Abstract
Differentiation of human embryonic stem cells into mesendoderm (ME) is directed by extrinsic signals and intrinsic epigenetic modifications. However, the dynamics of these epigenetic modifications and the mechanisms by which extrinsic signals regulate the epigenetic modifications during the initiation of ME differentiation remain elusive. In this study, we report that levels of histone H3 Lys-27 trimethylation (H3K27me3) decrease during ME initiation, which is essential for subsequent differentiation induced by the combined effects of activin and Wnt signaling. Furthermore, we demonstrate that activin mediates the H3K27me3 decrease via the Smad2-mediated reduction of EZH2 protein level. Our results suggest a two-step process of ME initiation: first, epigenetic priming via removal of H3K27me3 marks and, second, transcription activation. Our findings demonstrate a critical role of H3K27me3 priming and a direct interaction between extrinsic signals and epigenetic modifications during ME initiation.
Keywords: activin, chromatin modification, embryonic stem cell, SMAD transcription factor, Wnt signaling, H3K27me3, Mesendoderm, PRC2
Introduction
Differentiation of human embryonic stem cells (hESCs)4 into endoderm and mesoderm involves an intermediate state called mesendoderm (ME), equivalent to the primitive streak during gastrulation in early embryo development (1). Multiple signals have been demonstrated to induce ME differentiation of hESCs. Among them, Wnt and the transforming growth factor-β (TGF-β) family member activin appear to play critical roles in this process (reviewed in Refs. 2–4). The correct state of chromatin and histone modification is also a prerequisite for initiation and activation of ME differentiation. In ES cells, many genes involved in differentiation are kept poised for activation by the bivalent histone modifications H3K4 trimethylation (H3K4me3, an active mark) and H3K27 trimethylation (H3K27me3, a repressive mark). These modifications help to maintain pluripotency and make the genes ready for activation in response to differentiation signals (5–8).
H3K27me3 levels are determined by the balance between the methyltransferase polycomb repressive complex 2 (PRC2) (9, 10) and the demethylases JMJD3/KDM6B and UTX/KDM6A (11, 12). PRC2, which contains three core components (EZH2, SUZ12, and EED), is one of the most extensively studied chromatin regulators involved in gastrulation and ME differentiation. Gene targeting studies have revealed that mouse embryos lacking Ezh2, the catalytic subunit of PRC2, cannot complete the process of gastrulation to form the primitive streak (13), and ablation of Suz12, which is required for EZH2-methyltransferase activity, yields phenotypes similar to those observed in Ezh2-null mouse embryos (14, 15). Eed mutant mice also exhibit mesoderm defects during gastrulation (16). These observations demonstrate an essential role of PRC2 in early mouse development. Accumulating evidence also suggests that PRC2 plays an important role in ESC differentiation toward mesoderm and endoderm lineages (8, 17, 18).
Interactions between extrinsic signals and epigenetic modifications to determine ME specification have started to emerge. The activin/Nodal signaling mediators Smad2/3 have been shown to recruit JMJD3 to the promoters of the T and Nodal genes in mouse ESCs, leading to removal of the repressive H3K27me3 mark and thereby initiating ME differentiation (19). JMJD3/UTX can also remove H3K27me3 from the WNT3 gene and activate its expression, which, in turn, stimulates β-catenin-mediated gene expression and promotes definitive endoderm differentiation (20). In response to TGF-β/Nodal signals, the PHD-Bromo-containing protein TRIM33 facilitates the binding of Smad2/3 to trimethylated histone H3 Lys-9 and acetylated histone H3 Lys-18 on the promoters of the ME regulators Gsc and MixL1, thus leading to displacement of the chromatin-compacting factor HP1γ and gene activation in mouse ESCs (21).
Although various extrinsic signals have been shown to govern ME specification of hESCs and the importance of epigenetic and transcriptional regulation has been recognized, most studies to date have focused on the late stages of ME differentiation. Furthermore, ME differentiation has been induced by small molecular inhibitors or relatively high doses of growth factors in these studies, which may not accurately reflect the physiological process of early embryo development and may have missed important molecular events in the early stages of differentiation. Although genome-wide analyses have shown little change in H3K27me3 levels between the pluripotent state and differentiated lineage states of ES cells (22, 23), changes in H3K27me3 levels at the onset of ME differentiation and the possible regulation of H3K27me3 levels by extrinsic signals during ME initiation of hESCs remain obscure. In this study, we demonstrate that activin A and Wnt3a at low concentrations can induce ME differentiation of hESCs, and ME differentiation is initiated by a two-step process of epigenetic priming followed by transcription activation. We further show that signaling by activin, but not Wnt, impairs PRC2 activity, reducing global H3K27me3 levels during ME initiation, and establishes an appropriate chromatin state for subsequent activation of ME gene transcription through the combined effects of activin and Wnt signaling. These results demonstrate a direct interaction between extrinsic signals and intrinsic epigenetic modification via the activin/Smad2-PRC2-H3K27me3 regulatory axis that modulates initiation of ME differentiation of hESCs.
Results
Activin A and Wnt3a at Low Doses Efficiently Induce ME Differentiation
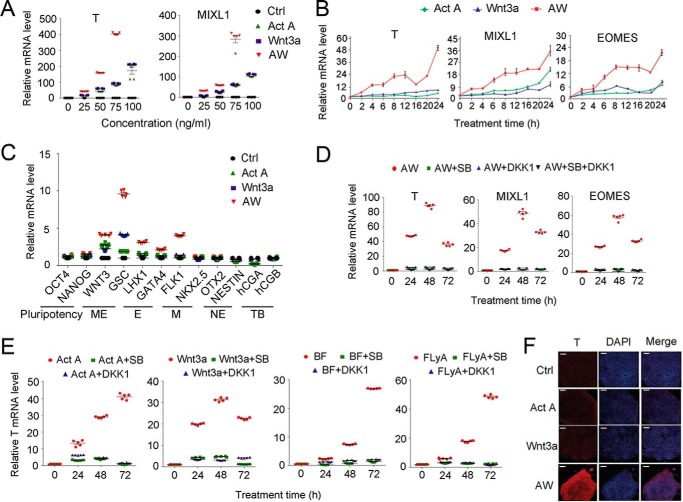
Various protocols have been used to induce the differentiation of human pluripotent stem cells toward ME and its derivative lineages (4). Activin and Wnt have been suggested as the essential extrinsic signals for differentiation of ESCs into ME (2, 4). Through a series of experiments investigating ME differentiation of H1 hESCs with activin A and Wnt3a, we established that a combination of activin A and Wnt3a (AW) at low concentrations (25 ng/ml each) is sufficient to induce ME differentiation, which was demonstrated by the increased mRNA expression of the ME marker genes T (BRACHYURY), MIXL1, EOMES, WNT3, and GSC (Fig. 1, A–C). Inhibition of either activin signaling with its type I receptor inhibitor SB431542 or Wnt signaling with its antagonist DKK1 impaired ME specification (Fig. 1, D and E). Immunostaining also revealed the expression of T upon AW treatment for 24 h (Fig. 1F), which was confirmed by FACS analysis (data not shown). In contrast, induction of trophoblast markers (CGA and CGB) or neuroectoderm markers (NESTIN and OTX2) was not observed (Fig. 1C). Expression of the pluripotency markers OCT4 and NANOG as well as of mesoderm and endoderm markers was slightly increased (Fig. 1C).
FIGURE 1.
Activin A and Wnt3a are necessary and sufficient to drive ME differentiation. A, H1 cells cultured on a Matrigel-coated plate were treated with various concentrations of activin A, Wnt3a, or AW for 48 h in B27 medium before they were harvested for qPCR. B, H1 cells were treated with 25 ng/ml activin A, 25 ng/ml Wnt3a, or AW for the indicated times before they were harvested for qPCR. C, H1 cells were treated with 25 ng/ml activin A, 25 ng/ml Wnt3a, and 25 ng/ml AW for 48 h before they were harvested for qPCR. E, endoderm; M, mesoderm; NE, neuroectoderm; TB, trophoblast. D, H1 cells were treated with AW, together with 10 μm SB431542 or 100 ng/ml DKK1 as indicated, and harvested at 24, 48, or 72 h for qPCR. E, H1 cells were treated with BF (5 ng/ml BMP4 and 40 ng/ml bFGF), FLyA (20 ng/ml bFGF, 10 μm Ly294002, and 20 ng/ml activin A), 100 ng/ml activin A, or 100 ng/ml Wnt3a in the presence or absence of 10 μm SB431542 or 100 ng/ml DKK1 for the indicated times and then harvested for qPCR. F, H1 cells cultured on a Matrigel-coated plate were treated with 25 ng/ml activin A, 25 ng/ml Wnt3a, and both for 24 h in B27 medium and then harvested for anti-T immunofluorescence. The nucleus was counterstained with DAPI. Scale bar, 100 μm. The data in A–E are shown as mean ± S.E. (error bars) (n = 15, including 5 biological replicates and 3 technical replicates). A two-way ANOVA test was used.
