Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
ZEB2 is a top hit of 2 short hairpin RNA screens for novel AML dependencies.
ZEB2 regulates differentiation in AML.
Abstract
Acute myeloid leukemia (AML) is a heterogeneous disease with complex molecular pathophysiology. To systematically characterize AML’s genetic dependencies, we conducted genome-scale short hairpin RNA screens in 17 AML cell lines and analyzed dependencies relative to parallel screens in 199 cell lines of other cancer types. We identified 353 genes specifically required for AML cell proliferation. To validate the in vivo relevance of genetic dependencies observed in human cell lines, we performed a secondary screen in a syngeneic murine AML model driven by the MLL-AF9 oncogenic fusion protein. Integrating the results of these interference RNA screens and additional gene expression data, we identified the transcription factor ZEB2 as a novel AML dependency. ZEB2 depletion impaired the proliferation of both human and mouse AML cells and resulted in aberrant differentiation of human AML cells. Mechanistically, we showed that ZEB2 transcriptionally represses genes that regulate myeloid differentiation, including genes involved in cell adhesion and migration. In addition, we found that epigenetic silencing of the miR-200 family microRNAs affects ZEB2 expression. Our results extend the role of ZEB2 beyond regulating epithelial–mesenchymal transition (EMT) and establish ZEB2 as a novel regulator of AML proliferation and differentiation.
Introduction
Acute myeloid leukemia (AML) is a complex, heterogeneous disorder with poor prognoses. Treatment strategies against AML have remained largely unchanged for the last 3 decades, with the majority of patients eventually succumbing to relapse after induction chemotherapy.1,2 The development of effective next-generation therapeutic options against AML relies on mechanistic understanding of AML biology, especially molecular regulators of AML pathogenesis and genetic dependencies of AML proliferation and differentiation.
Recent advances in genomic technologies have led to the generation of large-scale cancer data sets, such as the Cancer Cell Line Encyclopedia (CCLE)3 and The Cancer Genome Atlas (TCGA).4 The former provides copy number, mutation, gene expression, and pharmacologic profiling of >1000 cancer cell lines, including 34 AML cell lines. The latter profiles copy number, mutation, mRNA/microRNA expression, and methylation in >30 types of cancer samples, including 200 AML samples. These studies have revealed numerous genes and microRNAs that are altered in AML. However, a major challenge to interpreting these findings is to establish the functional relevance of these genes for AML and other cancers.
To address this challenge, we combined cancer genomic data with in vitro and in vivo RNA interference (RNAi) screens to systematically interrogate the genetic dependencies of AML. Massively parallel pooled short hairpin RNA (shRNA) screens coupled with next-generation sequencing deconvolution have yielded critical insights into a wide range of cancers and have demonstrated the value and feasibility of loss-of-function screening in cancer models. For example, such screens have led to the discovery of genotype-specific dependency such as ARID1B in ARID1A-mutant cell lines5 and lineage-specific dependency such as PAX8 in ovarian cancer.6 Corroborating in vitro screens, Zuber et al7 screened 824 inducible shRNAs in a murine MLL-AF9/NrasG12D model, and confirmed the in vivo requirement of an essential gene, Rpa3.
In this study, we identified novel genes regulating AML proliferation using in vitro genome-scale shRNA screens in multiple AML cell lines with diverse subtypes, in addition to an in vivo secondary screen in a syngeneic murine AML model driven by the MLL-AF9 oncogenic fusion protein. We established the transcription factor ZEB2 as a previously unknown regulator of AML proliferation and differentiation. Although ZEB2 is best known for its role in repressing E-cadherin and promoting epithelial–mesenchymal transition (EMT),8 our investigation uncovered its critical roles in regulating AML development.
Methods
shRNA screens
The human cell line screens were performed as described.9,10 The Probablility Analysis by Ranked Information Score (PARIS) module of GenePattern (http://genepattern.broadinstitute.org/gp) was used to analyze Project Achilles data v2.4.3. The in vivo screen was performed as described.11,12 All experiments and procedures were conducted in the Children’s Hospital Boston animal facility and were approved by the Children’s Hospital Institutional Animal Care and Use Committee on protocol 13-04-2393R. Briefly, 2 million quaternary transplanted mouse leukemia cells were infected by shRNA lentivirus pools in 5 replicates, and ∼1.2 million cells were injected into sublethally irradiated mice 1 day after infection. Infected cells were harvested at day 1 after infection, and bone marrows were harvest at day 14 after transplant (see supplemental Methods, available on the Blood Web site).
Cell culture, virus production, and infection
HL-60, THP-1, MOLM-13, THP-1, SKM-1, U-937, and KASUMI-1 cells were cultured in RPMI-1640 supplemented with 10% fetal bovine serum and Pen/Strep. Mouse leukemia cells were cultured in RPMI-1640, 10% fetal bovine serum, 10 ng/mL interleukin 3 (IL-3), and Pen/Strep. Phorbol myristate acetate treatment was carried out at a concentration of 100 ng/mL and a duration of 4 days. For lentivirus production, 293T cells were cotransfected with shRNA or single guide RNA (sgRNA) plasmids, psPAX2 (Addgene), and pMD2.G (Addgene). Virus was harvested 48 and 72 hours after transfection and concentrated by PEG-it Virus Precipitation Solution (System Biosciences). Spin infection was performed at 2500 rpm for 2 hours at 30°C. Polybrene (8 µg/mL) was used for human cell lines, and 5 µg/mL polybrene, 10 ng/mg IL-3, 10 ng/mL IL-6, and 20 ng/mL stem cell factor was used for mouse leukemia cells during infection. See supplemental Methods for shRNA/sgRNA sequences.
