection in primary placental fibroblast cells (Figure 7D)
Figure 7.
Nanchangmycin blocks ZIKV infection in primary cells. (A–B) HUVEC, (C) UtMEC, (D) or primary placental fibroblasts were treated with the indicated drugs and infected by ZIKV (FSS13025, MOI=1) and processed for immunofluorescence at 48 hpi. The percentage of infected cells was calculated and mean±SD shown; *p<0.05. (E–F) Murine mixed neuronal cultures were infected with ZIKA (MR766, MOI=2) for 24hr and processed for immunofluorescence. (E) Representative images shown for 3 independent experiments quantified in (F) with mean±SD shown; *p<0.05.
Given that ZIKV targets the fetal brain, we generated primary midbrain neuron-glia mixed cultures from embryonic mice (Gao, 2012; Gao et al., 2002) and validated that these cultures contained both neurons and glia (Figure S4A). Furthermore, we found that they are permissive to infection by ZIKV (Mex2-81) with infection largely of neurons (Figure S4B). We found that nanchangmycin efficiently blocks infection of ZIKV (Figure 7E–F). We also found that DENV (NGC) and CHIKV (181/25) infect these cultures and that infection is blocked by nanchangmycin (Figure S5) These data suggest that nanchangmycin is a potent inhibitor of flavivirus infection in diverse and primary cell types.
Discussion
The ZIKV pandemic is continuing to have a global impact, yet there are a dearth of therapeutic options available due to both the lack of anti-flavivirals and the fact that vaccine trials are in their infancy. Here we developed and performed three high-throughput screening assays using human cells to identify inhibitors of ZIKV infection and replication. We screened ~2000 compounds and identified 38 small molecules that block ZIKV infection many with potencies in the sub-micromolar range.
AXL is known to promote flavivirus infection and we identified inhibitors of AXL that block infection in U2OS cells as well as multiple primary cell types that likely express high levels of AXL. Some of these drugs, including cabozantinib, are in clinical use for cancer and are contraindicated during pregnancy. Nevertheless, it is striking that diverse primary cells such as HUVEC, UtMEC and placental fibroblasts could all be protected by AXL inhibitors. However, we also found that cells that do not express detectable levels of AXL, such as Jeg-3 cells, are not protected by these drugs. Moreover, a recent study showed no change in ZIKV titers in adult tissues of AXL knock out mice (Miner et al.). Collectively, these studies raise important questions as to the nature of the contribution of AXL to infection in vivo, and whether cell types that do not express AXL play important roles in pathogenesis.
While the majority of the compound collection that we screened has known targets, the most efficacious inhibitor we identified was nanchangmycin which inhibits ZIKV infection in every cell type we tested. Nanchangmycin is a polyether produced by Streptomyces nanchangensis (Sun et al., 2002). Nanchangmycin is structurally related to dianemycin and has been found to inhibit gram-positive bacteria and can be used as a growth promotant in poultry and to cure coccidiosis in chickens (Hamill et al., 1969). We do not know what the cellular target for nanchangmycin is. Our mechanistic studies found that nanchangmycin blocks an early step in the entry process of ZIKV. Since ZIKV, DENV, WNV, CHIKV and SINV use clathrin-mediated endocytosis for internalization, our data suggest that nanchangmycin may be blocking an early step in this entry process. However, when we tested the canonical endogenous cargo transferrin, we found that nanchanmycin did not block its uptake. This suggests that nanchangmycin may be selectively inhibiting the uptake of larger cargo or the particular protein receptors used by these viruses, which are largely unknown. Future studies will reveal the full spectrum of viruses that can be inhibited by nanchangmycin and whether entry inhibitors, which would prevent spread but not block replication in cells that were already infected, would be effective to treat these infections. Nevertheless, our discovery of an antiviral active against this large number ofimportant emerging viruses suggest that we may be able to block infection with such a therapeutic. This drug has not been tested in animals and thus future studies in animal models of infection are critical to test the efficacy and toxicity in vivo.