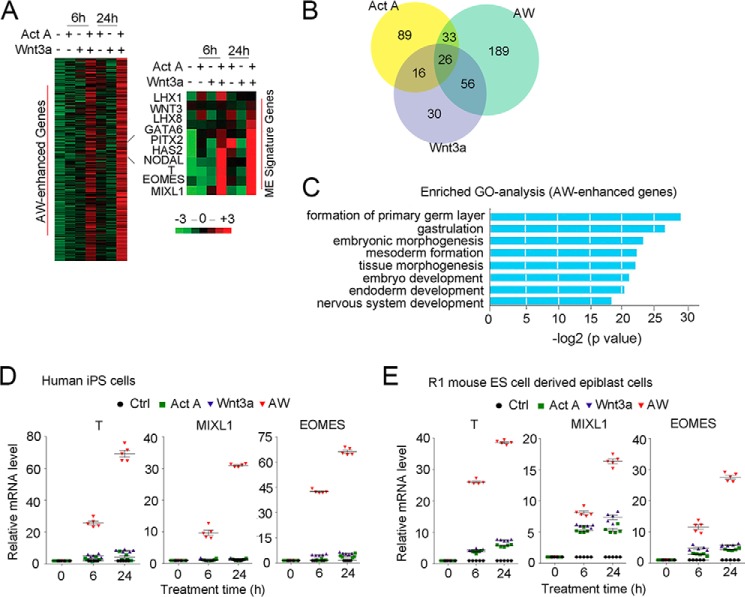
RNA-seq analysis of H1 cells treated with 25 ng/ml activin A or Wnt3a alone or in combination (AW) for 6 and 24 h revealed that a group of 304 genes was significantly up-regulated with AW treatment and exhibited expression patterns similar to those of the ME marker genes (T, MIXL1, and EOMES) (Fig. 2A and supplemental data sets 1 and 2). We defined these genes as AW-enhanced genes, among which 189 genes were induced only by AW (Fig. 2B and supplemental data set 3), indicating the cooperative effect of activin A and Wnt3a. DAVID gene ontology analysis indicated the functions of the AW-enhanced genes in gastrulation and in endoderm and mesoderm differentiation (Fig. 2C). In addition, we found that 25 ng/ml AW also induced ME differentiation of human induced pluripotent stem cells derived from CD34+ cord blood cells and R1 mouse ESCs (Fig. 2, D and E). Taken together, these results indicate that activin and Wnt signaling are necessary and sufficient to drive ME differentiation of pluripotent stem cells.
FIGURE 2.
Activin A and Wnt3a cooperate to induce ME signature genes. A, H1 cells were treated with 25 ng/ml activin A, 25 ng/ml Wnt3a, or AW for 6 or 24 h before they were harvested for RNA-seq. A heat map was generated to show gene expression. Right, genes showing a similar expression pattern as T were selected by PCC > 0.8 from the RNA-seq data. B, Venn plot showing overlapped DEGs after treatment with activin A, Wnt3a, or both. DEGs were obtained by the method described under “Experimental Procedures.” Genes expressed at both 6 and 24 h of each treatment were selected (supplemental data set 3). C, DAVID gene annotation enrichment analysis within AW-enhanced genes. Analysis was done with HOMER, requiring a p value <1e−5. D, human induced pluripotent stem cells derived from CD34+ cord blood cells were treated with 25 ng/ml activin A, 25 ng/ml Wnt3a, or both in B27 medium for 6 or 24 h before they were harvested for qPCR to examine the mRNA expression of T and MIXL1. E, mouse epiblast cells derived from R1 mouse ES cells were treated with 25 ng/ml activin A, 25 ng/ml Wnt3a, or both in B27 medium for 6 and 24 h before they were harvested for qPCR to examine the mRNA expression of T and MIXL1. The statistical data are shown as mean ± S.E. (error bars) (n = 15, including 5 biological replications and 3 technical replications for D and E).
H3K27me3 Reduction Facilitates ME Initiation
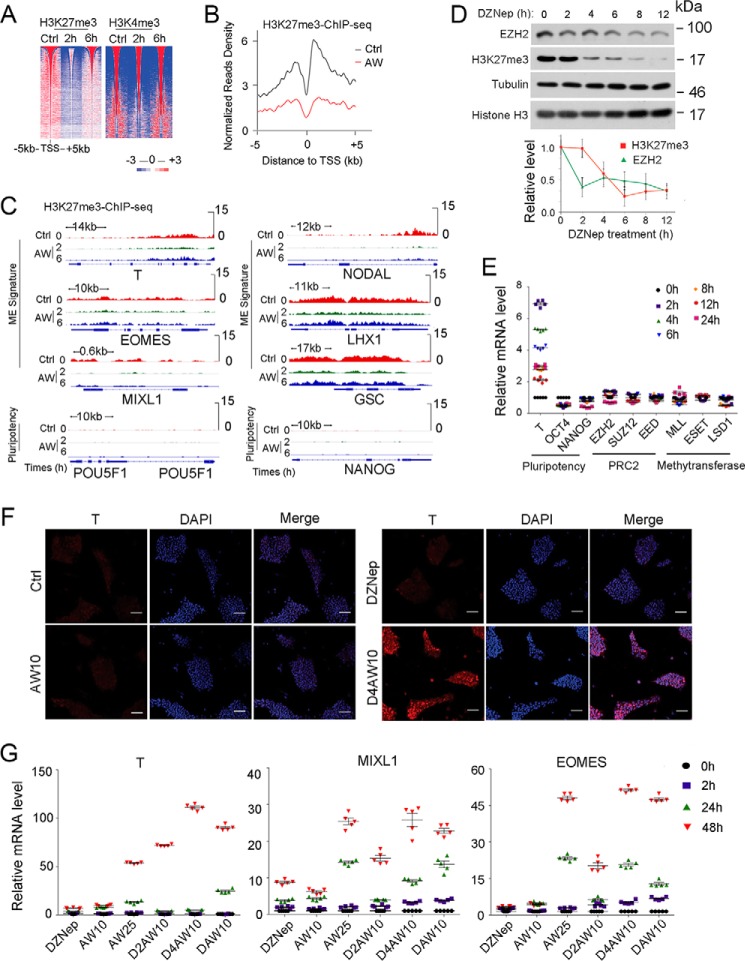
We then carried out a chromatin immunoprecipitation assay followed by high throughput sequencing (ChIP-seq) to assess H3K4me3 and H3K27me3 changes during AW-induced ME differentiation. As shown in Fig. 3A, the global H3K27me3 level was dramatically decreased at 2 h of AW treatment and then partially recovered at 6 h, indicating a dynamic change in H3K27me3 modification at ME initiation. Mean profile analysis also showed a significant decrease of H3K27me3 around the transcription start sites of AW-enhanced genes (Fig. 3B). In addition, the decreased H3K27me3 level at 2 h was observed for genes related to ME lineage specification (T, MIXL1, EOMES, NODAL, LHX1, and GSC) (Fig. 3C). Taken together, these results reveal for the first time a reduction in H3K27me3 levels during ME initiation. In contrast, there was little change for H3K4me3 at ME initiation (Fig. 3A).
FIGURE 3.