Cell proliferation, apoptosis, and differentiation assays
For shRNAs in vectors encoding puromycin resistance, cells were selected with puromycin for 72 hours, 48 hours after infection. Cells were subsequently seeded in 96-well plates for serial passage. An aliquot of cells was taken at different time points to analyze viability by the CellTiter-Glo assay (Promega). For shRNAs or sgRNAs in green fluorescent protein (GFP) or RFP657 vector, cells were infected at <70% efficiency, and the proportion of infected cells was monitored by flow cytometry. At day 6 after infection, apoptosis was measured by the allophycocyanin (APC) Annexin V Apoptosis Detection Kit (eBioscience), and BrdU incorporation was determined by the APC or fluorescein isothiocyanate 5-bromo-2′-deoxyuridine (BrdU) Flow Kit (BD Pharmingen) according to the manufacturer’s instructions. CD11b and other cell-surface markers were stained with fluorophore-conjugated antibodies (BD Pharmingen) and analyzed by flow cytometry at day 6 after infection. May-Grunwald-Giemsa staining was performed in HL-60 cells 10 days after infection (see supplemental Methods).
RNA sequencing and gene set enrichment analysis
THP-1 and U-937 cells were infected with shRNA lentivirus in GFP vector, and GFP+ cells were sorted at 72 hours after infection. Immediately after sorting, cells were pelleted and RNA was extracted with the RNeasy Plus Mini Kit (Qiagen). A library was prepared with the Illumina TruSeq mRNA Sample Preparation Kit using poly-A selection and strand-specific cDNA synthesis and sequenced with 50-bp paired-end sequencing on an Illumina HiSeq. Differentially expressed genes were analyzed by DESeq.13 Gene sets were created for genes upregulated and downregulated by ZEB2 knockdown and incorporated into the C6 collection of MsigDB (http://www.broadinstitute.org/gsea/msigdb/). Sample permutation was used for microarrays with >7 samples in ≥1 group, and gene set permutation was used for other microarrays (see supplemental Methods).
Motif analysis
Motif analysis using MsigDB was performed by the Investigate Gene Sets tool. For CisFinder and Regulatory Sequence Analysis Tools (RSAT) analysis, promoter sequences of all human genes were downloaded from The Eukaryotic Promoter Database (http://epd.vital-it.ch/), and promoters for ZEB2-regulated genes were retrieved. Both analyses used sequences of ZEB2-regulated genes as query and the same region of all human genes as background.
AML genomic data
Mutation, microRNA expression, and methylation data of TCGA AML samples were downloaded from The Broad Institute TCGA GDAC Firehose (http://gdac.broadinstitute.org/) or University of California, Santa Cruz Cancer Genome Browser (https://genome-cancer.ucsc.edu). mRNA expression data of CCLE was downloaded from the CCLE data portal (http://www.broadinstitute.org/ccle/home).
Results
Genome-scale shRNA screens identify genetic dependencies of AML cell lines
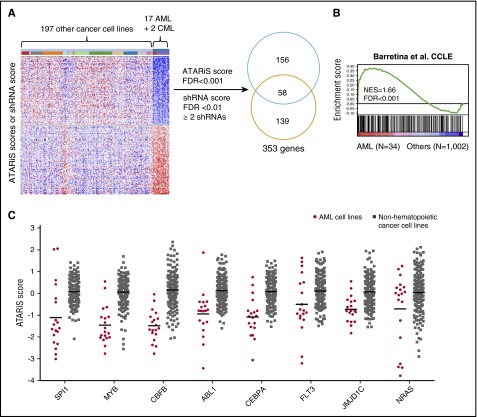
To explore novel genetic dependencies in AML, we performed genome-scale shRNA screens in 17 AML cell lines (supplemental Table 1) in parallel with screens in 199 cancer cell lines from other tissue origins (Project Achilles),9 including 2 chronic myeloid leukemia blast crisis (CML-BC) cell lines. To identify genes that were uniquely required by AML cell lines for survival, we compared the ATARiS (Analytic Technique for Assessment of RNAi by Similarity) scores14 and individual shRNA fold depletions of the 19 AML and 2 CML-BC cell lines (hereafter referred to as AML) to 197 other cancer cell lines and identified 353 genes, including 214 genes with significantly smaller ATARiS scores (false discovery rate [FDR] <0.001) and 197 genes targeted by ≥2 significantly depleted shRNAs (FDR <0.01; Figure 1A; supplemental Table 2). Notably, these candidates were expressed at higher levels in AML cell lines compared with other cancer cell lines from the Cancer Cell Line Encyclopedia (CCLE)3 (Figure 1B) and were enriched for genes involved in hematopoietic cell development (supplemental Figure 1A), which suggest their functional relevance for AML. Moreover, we found several genes that are frequently mutated or translocated in AML, such as NRAS, FLT3, CEBPA, PTPN11, CBFB, and ABL1,4 or genes known to be required for AML, such as SPI1,15,16 MYB,17 HOXA9,18 JMJD1C,19 EED,20 and KDM1A21,22 (Figure 1C; supplemental Table 2). The screen also unveiled the specific dependency of FLT3 in MLL-rearranged AML, and NRAS in NRAS mutant AML (supplemental Figure 1B). These observations identify a set of candidate genes uniquely required for the proliferation and survival of AML cell lines.
Figure 1.
shRNA screens for genes uniquely required by AML cell lines for survival. (A) Schematic representation of data analysis workflow. One hundred ninety-seven genes with ≥2 significant shRNAs were selected, and 214 genes with significant ATARiS scores were selected. The 2 lists have 58 overlapping genes and 353 genes in total. (B) The 353 screen hits are enriched of genes highly expressed in CCLE AML cell lines based on GSEA.30 (C) Examples of screen hits that are frequently altered in AML or required by AML cells for survival compared with non-AML cancer cell lines. Lines indicate mean. FDR, false discovery rate; NES, normalized enrichment score.
