Abstract
Drosophila melanogaster is able to thrive in harsh northern climates through adaptations in life-history traits and physiological mechanisms that allow for survival through the winter. We examined the genetic basis of natural variation in one such trait, female virgin egg retention, which was previously shown to vary clinally and seasonally. To further our understanding of the genetic basis and evolution of virgin egg retention, we performed a genome-wide association study (GWAS) using the previously sequenced Drosophila Genetic Reference Panel (DGRP) mapping population. We found 29 single nucleotide polymorphisms (SNPs) associated with virgin egg retention and assayed 6 available mutant lines, each harboring a mutation in a candidate gene, for effects on egg retention time. We found that four out of the six mutant lines had defects in egg retention time as compared with the respective controls: mun, T48, Mes-4, and Klp67A. Surprisingly, none of these genes has a recognized role in ovulation control, but three of the four genes have known effects on fertility or have high expression in the ovaries. We also found that the SNP set associated with egg retention time was enriched for clinal SNPs. The majority of clinal SNPs had alleles associated with longer egg retention present at higher frequencies in higher latitudes. Our results support previous studies that show higher frequency of long retention times at higher latitude, providing evidence for the adaptive value of virgin egg-retention.
Keywords: Drosophila Genetics Reference Panel, virgin egg retention, GWAS, cline
Various aspects of temperate environments challenge ectotherm populations, and many species exhibit patterns of local adaptation in direct response to spatial variation in such selection pressures. Spatially varying selection pressures often generate patterns of clinal variation in a variety of life-history traits, such as reproductive diapause, fecundity, longevity, and stress tolerance (Boulétreau-Merle et al. 1982; Mitrovski and Hoffmann 2001; Schmidt et al. 2005; Umina et al. 2005; Arthur et al. 2008; Schmidt and Paaby 2008). In addition to latitudinal clines, life history traits adaptively evolve over seasonal time scales in direct or indirect response to seasonal fluctuations in climate (Boulétreau-Merle et al. 1987, 1992; Schmidt and Conde 2006; Behrman et al. 2015). In the current study, we focus on Drosophila melanogaster as a model species for climatic adaptation because of the large span of its habitat and wealth of genetic tools that can be used to dissect genetic variation in life-history traits.
Virgin egg retention is a life-history trait that varies clinally, but has not yet been investigated with fine-scale mapping of natural genetic variation. The egg retention time of Drosophila melanogaster virgin females, or delay in the laying of unfertilized eggs, varies both with seasons and along latitudinal gradients (Boulétreau-Merle 1990; Boulétreau-Merle et al., 1992). Genotypes displaying longer egg retention in French D. melanogaster populations are more prevalent in spring and autumn and at higher latitudes (Boulétreau-Merle et al. 1992). Furthermore, long egg retention genotypes are associated with higher viability of pupa, longer life span even upon insemination, and higher fecundity at low winter temperatures than short retention genotypes (Boulétreau-Merle et al. 1992; Boulétreau-Merle and Fouillet 2002). A possible explanation is that a long egg retention phenotype minimizes pointless loss of reproductive potential, as both virgin and mated long retention females retain eggs longer at low temperature than short retention females (Boulétreau-Merle 1990; Boulétreau-Merle and Fouillet 2002). This is evidence that longer retention has adaptive value for overwintering survival, as the capacity to restart the population once spring arrives would be mostly dependent on females that were fertilized in autumn (Bouletreau-Merle et al. 1989; Santiago et al. 1989). Virgin egg retention is genetically determined, with major genetic components on the third chromosome and minor modifiers on the X chromosome, although significant interactions between developmental temperature and chromosomal origin implicate genetic components on the first and second chromosome as well (Bouletreau-Merle et al. 1989; 1998). Until now, naturally segregating genetic variants responsible for this trait have not been mapped at a finer scale.
Here, we investigate the genes and genetic variants underpinning natural variation in virgin egg retention in North American D. melanogaster through a genome-wide association study using inbred lines. Subsequent investigation of a set of candidate genes was performed through experiments using null mutants, gene expression analysis, correlation with other life-history traits, and enrichment analyses for clinality and seasonality. In general, we were able to verify that a majority of tested candidate genes affect virgin egg retention time and that some of the natural polymorphisms underlying virgin egg retention time may adaptively evolve in response to spatially varying selection pressures.
Methods
Drosophila Stocks and Rearing Conditions
We employed 90 lines from the Drosophila Genetic Reference Panel (DGRP; Mackay et al. 2012) for our initial association mapping study; a subset of the full 205 DGRP lines were used for logistic reasons. We note that the limited number of lines used restricts our analysis to the identification of common variants of large effect. Flies were reared on media containing 6.25% unsulfured molasses, 7.8% cornmeal, 3% yeast, and 1.25% agar on a 12L:12D light cycle at 25°C. Classical mutant flies for validation experiments were obtained from the Bloomington Drosophila Stock Center (Bloomington, IN).
Screening DGRP Lines for Egg Retention Phenotype
Ninety DGRP lines were screened in three blocks of ∼30 lines over the course of 3 mo, with each block taking 1 mo to screen. Ten virgin females were collected from each DGRP line and placed into individual vials for the assay. The females were kept on 2% yeast media until the first egg was laid or the fly died. The vials were kept on a 12L:12D light cycle at 25ºC over the course of the assay. The vials were checked for eggs in the morning and in the afternoon by examining the media in the vial under a dissection microscope. Egg retention time was measured by recording the date of initial placement in the vial and the date of the first egg. Vials were kept for an extra 3–4 d after the first egg was deposited to check for presence of larva, which would indicate that the female had been fertilized. Vials containing larva or vials where the flies died before laying the first egg were excluded from the analysis. Overall, 71 vials were removed from the 796 vials in the screen due to fly death or fertilization of the virgin female.
