Abstract
Human retinoid X receptor α (hRXRα) plays a critical role in DNA binding and transcriptional activity through heterodimeric association with several members of the nuclear receptor superfamily, including the human vitamin D receptor (hVDR). We previously showed that hRXRα phosphorylation at serine 260 through the Ras-Raf-MAPK ERK1/2 activation is responsible for resistance to the growth inhibitory effects of 1α,25-dihydroxyvitamin D3 (1α,25(OH)2D3), the biologically active metabolite of vitamin D3. To further investigate the mechanism of this resistance, we studied intranuclear dynamics of hVDR and hRXRα-tagged constructs in living cells together with endogenous and tagged protein in fixed cells. We find that hVDR-, hRXRα-, and hVDR-hRXRα complex accumulate in the nucleus in 1α,25(OH)2D3-treated HPK1A cells but to a lesser extent in HPK1ARas-treated cells. Also, by using fluorescence resonance energy transfer (FRET), we demonstrate increased interaction of the hVDR-hRXRα complex in 1α,25(OH)2D3-treated HPK1A but not HPK1ARas cells. In HPK1ARas cells, 1α,25(OH)2D3-induced nuclear localization and interaction of hRXRα are restored when cells are treated with the MEK1/2 inhibitor UO126 or following transfection of the non-phosphorylatable hRXRα Ala-260 mutant. Finally, we demonstrate using fluorescence loss in photobleaching and quantitative co-localization with chromatin that RXR immobilization and co-localization with chromatin are significantly increased in 1α,25(OH)2D3-treated HPK1ARas cells transfected with the non-phosphorylatable hRXRα Ala-260 mutant. This suggests that hRXRα phosphorylation significantly disrupts its nuclear localization, interaction with VDR, intra-nuclear trafficking, and binding to chromatin of the hVDR-hRXR complex.
Keywords: fluorescence resonance energy transfer (FRET), mitogen-activated protein kinase (MAPK), phosphorylation, retinoid, vitamin D, fluorescence loss in photobleaching (FLIP), retinoid X receptor, vitamin D receptor
Introduction
1α,25-Dihydroxyvitamin D3 (1α,25(OH)2D3),3 the hormonally active metabolite of vitamin D, along with its effect on bone and mineral homeostasis (1–5) is also known to affect immuno-modulation (6–8), to promote cellular differentiation, and to inhibit cell proliferation (9). The majority of the actions of 1α,25(OH)2D3 are mediated through the vitamin D receptor (VDR), a member of the nuclear receptor steroid/thyroid superfamily of transcriptional regulators. 1α,25(OH)2D3 directly modulates the transcription of several target genes by binding to the VDR. The ligand-bound activated complex functions as a heterodimer by interaction with the retinoid X receptor (RXR). This heterodimer (VDR/RXR) is capable of binding to the vitamin D response elements in the promoter regions of target genes resulting in activation or repression of transcription via interaction with transcriptional cofactors and the basal transcriptional machinery (1, 10–19).
Although in vitro studies have shown that 1α,25(OH)2D3 promotes differentiation of myeloid cells (20) and inhibits proliferation of breast (9, 20–25), colon (26), prostate (27, 28), and other cancer cells (29–37), it is now apparent that several human cancer cell lines are resistant to the growth inhibitory action of 1α,25(OH)2D3 (32, 34, 36–39). Attempts to treat cancer patients using vitamin D and its analogs have not yet translated into effective and approved therapies (40–45). Furthermore, epidemiological studies remain controversial (46–49). Thus, an understanding of the mechanism of that resistance could pave the way to improve the efficacy of vitamin D therapies in cancer.
Studies in our laboratory indicate that phosphorylation of the hRXRα is an important mechanism underlying the resistance to the growth inhibitory action of vitamin D in malignant keratinocytes secondary to its phosphorylation at serine 260, a critical site located in close spatial proximity to regions of potential co-activator and co-repressor interactions with the RXR (33–36). These earlier studies demonstrated that phosphorylation of hRXRα not only disrupts its vitamin D signaling but also co-activator recruitment perhaps by altering the conformation of the VDR-RXR-co-modulator complex and its biological activity. We therefore hypothesize that RXR phosphorylation at Ser-260 is a central regulator of 1α,25(OH)2D3 action in cancer cells that could be targeted therapeutically. In this study, we use in vitro imaging with live and fixed cells, including fluorescent resonance energy transfer (FRET) and fluorescence loss in photobleaching (FLIP), to examine whether phosphorylation at serine 260 altered VDR and RXR interaction, nuclear localization, and intranuclear kinetic of the VDR-RXR complex.