While the treatment of pregnant women is a major goal to block the vertical transmission of ZIKV, this is a difficult population to treat. However, non-pregnant individuals such as those at high risk for complications may also benefit from the identification of anti-ZIKV drugs. For example, an emerging literature demonstrates that ZIKV is sexually transmitted with a reservoir in the testes that remains active for long periods of time (Barzon et al., 2016; Nicastri et al., 2016). It is important to note that although ZIKV may be vertically transmitted by the hematogenous route, it is also possible that ascending infection following sexual intercourse with an infected male may transfer the virus to the developing fetus. Thus, the identification of antivirals that could be used in the non-pregnant population to suppress viral loads is also a vital component in the public health response to ZIKV. Furthermore, since nanchangmycin is a broad spectrum antiviral, it may ultimately be used to treat these other infections in non-pregnant individuals.
We identified a number of additional antivirals, including a number of nucleoside analogs, which are known to inhibit flavivirus infection (Diamond et al., 2002; Yin et al., 2009). Indeed, recent studies have shown that nucleoside inhibitors are active against ZIKV (Barrows et al., 2016; Hamel et al., 2015; Shan et al., 2016; Zmurko et al., 2016). In addition, we identified a number of other drugs that target diverse proteins and pathways and are active against ZIKV. However, most of these showed cell type specificity, thus making them potentially less useful as therapeutics, but informative about the biology of ZIKV infection, tropism and pathogenesis. For example, we have identified the group of antihelmithic agents of the benzimadole group including albendezole and oxibendazole that prevent ZIKV infection in U2OS and HBMEC cells but not Jeg-3 cells. Understanding the mechanism of action of these drugs will reveal important insights into the host factor dependencies during infection. This could also highlight additional targets of these drugs, that if understood may facilitate development of additional analogs to prevent these interactions with host proteins to decrease the toxicity of these drugs for use during helminth infections.
An understanding of the cell type specificity of antiviral drugs that target cellular factors is important. Indeed, a recent study in the human hepatocyte cell line Huh-7 using a similar assay at 10uM, identified 24 compounds that inhibited ZIKV (Barrows et al., 2016). Our screening set included 16 of these compounds, and we observed antiviral activity with the 4 nucleoside analogs. However, we did not observe antiviral activity for the other 12 drugs which is likely due to differences in the concentrations of drugs used. Barrows et al found that most of their inhibitors were active only at 10uM, a concentration well above our studies and thus would be missed in our primary screen. Another recently published screen for inhibitors of ZIKV-induced cell death identified two drugs that attenuated ZIKV replication, niclosamide and a CDK inhibitor PHA-690509 (Xu et al., 2016). We found that niclosamide as well as a panel of CDK inhibitors present in our screening library were toxic in the cell types we tested and thus were excluded from our analysis. Collectively, these studies suggest that host-targeted inhibitors may be active but that they must be tested across cell types in dose response to determine both efficacy and toxicity.
We identified 38 compounds from a wide variety of classes that inhibit ZIKV infection. While a number of these drugs have been shown to have antiviral activity previously, many of these drugs have not been previously implicated as antivirals, including our most potent candidate nanchangmycin. Therefore, while we feel that there should indeed be a call for open drug discovery efforts for ZIKV, it may be too narrow to only focus on known antiviral drugs or known antiviral drug classes (Ekins et al., 2016). Indeed, our findings provide a rich resource for future studies on the host pathways required for infection, the specific pathways engaged in particular cell lineages as well as compounds that can be used as a starting point for therapeutic interventions.