Genome-Wide Association Mapping for Egg Retention Phenotype
Random effect terms for each DGRP line were calculated using a generalized linear mixed effects model in R, using the LME4 package (Bates et al. 2015). The model employed was Yab = μ+ linea + dateb + εab, where Y is the age of females when eggs are first laid, linea represents the random effect of DGRP line, dateb is the random effect of date of virgin female collection for the DGRP line, and εab is the error term. We assumed a Poisson distribution of waiting times. Heritability was calculated as the variance explained by line divided by the sum of all sources of variance but for this analysis, we used the package MCMCglmm (Hadfield 2010) to calculate these variances. The values for each DGRP line were uploaded to the DGRP2 webtool (Huang et al. 2014), which performs mapping taking into account inversion and Wolbachia infection status. We chose a significance threshold of P < 10−5 to identify polymorphism for further consideration. While this is a liberal threshold that may include false positives, it has become the standard for DGRP studies (Mackay et al. 2012) and we view association mapping as a means of generating hypotheses about genes involved in the natural variation for the phenotype and will functionally validate the associations below. We also note that limited number of DGRP lines tested means that our experimental design has the strongest power to detect common variants.
Functional Validation of Genes Containing Associated SNPs
Classical null mutant lines were obtained for all genes (where available) containing SNPs significant at a threshold of P < 10−5. The mutant lines used were: Mi{ET1}GfrlMB00064, P{EPgy2}T48EY07311, P{EPgy2}Mes-4EY01950, Mi{ET1}stnBMB04192, P{EPgy2}Acph-1EY19501, and P{EP}Klp67AEP3516 (Supp Table 1 [online only]). Ten to fifteen virgin females were collected from each genotype (six mutants plus two controls, y1w67c23 and w1118) and placed individually in vials. Vials were checked daily for eggs and females were put on new vials every 5 d. After females laid eggs, vials were kept for an additional week to be sure that no eggs were fertilized because a mated female had been mistaken for a virgin. This design was replicated in three complete blocks for between 30 and 40 observations (individual virgin female flies) per genotype. Differences in egg retention between mutants and controls were ascertained by performing an ANOVA with genotype and block as factors and egg retention days as the response variable. Since all mutants were found on one of two backgrounds a Dunnett’s test was performed after the ANOVA to determine which mutant(s) differed significantly from the control (Hothorn et al. 2008). Note that Stoned-A, Mes-4, T48, and their proper controls were tested in a separate set of experimental blocks than the others and are, therefore, presented separately from the other genes.
Power was calculated by assuming a Poisson distribution of egg retention times with observed means for the control and the mutant. We performed 10,000 random draws of the appropriate sample size from those distributions determined what proportion of those 10,000 draws resulted in a significant difference in egg retention time at P < 0.05. All analysis was done in R (R Core Team 2015).
Analysis of Enrichment of Clinal SNPs
We examined whether our top GWAS hits were enriched among sets of previously identified clinal and seasonal SNPs from samples collected along the east coast of North America (five populations spanning from Florida to Maine) or among seasons (spring and fall over three years) in a focal North American population (Bergland et al. 2014; Machado et al. 2016). A dataset of allele frequencies at 550,895 SNPs sampled from five populations along the east coast of North America was used to assess enrichment of clinal SNPs in the GWAS results (Bergland et al. 2014; Machado et al. 2016). Seven of the SNPs from our GWAS results (P < 10−5) matched the SNPs from Bergland et al. and Machado et al. datasets (Table 1: 3L:1696552, 3L:16448208, 3R:22722373, 3R:23759237, 3R:27115805, X:3655147, X:22397621). These seven SNPs were used to assess enrichment of clinal and seasonal SNPs in our results. One thousand sets of seven control SNPs were generated from the Bergland et al. and Machado et al. datasets by matching the GWAS SNPs by chromosome, minor allele frequency, and whether the SNP was associated with the UTR, intronic, or coding sequences of known genes vs the intergenic regions. The sets of control SNPs were used to create distributions of expected numbers of clinal (seasonal) and non-clinal (non-seasonal) SNPs, to which the observed count of clinal (seasonal) and non-clinal (non-seasonal) SNPs was compared. All analysis was done in R (R Core Team 2015).
Table 1.
SNPs significantly associated with virgin egg retention (P < 10 − 5). MAF is minor allele frequency, Del, deletion, for non-synonymous SNPs, A55G is interpreted as a mutation that causes a change from alanine to glycine in the 55th position of the protein, SNPs downstream of the gene are denoted as “Down (position)”.
| Chr | Pos | MAF | Effect | P-value | Gene | Class |
|---|---|---|---|---|---|---|
| X | 22394286 | 0.384 | −0.120 | 5.86E−07 | StnA/B | Del (78bp—intron) |
| X | 22385304 | 0.333 | −0.130 | 5.93E−07 | StnA/B | Intron |
| 3L | 9356607 | 0.180 | −0.158 | 7.53E−07 | Klp67A | Nonsyn (S691N) |
| X | 22386644 | 0.356 | −0.121 | 1.08E−06 | StnA/B | Intron |
| 2R | 16443531 | 0.227 | −0.143 | 1.32E−06 | lms | Up(833) |
| 3L | 1696552 | 0.218 | −0.148 | 1.38E−06 | CG7991 | Intron |
| 3L | 9356604 | 0.191 | −0.151 | 1.44E−06 | Klp67A | Nonsyn (T692S) |
| 3L | 9356609 | 0.171 | −0.156 | 1.60E−06 | Klp67A | Syn |
| 3R | 25293837 | 0.165 | −0.157 | 2.24E−06 | Ptp99A | Intron |
| 3R | 22722373 | 0.446 | −0.105 | 2.56E−06 | T48 | Intron |
| 3R | 23759237 | 0.302 | −0.124 | 3.95E−06 | Mes-4 | Intron |
| 3R | 16293867 | 0.140 | −0.166 | 4.11E−06 | mun | Intron |
| X | 11867221 | 0.253 | −0.132 | 4.13E−06 | cac | Intron |
| X | 3655147 | 0.144 | −0.153 | 4.36E−06 | tlk | Intron |
| 3L | 12152503 | 0.130 | −0.164 | 4.57E−06 | CG9760 | Syn. |
| X | 22385502 | 0.366 | −0.117 | 5.89E−06 | StnA/B | Intron |
| 3R | 24327835 | 0.193 | −0.141 | 6.18E−06 | Dhc98D | Intron |
| X | 22397621 | 0.364 | −0.112 | 6.42E−06 | StnA/B | 5’ UTR |
| 3R | 16259969 | 0.056 | −0.236 | 6.45E−06 | mun | Intron |
| 3L | 9356600 | 0.227 | −0.123 | 6.70E−06 | Klp67A | Syn. |
| 3L | 16448208 | 0.216 | −0.130 | 6.75E−06 | CG33158 | Intron |
| 3R | 9294054 | 0.057 | −0.229 | 6.83E−06 | NA | NA |
| 3R | 25816726 | 0.112 | −0.178 | 7.05E−06 | Acph-1 | Down (140) |
| X | 3655346 | 0.148 | −0.151 | 7.27E−06 | tlk | Intron |
| 3R | 23719399 | 0.250 | −0.135 | 8.61E−06 | CG34353 | Intron |
| 3R | 6329823 | 0.229 | 0.137 | 8.66E−06 | NA | NA |
| 3R | 25816740 | 0.122 | −0.167 | 9.23E−06 | Acph-1 | Down (126) |
| X | 22382756 | 0.345 | −0.112 | 9.34E−06 | StnA/B | 3’ UTR |
| 3R | 27115805 | 0.333 | 0.109 | 9.86E−06 | NA | NA |
Phenotypic Correlations With Other Traits and Gene Expression
Gene expression data using the entire DGRP mapping population for the genes used in the validation experiments was compiled from Huang et al. (2015). We first tested the association between SNP status and expression of the gene associated with that SNP using the Huang et al. dataset. For the 90 DGRP lines tested for egg retention, we calculated the Pearson correlation between gene expression for and virgin egg retention. In addition, we examined the correlation in gene expression among genes containing significantly associated SNPs.