Validation of novel AML regulators by an in vivo secondary screen
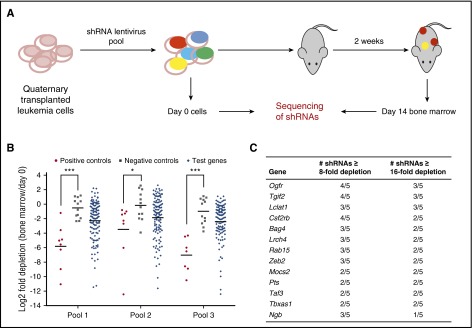
To validate the cell line screening results, we performed an in vivo secondary screen with pooled shRNAs targeting the murine homologs of the 37 top primary screen hits in a recently established MLL-AF9 xenotransplantation AML model11 (Figure 2A). These 37 genes were among the top depleting genes in the primary screen (Project Achilles; see supplemental Methods), prioritized for those that are highly expressed in AML cell lines, mutated in cancer, or having viable knockout mice. The in vivo screen was very sensitive given its high read depth (supplemental Figure 2A) and high consistency between replicates (supplemental Figure 2B-D). In all 3 pools, shRNAs targeting positive control genes were significantly more depleted than those targeting negative control genes (Figure 2B), suggesting that the screen was capable of detecting genetic dependencies in mouse AML. We selected genes that have ≥3 shRNAs depleted by >8-fold or ≥2 shRNAs depleted by >16-fold as hits. Four positive controls (Hoxa9, Ctnnb1, Mef2c, and Myc) but none of the negative controls were selected (supplemental Figure 2E). Among the 37 genes tested, 13 were selected as hits (Figure 2C; supplemental Table 3). We focused on ZEB2, a transcription factor that is known to promote EMT by repressing epithelial cell–cell junction genes.23,24
Figure 2.
Secondary shRNA screen for genes required by MLL-AF9 AML in vivo. (A) Schematic of secondary screen. MLL-AF9 transduced mouse granulocyte monocyte progenitors were serially transplanted into recipient mice. Quaternary transplanted leukemia cells (c-KithiCD34+FCRγRIIhi) were sorted and infected with 3 pools of shRNAs in 5 replicates and transplanted into sublethally irradiated mice. Pretransplant cells (day 0 sample) and day 14 bone marrow samples were sequenced to determine shRNA representation. (B) Positive control shRNAs are significantly more depleted than negative control shRNAs. Shown is the median log2 fold depletion of 5 replicates. Lines indicate mean. ***P < .001, *P < .05, t test. (C) List of 13 screen hits.
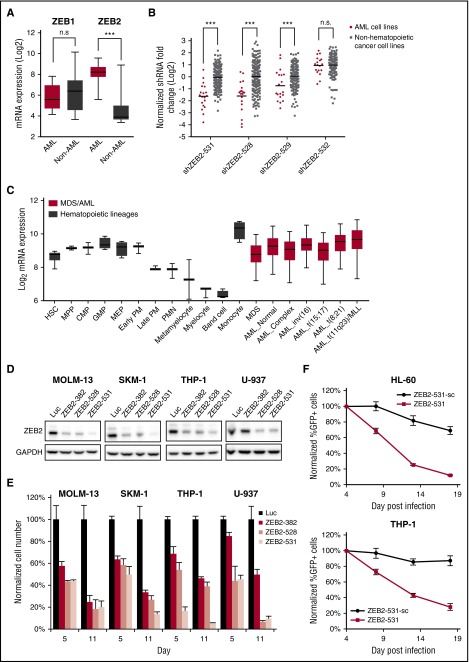
AML cells are sensitive to ZEB2 depletion
ZEB2 is highly expressed in AML cell lines compared to non-AML cancer cell lines (Figure 3A), and is specifically required for the proliferation of AML cell lines compared to non-hematopoietic cancer cell lines (Figure 3B). It is expressed at similar levels in MDS, major genetic subtypes of AML,4 and normal human stem and progenitor cells but is downregulated in most differentiated myeloid cells25,26 (Figure 3C). These findings suggested to us that ZEB2 might be essential in AML due to a role in the maintenance of hematopoietic cells in a less differentiated state.
Figure 3.
Human AML cell lines are sensitive to ZEB2 inhibition. (A) Box plot showing mRNA expression levels of ZEB1 and ZEB2 in CCLE3 cell lines, comparing AML cell lines vs non-AML cancer cell lines. ***P < .001; n.s., not significant, t test. (B) shRNA fold depletions of ZEB2 shRNAs in the human cell line screen. ***P < .001; n.s., not significant, PARIS analysis.9 (C) ZEB2 expression in hematopoiesis25 and AML.4 Error bars indicate 5% to 95% percentile. (D) Immunoblotting of ZEB2 at day 6 after shRNA lentivirus infection. (E) Viability of AML cells after ZEB2 knockdown. Error bars, standard deviation (SD) (n = 3). (F) Competitive proliferation assays of AML cells infected with ZEB2 shRNA (ZEB2-531) and its seed control (ZEB2-531-sc). Graph shows normalized cell number as a percentage of the first time point (4 days after infection). Error bars, SD (n = 3). AML_Complex, complex karyotype; AML_Normal, normal karyotype; CMP, common myeloid progenitor cell; GMP, granulocyte monocyte progenitors; HSC, hematopoietic stem cell; Luc, shRNA targeting firefly luciferase; MDS, myelodysplastic syndrome; MEP, megakaryocyte-erythroid progenitor cell; MPP, multipotential progenitors; PM, promyelocyte; PMN, polymorphonuclear cells; ZEB2-382, shRNA targeting ZEB2.
To validate ZEB2’s requirement in human AML cell lines, we suppressed ZEB2 using 3 shRNAs, including one that was not used in the screen (ZEB2-382). ZEB2 depletion, as determined by immunoblotting (Figure 3D), strongly impaired the growth of 4 different AML cell lines, including 2 that were not in the screen (SKM-1 and U-937) (Figure 3E). Similar growth inhibitory effects were observed in GFP-labeled competitive proliferation assays for all 3 ZEB2 shRNAs (supplemental Figure 3A).
shRNA screens are frequently confounded by seed sequence-based off-target effects.27 To ensure the specificity of the shRNAs, we generated “seed control”28 for the shRNA that showed the highest knockdown efficiency (ZEB2-531), by introducing a 3-bp substitution in the middle of the shRNA. As expected, the seed control (ZEB2-531-sc) was no longer able to suppress ZEB2 (supplemental Figure 2B). ZEB2-531 showed significantly more proliferation–inhibitory effects compared with ZEB2-531-sc (Figure 3F; supplemental Figure 3C), indicating that ZEB2 knockdown is sufficient to impair cell proliferation. Further supporting these findings, we showed that CRISPR-Cas9–mediated depletion of ZEB2 consistently impaired the proliferation of AML cell lines (supplemental Figure 3E-F). These results establish ZEB2 as a critical requirement for the proliferation of multiple human AML cell lines.