Methods
Cells
Human U2OS, BHK, Vero (CCL-81) and Jeg-3 were from ATCC and maintained in DMEM plus 10% FBS for all but Jeg-3 that were maintained in MEM plus 10% FBS. HBMECs have been described previously (Bayer et al., 2016). Primary HUVEC and UtMEC were purchased from Lonza and were cultured in endothelial growth media (EndoGRO, Millipore). Isolation of primary term human fibroblasts was performed during standard placental cell isolation from full-term placentas under an exempt protocol approved by the institutional review board at the University of Pittsburgh (Bayer et al., 2016). Patients provided written consent for the use of de-identified and discarded tissues for research purposes upon admission to the hospital. Briefly, fibroblasts were isolated using the LD Column, (Miltenyi Biotec # 130--042--901) and a MACS Separation Unit. The cells were incubated with a mouse anti--vimentin, (1:20 dilution, Clone V9, Dako # M--0725), for 10 min at 4°C, washed by centrifugation at 300 xg, and resuspended in the PBS column with added MACS Microbeads coated with goat anti--mouse IgG (Miltenyi Biotec # 120--000--288). After incubation for 15 min at 4°C the bound cells were washed three times, removed from the column, and washed in Ca/Mg free PBS supplemented with EDTA and 0.5% BSA. The purity of isolated cells has been repeatedly validated using flow cytometry. Primary mixed neuronal cultures were derived from mice as described (Gao, 2012).
Virus stocks
ZIKV (mR766 and Mex2-81) and WNV (Kunjin) were obtained from Dr. R. Tesh (The World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) at UTMB) and M. Diamond (Washington University), ZIKV from Cambodia (FSS13025) was obtained from Dr. C. Coyne (University of Pittsburgh). CHIKV-mKate was obtained from Dr. M. Heise (University of North Carolina). DENV-2 (New Guinea C) was purchased (ATCC). PIV5 was from ATCC, and SINV-GFP was from Dr. R. Hardy (University of Indiana). All viruses were propagated in C636 and tittered on Vero cells.
Antibodies
Monoclonal antibodies against flavivirus glycoproteins (4G2) was provided by Dr. M. Diamond (Washington University) or anti-dsRNA (J2) from Dr. A. Brass (UMASS). Fluorescent secondary antibodies were from Life technologies. Hoechst 33342 was from Sigma-Aldrich. Anti-AXL was from Bethyl Laboratories (A302-168A), anti-alpha-tubulin was from Sigma-Aldrich (T6199) anti-NeuN was from Millipore (MAB377) and anti-GFAP was from Dako (Z0334).).
Drugs
The Selleckchem bioactive compound library was obtained in DMSO at 10mM. Candidates were re-purchased from Selleckchem and dissolved in DMSO at 10mM and diluted in Optimem prior to use. Other compounds were purchased from Sigma.
Drug Screening, IC50s, CC50s
For the primary screen, 5000 cells were plated in 25ul media per well in 384-well plates, using a Well Mate in duplicate plates. The next day, 50 nl of drugs were transfered to each well with a Janus MDT 50 nl pintool for a final concentration of 0.7uM/0.2% DMSO in U2OS and HBMECs. For Jeg-3 cells, 200 nl of drugs were transferred to each well of assay plates for a final concentration of 2.8uM/4 % DMSO 1 hour prior to infection. The drug treated U2OS cells were infected with Zika MR766, MOI=0.125 and fixed at 48 hours post infection (hpi). The drug treated HBMEC cells were infected with Zika MR766, MOI= 0.125 and fixed at 24 hpi. The drug treated Jeg-3 cells were infected by adding Zika Mex2-81, MOI=0.5 and fixed at 36 hpi. For IC50 analysis, 5000 cells were plated and the next day 50nL of serial dilutions of the drugs were transferred to assay plates and 1 h later, cells were infected with ZIKV Mex2-81: MOI=1.5 for U2OS and HBMEC for 24h and MOI=5 for Jeg-3 for 36h. For CC50 analysis, 5000 cells were plated and the next day 50nL of serial dilutions of the drugs were transferred to assay plates and 24h later for U2OS and HBMEC, or 36h for Jeg-3, 5uL of 0.5mM resazurin was added and 4h later analyzed by Envision reader (AlamarBlue_Ex560_Em590) and normalized to DMSO control wells.
Drug re-tests across cell types and viruses
U2OS, HBMEC or Jeg-3 cells were plated in 96-well plates. The next day, 1 hour prior to infection drugs were added at a final concentration of 2uM and infected at the following conditions: ZIKV mR766 (MOI 0.3, 48h), ZIKV Mex2-81 (MOI 10, 24h), ZIKV Cambodia (MOI 0.33, 24), DENV2-NGC (MOI 2.25, 24h).