Line means for traits from several other studies (Mackay et al. 2012; Durham et al. 2014; Unckless et al. 2015) were compiled and correlation was assessed for each trait against virgin egg retention time. All analysis was done in R (R Core Team 2015).
Results
Genome-Wide Association Mapping for Egg Retention
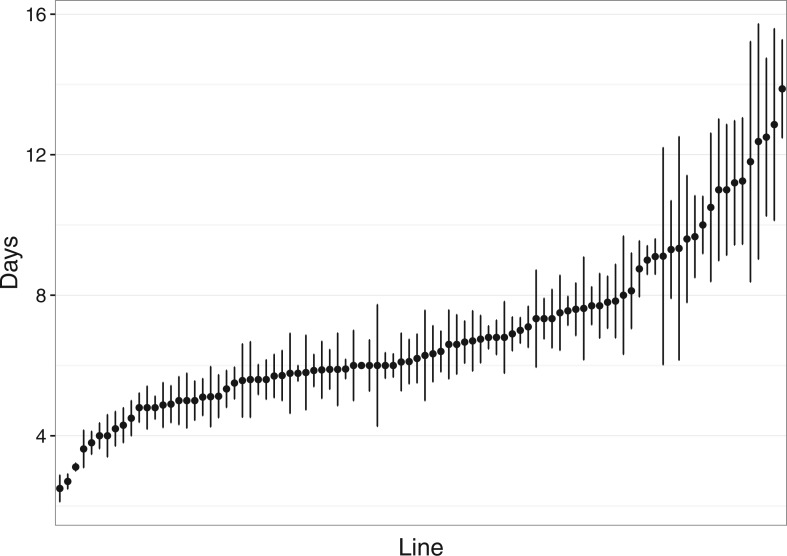
Initial analysis of the egg retention phenotype in the DGRP lines tested showed inter-line variability that is comparable with previous studies (Fig. 1; Bouletreau-Merle et al. 1989). In our study, heritability was estimated to be 0.596 (highest posterior density interval: 0.451–0.727). Twenty-nine of the 1,896,196 variant sites met our threshold significance cutoff of P < 10−5 for association with virgin egg retention time. SNPs significantly associated with egg retention time were distributed across the genome and inspection of the distribution of P-values compared to the expected values via the QQ plot reveals that the model performed relatively well (Table 1; Supp Figs 1A and 1B [online only]). Only one SNP on the second chromosome met our significance threshold of P < 10−5, whereas 19 and 9 met the threshold for the third and X chromosomes, respectively (Fisher’s exact test (FET), P < 0.0001 for both comparisons). SNPs showing significant association with virgin egg retention were more likely to be in or within 1,000 bp of genes, with 26 of 29 (∼90%) of egg retention associated SNPs vs 1,509,979 of 1,896,196 (∼80%) of genome-wide SNPs, although this difference was not significant (FET, P = 0.249). Non-synonymous SNPs (n = 2) were also more likely to be significantly associated with the egg retention phenotype than synonymous SNPs (n = 3) when compared to the rest of the genome (48,720 non-synonymous SNPs, 167,361 synonymous SNPs), but the likelihood was not statistically significant (FET, P = 0.316). Spurious linkage disequilibrium (LD) among physically unlinked SNPs due to the small size of the mapping panel does not appear to be a problem as only tightly physically linked SNPs are in LD with each other (Supp Fig. 2 [online only]; Houle and Marquez 2015; Skelly et al. 2016).
Fig. 1.
Plot of mean egg retention time (± 1 SE) in ascending order for the DGRP lines screened.