H3K27me3 reduction facilitates ME initiation. A, heat map of H3K27me3 and H3K4me3 intensity centered at all of the detected peaks after H1 cells were treated with 25 ng/ml AW for 2 or 6 h before they were harvested for anti-H3K27me3 and anti-H3K4me3 ChIP-seq. B, mean ChIP-seq intensity profile showed the decrease of H3K27me3 around the TSS of AW enhanced genes. Valuable peaks were sorted by peak size. Pluripotent H1 cells were used as a control. C, IGV showed H3K27me3 enrichment at 2 and 6 h on ME and pluripotency genes as revealed by H3K27me3 ChIP-seq upon AW treatment. D, H1 cells were treated with 10 ng/ml DZNep in B27 medium for the indicated times before they were harvested for anti-EZH2 immunoblotting or for histone extraction followed by anti-H3K27me3 and anti-histone H3 immunoblotting. The relative levels of H3K27me3 and EZH2 were shown after normalization with histone H3 or tubulin, respectively. E, H1 cells were treated with 10 ng/ml DZNep for indicated times before they were harvested for qPCR. F, H1 cells were pretreated with 10 ng/ml DZNep for 4 h (D4) before they were treated with 10 ng/ml both activin A and Wnt3a (AW10) for another 24 h, and then cells were harvested for anti-T immunofluorescence. The nucleus was counterstained with DAPI. Scale bar, 100 μm. G, H1 cells were pretreated with or without 10 ng/ml DZNep for 2 h (D2) or 4 h (D4) before 10 ng/ml both activin A and Wnt3a were added for another 2, 24, and 48 h. Then the cells were harvested for qPCR. DAW10, 10 ng/ml DZNep plus AW10. 25 ng/ml AW (AW25) was used as the positive control for ME differentiation. The statistical data are shown as mean ± S.E. (error bars) (n = 15, including 5 biological replicates and 3 technical replicates for E and G; n = 9, including 3 biological replicates and 3 technical replicates for D; n = 3, including 3 biological replicates for F). The Friedman test was performed in D.
H3K27me3 is maintained by the methyltransferase PRC2 (24, 25). To elucidate the functional significance of the H3K27me3 reduction during ME initiation, we used the EZH2 inhibitor DZNep (26) to assess whether reducing H3K27me3 levels by inhibiting PRC2 activity would facilitate ME differentiation. As shown in Fig. 3D, DZNep treatment of H1 cells for 2 h resulted in a decrease in EZH2 protein levels, followed by the reduction of H3K27me3 levels at 4 h. There was no significant change in mRNA levels of PRC2 components as well as other lysine methyltransferases, such as ESET, MLL, and LSD1, in the 48 h after DZNep treatment (Fig. 3E). Treatment with DZNep alone increased ME marker gene expression slightly (Fig. 3, E and F). The mRNA levels of the pluripotent marker OCT4 decreased, and NANOG mRNA levels showed no significant change in the 24 h after DZNep treatment (Fig. 3E). However, pretreatment of DZNep for 2–4 h enabled ME differentiation at low concentrations of activin A and Wnt3a (10 ng/ml each), which otherwise had little effect (Fig. 3, F and G). These results together indicate that the reduction of H3K27me3 levels primes cells for subsequent differentiation to ME.
Reduction of H3K27me3 Levels Is Specific during ME Initiation
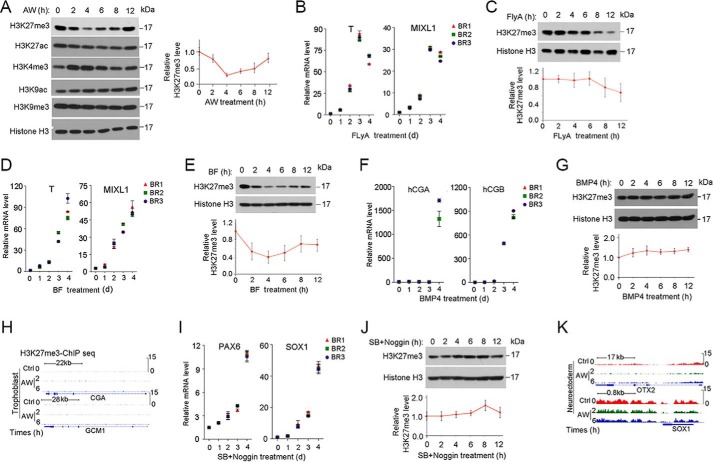
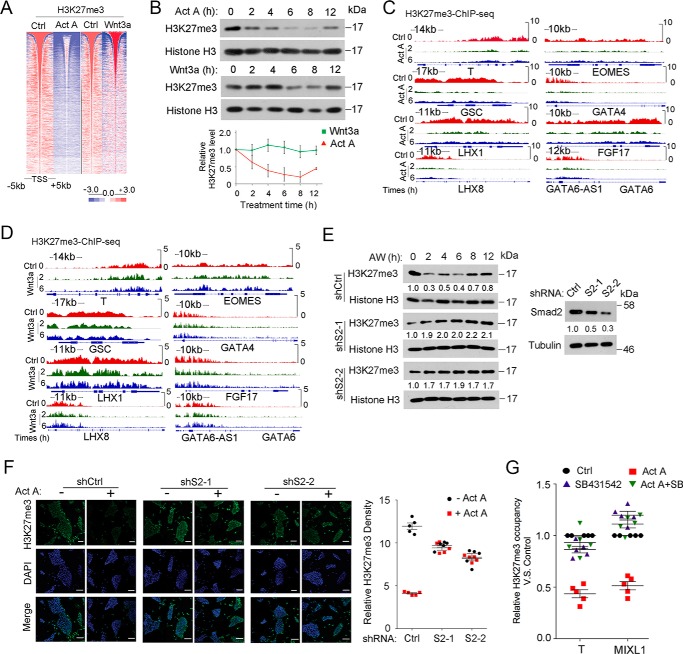
To further assess the changes in H3K27me3 levels during initiation of ME differentiation, we examined the global pattern of changes by immunoblotting. Consistent with the ChIP-seq results, immunoblotting analysis revealed that the global H3K27me3 level was reduced at the beginning (2–8 h) of AW-induced ME specification and then recovered at 12 h (Fig. 4A). In contrast, the global H3K4me3 level was slightly increased at 2–6 h. Additionally, levels of trimethylated histone H3 Lys-9 and acetylated histone H3 Lys-27 remained unchanged during this period, whereas levels of acetylated histone H3 Lys-9 slightly increased along the time (Fig. 4A). To determine whether the H3K27me3 reduction is ME-specific, we confirmed H3K27me3 levels with other ME differentiation protocols as well as other cell fate differentiation systems. Several protocols have been used to induce ME differentiation, including bone morphogenetic protein 4 (BMP4) + FGF2 (BF) (27) and FLyA (28). A reduction in H3K27me3 levels was observed in BF- and FLyA-induced ME specification, albeit at a delayed time point (Fig. 4, B–E). In contrast, the H3K27me3 level was little changed in the early stage of trophoblast differentiation (27, 29, 30) (Fig. 4, F–H) and neuroectoderm differentiation (31, 32) (Fig. 4, I–K). These data together suggest that reduction of H3K27me3 specifically occurs in the early stage of ME differentiation.
FIGURE 4.
H3K27me3 is specifically reduced during ME initiation. A, H1 cells were treated with 25 ng/ml AW in B27 medium for the indicated times before they were harvested for histone extraction, followed by immunoblotting with the indicated antibodies. B and C, H1 cells were treated with FLyA (20 ng/ml bFGF, 10 μm Ly294002, and 20 ng/ml activin A) for the indicated times before they were harvested for qPCR (B) or for histone extraction followed anti-H3K27me3 immunoblotting (C). D and E, H1 cells treated with BF (5 ng/ml BMP4 and 40 ng/ml bFGF) for the indicated times before they were harvested for qPCR (D) or for histone extraction and anti-H3K27me3 immunoblotting (E). F and G, H1 cells treated with 5 ng/ml BMP4 in B27 medium for the indicated times before they were harvested for qPCR (F) or for histone extraction and then anti-H3K27me3 immunoblotting (G). H, IGV showed H3K27me3 enrichment at 2 and 6 h on the specific trophoblast genes CGA and GCM1, as revealed by H3K27me3 ChIP-seq upon AW treatment. I and J, H1 cells were treated with 10 μm SB431542 and 100 ng/ml Noggin in B27 medium for the indicated times before they were harvested for qPCR (I) or for histone extraction and then anti-H3K27me3 immunoblotting (J). K, IGV showed H3K27me3 enrichment at 2 and 6 h on the specific neuroectoderm genes OTX2 and SOX1, as revealed by H3K27me3 ChIP-seq upon AW treatment. H3K27me3 bands were quantified, and the relative levels were shown after normalization to histone H3, and the statistical data are shown as mean ± S.E. (error bars) (n = 9, including 3 biological replicates and 3 technical replicates for A, C, E, G, and J). A Friedman test was performed in A, C, E, G, and J.