Automated imaging
Briefly, for immunofluorescence infection experiments cells were fixed and stained for ZIKV (4G2) and counterstained with Hoescht. The cells were imaged at 10X (ImageXpress Micro, Molecular Devices) with 4 sites per well in a 384-well plate or 9 sites per well in a 96-well plate. The number of cells and the number of infected cells was measured using MetaXpress software and the percentage of infected cells calculated. For screening, percent-of-control (POC) and z-scores relative to DMSO control wells were calculated. For IC50 and CC50 analysis we used CeuticalSoft, Syndeo to calculate values.
Viral replication by RT-qPCR
U2OS or HBMEC cells were treated with the indicated drugs at 2uM, infected with ZIKV (Mex-2-81, MOI=2.5), or WNV (Kunjin, MO1=1), DENV (NGC, MOI=1), CHIKV (181/25, MOI=1), SINV (HRsp, MOI=1), PIV5 (MOI=1) and processed for RNA analysis at 24 hpi. Total RNA was extracted using Trizol reagent (Invitrogen). cDNA was generated from random hexamers to prime reverse transcription reactions using M-MLV reverse transcriptase. Quantitative PCR (qPCR) was performed with the cDNA using Power SYBR Green PCR Master Mix and a StepOnePlus RT-PCR system (Applied Biosystems). Primer sequences used in qPCR reactions are in Table S1. Reactions were initially run at initial 95°C for 5min, then 40 cycles of 95°C for 20 sec, 52°C for 30 sec, and 72°C for 30 sec. Relative viral copy numbers were generated by normalizing to cells treated with vehicle. ZIKV CC1-F: GTGTCATACTGTGGGCCTTG; ZIKV CC1-R: AATATTCCAGGCAGGGTCTG, WNV F: CCTGTGTGAGCTGACAAACTTAGT, WNV-R: GCGTTTTAGCATATTGACAGCC, DENV F: TGAGGACTACATGGGCTCTG DENV R: AAACCTCCCTGGATTTCCTT, CHIKV F: GGCAGTGGTCCCAGATAATTCAAG, CHIKV R: ACTGTCTAGATCCACCCCATACATG, SINV F: GCTGAAACACCATCGCTCTGCTTT, SINV R: TGGTGTCGAAGCCAATCCAGTACA, PIV5 F: TCCATCCTGTTCTCCAATCA, PIV5 R: TGATTCATCCGAGAGTCCAA, GAPDH-F: ACCAAATCCGTTGACTCCGACCTT; qGAPDH-R: TCGACAGTCAGCCGCATCTTCTTT.
Viral titers by TCID50s
U2OS, Vero CCL81, or Jeg-3 cells were plated in 96-well plates. The next day cells were pretreated with drugs 1hr prior to infection with 10-fold dilutions of ZIKV Mex2-81 strain. Jeg-3 cells were infected for 24hrs. U2OS and Vero CCL81 were infected for 48hrs prior to fixation and assaying by automated microscopy.
Time of addition experiments
U2OS cells were pretreated with drugs at a final concentration of 2uM either 1 h prior to or 4 hpi with ZIKV (Mex2-81, MOI=5). In addition, 20mM ammonium chloride was added 4hpi and the cells were processed 24 hpi. Alternatively, cells were pretreated and infected as above, but the drugs were washed out 4 hpi and processed for automated immunofluorescence microscopy 24 hpi.
Microscopy-based viral uptake assay
U2OS cells were pretreated with drugs for a final concentration of 2uM 1h prior to infection with ZIKV (Mex2-81, MOI=100) in the cold and centrifuged at 4°C (1500rpm, 30min) for binding (t=0) or released at 37°C for 3h (t=3). Cells were washed twice with PBS, fixed in 4% formaldehyde (10min, RT) and then blocked in PBS/2%BSA for 30min (4°C) prior to processing for confocal microscopy staining for ZIKV envelope (4G2) and DAPI. Confocal microscopy images were taken at 63X for 3 independent experiments.