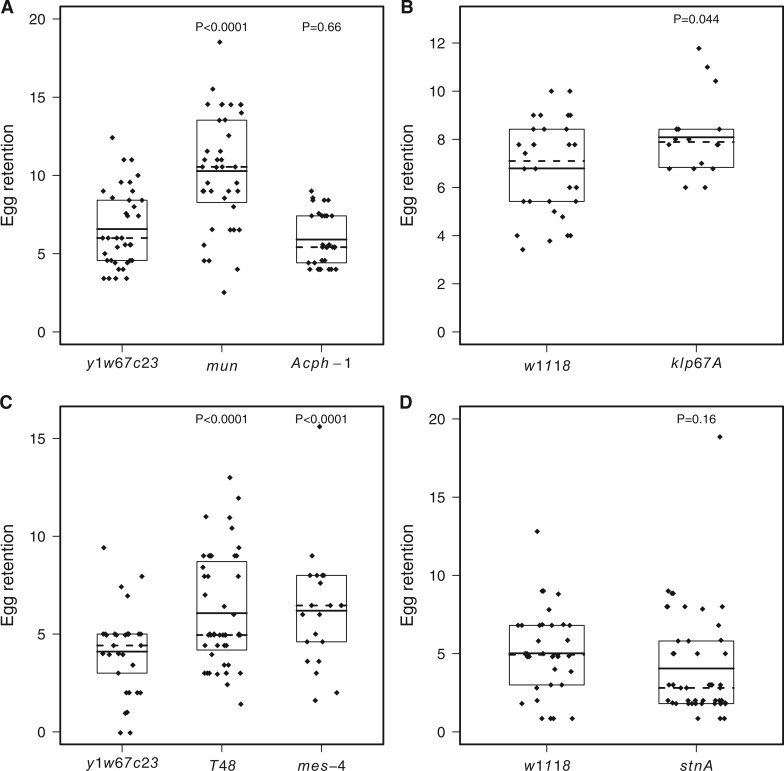
Functional Validation of Genes Containing Associated SNPs
Four of six candidate gene mutants showed a significant difference in egg retention time compared to their controls (Supp Table 2 [online only]; Fig. 2). In all cases where differences were significant, mutant virgins retained eggs longer than their wild-type controls. Below we briefly summarize these results and putative functional roles of these genes.mun: An intronic SNP (3R.16259867, P = 4.11e−06) in mun (also known as Gfrl) was significantly associated with virgin egg retention in our genome-wide analysis. A second SNP (3R:16293969, P = 6.45e−06) was also significantly associated. The mutant allele for mun resulted in significantly longer virgin egg retention (P < 0.0001, mean (control) = 6.00 d, mean (mun mutant) = 10.28 d).
Fig. 2.
Classical mutants in candidate genes show differences in egg retention times compared to their control genotypes. Each panel represents a different control (y1w67c23 or w1118) or experiment: A) first validation experiment with y1w67c23 control, B) first validation experiment with w1118 control, C) second validation experiment with y1w67c23 control, D) second validation experiment with w1118 control. Each point represents a single female corrected for block effects, boxes represent the middle 50th percentile, solid horizontal lines represent means and dashed horizontal lines represent medians. P values are given for comparison of each mutant to the control genotype using a Dunnett’s Test.
T48: An intronic SNP in T48 was significantly associated with virgin egg retention (3R:22722373, P = 2.56e−06). The mutant allele for T48 resulted in significantly longer virgin egg retention (P < 0.0001, mean (control) = 4.10 d, mean (T48 mutant) = 6.07 d).
Mes-4: A single intronic SNP in Mes-4 met our significance threshold (3R: 23759237, P = 3.95e−06) and the mutant allele confirmed a role of Mes-4 in virgin egg retention (P < 0.0001, mean (control) = 4.10 d, mean (Mes-4 mutant) = 6.19 d).
Klp67A: Three SNPs in Klp67A met our significance threshold of P < 10−5: 3L:9356607 (non-synonymous), P = 7.53e−07; 3L:9356604 (non-synonymous), P = 1.44e−06; 3L:9356609 (synonymous), P = 1.60e−06). The mutant allele for Klp67A resulted in significantly longer virgin egg retention compared with the wild-type control (P = 0.0438, mean (control) = 6.84 d, mean (Klp67A mutant) = 8.15 d), although this level of significance is marginal.
Genes failing to validate with mutant lines: Two mutant genes, StnA and Acph-1, failed to show a phenotypic effect in our validation experiments. StnA and StnB are dicistronic genes. Three intronic SNPs, one in the 3′UTR, one in the 5′UTR, one 78-bp deletion in an intron were significantly associated with virgin egg retention (Table 1). Although not significant, mutants for StnA/B exhibited a shorter duration of egg retention (P = 0.164, mean (control) = 5.02 d, mean (StnA/B mutant) = 4.05 d). Both proteins influence neurotransmission in a calcium dependent manner (Soekmadji et al. 2012). Two SNPs (3R.25816726, P = 7.05e−06; 3R.25816740, P = 9.22e−06), 140 and 126 bp downstream of Acph-1, respectively, were significantly associated with virgin egg retention in our genome-wide analysis. The mutant allele for Acph-1 led to a non-significant reduction in virgin egg retention compared to the wild-type control (P = 0.66, mean (control) = 6.53 d, mean Acph-1 (mutant) = 5.82 d). Acph-1 is an acid phosphatase primarily with no obvious connection to virgin egg retention (Tweedie et al. 2009). Note that both these analyses may have suffered from a lack of power since values are small. If we assume the means listed above and a Poisson distribution of egg retention times, then the power to detect a significant difference (assessed by simulation) at P < 0.05 is ∼0.55 for StnA/B and ∼0.19 for Acph-1. This means that if a mutation at StnA/B decreased virgin egg retention time, ∼45% of the time the analysis would not mark it a significant difference. The same can be said for Acph-1, where ∼81% of the time the difference in egg retention times would not be deemed significant.
Significantly associated SNPs in genes not tested: SNPs in several other genes were significantly associated with virgin egg retention but were not functionally validated due to a lack of available mutant stock, proper control, or both (Table 1).