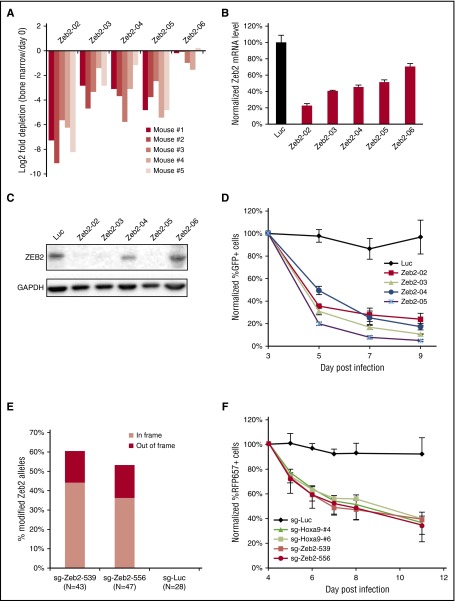
We next sought to validate ZEB2’s requirement in mouse MLL-AF9 AML cells. Four of 5 shRNAs targeting Zeb2 were significantly depleted in the in vivo screen (Figure 4A), and the effects of shRNAs correlated well with the knockdown efficiency at mRNA and protein level (Figures 4B-C), demonstrating the specificity of the screen’s results. In vivo depletion of AML cells might be due to impaired cell proliferation or impaired homing and engraftment to the bone marrow. To evaluate whether Zeb2 depletion affects cell autonomous growth, we cultured MLL-AF9 AML cells with IL-3 in vitro and infected them with 4 shRNAs that efficiently knockdown Zeb2. Indeed, all shRNAs quickly decreased cell proliferation (Figure 4D). To corroborate the shRNA results, we infected MLL-AF9 AML cells stably expressing Cas9 with 2 Zeb2 sgRNAs. As expected, both Zeb2 sgRNAs quickly suppressed cell proliferation, whereas the control sgRNA targeting firefly luciferase did not (Figure 4F). The sgRNAs generated mutations at ∼55% to 60% of Zeb2 genomic loci (Figure 4E; supplemental Figure 4A-B), of which only ∼20% were frameshift. The lower-than-expected rate of frameshift mutations suggests that functional in-frame mutations were selected for, providing additional evidence that functional ZEB2 is required for AML survival. Importantly, the inhibitory effects of Zeb2 sgRNAs were comparable to 2 Hoxa9 sgRNAs tested in parallel, highlighting the critical dependency of mouse MLL-AF9 AML cells on Zeb2.
Figure 4.
Mouse AML cells are sensitive to ZEB2 inhibition. (A) Log2 fold depletions of Zeb2 shRNA in the in vivo screen. (B) qPCR of Zeb2 mRNA level at day 6 after infection. (C) Immunoblotting of ZEB2 at day 6 after infection. (D) Competitive proliferation assay of leukemia cells infected with Zeb2 shRNA and control shRNA. Error bars indicate SD (n = 3). (E) Frequency of mutated Zeb2 genomic alleles at 4.5 days after sgRNA infection. (F) Competitive proliferation assay of leukemia cells stably expressing Cas9 after infection with sgRNAs. Error bars indicate SD (n = 3).
ZEB2 suppression induces aberrant differentiation in human AML cells
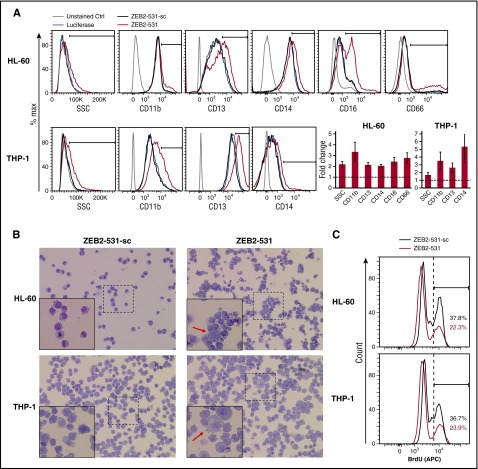
Based on the differential expression of ZEB2 in AML and normal hematopoietic stem and progenitor cells, compared with more differentiated cells, we hypothesized that ZEB2 is required to maintain hematopoietic cells in a stem/progenitor state and that its loss depletes AML cells through differentiation. To investigate this, we transduced well-studied AML models known for their differentiation potential, HL-60, THP-1, and U937 cells, with shRNA to ZEB2 (ZEB2-531) and negative controls (ZEB2-531-sc and Luciferase).
Compared with the more “physiologic” differentiation induced by phorbol myristate acetate treatment (supplemental Figure 6),29 ZEB2 knockdown led to aberrant differentiation in the AML cell lines tested, as shown by increased expression of cell surface differentiation markers such as CD11b, CD13, CD14, CD16, and CD66, as well as moderately altered side-scatter profiles (Figure 5A; supplemental Figure 6). Specifically, ZEB2-531 caused induction of CD11b, CD13, and CD14 in both HL-60 and THP-1 cells, as well as an increase of CD16 and CD66 in HL-60 cells and CD11b and CD13 in U937 cells. These changes corresponded with morphological alterations, with cells becoming larger and more granulated (Figure 5B), but not clearly differentiating into macrophages or neutrophils. Consistent with differentiation, these cells had decreased proliferation based on BrdU incorporation (Figure 5C). In addition, in MOLM-13 and SKM-1 cells, the frequency of CD11blowBrdU+ cells decreased, whereas CD11bhighBrdU- cells increased, suggesting decreased proliferation was coupled with increased differentiation (supplemental Figure 5B). In contrast, we did not observe a significant difference in apoptosis between cells infected by ZEB2-531 and its seed control (supplemental Figure 5A).
Figure 5.