Transferrin uptake
U2OS cells were pretreated with drugs at a final concentration of 2uM for 1h and 50 μg/mL AlexaFluor594-labeled transferrin was prebound in the cold for 1h. Cells were then incubated at 37°C for 15min. After washing, cells were placed in acid strip buffer to remove surface bound transferrin and cells were imaged by confocal microscopy at 63X for 3 independent experiments.
Primary cell infections
Primary neurons were plated in 96 well plates and the next day treated with the indicated drug at 2uM and 1h later infected with ZIKV (MR766) or DENV-2 (NGC) at MOI=2 for 24h and processed for automated microscopy. Representative images shown for 3 independent experiments. For other primary cells, the cells were plated in 24-well plates at a density of 5×105 cells per well and infected with ZIKV (FSS13025) at MOI=1 for 48hrs and processed for microscopy. Cells were fixed with ice-cold methanol and incubated with anti-dsRNA antibody (J2, Scion) and images were captured using an inverted IX81 Olympus microscope and image analysis performed using Fiji.
Statistical analysis
The primary screen was performed in duplicate and percent-of-control (POC) and z-scores relative to DMSO control wells were calculated based on standard deviation across each plate. For IC50 and CC50 analysis we used CeuticalSoft, Syndeo to calculate values. In additional studies a student’s t test was used to determine significance.
Supplementary Material
Acknowledgments
We thank Drs. R. Tesh, S. Hensley, S. Weiss, M. Heise and M. Diamond for viruses. We thank Antonia Bass for initial RT-qPCR studies, Yuan Liu for Chinese translations, Stephen Goldstein for technical advice and Qun Fang for generating neuronal cultures. In addition, we thank members of Cherry, Coyne and High-throughput screening core for technical support and advice. This work was supported by National Institute of Health grants R01AI074951, RO1AI122749, and R01AI095500 to SC. Support was provided by the Linda Montague Investigator Award, ITMAT Pilot and the Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award to S.C.
Footnotes
Author Contributions
Conception of the work: S.C. Data collection: K.R., B.H., N.W., S.R., D.C.S. Methodology and critical reagents: K.R., B.H., N.W., S.R., Y.S., C.H., D.C.S, C.C. Data analysis and interpretation: K.R., B.H., N.W., D.C.S, C.C., S.C. Drug Screening: K.R. and D.C.S. Validation studies: K.R., B.H., N.W., D.C.S, C.C., S.C Drafting the article: S.C. with assistance from all co-authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Araujo AQ, Silva MT, Araujo AP. Zika virus-associated neurological disorders: a review. Brain. 2016;139:2122–2130. doi: 10.1093/brain/aww158. [DOI] [PubMed] [Google Scholar]
- Barrows NJ, Campos RK, Powell ST, Prasanth KR, Schott-Lerner G, Soto-Acosta R, Galarza-Munoz G, McGrath EL, Urrabaz-Garza R, Gao J, et al. A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection. Cell Host Microbe. 2016;20:259–270. doi: 10.1016/j.chom.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzon L, Trevisan M, Sinigaglia A, Lavezzo E, Palu G. Zika virus: From pathogenesis to disease control. FEMS Microbiol Lett. 2016 doi: 10.1093/femsle/fnw202. [DOI] [PubMed] [Google Scholar]