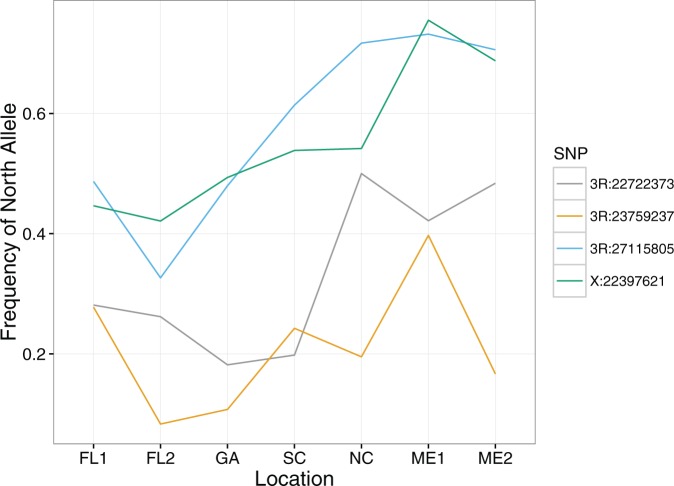
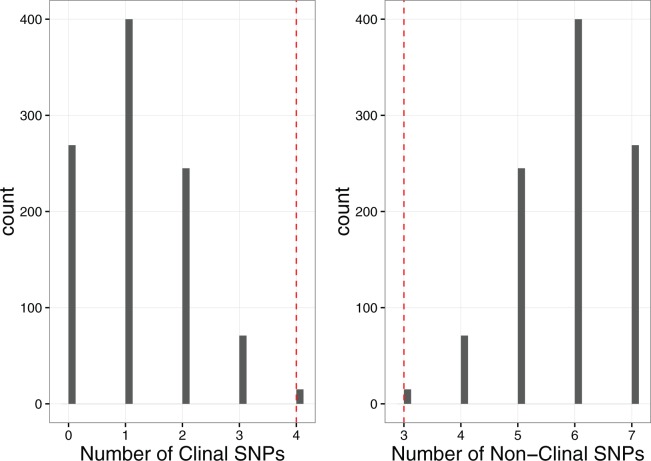
Enrichment for clinality in significantly associated SNPs: Four SNPs out of seven significantly associated with egg retention time (P < 10−5, Table 2) were found to have a significant change in frequency over a North American cline (Fig. 3, FDR < 0.05; Bergland et al. 2014). None of the SNPs were found to have significant change in frequency over seasons (Table 2). To determine if our set of seven SNPs was enriched for clinal SNPs, we used 1,000 sets of seven matched control SNPs to generate distributions of the number of clinal and non-clinal SNPs expected by chance alone (Fig. 4). The mean number of clinal SNPs expected by chance is 1.09± 0.92 (SD) and mean number of non-clinal SNPs expected by chance is 5.91 ± 0.92. Our data show an average excess of 2.91 clinal SNPs, which corresponds to an average odds ratio of 5.86. Therefore, we conclude that there is a significant enrichment of clinal SNPs in our dataset. In addition, we found that three of the four clinal SNPs (3R: 22722373, 3R:23759237, and 3R:27115805), of which 3R:22722373 and 3R:23759237 are in the functionally validated genes T48 and Mes-4, respectively, have the allele found more frequently in the northern latitudes coincide with a longer retention time (Fig. 3; Table 2).
Table 2.
Significance of clinal and seasonal frequency changes of SNPs that are associated with egg retention (P < 10 − 5). The effect (N–S) column shows the effect of each SNP on egg retention time, calculated as ½ (North Allele mean – South Allele mean). If effect (N–S) is greater than 0, then egg retention time is longer in the north vs. south.
| Chr | Pos | MAF | Gene | Class | Season q-value | Clinal q-value | North allele | South allele | Effect (N–S) |
|---|---|---|---|---|---|---|---|---|---|
| 3R | 22722373 | 0.446 | T48 | Intronic | 0.953 | 0.025 | T | C | 0.105 |
| 3R | 23759237 | 0.302 | Mes-4 | Intronic | 0.947 | 0.016 | A | T | 0.124 |
| 3R | 27115805 | 0.333 | NA | NA | 0.788 | 0.001 | C | T | 0.109 |
| 3L | 16448208 | 0.216 | CG33158 | Intronic | 0.939 | 0.665 | – | – | – |
| 3L | 1696552 | 0.218 | CG7991 | Intronic | 0.992 | 0.236 | – | – | – |
| X | 3655147 | 0.144 | tlk | Intronic | 0.966 | 0.799 | – | – | – |
| X | 22397621 | 0.364 | StnA/B | 5’ UTR | 0.954 | 0.044 | T | C | −0.112 |
Fig. 3.
Frequency of the ‘north allele’ along a cline for four clinal SNPs associated with variation in egg retention time. A ‘north allele’ is determined as the most frequent allele for a particular SNP in the northern populations. The location abbreviations correspond to the USA state from which the flies were collected and are ordered from most southern state (FL) to most northern state (ME).
Fig. 4.
SNPs significantly associated with egg retention time are enriched for clinality. The histograms present the distribution of the number of (non-) clinal SNPs from sets of control SNPs matched to our observed SNPs on the basis of chromosome and MAF. The red dashed line is the number of observed (non-) clinal SNPs from our GWAS dataset.
Correlations Among Gene Expression, SNPs and Other Traits
We examined whether variation in gene expression of genes used for validation (a) is correlated with virgin egg retention and (b) whether the state at a given SNP is associated with expression of the gene in which it falls.
Of the five genes we used for validation (no expression data was available for StnA/B), three genes exhibited strong positive correlation in gene expression, Acph-1, Klp67A, and Mes-4 (Supp Fig. 3 [online only]). Pairwise correlation coefficients for these three genes ranged from 0.43 to 0.68, all of which were highly significant (P < 0.001). Expression of CG34356, on the other hand, was significantly negatively correlated with expression of the four other genes, T48, Acph-1, Klp67A, and Mes-4 (Supp Fig. 3 [online only]). To assess the biological significance of these correlations, we calculated pairwise correlations among 1,000 randomly chosen pairs of genes from the rest of the genome. Supp Table 3 [online only] lists the percentile in pairwise correlation of each of the correlation coefficients presented in Supp Fig. 3 [online only]. Four of ten are in the top or bottom five percent of correlations and all but one are in the top or bottom twenty percent.
No genes were significantly correlated with the virgin egg retention phenotype, though Acph-1 expression showed a moderate negative correlation. Only two downstream SNPs near Acph-1 were significantly associated with Acph-1 gene expression (Table 3).
Table 3.
Association between significantly associated SNPs and expression of those genes.
| Chr | Pos | MAF | Gene | Class | t-score/df | P value |
|---|---|---|---|---|---|---|
| 3L | 9356600 | 0.227 | Klp67A | Syn. | 0.667/27.65 | 0.510 |
| 3L | 9356604 | 0.191 | Klp67A | Nonsyn. | 1.561/22.48 | 0.132 |
| 3L | 9356607 | 0.180 | Klp67A | Nonsyn. | 1.469/20.19 | 0.157 |
| 3L | 9356609 | 0.191 | Klp67A | Syn. | 1.489/18.53 | 0.153 |
| 3R | 22722373 | 0.446 | T48 | Intronic | 0.052/75.89 | 0.959 |
| 3R | 23759237 | 0.302 | Mes-4 | Intronic | 0.794/51.14 | 0.431 |
| 3R | 25816726 | 0.112 | Acph-1 | Downstream | 3.179/15.84 | 0.006* |
| 3R | 25816740 | 0.122 | Acph-1 | Downstream | 3.385/19.43 | 0.003* |
*Refers to P < 0.05.
