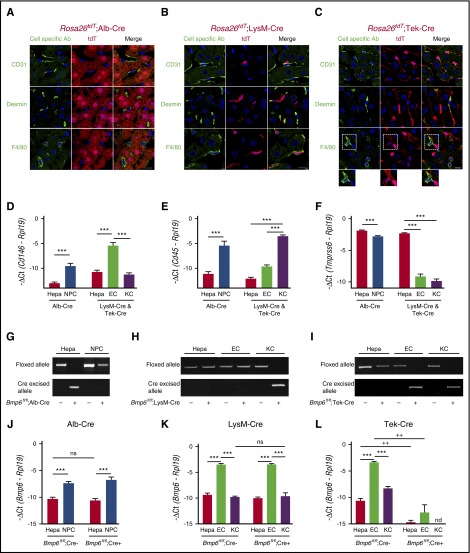
Figure 2.
Validation of conditional Bmp6 knockout mice. (A-C) Immunofluorescence microscopy of liver from offspring of Rosa26tdT Cre-reporter mice mated with (A) Alb-Cre, (B) LysM-Cre, and (C) Tek-Cre mice. Cells expressing Cre recombinase are labeled red throughout the cytoplasm because of the removal of a stop codon upstream of the tdT fluorescent protein. Other NPC populations are labeled green by using CD31 (ECs), desmin (HSCs), and F4/80 (KCs) antibodies. Nuclei are labeled blue with 4′,6-diamidino-2-phenylindole (DAPI). Dotted areas are shown with increased brightness below the right panel. Scale bars represent 10 μM. One representative mouse of 4 to 7 per group is shown. (D-F,J-L) qRT-PCR of (D) the EC marker Cd146, (E) the leukocyte/KC marker Cd45, (F) the hepatocyte (Hepa) marker Tmprss6, and (J-L) Bmp6 relative to Rpl19 mRNA from isolated hepatocytes, NPCs, ECs, or KCs from Bmp6 conditional knockout and littermate control Bmp6fl/fl;Cre– mice (n = 3-5 per group). Results are reported as mean ± SEM of –ΔCt as in Figure 1. (G-I) PCR of genomic DNA for floxed and cre-excised Bmp6 alleles from isolated hepatocytes, NPCs, ECs, or KCs from Bmp6 conditional knockout and littermate control Bmp6fl/fl;Cre– mice (n = 3-5 per group). Representative gels are shown. ***P < .001 for NPCs relative to hepatocytes by Student t test or for ECs relative to hepatocytes or KCs by one-way ANOVA with Tukey post hoc test for mice of the same genotype. ++P < .01 in Bmp6fl/fl;Cre+ relative to Bmp6fl/fl;Cre– mice for the same cell type by Student t test. nd, not detectable; ns, not significant.