- Bayer A, Lennemann NJ, Ouyang Y, Bramley JC, Morosky S, Marques ET, Jr, Cherry S, Sadovsky Y, Coyne CB. Type III Interferons Produced by Human Placental Trophoblasts Confer Protection against Zika Virus Infection. Cell Host Microbe. 2016;19:705–712. doi: 10.1016/j.chom.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitvich BJ, Firth AE. Insect-specific flaviviruses: a systematic review of their discovery, host range, mode of transmission, superinfection exclusion potential and genomic organization. Viruses. 2015;7:1927–1959. doi: 10.3390/v7041927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu JJ, Yang PL. c-Src protein kinase inhibitors block assembly and maturation of dengue virus. Proc Natl Acad Sci U S A. 2007;104:3520–3525. doi: 10.1073/pnas.0611681104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne CB, Lazear HM. Zika virus - reigniting the TORCH. Nat Rev Microbiol. 2016 doi: 10.1038/nrmicro.2016.125. [DOI] [PubMed] [Google Scholar]
- D’Ortenzio E, Matheron S, Yazdanpanah Y, de Lamballerie X, Hubert B, Piorkowski G, Maquart M, Descamps D, Damond F, Leparc-Goffart I. Evidence of Sexual Transmission of Zika Virus. The New England journal of medicine. 2016;374:2195–2198. doi: 10.1056/NEJMc1604449. [DOI] [PubMed] [Google Scholar]
- de Wispelaere M, LaCroix AJ, Yang PL. The small molecules AZD0530 and dasatinib inhibit dengue virus RNA replication via Fyn kinase. J Virol. 2013a;87:7367–7381. doi: 10.1128/JVI.00632-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wispelaere M, LaCroix AJ, Yang PL. The small molecules AZD0530 and dasatinib inhibit dengue virus RNA replication via Fyn kinase. J Virol. 2013b;87:7367–7381. doi: 10.1128/JVI.00632-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Zachariah M, Harris E. Mycophenolic acid inhibits dengue virus infection by preventing replication of viral RNA. Virology. 2002;304:211–221. doi: 10.1006/viro.2002.1685. [DOI] [PubMed] [Google Scholar]
- Ekins S, Mietchen D, Coffee M, Stratton TP, Freundlich JS, Freitas-Junior L, Muratov E, Siqueira-Neto J, Williams AJ, Andrade C. Open drug discovery for the Zika virus. F1000Res. 2016;5:150. doi: 10.12688/f1000research.8013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H. Midbrain Neuron-glia Mixed Cultures. bio-protocol. 2012;2:e148. [Google Scholar]
- Gao HM, Hong JS, Zhang W, Liu B. Distinct role for microglia in rotenone-induced degeneration of dopaminergic neurons. J Neurosci. 2002;22:782–790. doi: 10.1523/JNEUROSCI.22-03-00782.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett BA, Yasunaga A, Panda D, Tartell MA, Hopkins KC, Hensley SE, Cherry S. RNASEK is required for internalization of diverse acid-dependent viruses. Proc Natl Acad Sci U S A. 2015;112:7797–7802. doi: 10.1073/pnas.1424098112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel R, Dejarnac O, Wichit S, Ekchariyawat P, Neyret A, Luplertlop N, Perera-Lecoin M, Surasombatpattana P, Talignani L, Thomas F, et al. Biology of Zika Virus Infection in Human Skin Cells. J Virol. 2015;89:8880–8896. doi: 10.1128/JVI.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel R, Liegeois F, Wichit S, Pompon J, Diop F, Talignani L, Thomas F, Despres P, Yssel H, Misse D. Zika virus: epidemiology, clinical features and host-virus interactions. Microbes and infection/Institut Pasteur. 2016;18:441–449. doi: 10.1016/j.micinf.2016.03.009. [DOI] [PubMed] [Google Scholar]
- Hamill RL, Hoehn MM, Pittenger GE, Chamberlin J, Gorman M. Dianemycin, an antibiotic of the group affecting ion transport. J Antibiot (Tokyo) 1969;22:161–164. doi: 10.7164/antibiotics.22.161. [DOI] [PubMed] [Google Scholar]