Virgin egg retention was not significantly correlated with any other trait examined including measures of nutritional indices, body mass, and fecundity (Supp Table 4 [online only]). The only phenotype that came close to a significant correlation was a weak negative correlation with chill coma recovery time (R2 = 0.038, P = 0.095).
Discussion
Virgin egg retention is associated with female overwintering success, with long retention genotype females being more likely to survive winter than short retention genotype females (Boulétreau-Merle and Fouillet 2002). Here we examined natural genetic variation in virgin egg retention using a set of inbred Drosophila melanogaster lines. We find genetically based phenotypic variation among lines and a genome-wide association study revealed several SNPs significantly associated with the virgin egg retention phenotype. We were able to confirm a role of four out of six tested candidate genes using classical mutants. The four validated genes were mun, T48, Mes-4, and Klp67A. We also find correlation in gene expression among these genes, though the correlation between gene expression of these genes and virgin egg retention was not significantly different from zero.
Three of the four genes validated have a role in female fertility or have high expression in ovaries. GfrI (mun) and Klp67A mutants show fertility defects (Gandhi et al. 2004; Kallijarvi et al. 2012). GfrI has been shown bind FasII, a neural cell adhesion molecule – the male and female fertility effects were hypothesized to be evolutionary ancient roles for this gene (Kallijarvi et al. 2012). Klp67A is a kinesin-like protein that affects spindle polymerization in mitosis and meiosis, leading to reduced fertility and also defects in embryonic cell divisions (Gandhi et al. 2004). Mes-4 is a histone methyltransferase that is highly expressed in ovaries (Tweedie et al. 2009; Alekseyenko et al. 2014). In C. elegans, Mes-4 is important for proper development of the germline, but no reproductive role for Mes-4 has been identified in Drosophila (Fong et al. 2002). There do not seem to be direct roles of the above genes in ovulation control, but there is a significant effect on virgin egg retention. A possible explanation is that GfrI (mun) and Klp67A have unidentified pleiotropic mechanisms that control both fertility and virgin egg retention. No role in female reproduction or other life-history traits related to overwintering was found for T48. It may be that there are other unidentified pleiotropic effects of this gene.
Temperate populations of D. melanogaster can survive harsh winter conditions in place, whereas the D. simulans sister species likely recolonizes the area through migration from the south or local refugia (Boulétreau-Merle et al. 2003; Bergland et al. 2014; Machado et al. 2016). Clinal and seasonal allele changes associated with life-history traits such as fecundity, reproductive timing and potential, longevity, and reproductive diapause show that selection pressures exerted by the environment alter life-history to favor overwintering (Boulétreau-Merle et al. 1987, 1992; Mitrovski and Hoffmann 2001; Boulétreau-Merle et al. 2003; Hoffmann et al. 2003; Schmidt and Conde 2006). Virgin egg retention phenotypes not only vary clinally and seasonally (Boulétreau-Merle et al. 1992), but are also associated with other life-history traits that favor either overwintering or summertime proliferation (Boulétreau-Merle 1990; Boulétreau-Merle and Fouillet 2002). For example, long retention phenotypes have increased longevity whether virgin or mated vs. short retention phenotypes at 14ºC, long enough for individuals to survive to re-establish the population under more favorable temperatures (Boulétreau-Merle et al. 1992; Boulétreau-Merle and Fouillet 2002).
In the current study, the SNPs that were significantly associated with egg retention time were enriched for clinally varying SNPs, although not for seasonally varying SNPs. All of the clinally varying SNPs were located on the third or X chromosomes, which is expected as the predicted genetic determinants of egg retention were mapped on to the 3rd and X chromosomes (Bouletreau-Merle et al. 1989). In addition, two of the clinal SNPs are located in the functionally validated T48 and Mes-4 genes. The clinal SNPs in these genes are not associated with changes in gene expression, though T48 and Mes-4 increase egg retention time if they are knocked out. Moreover, the most frequent clinal SNP alleles of the T48 and Mes-4 genes in the northern populations are associated with longer egg retention. This coincides with previous findings that northern populations of D. melanogaster have longer egg retention (Boulétreau-Merle et al. 1992). T48 and Mes-4 are good candidates for further study on the mechanisms of how these variants affect egg retention, and perhaps other life history traits that vary clinally. In contrast to previous work (Boulétreau-Merle et al. 1992), we did not find any enrichment of seasonal SNPs, even at liberal thresholds. Association studies using more SNPs may be needed to find the seasonally varying SNPs for the virgin egg retention phenotype.
The expression levels of Klp67A, and Mes-4, genes that were validated in this study, are strongly positively correlated with each other and with Acph-1, a gene whose mutant did not have a significant effect on virgin egg retention, across the DGRP mapping population. The rank correlations in the expression levels of these genes fall into the top five percent of genetic pairwise correlations. T48 has a weak positive correlation with these three genes. Klp67A, Mes-4, Acph-1, and T48 are all negatively correlated with CG34356, with the rank correlations falling into the top or bottom twenty percent of the genetic pairwise correlations. The strong correlations in gene expression suggest that these five genes might be under similar regulatory control and that the cis-regulatory variants in each influence virgin egg retention. Global changes in the expression of several genes could produce the life-history changes associated with the long retention phenotype (Boulétreau-Merle et al. 1992; Boulétreau-Merle and Fouillet 2002). This is similar to the variety of changes in gene expression associated with onset of reproductive diapause (Zhao et al. 2016). Like virgin egg retention, a higher incidence of reproductive diapause is associated with predictable changes to several life history traits, such as increased life span, decreased per capita fecundity, and decreased mortality, and diapause expression is associated with overwintering survival (Schmidt and Paaby 2008; Tatar et al. 2001; Schmidt et al. 2005; Schmidt and Conde 2006). Our results indicate that virgin egg retention and reproductive diapause have different genetic bases, as the causal SNP for diapause variation did not show up in our top hits (Schmidt et al. 2008).