Results
Effects of 1α,25(OH)2D3 Treatment on Subcellular Localization of hVDR and hRXRα in Non-transformed HPK1A and Ras-transformed HPK1ARas Cells
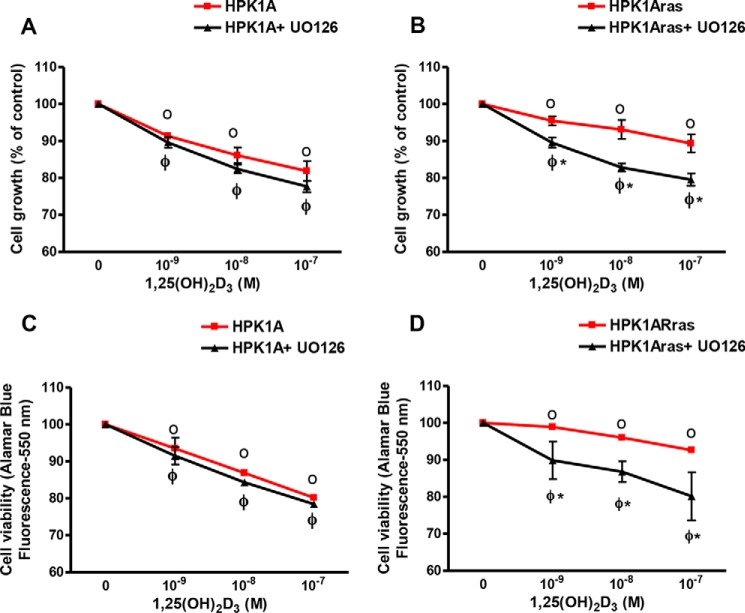
First, we established the conditions for reversal of 1α,25(OH)2D3 resistance with the MAPK inhibitor UO126 in Ras-transformed HPK1ARas cells. To test whether the non-transformed HPK1A and the Ras-transformed (HPK1ARas) cells were sensitive to the growth inhibitory action of 1α,25(OH)2D3, we treated both cell lines with UO126 alone, various concentrations of 1α,25(OH)2D3 alone, or in combination with UO126. In HPK1A cells, we observed a dose-dependent growth inhibition with 1α,25(OH)2D3 but no further growth inhibition when UO126 was added (Fig. 1A, p < 0.05). In contrast HPK1ARas-transformed cells were less responsive to treatment with 1α,25(OH)2D3 alone (Fig. 1B, p > 0.05), but pre-treatment for 30 min with the MEK inhibitor UO126, restored 1α,25(OH)2D3 response (p < 0.05). Similar effects were observed on cell viability in both cell lines using the Alamar blue viability assay (Fig. 1, C and D). We used flow cytometry to determine the effects of 1α,25(OH)2D3 on the cell cycle. HPK1A and HPK1ARas cells were treated with either vehicle, 1α,25(OH)2D3 alone, or in combination with UO126 for 72 h before flow cytometric analysis. In HPK1A cells, 1α,25(OH)2D3 addition or pre-treatment with MEK inhibitor UO126 significantly increased the percentage of cells in G0/G1 phase from 70.6 to 81.2% when compared with vehicle control (p < 0.05). Although UO126 significantly increased the percentage of cells in G0/G1 when added alone, it did not enhance 1α,25(OH)2D3 effects (Table 1, p > 0.05). In HPK1ARas cells, treatment with 1α,25(OH)2D3 slightly increased the percentage of cells in G0/G1 phase (64.9–69.2%) when compared with vehicle (p < 0.05). However, pre-treatment with the MEK inhibitor UO126 followed by addition of 1α,25(OH)2D3 significantly increased the percentage of cells in G0/G1 phase (69.2-81.0%) as compared with 1α,25(OH)2D3 treatment alone (Table 1, p < 0.05).