ZEB2 suppression induces aberrant differentiation of human AML cells. (A) (Left) Flow cytometric analysis of side-scatter profiles and the expression of CD11b, CD13, CD14, CD16, and CD16 in HL-60 and THP-1 cells infected with ZEB2-531 and its seed control (ZEB2-531-sc) at day 6. (Right) Quantification of results from the left shown as average fold change of marker expression comparing ZEB2 shRNA (ZEB2-531) and seed control (ZEB2-531-sc). Error bars, SD (n = 3). (B) Representative May-Grunwald-Giemsa staining of HL-60 and THP-1 cells at day 10 after infection. Original magnification ×200; original magnification for insets ×400. (C) Flow cytometric analysis of BrdU incorporation in HL-60 and THP-1 cells at day 6 after infection.
To further understand ZEB2’s role in differentiation, we investigated the transcriptional program regulated by ZEB2. To this end, we performed RNA sequencing in THP-1 and U-937 cells at 72 hours after shRNA infection. We identified 438 genes that are significantly differentially expressed in both cell lines (supplemental Figure 5C). Importantly, the log2 fold change of these 438 genes are highly correlated between the 2 cell lines (R2 = 0.69, P < 1×107), suggesting ZEB2 regulates a common transcriptional program in AML. Specifically, 283 genes were upregulated (ZEB2KDup) and 144 genes were downregulated (ZEB2KDdown) in both cell lines, whereas only 11 genes (2.5%) were affected in opposite directions (supplemental Figure 7A). Gene set enrichment analysis (GSEA)30 suggests that ZEB2KDup genes were significantly associated with granulocytic differentiation31,32 (supplemental Figure 7B), an unexpected finding in these monoblastic leukemia cell lines, which emphasizes the aberrant nature of differentiation due to ZEB2 loss.
ZEB2 directly represses genes involved in myeloid differentiation
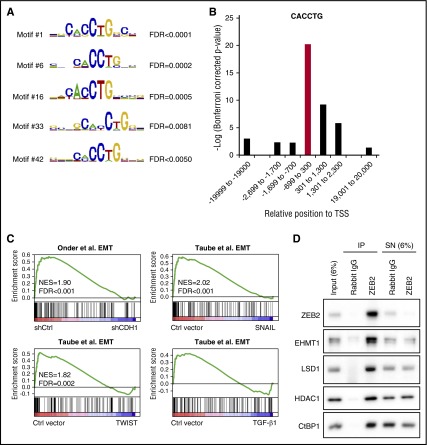
We hypothesized that ZEB2 might regulate the level of key myeloid transcription factors and modulate myeloid differentiation through these genes. However, known transcription factors regulating granulocytic differentiation, such as CEBPα/β/δ/ε/γ/ζ, PU.1, GFI-1, and LEF-1,33 were not among the 438 differentially expressed genes on ZEB2 knockdown. An alternative mechanism is that ZEB2 directly targets multiple downstream genes involved in differentiation. To test this possibility, we searched for the enrichment of transcription factor binding motifs in the promoters of genes regulated by ZEB2 using MsigDB.30 Intriguingly, the highest-ranked motif in promoters of ZEB2KDup genes is CACCTG, whereas CACCT(G) is the known binding motif of ZEB2 in the promoters of E-cadherin23 and other cell–cell junction genes.24 The CACCTG motif was not enriched in promoters of ZEB2KDdown genes (supplemental Figure 8A), suggesting that ZEB2 regulates its direct targets mainly by transcription repression, which is consistent with its known role as a repressor.34 We therefore focused on the promoters of ZEB2KDup genes and confirmed that the highest ranked motif cluster contains CACCTG by CisFinder35 (Figure 6A; supplemental Figure 8B). Moreover, using the oligo-diff tool of RSAT,36 we found that the CACCTG/CACCT/ACCTG motifs are enriched in regions close to the transcription start sites of ZEB2KDup genes, but not distant regions (Figure 6B; supplemental Figure 8C). Further supporting these results, we found that ZEB2KDup genes with ≥1 CACCTG sequence in their promoters were enriched of genes that are expressed in human mammary epithelial cells, but are downregulated during EMT induced by E-cadherin knockdown, or TWIST/SNAIL/TGF-β1 expression,37,38 which is consistent with ZEB2’s known role in repressing epithelial genes and promoting EMT. Finally, these putative ZEB2 direct targets were enriched of cell movement and adhesion genes, several of which are granulocytic differentiation markers, such as ITGAM (CD11b), MPO, and CEACAM6 (CD66c), or genes that are required for granulocytic differentiation, such as CSF3R39 and CORO1A40 (supplemental Figure 8D-E). Taken together, these observations suggest that ZEB2 may directly suppress multiple genes that are critical for myeloid differentiation.
Figure 6.
ZEB2 directly represses genes involved in myeloid differentiation. (A) Motifs with the CACCTG sequence are enriched in the promoters of ZEB2KDup genes based on CisFinder analysis. (B) Enrichment of CTCCTG sequence in different genomic regions of ZEB2KDup genes as analyzed by the oligo-diff tool of RSAT. (C) GSEA shows that ZEB2KDup genes with CACCTG sequence in their promoters are highly expressed in epithelial cells and downregulated during EMT triggered by E-cadherin knockdown or TWIST/SNAIL/TGF-β1 expression. (D) ZEB2 coimmunoprecipitate with CtBP1, LSD1, HDAC1, and EHMT1 in THP-1 cells. TSS, transcription start site; FDR, false discovery rate; NES, normalized enrichment score; IP, immunoprecipitate; SN, supernatant.
We next asked how ZEB2 represses these targets in AML cells. ZEB2 is a component of the CtBP corepressor complex41 and directly interacts with CtBP1.34 Several components of the CtBP complex, such as CtBP1,41 G9a,41 EuHMT,41 and LSD1,42 have been shown to mediate the transcription repression of E-cadherin. We therefore tested the interaction between endogenous ZEB2 and the CtBP complex. Indeed, in THP-1 cells, ZEB2 coimmunoprecipitated with CtBP1 and several other epigenetic enzymes of the CtBP complex, including LSD1, HDAC, and EHMT1 (EuHMT) (Figure 6D). Similar results were obtained in U-937 cells (supplemental Figure 9). Small molecule inhibitors of LSD1,21,22 HDAC,43 and G9a/EuHMT44 have all been shown to induce myeloid differentiation. Therefore, it is possible that ZEB2 recruits these epigenetic regulators to the promoters of genes involved in myeloid differentiation and repress their transcription.