The correlation between egg retention and other life-history traits is not always the case, as in D. melanogaster populations in eastern Australia, which lack a virgin egg retention cline, but have clines for fecundity, longevity, and diapause (Mitrovski and Hoffmann 2001; Sgrò et al. 2006; Lee et al. 2011). In this case, virgin egg retention phenotype should not be predictive of overwintering success. Therefore, long virgin egg retention alone may not be vitally important for overwintering survival, but rather is often linked with a suite of other life history changes that do provide an advantage. Our results point to a weak (but non-significant) correlation with chill coma recovery time, with longer egg retention associated with short chill coma recovery time, but no significant correlations with fecundity, lifespan, or stress resistance. Previous results establishing relationships between virgin egg retention and other life-history traits were done in natural populations or recently established lines (Boulétreau-Merle et al. 1992; Boulétreau-Merle and Fouillet 2002). It is possible that the inbred nature of the DGRP lines induced a general depression in fecundity, longevity, and stress resistance that destroyed any correlation between virgin egg retention and these life-history traits.
Our study sought to find out the genes responsible for variation in virgin egg retention. Using the DGRP panel to conduct a GWAS, four genes out of the top six candidates were validated. These genes, mun, T48, Mes-4, and Klp67A, have pleiotropic effects and their expression levels are highly correlated. These genes are likely under similar regulation and operate in concert to control the physiological changes that are associated with changes in virgin egg retention. Understanding the genes associated with overwintering adaptive traits in D. melanogaster adds to our current knowledge of how climactic adaptation happens in cosmopolitan invertebrates, such as mosquitoes, bees, and moths (Armbruster 2016; Pitts-Singer et al. 2014; Stuckas et al. 2014). Future studies should address the roles of these genes and genetic variants in controlling virgin egg retention and overwintering survival in natural populations of D. melanogaster.
Acknowledgments
This research is supported by the National Institutes of Health grants: NIH NIAID R01-AI083932 to B. Lazzaro; NIH NIGMS K99-GM114714 to R.L.U.; NIH NIGMS R01-GM089926 to P. Schmidt and D. Petrov; NIH NRSA NIGMS F32-GM097837 to A.O.B.
Supplementary Data
Supplementary data are available at Journal of Insect Science online.
References Cited
- Alekseyenko A. A., Gorchakov A. A., Zee B. M., Fuchs S. M., Kharchenko P. V., Kuroda M. I. 2014. Heterochromatin-associated interactions of Drosophila HP1a with dADD1, HIPP1, and repetitive RNAs. Genes Dev. 28: 1445–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster P. A. 2016. Photoperiodic diapause and the establishment of Aedes albopictus (Diptera: Culicidae) in North America. J. Med. Entomol. 53: 1013–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur A. L., Weeks A. R., Sgro C. M. 2008. Investigating latitudinal clines for life history and stress resistance traits in Drosophila simulans from eastern Australia. J. Evol. Biol. 21: 1470–1479. [DOI] [PubMed] [Google Scholar]
- Bates D., Mächler M., Bolker B., Walker S. 2015. Fitting linear mixed-effects models using lme4. 67: 48. [Google Scholar]
- Behrman E. L., Watson S. S., O'brien K. R., Heschel M. S., Schmidt P. S. 2015. Seasonal variation in life history traits in two Drosophila species. J. Evol. Biol. 28: 1691–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland A. O., Behrman E. L., O'brien K. R., Schmidt P. S., Petrov D. A. 2014. Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genet. 10: e1004775.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulétreau-Merle J. 1990. Genetic divergence in the kinetics of ovarian activity and brain control in virgin Drosophila melanogaster females. J. Insect Physiol. 36: 119–124. [Google Scholar]
- Boulétreau-Merle J., Allemand R., Cohet Y., David J. R. 1982. Reproductive strategy in Drosophila melanogaster: significance of a genetic divergence between temperate and tropical populations. Oecologia 53: 323–329. [DOI] [PubMed] [Google Scholar]
- Boulétreau-Merle J., Fouillet P. 2002. How to overwinter and be a founder: egg-retention phenotypes and mating status in Drosophila melanogaster. Evol. Ecol. 16: 309–332. [Google Scholar]
- Boulétreau-Merle J., Fouillet P., Terrier O. 1987. Seasonal variations and balanced polymorphisms in the reproductive potential of temperate D. melanogaster populations. Entomol. Exp. Appl. 43: 39–48. [Google Scholar]
- Boulétreau-Merle J., Fouillet P., Terrier O. 1992. Clinal and seasonal variations in initial retention capacity of virgin Drosophila melanogaster females as a strategy for fitness. Evol. Ecol. 6: 223–242. [Google Scholar]
- Boulétreau-Merle J., Fouillet P., Varaldi J. 2003. Divergent strategies in low temperature environment for the sibling species Drosophila melanogaster and D. simulans: overwintering in extension border areas of France and comparison with African populations. Evolutionary Ecol. 17: 523 [Google Scholar]
- Bouletreau-Merle J., Terrier O., Fouillet P. 1989. Chromosomal analysis of initial retention capacity in virgin Drosophila melanogaster females. Heredity (Edinb). 62 (Pt 2): 145–151. [DOI] [PubMed] [Google Scholar]
- Bouletreau-Merle J., Terrier O., Fouillet P. 1998. A chromosomal analysis of the phenotypic plasticity of some life history traits in relation to developmental temperature. Behav Genetics. 28:403–414. [DOI] [PubMed] [Google Scholar]
- Durham M. F., Magwire M. M., Stone E. A., Leips J. 2014. Genome-wide analysis in Drosophila reveals age-specific effects of SNPs on fitness traits. Nat. Commun. 5: 4338.. [DOI] [PubMed] [Google Scholar]