FIGURE 1.
Effects of 1,25(OH)2D3 and UO126 on cell growth (A and B) and viability (C and D). HPK1A (A) and HPK1ARas (B) cells were grown in DMEM containing 10% FBS. At 40% confluency the medium was changed, and cells were treated with increasing concentrations of 1,25(OH)2D3 in the absence or presence of UO126 (10−6 m). After 72 h, cells were collected, and cells were counted using a Coulter counter. HPK1A (C) and HPK1ARas (D) cells were seeded at a density of 5 × 103 cells/well. Twenty four hours later, cells were treated with vehicle, 1,25(OH)2D3 alone, or a combination of UO126 and 1,25(OH)2D3 for 72 h. Alamar blue metabolism was used to assess cell viability. Values are expressed as a percentage of vehicle-treated cells. All values represent means ± S.D. (bars) of at least three separate experiments conducted in triplicate. In the cell viability experiment (C and D) HPK1A and HPK1ARas showed very little variation in the absence of UO126, and error bars are less than the width of the plotted points. Closed circles indicate a significant growth inhibition in either HPK1A + UO126 or HPK1ARas + UO126 cells as compared with vehicle-treated cells. Open circles indicate a significant growth inhibition in either HPK1A or HPK1ARas cells as compared with vehicle-treated cells. Asterisks indicate a significant difference between vehicle-treated and drug-treated cells. A p value of p < 0.05 was considered significant.
TABLE 1.
Cell cycle analysis of HPK1A and HPK1ARas cells following treatment with 1,25(OH)2D3 in the absence or presence of UO126
After 24 h of serum starvation (5% FBS), cells were treated with 1,25(OH)2D3 at 10−7 m with or without UO126. Cells were trypsinized after 72 h, and cell cycle was analyzed by flow cytometry as described under “Experimental Procedures.” Results are expressed as percentage of cells in G0/G1 phase of the cell cycle. Asterisks indicate a significant difference in G0/G1 cell cycle phase as compared with vehicle-treated control. Open circle indicates a significant difference between 1,25(OH)2D3 treatment alone and combined treatment of UO126 and 1,25(OH)2D3. A triangle represents significant difference between UO126 treatment alone and combined treatment of UO126 and 1,25(OH)2D3. A p value compared with control was considered significant when p < 0.05.
| Percentage of cells in G0/G1 cell cycle phase | ||
|---|---|---|
| Treatment | HPK1A | HPK1ARas |
| Vehicle | 70.66 ± 3.22 | 64.88 ± 1.96 |
| 1,25(OH)2D3 (10−7 m) | 79.50 ± 3.59* | 69.23 ± 1.61* |
| UO126 | 80.92 ± 3.05* | 74.71 ± 01.21* |
| UO126 + 1,25(OH)2D3 (10−7 m) | 81.20 ± 3.81* | 81.03 ± 3.29*○▵ |
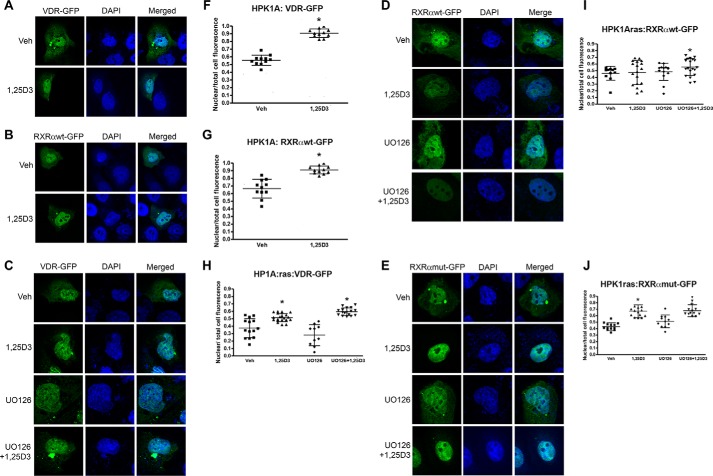
In HPK1A cells, hVDR distribution was both cytoplasmic and nuclear in the absence of the ligand (0.56 ± 0.07 nuclear). However, ligand addition significantly increased the nuclear localization of hVDR (0.91 ± 0.06; Fig. 2, A and F, p < 0.001). Similarly, in the absence of the ligand, hRXRα was both cytoplasmic and nuclear (0.66 ± 0.12), but nuclear translocation increased after ligand addition (0.91 ± 0.05; Fig. 2, B and G, p < 0.05). To determine the involvement of hRXRα phosphorylation on VDR localization in HPK1ARas cells, we similarly transfected the cells with VDR-GFP. VDR was found to be both cytoplasmic and nuclear in vehicle-treated cells (0.37 ± 0.13). Addition of 1α,25(OH)2D3 alone (0.52 ± 0.06) or a combination of the MEK inhibitor UO126 and 1α,25(OH)2D3 significantly increased VDR accumulation in the nucleus compared with control (0.60 ± 0.05) (Fig. 2, C and H, p < 0.05). However, addition of the MEK inhibitor UO126 alone did not increase the nuclear accumulation of VDR when compared with control (0.28 ± 0.14). The combination of UO126 and 1α,25(OH)2D3 increased nuclear accumulation of VDR by more than 1.5-fold compared with control and about 2-fold compared with 1α,25(OH)2D3 or UO126 treatment alone (0.60 ± 0.05) (Fig. 2, C and H, p < 0.005).