ZEB2-targeting miR-200 family miRNAs are epigenetically silenced in AML
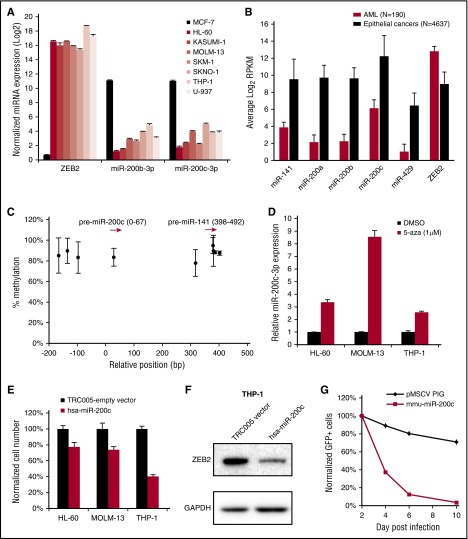
Given the critical role of ZEB2 in AML growth and differentiation, we hypothesized that negative regulators of ZEB2 might be repressed in AML. The major known negative regulators of ZEB2 are the microRNAs (miRNAs) of the miR-200 family, which include miR-141, miR-200a, miR-200b, miR-200c, and miR-429.8 miR-200 family members are expressed in epithelial cells but silenced in mesenchymal cells, whereas ZEB2 has opposite expression pattern.8 We quantified the expression of miR-200b and miR-200c in parallel with ZEB2 in a panel of AML cell lines by quantitative polymerase chain reaction (qPCR). As expected, ZEB2 was highly expressed in AML cell lines but almost absent in epithelial cell line MCF-7. Interestingly, the expression levels of miR-200b and miR-200c were 60- to 900-fold lower in AML cells than MCF-7 (Figure 7A). In fact, all 5 miRNAs in the miR-200 family were expressed at significantly lower levels in TCGA AML samples4 compared with samples of 10 types of carcinomas, whereas the opposite trend was observed for ZEB2 (Figure 7B).
Figure 7.
ZEB2-targeting miR-200 family miRNAs are epigenetically silenced in AML. (A) qPCR of ZEB2, miR-200b, and miR-200c in a panel of AML cell lines and epithelial cancer cell line MCF-7. (B) Expression of miR-200 family members and ZEB2 in cancer samples from TCGA RNA-Seq data; expression level is shown as log2 of reads per kilobase of transcript per million mapped reads. (C) Methylation of DNA sequences upstream of miR-200c and miR-141 in TCGA AML samples. Red arrows indicate the relative position of miR-200c and miR-141. Each dot indicates average level of methylation in all patients. (D) miR-200c is upregulated after 3 days of 5-aza treatment as measured by qPCR. (E) Viability of AML cell lines after miR-200c and empty vector infection. Cells were seeded at day 6 after infection, and cell viability was measured at day 9 by the CellTiter-Glo assay. (F) Immunoblot of ZEB2 in cells infected with miR-200c. (G) Competitive proliferation assay in mouse leukemia cells infected with mouse miR-200c or control vector. Error bars, SD. n = 190 for AML, n = 4837 for epithelial cancers in B; n = 193 for C; and n = 3 for D and E.
Repression of miR-200c and miR-141 in breast cancer cell lines is mediated by promoter methylation.45 We therefore analyzed the TCGA AML 450k methylation data and found high levels of DNA methylation in nearly all of the CpG probes upstream of miR-200 family. By contrast, the promoter of a highly expressed miRNA, miR-150, was not methylated in AML (Figure 7C; supplemental Figure 10). When treated with DNA demethylating agent 5-aza-2′-deoxycytidine, miR-200c-3p was upregulated by three- to eightfold in 3 different AML cell lines (Figure 7D). We next directly tested the role of miR-200 on AML survival and focused on miR-200c as it is the most abundant member of the family. Introduction of a pre–miR-200c reduced the expression of ZEB2 protein and inhibited the proliferation of HL-60, MOLM-13, and THP-1 cell lines compared with an empty vector (Figure 7E-F). Likewise, mouse miR-200c significantly impaired the proliferation of mouse leukemia cells (Figure 7G). Taken together, these data suggest miR-200 family members negatively regulate AML growth and are epigenetically silenced in AML, which supports the role of ZEB2 as a positive regulator of AML.
Discussion
Using an unbiased genome-wide RNA interface screen in human cell lines coupled with an in vivo validation screen in a murine AML model, we identified ZEB2 as a novel essential requirement in AML. Our genome-scale human cancer screening data have also suggested that ZEB2 plays critical roles in the pathogenesis of a variety of human leukemias including AML, CML-BC, and B-cell acute lymphoblastic leukemia.9 Indeed, ZEB2 overexpression was reported to enhance the development of murine B-cell acute lymphoblastic leukemia driven by the CALM-AF10 chimeric oncogene.46 ZEB2 has also been shown to drive the development of human early T-cell precursor acute lymphoblastic leukemia through enhanced cytokine signaling.47 Our results demonstrate that ZEB2 is an ongoing, critical AML dependency: specifically, ZEB2 depletion appears to terminate AML proliferation by triggering aberrant differentiation via ZEB2’s transcriptional repression of elements of a myeloid differentiation gene expression program.
It is interesting to consider the relationship between ZEB2’s role in AML and its better-studied role in promoting the EMT. EMT transcription factors such as SNAI1 and SNAI2 have been shown to induce AML in mouse models,48,49 although it is unclear whether their roles in AML are related to EMT pathways. Our data reveal that the transcriptional program regulated by ZEB2 in AML significantly overlaps with the genes that are known to be downregulated by ZEB2 during EMT24 (Figure 6C). In AML cells, we have shown that ZEB2 depletion results in upregulation of multiple cell adhesion and migration genes, such as integrins (ITGAM, ITGB5, ITGB7), Toll-like receptors (TLR6, TLR9), and other neutrophil adhesion molecules (FPR2, SELPLG, ICAM3) (supplemental Figure 6D-E). Consistent with our findings, in the murine hematopoietic system, Zeb2 knockout induces cell adhesion genes integrin β1 and Cxcr4, resulting in the retention of hematopoietic stem cells in the fetal liver.26 Thus, much of the ZEB2 regulatory circuit in epithelial cells is preserved in AML cells.