- Fong Y., Bender L., Wang W., Strome S. 2002. Regulation of the different chromatin states of autosomes and X chromosomes in the germ line of C. elegans. Science. 296: 2235–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R., Bonaccorsi S., Wentworth D., Doxsey S., Gatti M., Pereira A. 2004. The Drosophila kinesin-like protein KLP67A is essential for mitotic and male meiotic spindle assembly. Mol. Biol. Cell. 15: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J. 2010. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J. Stat. Software. 33: 1–22. [Google Scholar]
- Hoffmann A. A., Scott M., Partridge L., Hallas R. 2003. Overwintering in Drosophila melanogaster: outdoor field cage experiments on clinal and laboratory selected populations help to elucidate traits under selection. J. Evol. Biol. 16: 614–623. [DOI] [PubMed] [Google Scholar]
- Hothorn T., Bretz F., Westfall P. 2008. Simultaneous inference in general parametric models. Biomet. J. 50: 346–363. [DOI] [PubMed] [Google Scholar]
- Houle D., Marquez E. J. 2015. Linkage disequilibrium and inversion-typing of the Drosophila melanogaster Genome Reference Panel. G3 (Bethesda). 5: 1695–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Carbone M. A., Magwire M. M., Peiffer J. A., Lyman R. F., Stone E. A., Anholt R. R., Mackay T. F. 2015. Genetic basis of transcriptome diversity in Drosophila melanogaster. Proc. Natl. Acad. Sci. U SA. 112: E6010–E6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Massouras A., Inoue Y., Peiffer J., Ramia M., Tarone A. M., Turlapati L., Zichner T., Zhu D., Lyman R. F., et al. 2014. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 24: 1193–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallijarvi J., Stratoulias V., Virtanen K., Hietakangas V., Heino T. I., Saarma M. 2012. Characterization of Drosophila GDNF receptor-like and evidence for its evolutionarily conserved interaction with neural cell adhesion molecule (NCAM)/FasII. PLoS One. 7: e51997.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. F., Sgro C. M., Shirriffs J., Wee C. W., Rako L., Van Heerwaarden B., Hoffmann A. A. 2011. Polymorphism in the couch potato gene clines in eastern Australia but is not associated with ovarian dormancy in Drosophila melanogaster. Mol. Ecol. 20: 2973–2984. [DOI] [PubMed] [Google Scholar]
- Machado H. E., Bergland A. O., O'brien K. R., Behrman E. L., Schmidt P. S., Petrov D. A. 2016. Comparative population genomics of latitudinal variation in Drosophila simulans and Drosophila melanogaster. Mol. Ecol. 25: 723–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., Zhu D., Casillas S., Han Y., Magwire M. M., Cridland J. M., et al. 2012. The Drosophila melanogaster Genetic Reference Panel. Nature. 482: 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovski P., Hoffmann A. A. 2001. Postponed reproduction as an adaptation to winter conditions in Drosophila melanogaster: evidence for clinal variation under semi-natural conditions. Proc. Biol. Sci. 268: 2163–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts-Singer T. L., Cane J. H., Trostle G. 2014. Progeny of Osmia lignaria from distinct regions differ in developmental phenology and survival under a common thermal regime. J. Insect Physiol. 67: 9–19. [DOI] [PubMed] [Google Scholar]
- Santiago E., Dominguez A., Albornoz J., Pineiro R., Izquierdo J. I. 1989. Environmental sensitivity and heterosis for egg laying in Drosophila melanogaster. Theor. Appl. Genet. 78: 243–248. [DOI] [PubMed] [Google Scholar]
- Schmidt P. S., Conde D. R. 2006. Environmental heterogeneity and the maintenance of genetic variation for reproductive diapause in Drosophila melanogaster. Evolution. 60: 1602–1611. [PubMed] [Google Scholar]
- Schmidt P. S., Matzkin L., Ippolito M., Eanes W. F. 2005. Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evolution. 59: 1721–1732. [PubMed] [Google Scholar]
- Schmidt P. S., Paaby A. B. 2008. Reproductive diapause and life-history clines in North American populations of Drosophila melanogaster. Evolution 62: 1204–1215. [DOI] [PubMed] [Google Scholar]
- Schmidt P. S., Zhu C. T., Das J., Batavia M., Yang L., Eanes W. F. 2008. An amino acid polymorphism in the couch potato gene forms the basis for climatic adaptation in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 105: 16207–16211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgrò C. M., Magiafoglou A., Faine L., Hoffmann A. A. 2006. Absence of clinal variation in virgin retention capacity in Australian Drosophila melanogaster. Evol. Ecol. 20: 407–413. [Google Scholar]
- Skelly D. A., Magwene P. M., Stone E. A. 2016. Sporadic, global linkage disequilibrium between unlinked segregating sites. Genetics. 202: 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soekmadji C., Angkawidjaja C., Kelly L. E. 2012. Ca2+ regulates the Drosophila Stoned-A and Stoned-B proteins interaction with the C2B domain of Synaptotagmin-1. PLoS One 7: e38822.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckas H., Mende M. B., Hundsdoerfer A. K. 2014. Response to cold acclimation in diapause pupae of Hyles euphorbiae (Lepidoptera: Sphingidae): candidate biomarker identification using proteomics. Insect Mol. Biol. 23: 444–456. [DOI] [PubMed] [Google Scholar]
- Tatar M., Chien S. A., Priest N. K. 2001. Negligible senescence during reproductive dormancy in Drosophila melanogaster. Am. Nat. 158: 248–258. [DOI] [PubMed] [Google Scholar]
- Tweedie S., Ashburner M., Falls K., Leyland P., Mcquilton P., Marygold S., Millburn G., Osumi-Sutherland D., Schroeder A., Seal R., et al. 2009. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 37: D555–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umina P. A., Weeks A. R., Kearney M. R., Mckechnie S. W., Hoffmann A. A. 2005. A rapid shift in a classic clinal pattern in Drosophila reflecting climate change. Science 308: 691–693. [DOI] [PubMed] [Google Scholar]
- Unckless R. L., Rottschaefer S. M., Lazzaro B. P. 2015. A genome-wide association study for nutritional indices in Drosophila. G3 (Bethesda). 5: 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Bergland A. O., Behrman E. L., Gregory B. D., Petrov D. A., Schmidt P. S. 2016. Global transcriptional profiling of diapause and climatic adaptation in Drosophila melanogaster. Mol. Biol. Evol. 33: 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]