Activin Reduces H3K27me3 Levels
To explore the mechanism underlying the decline of H3K27me3 during AW-induced ME differentiation, we assessed the respective roles of activin and Wnt signaling during ME initiation. As shown in Fig. 5A, heat map analysis of H3K27me3 ChIP-seq showed that activin decreased the H3K27me3 level at 2 h, which was similar to AW treatment, but Wnt had little effect at this time point. When the respective effect of activin and Wnt on H3K27me3 levels by immunoblotting was assessed, we found that activin decreased the H3K27me3 levels between 2 and 8 h, but Wnt3a slightly reduced H3K27me3 levels at 6–8 h (Fig. 5B). Consistently, H3K27me3 occupancy of ME signature genes decreased significantly upon 2-h activin A treatment (Fig. 5C), whereas there was little change by Wnt3a (Fig. 5D). Furthermore, the reduction of H3K27me3 by AW was blocked with Smad2 depletion (Fig. 5, E and F). These results were further confirmed by the observation that the activin type I receptor inhibitor SB431542 blocked the activin A-induced reduction of H3K27me3 on the ME signature genes T and MIXL1 (Fig. 5G).
FIGURE 5.
Activin/Smad2 signaling is critical for H3K27me3 reduction. A, heat map of H3K27me3 and H3K4me3 intensity centered at all of the detected peaks after H1 cells were treated with 25 ng/ml activin A or 25 ng/ml Wnt3a for 2 h before they were harvested for the ChIP assay followed by high throughput sequencing against H3K27me3 and H3K4me3 antibody. B, H1 cells treated with 25 ng/ml activin A or 25 ng/ml Wnt3a for the indicated times before they were harvested for histone extraction followed by anti-H3K27me3 immunoblotting. H3K27me3 bands were quantified, and the relative levels were shown after normalization to histone H3. A Friedman test was performed, and the statistical data are shown as mean ± S.E. (error bars) (n = 9, including 3 biological replicates and 3 technical replicates). C and D, IGV showed examples of H3K27me3 enrichment at 2 and 6 h on specific ME genes as revealed by H3K27me3 ChIP-seq upon activin A (C) or Wnt3a treatment (D). E, Smad2 knockdown H1 cells were treated with 25 ng/ml AW in B27 medium for the indicated times before they were harvested for histone extraction followed by anti-H3K27me3 immunoblotting. Nonspecific target shRNA was used as control. Knockdown efficiency of Smad2 was shown in the left panel. Smad2 and H3K27me3 bands were quantified. F, Smad2 knockdown H1 cells treated with 25 ng/ml activin A for 2 h before they were harvested for anti-H3K27me3 immunofluorescence. H3K27me3 densities were quantified with ImageJ. The nucleus was counterstained with DAPI. Scale bar, 100 μm. G, H1 cells treated with 25 ng/ml activin A, 10 μm SB431542, or both for 2 h before they were harvested for anti-H3K27me3 ChIP followed by qPCR. The amplified promoter regions for ChIP-qPCR were as follows: T at about 1 kb upstream of TSS; MIXL1 at about 0.8 kb upstream of the TSS. The statistical data are shown as mean ± S.E. (F, n = 5, including 5 biological replicates; G, n = 15, including 5 biological replicates and 3 technical replicates). A two-way ANOVA test was used in F and G.
Activin/Smad2 Decreases H3K27me3 Level via Reducing EZH2 Protein Level
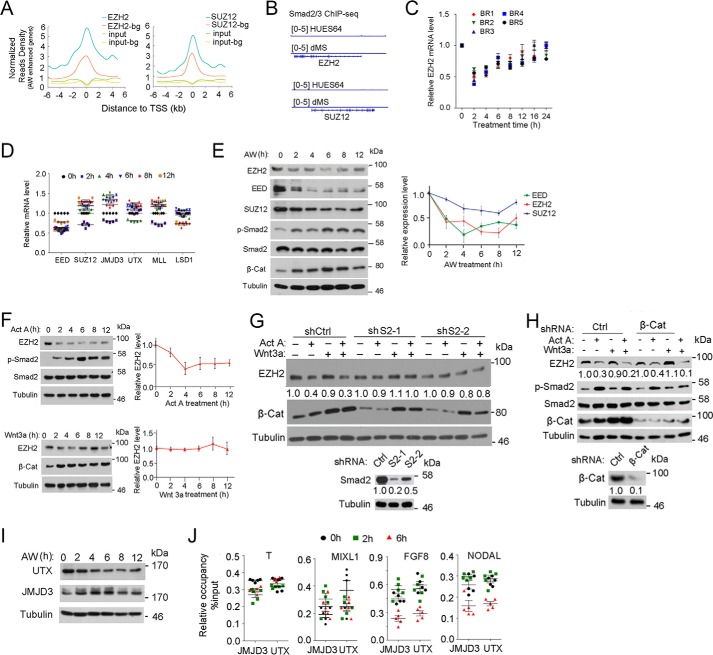
H3K27me3 levels are determined by balanced activities of the methyltransferase PRC2 and the demethylases JMJD3/KDM6B and UTX/KDM6A (24, 25). By analyzing transcription factor-binding enrichment in H1 cells with the published EZH2 and SUZ12 ChIP-seq data (21), we found a significant enrichment of both EZH2 and SUZ12 binding on AW-enhanced genes (Fig. 6A and supplemental data set 4). To explore whether activin/Smad2 signaling reduces H3K27me3 levels via the PRC2 complex, we first examined whether Smad2 directly regulated the mRNA expression of EZH2 or SUZ12. By scrutinizing the published Smad2/3 ChIP-seq data (22, 33), we found that Smad2 does not bind to the EZH2 and SUZ12 genes (Fig. 6B). In agreement, AW treatment did not significantly change the mRNA expression of EZH2 and SUZ12 (Fig. 6, C and D). Then we assessed the protein level of PRC2 complex proteins and found that EZH2 protein levels were decreased under AW or activin A treatment with a dynamic similar to the pattern of changes in H3K27me3 levels, whereas Wnt3a did not alter EZH2 protein level (Fig. 6, E and F). The protein levels of SUZ12 and EED were also reduced after AW treatment (Fig. 6E). In agreement with the minimal change observed in H3K4me3 levels (Figs. 3A and 4A), the mRNA levels of MLL and LSD1 did not change significantly at the early time points (Fig. 6D).
FIGURE 6.
Activin/Smad2 signaling decreases EZH2 protein levels. A, mean ChIP-seq profile for EZH2 and SUZ12 in H1 cells (GSE32509) at ±5 kb around the TSS of AW-enhanced genes together with background (bg) genes. 1477 background genes that show random response were selected as having −0.05 < PCC < 0.05 with T, EOMES, and MIXL1. B, IGV was performed on the Smad2/3 ChIP-seq data to show the occupancy of Smad2/3 on EZH2 and SUZ12 promoters. C, H1 cells were treated with 25 ng/ml AW for the indicated times before they were harvested for qPCR to assess EZH2 mRNA expression. BR, biological replicate. D, H1 cells were treated with 25 ng/ml AW for the indicated times before they were harvested for qPCR. E, H1 cells treated with 25 ng/ml AW for they indicated times before they were harvested for immunoblotting against EZH2, SUZ12, and EED. Phospho-Smad2 and β-catenin levels indicate activin and Wnt signaling activities, respectively. EZH2, SUZ12, and EED bands were quantified, and the relative levels were shown after normalization to tubulin. F, H1 cells treated with 25 ng/ml activin A or 25 ng/ml Wnt3a for the indicated times before they were harvested for anti-EZH2 immunoblotting. EZH2 bands were quantified, and the relative levels were shown after normalization to tubulin. G, Smad2 knockdown H1 cells were treated with 25 ng/ml activin A, 25 ng/ml Wnt3a, or both for 2 h before they were harvested for anti-EZH2 immunoblotting. EZH2 and Smad2 bands were quantified. Knockdown efficiency of Smad2 is shown in the bottom panels. H, β-catenin knockdown H1 cells were treated with 25 ng/ml activin A, 25 ng/ml Wnt3a, or both for 2 h before they were harvested for anti-EZH2 immunoblotting. Phospho-Smad2 and β-catenin levels indicate activin and Wnt signaling activities, respectively; tubulin was used as a loading control. EZH2 and β-catenin bands were quantified and normalized to tubulin. Knockdown efficiency of β-catenin is shown in the bottom panels. I, H1 cells were treated with 25 ng/ml AW for the indicated times before they were harvested for anti-UTX or anti-JMJD3 immunoblotting. Tubulin served as a loading control. J, H1 cells were treated with 25 ng/ml AW for 2 or 6 h before they were harvested for anti-JMJD3 and anti-UTX ChIP followed by qPCR of the ME signature genes. The statistical data are shown as mean ± S.E. (error bars) (n = 15, including 5 biological replicates and 3 technical replicates for C, D, and J; n = 9, including 3 biological replicates and 3 technical replicates for E and F). A Friedman test was performed in E and F.