- Jurado KA, Simoni MK, Tang Z, Uraki R, Hwang J, Householder S, Wu M, Lindenbach BD, Abrahams VM, Guller S, et al. Zika virus productively infects primary human placenta-specific macrophages. JCI Insight. 2016;1 doi: 10.1172/jci.insight.88461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Diamond MS. Zika Virus: New Clinical Syndromes and its Emergence in the Western Hemisphere. J Virol. 2016 doi: 10.1128/JVI.00252-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessler J, Chaisson LH, Kucirka LM, Bi Q, Grantz K, Salje H, Carcelen AC, Ott CT, Sheffield JS, Ferguson NM, et al. Assessing the global threat from Zika virus. Science. 2016;353:aaf8160. doi: 10.1126/science.aaf8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meertens L, Carnec X, Lecoin MP, Ramdasi R, Guivel-Benhassine F, Lew E, Lemke G, Schwartz O, Amara A. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe. 2012;12:544–557. doi: 10.1016/j.chom.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner Jonathan J, Sene A, Richner Justin M, Smith Amber M, Santeford A, Ban N, Weger-Lucarelli J, Manzella F, Rückert C, Govero J, et al. Zika Virus Infection in Mice Causes Panuveitis with Shedding of Virus in Tears. Cell Rep. doi: 10.1016/j.celrep.2016.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan AJ, Morin CW, Steinhoff DF, Wilhelmi O, Hayden M, Quattrochi DA, Reiskind M, Lloyd AL, Smith K, Schmidt CA, et al. On the Seasonal Occurrence and Abundance of the Zika Virus Vector Mosquito Aedes Aegypti in the Contiguous United States. PLoS Curr. 2016;8 doi: 10.1371/currents.outbreaks.50dfc7f46798675fc63e7d7da563da76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy RH, Gold B, Molleston JM, Schad V, Yanger K, Salzano MV, Yagi Y, Fitzgerald KA, Stanger BZ, Soldan SS, et al. Antiviral autophagy restrictsRift Valley fever virus infection and is conserved from flies to mammals. Immunity. 2014;40:51–65. doi: 10.1016/j.immuni.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastri E, Castilletti C, Liuzzi G, Iannetta M, Capobianchi MR, Ippolito G. Persistent detection of Zika virus RNA in semen for six months after symptom onset in a traveller returning from Haiti to Italy, February 2016. Euro Surveill. 2016;21 doi: 10.2807/1560-7917.ES.2016.21.32.30314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira Melo AS, Malinger G, Ximenes R, Szejnfeld PO, Alves Sampaio S, Bispo de Filippis AM. Zika virus intrauterine infection causes fetal brain abnormality and microcephaly: tip of the iceberg? Ultrasound in obstetrics & gynecology: the official journal of the International Society of Ultrasound in Obstetrics and Gynecology. 2016;47:6–7. doi: 10.1002/uog.15831. [DOI] [PubMed] [Google Scholar]
- Ouyang LTG, Gao Y, Zhang P, Xie X. Two insecticidal antibiotics produced by Streptomyces nanchangensis. J Jiangxi AgricUniv. 1993;15:148–153. [Google Scholar]
- Panda D, Rose PP, Hanna SL, Gold B, Hopkins KC, Lyde RB, Marks MS, Cherry S. Genome-wide RNAi screen identifies SEC61A and VCP as conserved regulators of Sindbis virus entry. Cell Rep. 2013;5:1737–1748. doi: 10.1016/j.celrep.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson TC, Kielian M. Flaviviruses: braking the entering. Curr Opin Virol. 2013;3:3–12. doi: 10.1016/j.coviro.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage H, Cherry S. Virus-Host Interactions: From Unbiased Genetic Screens to Function. Annu Rev Virol. 2015;2:497–524. doi: 10.1146/annurev-virology-100114-055238. [DOI] [PubMed] [Google Scholar]
- Randolph VB, Winkler G, Stollar V. Acidotropic amines inhibit proteolytic processing of flavivirus prM protein. Virology. 1990;174:450–458. doi: 10.1016/0042-6822(90)90099-d. [DOI] [PubMed] [Google Scholar]
- Savidis G, McDougall WM, Meraner P, Perreira JM, Portmann JM, Trincucci G, John SP, Aker AM, Renzette N, Robbins DR, et al. Identification of Zika Virus and Dengue Virus Dependency Factors using Functional Genomics. Cell Rep. 2016;16:232–246. doi: 10.1016/j.celrep.2016.06.028. [DOI] [PubMed] [Google Scholar]