Indeed, we found that the regulatory control of ZEB2 by microRNAs (miRNAs) is also preserved in AML. ZEB2 is regulated by the miR-200 family miRNAs during EMT: miR-200 miRNAs are highly expressed in epithelial cells where they block ZEB2 expression, but are silenced in mesenchymal cells where high ZEB2 levels directly repress miR-200 family miRNAs, potentially leading to their promote methylation.8,45 Consistent with this double-inhibitory feedback loop, we showed that ZEB2 is expressed at high levels in AML and miR-200 at low levels (Figure 3A). Notably, this reciprocal expression pattern of ZEB2 and miR-200 is also conserved in normal mouse hematopoiesis.26,50
ZEB2 is required for embryonic hematopoiesis with embryonic lethality,26 but remarkably, mice with Zeb2 knockout in adult hematopoiesis survive for many months, albeit with differentiation defects that lead to cytopenias (V. Janzen, personal communication, 19 April 2016). This complementary work both confirms ZEB2's important role in hematopoietic differentiation yet also raises the possibility that a meaningful therapeutic window exists as our studies show that there is a rapid differentiation of AML cells with Zeb2 knockdown within days to weeks. Determining the extent to which there is a window for therapeutic Zeb2 inhibition needs to be rigorously evaluated in future work.
Although transcription factors represent historically difficult drug targets, recent reports have demonstrated that they can be selectively targeted for degradation through specific recruitment of E3 ubiquitin ligases. Lenalidomide, a small-molecule drug with clinical efficacy in multiple myeloma, has been shown to cause selective degradation of lymphoid transcription factors IKZF1 and IKZF3.51,52 As a further proof of concept, this strategy has been used to rationally target and degrade the leukemia essential protein, BRD4, via the conjugation of the small-molecule BRD4 inhibitor JQ1 to the aryl ring of thalidomide. This compound, dBET1, was shown to selectively target BRD4 for proteasomal degradation by recruiting E3 ubiquitin ligases.53
In summary, using unbiased screens, we identified ZEB2 as an essential gene in AML, with its loss leading to aberrant differentiation and a block in proliferation. ZEB2 appears to directly suppress genes critical for myeloid differentiation and itself is repressed by the miR-200 family of microRNAs. This and other studies demonstrate that ZEB2 is a critical regulator of normal and malignant hematopoiesis, and further studies are needed to study if a therapeutic window for targeting may exist.
Acknowledgments
The authors thank the Broad Institute RNAi Platform for shRNA plasmids, William McConaughy for assistance with microRNA cloning, Alexandre Puissant for assistance with May-Grunwald-Giemsa staining, Xing Zeng for help with immunoprecipitation, and Cigall Kadoch for guidance on RNA sequencing.
This work is supported by National Institutes of Health (NIH) National Cancer Institute grants U01 CA176058 (W.C.H.), P01 CA066996 (B.E.), and K08 CA184419 (B.G.M.); NIH National Heart, Lung, and Blood Institute grant R01 HL082945 (B.E.); NIH National Institute of General Medical Sciences grant R01 GM083299 (D.P.); St. Baldrick's Foundation Scholar grant and Alex's Lemonade Stand Foundation Young Investigator grant (B.G.M.); and by the Project Achilles Consortium. D.P. is a Howard Hughes Medical Institute Investigator.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: H.L., W.C.H., B.E., and D.P. designed the studies; H.L., B.G.M., H.Z., R.V.P., F.V., and B.A.W. performed experiments; and H.L., B.G.M., and H.Z. analyzed data and wrote the manuscript with input and editing from all authors.
Conflict-of-interest disclosure: W.C.H. and D.P. are consultants for Novartis. The remaining authors declare no competing financial interests.
Correspondence: David Pellman, Howard Hughes Medical Institute, Dana-Farber Cancer Institute and Children's Hospital, Harvard Medical School, 450 Brookline Ave, Boston, MA 02215; e-mail: david_pellman@dfci.harvard.edu.
References
- 1.National Cancer Institute. SEER (Surveillance, Epidemiology and End Results) Cancer Statistics Review, 1975-2010; 2013.
- 2.Saultz JN, Garzon R. Acute myeloid leukemia: a concise review. J Clin Med. 2016;5(3):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barretina J, Caponigro G, Stransky N, et al. . The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helming KC, Wang X, Wilson BG, et al. . ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat Med. 2014;20(3):251-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung HW, Cowley GS, Weir BA, et al. . Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci USA. 2011;108(30):12372-12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuber J, McJunkin K, Fellmann C, et al. . Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol. 2011;29(1):79-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill L, Browne G, Tulchinsky E. ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. Int J Cancer. 2013;132(4):745-754. [DOI] [PubMed] [Google Scholar]
- 9.Cowley GS, Weir BA, Vazquez F, et al. . Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data. 2014;1(140035):140035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo B, Cheung HW, Subramanian A, et al. . Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci USA. 2008;105(51):20380-20385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller PG, Al-Shahrour F, Hartwell KA, et al. . In Vivo RNAi screening identifies a leukemia-specific dependence on integrin beta 3 signaling. Cancer Cell. 2013;24(1):45-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puram RV, Kowalczyk MS, de Boer CG, et al. . Core Circadian Clock Genes Regulate Leukemia Stem Cells in AML. Cell. 2016;165(2):303-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shao DD, Tsherniak A, Gopal S, et al. . ATARiS: computational quantification of gene suppression phenotypes from multisample RNAi screens. Genome Res. 2013;23(4):665-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aikawa Y, Katsumoto T, Zhang P, et al. . PU.1-mediated upregulation of CSF1R is crucial for leukemia stem cell potential induced by MOZ-TIF2. Nat Med. 2010;16(5):580-585. [DOI] [PMC free article] [PubMed]
- 16.Zhou J, Wu J, Li B, et al. . PU.1 is essential for MLL leukemia partially via crosstalk with the MEIS/HOX pathway. Leukemia. 2014;28(7):1436-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuber J, Rappaport AR, Luo W, et al. . An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 2011;25(15):1628-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faber J, Krivtsov AV, Stubbs MC, et al. . HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113(11):2375-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sroczynska P, Cruickshank VA, Bukowski JP, et al. . shRNA screening identifies JMJD1C as being required for leukemia maintenance. Blood. 2014;123(12):1870-1882. [DOI] [PubMed] [Google Scholar]