To explore whether activin decreases EZH2 expression via Smad2, we examined EZH2 protein level in Smad2 knockdown H1 cells. Depletion of Smad2 attenuated AW-induced EZH2 degradation at ME initiation (Fig. 6G). In contrast, knockdown of β-catenin had no effect on AW-induced EZH2 degradation (Fig. 6H), indicating that activin/Smad2 signaling, but not Wnt/β-catenin signaling, controls EZH2 stability, decreasing its methyltransferase activity and thus the H3K27me3 level during ME initiation.
It has been reported that JMJD3 can be recruited to the T and NODAL loci via Smad2/3 and thus decrease H3K27me3 levels, which is critical for transcriptional activation in mouse ESCs (19, 34). Upon AW treatment, there was a transient decrease of UTX protein levels at 2 h, whereas JMJD3 protein levels were slightly increased (Fig. 6I). However, we did not detect enrichment in the occupancy of either JMJD3 or UTX in the ME signature genes T, MIXL1, NODAL, or FGF8 (Fig. 6J). Moreover, no interaction was observed between JMJD3 and Smad2/3 or JMJD3 and β-catenin at 2 h of AW treatment (data not shown). These data together suggest that the demethylases JMJD3 and UTX may not play a role in the AW-induced reduction in H3K27me3 levels that occurs during ME initiation.
Smad2 Recruits β-Catenin to Activate Transcription after H3K27me3 Priming
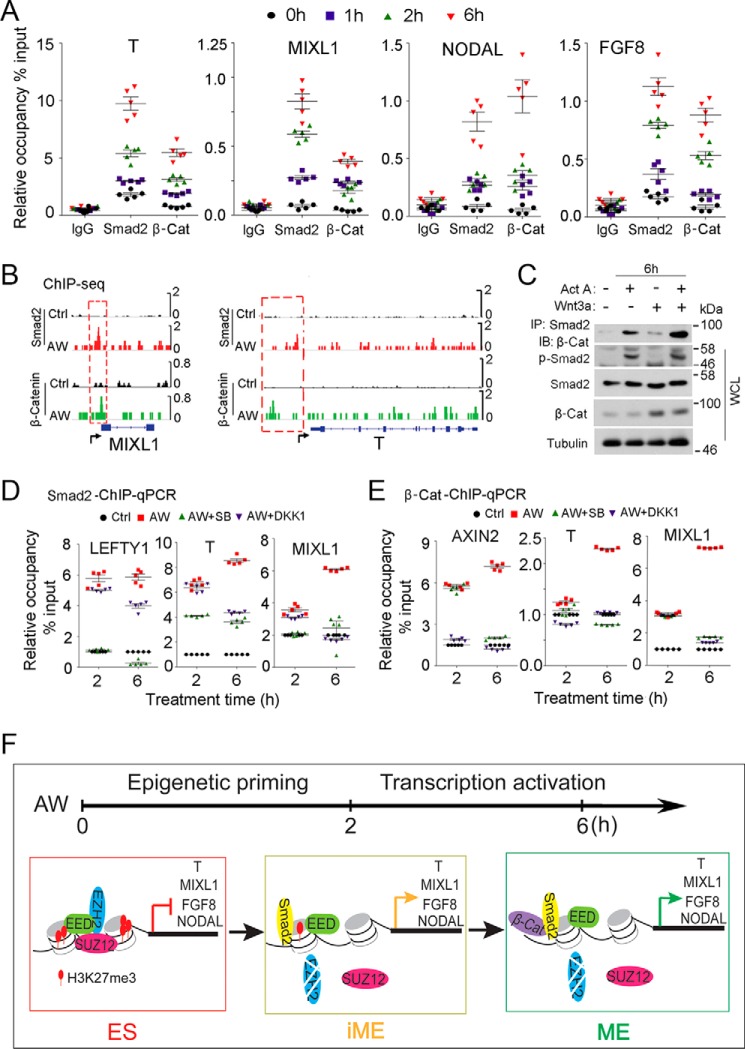
Because low activin A (25 ng/ml) also cannot drive ME differentiation (Fig. 1A), cooperation between activin and Wnt signaling is important even after H3K27me3 reduction. Both Smad2 and Smad3 can transduce activin signaling to control gene expression, but Smad2 is critical for embryogenesis, whereas Smad3 is important for tissue homeostasis (35, 36). Although the full-length Smad2 cannot bind to DNA directly, it can do so by forming a complex with Smad4 or via its short splicing form that has been shown to directly associate with DNA (37). To further investigate the effect of combined activin and Wnt signaling to promote ME differentiation, we thus examined the binding of Smad2 and β-catenin to the promoters of ME signature genes. Binding of Smad2 and β-catenin to ME signature genes, such as T, MIXL1, NODAL, and FGF8, began to increase within 2 h of AW treatment, and significant enrichment was observed at 6 h (Fig. 7A). Furthermore, we observed overlaps between Smad2 and β-catenin binding on certain ME signature genes (Fig. 7B), and AW treatment enhanced the interaction between Smad2 and β-catenin (Fig. 7C). In addition, inhibition of activin and Wnt signaling with SB431542 and DKK1 decreased the occupancy of Smad2 and β-catenin on those ME signature genes at 6 h (Fig. 7, D and E). LEFTY1 and AXIN2, the known targets of activin and Wnt, respectively, were used as controls. These results indicate a cooperative effect of Smad2 and β-catenin on transcription levels. Interestingly, SB431542 only inhibited Smad2 occupancy, whereas DKK1 only decreased β-catenin bindings at 2 h of AW treatment, indicating that the cooperation between Smad2 and β-catenin might occur after this time point.
FIGURE 7.
Smad2 collaborates with β-catenin to activate ME genes after H3K27me3 priming. A, H1 cells were treated with 25 ng/ml AW for the indicated times before they were harvested for anti-Smad2 or anti-β-catenin ChIP followed by qPCR to assess the occupancy of Smad2 or β-catenin on the promoters of T, MIXL1, FGF8 and NODAL. The statistical data are shown as mean ± S.E. (error bars) (n = 15, including 5 biological replicates and 3 technical replicates). B, H1 cells treated with 25 ng/ml activin A and 25 ng/ml Wnt3a for 6 h before they were harvested for an anti-Smad2 or anti-β-catenin ChIP-seq assay. IGV was performed to show the binding of Smad2 and β-catenin on ME signature genes T and MIXL1. C, H1 cells were treated with 25 ng/ml activin A, 25 ng/ml Wnt3a, and both for 6 h before they were harvested for anti-Smad2 immunoprecipitation (IP) followed by anti-β-catenin immunoblotting (IB). Phospho-Smad2 and β-catenin levels indicate activin and Wnt signaling activities, respectively. Tubulin was used as loading control. The statistical data are shown as mean ± S.E. (n = 6, including 3 biological replicates and 2 technical replicates). D and E, H1 cells treated with 25 ng/ml AW with or without 10 μm SB431542 or 100 ng/ml DKK1 for 2 or 6 h before they were harvested for anti-Smad2 (D) or anti-β-catenin (E) ChIP-qPCR. H1 cells in the self-renewal condition were used as control. F, working model for AW-induced ME initiation of human ES cells. AW-induced ME specification involves two steps. During the early initiation stage, activin/Smad2 signaling reduces the global H3K27me3 level by decreasing EZH2 protein level, thus leading to a transient “intermediate” state (iME), and then the intermediate state cells are directed toward ME cell fates by the collaborative effects of activin/Smad2 and Wnt/β-catenin signaling on transcription.
Discussion
Various extrinsic signals have been demonstrated to induce ME specification, among which activin and Wnt play critical roles in inducing ME differentiation of ESCs (2, 4). Large scale ChIP-seq analyses have revealed the epigenetic landscape of ME (22, 23), and connections among extrinsic signals, epigenetic modifications, and chromatin remodeling have also been explored (38–42). However, these studies mainly focused on the later stages of ME differentiation, when expression of ME marker genes has reached a high level. The early events in the initiation stage of ME specification remain largely unknown. In the present study, we provide compelling evidence that reduction of H3K27me3 levels is critical for initiation of ME differentiation. Our data suggest that ME differentiation can be divided into two stages: first, epigenetic priming and, second, transcription activation (Fig. 7F). During the priming stage (iME, for intermediate ME), activin signaling via Smad2 attenuates PRC2 activity and thus decreases global H3K27me3 levels. The low H3K27me3 level then enables subsequent transcriptional activation of ME genes by the collaborative effects of activin/Smad2 and Wnt/β-catenin signaling.