- Schuler-Faccini L, Ribeiro EM, Feitosa IM, Horovitz DD, Cavalcanti DP, Pessoa A, Doriqui MJ, Neri JI, Neto JM, Wanderley HY, et al. Possible Association Between Zika Virus Infection and Microcephaly - Brazil, 2015. MMWR Morbidity and mortality weekly report. 2016;65:59–62. doi: 10.15585/mmwr.mm6503e2. [DOI] [PubMed] [Google Scholar]
- Shan C, Xie X, Muruato AE, Rossi SL, Roundy CM, Azar SR, Yang Y, Tesh RB, Bourne N, Barrett AD, et al. An Infectious cDNA Clone of Zika Virus to Study Viral Virulence, Mosquito Transmission, and Antiviral Inhibitors. Cell Host Microbe. 2016;19:891–900. doi: 10.1016/j.chom.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Zhou X, Liu J, Bao K, Zhang G, Tu G, Kieser T, Deng Z. ‘Streptomyces nanchangensis’, a producer of the insecticidal polyether antibiotic nanchangmycin and the antiparasitic macrolide meilingmycin, contains multiple polyketide gene clusters. Microbiology. 2002;148:361–371. doi: 10.1099/00221287-148-2-361. [DOI] [PubMed] [Google Scholar]
- Tabata T, Petitt M, Puerta-Guardo H, Michlmayr D, Wang C, Fang-Hoover J, Harris E, Pereira L. Zika Virus Targets Different Primary Human Placental Cells, Suggesting Two Routes for Vertical Transmission. Cell Host Microbe. 2016;20:155–166. doi: 10.1016/j.chom.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetro JA. Microbes and infection/Institut Pasteur. 2016. Zika and microcephaly: Causation, correlation, or coincidence? [DOI] [PubMed] [Google Scholar]
- Turmel JM, Abgueguen P, Hubert B, Vandamme YM, Maquart M, Le Guillou-Guillemette H, Leparc-Goffart I. Late sexual transmission of Zika virus related to persistence in the semen. Lancet. 2016;387:2501. doi: 10.1016/S0140-6736(16)30775-9. [DOI] [PubMed] [Google Scholar]
- Xu M, Lee EM, Wen Z, Cheng Y, Huang WK, Qian X, Tcw J, Kouznetsova J, Ogden SC, Hammack C, et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med. 2016 doi: 10.1038/nm.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasunaga A, Hanna SL, Li J, Cho H, Rose PP, Spiridigliozzi A, Gold B, Diamond MS, Cherry S. Genome-wide RNAi screen identifies broadly-acting host factors that inhibit arbovirus infection. PLoS Pathog. 2014;10:e1003914. doi: 10.1371/journal.ppat.1003914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Chen YL, Schul W, Wang QY, Gu F, Duraiswamy J, Kondreddi RR, Niyomrattanakit P, Lakshminarayana SB, Goh A, et al. An adenosine nucleoside inhibitor of dengue virus. Proc Natl Acad Sci U S A. 2009;106:20435–20439. doi: 10.1073/pnas.0907010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Miner JJ, Gorman MJ, Rausch K, Ramage H, White JP, Zuiani A, Zhang P, Fernandez E, Zhang Q, et al. A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature. 2016;535:164–168. doi: 10.1038/nature18625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kwiatkowski N, Zeng H, Lim SM, Gray NS, Zhang W, Yang PL. Leveraging kinase inhibitors to develop small molecule tools for imaging kinases by fluorescence microscopy. Mol Biosyst. 2012;8:2523–2526. doi: 10.1039/c2mb25099c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmurko J, Marques RE, Schols D, Verbeken E, Kaptein SJ, Neyts J. The Viral Polymerase Inhibitor 7-Deaza-2′-C-Methyladenosine Is a Potent Inhibitor of In Vitro Zika Virus Replication and Delays Disease Progression in a Robust Mouse Infection Model. PLoS Negl Trop Dis. 2016;10:e0004695. doi: 10.1371/journal.pntd.0004695. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.