- 20.Neff T, Sinha AU, Kluk MJ, et al. . Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc Natl Acad Sci USA. 2012;109(13):5028-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris WJ, Huang X, Lynch JT, et al. . The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 2012;21(4):473-487. [DOI] [PubMed] [Google Scholar]
- 22.Schenk T, Chen WC, Göllner S, et al. . Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med. 2012;18(4):605-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Comijn J, Berx G, Vermassen P, et al. . The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7(6):1267-1278. [DOI] [PubMed] [Google Scholar]
- 24.Vandewalle C, Comijn J, De Craene B, et al. . SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res. 2005;33(20):6566-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rapin N, Bagger FO, Jendholm J, et al. . Comparing cancer vs normal gene expression profiles identifies new disease entities and common transcriptional programs in AML patients. Blood. 2014;123(6):894-904. [DOI] [PubMed] [Google Scholar]
- 26.Goossens S, Janzen V, Bartunkova S, et al. . The EMT regulator Zeb2/Sip1 is essential for murine embryonic hematopoietic stem/progenitor cell differentiation and mobilization. Blood. 2011;117(21):5620-5630. [DOI] [PubMed] [Google Scholar]
- 27.Jackson AL, Burchard J, Schelter J, et al. . Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12(7):1179-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buehler E, Chen YC, Martin S. C911: A bench-level control for sequence specific siRNA off-target effects. PLoS One. 2012;7(12):e51942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daigneault M, Preston JA, Marriott HM, Whyte MK, Dockrell DH. The identification of markers of macrophage differentiation in PMA-stimulated THP-1 cells and monocyte-derived macrophages. PLoS One. 2010;5(1):e8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stegmaier K, Ross KN, Colavito SA, O’Malley S, Stockwell BR, Golub TR. Gene expression-based high-throughput screening(GE-HTS) and application to leukemia differentiation. Nat Genet. 2004;36(3):257-263. [DOI] [PubMed] [Google Scholar]
- 32.Novershtern N, Subramanian A, Lawton LN, et al. . Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144(2):296-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borregaard N. Neutrophils, from marrow to microbes. Immunity. 2010;33(5):657-670. [DOI] [PubMed] [Google Scholar]
- 34.Postigo AA, Dean DC. Differential expression and function of members of the zfh-1 family of zinc finger/homeodomain repressors. Proc Natl Acad Sci USA. 2000;97(12):6391-6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharov AA, Ko MS. Exhaustive search for over-represented DNA sequence motifs with CisFinder. DNA Res. 2009;16(5):261-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas-Chollier M, Defrance M, Medina-Rivera A, et al. . RSAT 2011: regulatory sequence analysis tools. Nucleic Acids Res. 2011;39(Web Server issue):W86-W91. [DOI] [PMC free article] [PubMed]
- 37.Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68(10):3645-3654. [DOI] [PubMed] [Google Scholar]
- 38.Taube JH, Herschkowitz JI, Komurov K, et al. . Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA. 2010;107(35):15449-15454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ward AC. The role of the granulocyte colony-stimulating factor receptor (G-CSF-R) in disease. Front Biosci. 2007;12:608-618. [DOI] [PubMed] [Google Scholar]
- 40.Federzoni EA, Humbert M, Valk PJ, et al. . The actin-binding protein CORO1A is a novel PU.1 (SPI1)- and CEBPA-regulated gene with significantly lower expression in APL and CEBPA-mutated AML patients. Br J Haematol. 2013;160(6):855-859. [DOI] [PubMed] [Google Scholar]
- 41.Shi Y, Sawada J, Sui G, et al. . Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422(6933):735-738. [DOI] [PubMed] [Google Scholar]
- 42.Ding J, Zhang ZM, Xia Y, et al. . LSD1-mediated epigenetic modification contributes to proliferation and metastasis of colon cancer. Br J Cancer. 2013;109(4):994-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bots M, Verbrugge I, Martin BP, et al. . Differentiation therapy for the treatment of t(8;21) acute myeloid leukemia using histone deacetylase inhibitors. Blood. 2014;123(9):1341-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehnertz B, Pabst C, Su L, et al. . The methyltransferase G9a regulates HoxA9-dependent transcription in AML. Genes Dev. 2014;28(4):317-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neves R, Scheel C, Weinhold S, et al. . Role of DNA methylation in miR-200c/141 cluster silencing in invasive breast cancer cells. BMC Res Notes. 2010;3:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caudell D, Harper DP, Novak RL, et al. . Retroviral insertional mutagenesis identifies Zeb2 activation as a novel leukemogenic collaborating event in CALM-AF10 transgenic mice. Blood. 2010;115(6):1194-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goossens S, Radaelli E, Blanchet O, et al. . ZEB2 drives immature T-cell lymphoblastic leukaemia development via enhanced tumour-initiating potential and IL-7 receptor signalling. Nat Commun. 2015;6:5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pérez-Mancera PA, Pérez-Caro M, González-Herrero I, et al. . Cancer development induced by graded expression of Snail in mice. Hum Mol Genet. 2005;14(22):3449-3461. [DOI] [PubMed] [Google Scholar]
- 49.Pérez-Mancera PA, González-Herrero I, Pérez-Caro M, et al. . SLUG in cancer development. Oncogene. 2005;24(19):3073-3082. [DOI] [PubMed] [Google Scholar]
- 50.Petriv OI, Kuchenbauer F, Delaney AD, et al. . Comprehensive microRNA expression profiling of the hematopoietic hierarchy. Proc Natl Acad Sci USA. 2010;107(35):15443-15448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu G, Middleton RE, Sun H, et al. . The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343(6168):305-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krönke J, Udeshi ND, Narla A, et al. . Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343(6168):301-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winter GE, Buckley DL, Paulk J, et al. . DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348(6241):1376-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]