Previous studies demonstrated no significant changes in global H3K27me3 modification between the pluripotent cell state and differentiated lineage states (5, 6, 22, 43–45), but these studies were focused on the 12-h or later stage of differentiation. We found that with AW treatment, the global level of H3K27me3 significantly decreased between 2 and 6 h after the initiation of treatment, and recovered at 12 h (Figs. 3 (A and B) and 4A). The reduction in H3K27me3 levels was observed on ME signature genes, and expression of these genes was activated after 2–4 h of AW treatment, corresponding to the pattern of changes in H3K27me3 levels (Figs. 1B and 3C). In contrast, other epigenetic modifications, including H3K4me3 of ME genes, did not undergo significant changes at the early time point of ME differentiation. These data are in agreement with the notion that H3K27me3 functions together with H3K4me3 to keep differentiation genes in a poised state, and upon induction of differentiation, the low H3K27me3 level ensures the expression of these genes (5–7, 21). The importance of H3K27me3 reduction during ME initiation was further confirmed by the finding that a brief treatment with the EZH2 inhibitor DZNep for 2–4 h significantly potentiated ME differentiation induced by low AW (Fig. 3, F and G).
Our data indicate that cooperation of activin and Wnt signaling is necessary and sufficient for ME induction (Figs. 1 and 2), which is consistent with previous reports that activin and Wnt signaling combine to drive ME induction (40, 41, 46). However, we found that activin and Wnt function differently in the early stage of ME differentiation. Smad2-dependent activin signaling is crucial for epigenetic priming by reducing H3K27me3 levels (Fig. 5). We observed that activin/Smad2 signaling decreases EZH2 protein levels (Fig. 6), which is not consistent with the report that JMJD3 interacts with Smad2/3 and is important for H3K27me3 erasing in the T and NODAL genes in mouse ESCs (19). The helix-loop-helix protein HEB was reported to be a functional link between Nodal/Smad signaling and the derepression of the PRC2 complex-repressed developmental genes in mouse ESCs (17). The mechanism for direct regulatory networks between activin/Smad2 and PRC2 is still unclear and needs further exploration. Wnt/β-catenin signaling, in contrast, is not required for reduction of H3K27me3 levels. Instead, Wnt/β-catenin signaling in cooperation with activin/Smad signaling is necessary for efficient expression of ME genes. At this stage, the function of Wnt/β-catenin signaling in ME initiation awaits further investigation.
Activin signaling has been shown to regulate both self-renewal and mesendoderm-endoderm differentiation of human embryonic stem cells (3, 36, 38, 39, 42, 46). At low activin dose, Smad2 facilitates pluripotency of human embryonic stem cells by directly regulating NANOG expression (46) or by interacting with OCT4 to form a complex with TEAD to regulate expression of pluripotency genes (47). However, upon mesendoderm differentiation, Smad2/Smad4 can directly target mesendoderm signature genes and activate their expression, thus promoting the mesendoderm differentiation of human embryonic stem cells (3, 35, 40, 41, 46).
In summary, by focusing on the early stage of ME differentiation, we observed a reduction in H3K27me3 levels, which is critical for subsequent combined activin/Smad and Wnt/β-catenin signaling to effectively specify ME fate. We also demonstrated that attenuation of PRC2 activity is responsible for the activin/Smad-elicited H3K27me3 decrease. These findings advance our understanding of the complex signaling underlying cell fate determination of pluripotent stem cells and the molecular events in early embryogenesis.
Experimental Procedures
Cell Culture, Reagents, and Antibodies
H1 human embryonic stem cells (WiCell, 12-W0296) were cultured on feeder cells in DMEM/F-12 (Gibco) supplemented with 20% Knock-out Serum Replacement (KSR) (Gibco), 1 mm l-glutamine, 0.1 mm nonessential amino acids, 0.1 mm β-mercaptoethanol, and 10 ng/ml recombinant human bFGF. For feeder-free culture condition, H1 cells were cultured on a Matrigel (BD Biosciences)-coated plate with N2B27 chemically defined medium (DMEM/F-12 supplemented with N2, B27, 1 mm l-glutamine, 0.1 mm nonessential amino acids, 0.1 mm β-mercaptoethanol, and 20 ng/ml recombinant human bFGF). Feeder cells were generated and maintained in DMEM medium supplemented with 10% FBS (HyClone), and mouse manipulations followed the protocols approved by the Tsinghua Animal Ethics Committee. Reagents and antibodies used in this study are listed in supplemental Table S1.
Mesendoderm, Neuroectoderm, and Trophoblast Differentiation of ESCs
For mesendoderm differentiation, H1 cells were maintained on a Matrigel-coated plate with N2B27-bFGF chemically defined medium to about 40–70% confluence, and then differentiation was induced in B27 chemically defined basal medium (DMEM/F-12, B27, 1 mm l-glutamine, 0.1 mm nonessential amino acids) supplemented with activin A (25 ng/ml), Wnt3a (25 ng/ml), activin A plus Wnt3a (25 ng/ml each), BMP4 (5 ng/ml) plus bFGF (40 ng/ml), or bFGF (20 ng/ml) plus activin A (25 ng/ml) and Ly294002 (10 μm). Neuroectoderm was induced in B27 medium supplemented with SB431542 (10 μm) and Noggin (100 ng/ml), whereas trophoblast was induced in B27 medium supplemented with BMP4 (5 ng/ml). R1 cells cultured on a gelatin-coated plate were induced to form an embryoid body as described by Kurosawa (48). After 2 days of induction, embryoid bodies were digested into single cells with TrypleTM and replaced in gelatin-coated 6-well plates with N2B27 chemically defined medium overnight to form epiblast cells, which were then used for ME differentiation with activin A (25 ng/ml), Wnt3a (25 ng/ml), or both in B27 medium.
Derivation of Human Induced Pluripotent Stem Cells
CD34+ cells were purified from human cord blood using CD34 magnetic beads (MACS) and expanded in StemPro-34 medium (Invitrogen) for 7 days. Transgene-free induced pluripotent stem cells were generated using the CytoTuneTM-iPS reprogramming kit (Invitrogen) following the instructions. The induced pluripotent stem cell has a 46 XX normal karyotype and could differentiate to ectoderm, mesoderm, and endoderm tissues in an induced differentiation assay and teratoma assay (data not shown).
shRNA Knockdown
All shRNA plasmids were obtained from the Sigma TRC shRNA library, and non-targeting nucleotide plasmid served as control shRNA. shRNAs were packed into lentivirus following the instructions. H1 cells were infected with shRNA lentiviral supernatant. Puromycin (1 μg/ml; Invitrogen) was added into culture medium for 4–7 days to select cells for stable viral integration. shRNA sequences are showed in supplemental Table S2.
Chromatin Immunoprecipitation
A ChIP assay was performed as described previously (49). High throughput sequencing using Illumina HiSeq 2000 devices was carried out at Berry Genomics (Beijing, China). Specific PCR primers for ChIP are described in supplemental Table S3.
Histone Extraction
H1 cells were lysed with TEB buffer (PBS containing 0.5% Triton X-100, 2 mm phenylmethylsulfonyl fluoride, 0.02% NaN3) with proteinase inhibitor mixture (Roche Applied Science) for 10 min at 4 °C and then centrifuged at 2000 rpm for 10 min. After removing the supernatant and washing the pellets once with 300 μl of cold TEB buffer with proteinase inhibitors, the pellets were suspended with 0.2 n HCl and incubated at 4 °C overnight. Histones were collected with 100% trichloroacetic acid at a final concentration of 33% by incubation for 1–2 h on ice. After centrifugation at 13,000 rpm for 10 min at 4 °C, the supernatant was discarded, histone pellets were washed with ice-cold acetone, and pellets were air-dried for 20 min before being dissolved with a suitable volume of 150 mm NaCl.
RNA-seq Data Processing, Transcription Factor Binding Enrichment Analysis, Gene Ontology Term Enrichment Analysis, and Motif Analysis
Paired end reads were first trimmed of the first 15 bp from each end and then mapped to human genome (hg19) using STAR (version 2.4.0d) (50). Gene expression was estimated and normalized with Cuffnorm from the Cufflinks package (version 2.2.1) (51) into an FPKM matrix using default parameters for the annotation GTF file downloaded from GENCODE (version 19) (52). Genes were filtered if their expression in all samples was <0.3 FPKM, and the expression matrix was log2-transformed. Differentially expressed genes (DEGs) were obtained by Cuffdiff (version 2.2.1) at the gene level by the following strategies. (a) All other samples were compared with control. (b) We analyzed each treatment as a time series like (C, A6, A24), which implements the Cuffdiff -T option, and termed the result as A group, W group, and AW group. This set of genes was used in Fig. 2. (c) Samples (C, A6, W6, AW6), termed as 6 h, and (C, A24, W24, AW24), termed as 24 h, also use the Cuffdiff -T option. The DEGs were selected by p value <0.001. The expression matrix for the union set of DEGs (n = 439) was feed to Cluster version 3.0 (53), grouping k-means into 14 clusters. We defined a group of genes (n = 304) termed as AW enhanced genes, which showed an expression pattern similar to those of the mesendoderm markers BRACHYURY/T, MIXL1, and EOMES, with a Pearson correlation coefficient (PCC) >0.8. We also defined a set of genes termed as the background group (n = 1477) used for comparison with the AW enhanced genes because the expression pattern showed a random response to the treatment, selected by −0.05 < PCC < 0.05. Transcription factor binding data available in H1 cells were downloaded from ENCODE (54). The peaks (BED files) were assigned to a gene when the gene was targeted by a transcription factor and there were peaks around its transcription start site (TSS) at ±2 kb. The transcription factor binding enrichment analysis was carried out by a hypergeometric test as follows, similar to the method described previously (55).
ChIP-seq Mapping, Peak Calling, Heat Maps, Profile Plots, and Peak Visualization
Reads were mapped into the human genome (hg19) using bowtie (version 1.1.0) with the following options: −y −k 1 −m 1 −best −chunkmbs 200. MACS (version 2.1.0) was used to identity significant binding events for each ChIP-seq data with the following options: −nomodel −extsize 75 −keep-dup 1 −broad. The peaks were further filtered using the ENCODE blacklist regions. Peaks were annotated using annotatePeak.pl in HOMER. The bedGraph pileups were also generated by MACS and converted to bigWig for visualization in the integrative genomics viewer (IGV). The command annotatePeak.pl in HOMER was used to generate the normalized matrix for target genes' TSS and peaks, with the following options: TSS hg19 −size 10000 −hist 200 −ghist. The smoothed average signal for the matrix was used to plot the average profile, whereas the matrix was visualized by Treeview (53) to show a heat map. H1-hESC EZH2 and SUZ12 ChIP-seq data were obtained with GEO accession numbers GSM831028 and GSM831042. H3K27me3, H3K4me3, ChIP-seq, and RNA-seq data of activin A, Wnt3a, and AW treatments used in this study were shown in GEO 81617 and GSM2159741–2159756.
Statistical Analysis
All of the values are shown as mean ± S.E. with a two-way ANOVA test. The significance between groups was determined by Student's t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001. The Friedman test was used for immunoblotting protein level analysis.
Author Contributions
L. W. and Y.-G. C. designed the study and wrote the paper; L. W., X. X., Z. L., and G. Z. performed the experiments; Y. C., H. C., and J.-D. J. H. performed the bioinformatic analyses. F. D. and J. N. provided CD34+ human iPSCs. All authors analyzed the results.
Acknowledgments
We are grateful to Dr. Teng Fei for suggestions and Drs. Qiaoran Xi and Xiaohua Shen for critical reading of the manuscript.
This work was supported by National Natural Science Foundation of China Grants 91519310 and 31330049 and 973 Program Grant 2013CB933700 (to Y. G. C.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables S1 and S2 and data sets 1–4.
- hESC
- human embryonic stem cell
- bFGF
- basal fibroblast growth factor
- ChIP-seq
- chromatin immunoprecipitation-sequencing
- RNA-seq
- RNA-sequencing
- H3K4 and H3K27
- histone H3 Lys-4 and Lys-27, respectively
- H3K4me3
- H3K4 trimethylation
- H3K27me3
- H3K27 trimethylation
- ME
- mesendoderm
- PRC2
- polycomb repressive complex 2
- DEGs
- differentially expressed genes
- TSS
- transcription start site
- ANOVA
- analysis of variance
- qPCR
- quantitative PCR
- IGV
- integrative genomics viewer
- PCC
- Pearson correlation coefficient.
References
- 1. Tam P. P., and Behringer R. R. (1997) Mouse gastrulation: the formation of a mammalian body plan. Mech. Dev. 68, 3–25 [DOI] [PubMed] [Google Scholar]
- 2. Murry C. E., and Keller G. (2008) Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132, 661–680 [DOI] [PubMed] [Google Scholar]
- 3. Gaarenstroom T., and Hill C. S. (2014) TGF-β signaling to chromatin: how Smads regulate transcription during self-renewal and differentiation. Semin. Cell Dev. Biol. 32, 107–118 [DOI] [PubMed] [Google Scholar]
- 4. Wang L., and Chen Y. G. (2016) Signaling control of differentiation of embryonic stem cells toward mesendoderm. J. Mol. Biol. 428, 1409–1422 [DOI] [PubMed] [Google Scholar]
- 5. Li M., Liu G. H., and Izpisua Belmonte J. C. (2012) Navigating the epigenetic landscape of pluripotent stem cells. Nat. Rev. Mol. Cell Biol. 13, 524–535 [DOI] [PubMed] [Google Scholar]
- 6. Young R. A. (2011) Control of the embryonic stem cell state. Cell 144, 940–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Voigt P., Tee W. W., and Reinberg D. (2013) A double take on bivalent promoters. Gene Dev. 27, 1318–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shen X. H., Liu Y., Hsu Y. J., Fujiwara Y., Kim J., Mao X., Yuan G. C., and Orkin S. H. (2008) EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 32, 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., and Zhang Y. (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 10. Kirmizis A., Bartley S. M., Kuzmichev A., Margueron R., Reinberg D., Green R., and Farnham P. J. (2004) Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Gene Dev. 18, 1592–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hübner M. R., and Spector D. L. (2010) Role of H3K27 demethylases Jmjd3 and UTX in transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol. 75, 43–49 [DOI] [PubMed] [Google Scholar]
- 12. Swigut T., and Wysocka J. (2007) H3K27 demethylases, at long last. Cell 131, 29–32 [DOI] [PubMed] [Google Scholar]
- 13. O'Carroll D., Erhardt S., Pagani M., Barton S. C., Surani M. A., and Jenuwein T. (2001) The Polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 21, 4330–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen T., and Dent S. Y. R. (2014) Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat. Rev. Genet. 15, 93–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pasini D., Bracken A. P., Jensen M. R., Lazzerini Denchi E., and Helin K. (2004) Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 23, 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Faust C., Schumacher A., Holdener B., and Magnuson T. (1995) The Eed mutation disrupts anterior mesoderm production in mice. Development 121, 273–285 [DOI] [PubMed] [Google Scholar]
- 17. Yoon S. J., Foley J. W., and Baker J. C. (2015) HEB associates with PRC2 and SMAD2/3 to regulate developmental fates. Nat. Commun. 6, 6546. [DOI] [PubMed] [Google Scholar]
- 18. Pethe P., Nagvenkar P., and Bhartiya D. (2014) Polycomb group protein expression during differentiation of human embryonic stem cells into pancreatic lineage in vitro. BMC Cell Biol. 15, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dahle Ø., Kumar A., and Kuehn M. R. (2010) Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci. Signal. 3, ra48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang W., Wang J., and Zhang Y. (2013) Histone H3K27me3 demethylases KDM6A and KDM6B modulate definitive endoderm differentiation from human ESCs by regulating WNT signaling pathway. Cell Res. 23, 122–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xi Q., Wang Z., Zaromytidou A. I., Zhang X. H., Chow-Tsang L. F., Liu J. X., Kim H., Barlas A., Manova-Todorova K., Kaartinen V., Studer L., Mark W., Patel D. J., and Massagué J. (2011) A poised chromatin platform for TGF-beta access to master regulators. Cell 147, 1511–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dixon J. R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J. E., Lee A. Y., Ye Z., Kim A., Rajagopal N., Xie W., Diao Y., Liang J., Zhao H., Lobanenkov V. V., Ecker J. R., et al. (2015) Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xie W., Schultz M. D., Lister R., Hou Z., Rajagopal N., Ray P., Whitaker J. W., Tian S., Hawkins R. D., Leung D., Yang H., Wang T., Lee A. Y., Swanson S. A., Zhang J., et al. (2013) Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153, 1134–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hansen K. H., Bracken A. P., Pasini D., Dietrich N., Gehani S. S., Monrad A., Rappsilber J., Lerdrup M., and Helin K. (2008) A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 10, 1291–1300 [DOI] [PubMed] [Google Scholar]
- 25. Margueron R., and Reinberg D. (2011) The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miranda T. B., Cortez C. C., Yoo C. B., Liang G., Abe M., Kelly T. K., Marquez V. E., and Jones P. A. (2009) DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol. Cancer Ther. 8, 1579–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu P., Pan G., Yu J., and Thomson J. A. (2011) FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell 8, 326–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bernardo A. S., Faial T., Gardner L., Niakan K. K., Ortmann D., Senner C. E., Callery E. M., Trotter M. W., Hemberger M., Smith J. C., Bardwell L., Moffett A., and Pedersen R. A. (2011) BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell 9, 144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu R. H., Chen X., Li D. S., Li R., Addicks G. C., Glennon C., Zwaka T. P., and Thomson J. A. (2002) BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat. Biotechnol. 20, 1261–1264 [DOI] [PubMed] [Google Scholar]
- 30. Zhang P., Li J., Tan Z., Wang C., Liu T., Chen L., Yong J., Jiang W., Sun X., Du L., Ding M., and Deng H. (2008) Short-term BMP-4 treatment initiates mesoderm induction in human embryonic stem cells. Blood 111, 1933–1941 [DOI] [PubMed] [Google Scholar]
- 31. Chambers S. M., Fasano C. A., Papapetrou E. P., Tomishima M., Sadelain M., and Studer L. (2009) Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim D. S., Lee J. S., Leem J. W., Huh Y. J., Kim J. Y., Kim H. S., Park I. H., Daley G. Q., Hwang D. Y., and Kim D. W. (2010) Robust enhancement of neural differentiation from human ES and iPS cells regardless of their innate difference in differentiation propensity. Stem Cell Rev. 6, 270–281 [DOI] [PubMed] [Google Scholar]
- 33. Tsankov A. M., Gu H., Akopian V., Ziller M. J., Donaghey J., Amit I., Gnirke A., and Meissner A. (2015) Transcription factor binding dynamics during human ES cell differentiation. Nature 518, 344–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kartikasari A. E., Zhou J. X., Kanji M. S., Chan D. N., Sinha A., Grapin-Botton A., Magnuson M. A., Lowry W. E., and Bhushan A. (2013) The histone demethylase Jmjd3 sequentially associates with the transcription factors Tbx3 and Eomes to drive endoderm differentiation. EMBO J. 32, 1393–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brown S., Teo A., Pauklin S., Hannan N., Cho C. H., Lim B., Vardy L., Dunn N. R., Trotter M., Pedersen R., and Vallier L. (2011) Activin/Nodal signaling controls divergent transcriptional networks in human embryonic stem cells and in endoderm progenitors. Stem Cells 29, 1176–1185 [DOI] [PubMed] [Google Scholar]
- 36. Seuntjens E., Umans L., Zwijsen A., Sampaolesi M., Verfaillie C. M., and Huylebroeck D. (2009) Transforming growth factor type β and Smad family signaling in stem cell function. Cytokine Growth Factor Rev. 20, 449–458 [DOI] [PubMed] [Google Scholar]
- 37. Yagi K., Goto D., Hamamoto T., Takenoshita S., Kato M., and Miyazono K. (1999) Alternatively spliced variant of Smad2 lacking exon 3: comparison with wild-type Smad2 and Smad3. J. Biol. Chem. 274, 703–709 [DOI] [PubMed] [Google Scholar]
- 38. Mendjan S., Mascetti V. L., Ortmann D., Ortiz M., Karjosukarso D. W., Ng Y., Moreau T., and Pedersen R. A. (2014) NANOG and CDX2 pattern distinct subtypes of human mesoderm during exit from pluripotency. Cell Stem Cell 15, 310–325 [DOI] [PubMed] [Google Scholar]
- 39. Vallier L., Touboul T., Brown S., Cho C., Bilican B., Alexander M., Cedervall J., Chandran S., Ahrlund-Richter L., Weber A., and Pedersen R. A. (2009) Signaling pathways controlling pluripotency and early cell fate decisions of human induced pluripotent stem cells. Stem Cells 27, 2655–2666 [DOI] [PubMed] [Google Scholar]
- 40. Funa N. S., Schachter K. A., Lerdrup M., Ekberg J., Hess K., Dietrich N., Honoré C., Hansen K., and Semb H. (2015) β-Catenin regulates primitive streak induction through collaborative interactions with SMAD2/SMAD3 and OCT4. Cell Stem Cell 16, 639–652 [DOI] [PubMed] [Google Scholar]
- 41. Estarás C., Benner C., and Jones K. A. (2015) SMADs and YAP compete to control elongation of β-catenin: LEF-1-recruited RNAPII during hESC differentiation. Mol. Cell 58, 780–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bertero A., Madrigal P., Galli A., Hubner N. C., Moreno I., Burks D., Brown S., Pedersen R. A., Gaffney D., Mendjan S., Pauklin S., and Vallier L. (2015) Activin/Nodal signaling and NANOG orchestrate human embryonic stem cell fate decisions by controlling the H3K4me3 chromatin mark. Gene Dev. 29, 702–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Howard T. D., Ho S. M., Zhang L., Chen J., Cui W., Slager R., Gray S., Hawkins G. A., Medvedovic M., and Wagner J. D. (2011) Epigenetic changes with dietary soy in cynomolgus monkeys. PLoS One 6, e26791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Meissner A., Mikkelsen T. S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B. E., Nusbaum C., Jaffe D. B., Gnirke A., Jaenisch R., and Lander E. S. (2008) Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xie R., Everett L. J., Lim H. W., Patel N. A., Schug J., Kroon E., Kelly O. G., Wang A., D'Amour K. A., Robins A. J., Won K. J., Kaestner K. H., and Sander M. (2013) Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell Stem Cell 12, 224–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Singh A. M., Reynolds D., Cliff T., Ohtsuka S., Mattheyses A. L., Sun Y., Menendez L., Kulik M., and Dalton S. (2012) Signaling network crosstalk in human pluripotent cells: a Smad2/3-regulated switch that controls the balance between self-renewal and differentiation. Cell Stem Cell 10, 312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Beyer T. A., Weiss A., Khomchuk Y., Huang K., Ogunjimi A. A., Varelas X., and Wrana J. L. (2013) Switch enhancers interpret TGF-β and Hippo signaling to control cell fate in human embryonic stem cells. Cell Rep. 5, 1611–1624 [DOI] [PubMed] [Google Scholar]
- 48. Kurosawa H. (2007) Methods for inducing embryoid body formation: in vitro differentiation system of embryonic stem cells. J. Biosci. Bioeng. 103, 389–398 [DOI] [PubMed] [Google Scholar]
- 49. Fei T., Zhu S., Xia K., Zhang J., Li Z., Han J. D., and Chen Y. G. (2010) Smad2 mediates Activin/Nodal signaling in mesendoderm differentiation of mouse embryonic stem cells. Cell Res. 20, 1306–1318 [DOI] [PubMed] [Google Scholar]
- 50. Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T. R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J., and Pachter L. (2010) Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Harrow J., Frankish A., Gonzalez J. M., Tapanari E., Diekhans M., Kokocinski F., Aken B. L., Barrell D., Zadissa A., Searle S., Barnes I., Bignell A., Boychenko V., Hunt T., Kay M., et al. (2012) GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Eisen M. B., Spellman P. T., Brown P. O., and Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Boyle A. P., Araya C. L., Brdlik C., Cayting P., Cheng C., Cheng Y., Gardner K., Hillier L. W., Janette J., Jiang L., Kasper D., Kawli T., Kheradpour P., Kundaje A., Li J. J., et al. (2014) Comparative analysis of regulatory information and circuits across distant species. Nature 512, 453–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Auerbach R. K., Chen B., and Butte A. J. (2013) Relating genes to function: identifying enriched transcription factors using the ENCODE ChIP-seq significance tool. Bioinformatics 29, 1922–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]