FIGURE 2.
Effects of 1,25(OH)2D3 with or without UO126 treatment on nuclear localization of VDR and RXRα in HPK1A and HPK1ARas cells. HPK1A cells were transfected with VDR-GFP (A) or RXRα-WT-GFP (B) followed by treatment with either vehicle (veh) or 1,25(OH)2D3 (1,25D3). Similarly, HPK1ARas cells were transfected with either VDR-GFP (C), RXRα-WT-GFP (D), or RXRα-mut-GFP (E) followed by treatment with either vehicle, 1,25(OH)2D3, UO126 alone, or a combination of UO126 and 1,25(OH)2D3. Nuclear localization was assessed as under “Experimental Procedures.” Scatter plots show the quantitation of fluorescence of nuclear receptors normalized to total cell fluorescence (F–J). Values represent mean ± S.D. of at least 10 different cells. Asterisks indicate a significant difference in nuclear localization between 1,25(OH)2D3 treatment alone compared with vehicle-treated control. Open circles indicate a significant difference in nuclear localization of receptors in 1,25(OH)2D3-treated cells alone compared with combined UO126 and 1,25(OH)2D3 treatment. A p value of p < 0.05 was considered significant.
We also assessed the effects of phosphorylation on hRXRα subcellular localization. Similar to the protocol above, we transfected the cells with either RXRαWT-GFP or RXRαmut-GFP plasmids. In HPK1ARas cells transfected with RXRαWT-GFP plasmids, RXRα was found to be predominantly nuclear without treatment (vehicle) (0.46 ± 0.10). Treatment with 1α,25(OH)2D3 (0.47 ± 0.18) or MEK inhibitor UO126 (0.48 ± 0.13) alone did not increase nuclear accumulation of RXRα compared with control (0.46 ± 0.10) (Fig. 2, D and I, p > 0.05). However, a combination of UO126 and 1α,25(OH)2D3 (0.56 ± 0.12 versus 0.46 ± 0.10 p < 0.05) increased the nuclear accumulation of RXRα compared with vehicle, 1α,25(OH)2D3, and UO126 alone treated cells (Fig. 2, D and I, p < 0.05). In cells transfected with RXRαmut-GFP plasmids, 1α,25(OH)2D3 (0.67 ± 0.11) or a combination of UO126 and 1α,25(OH)2D3 (0.68 ± 0.11) significantly increased nuclear accumulation of RXRα compared with vehicle (0.43 ± 0.06) or MEK inhibitor UO126 alone (0.51 ± 0.10, Fig. 2, E and J, p < 0.05). Furthermore, treatment with MEK UO126 alone (0.51 ± 0.10) did not increase nuclear accumulation of RXRα when compared with vehicle (0.43 ± 0.06, p < 0.05).
Effects of 1α,25(OH)2D3 Treatment on Subcellular Localization of Endogenous hVDR and hRXRα in Non-transformed HPK1A and Ras-transformed HPK1ARas Cells
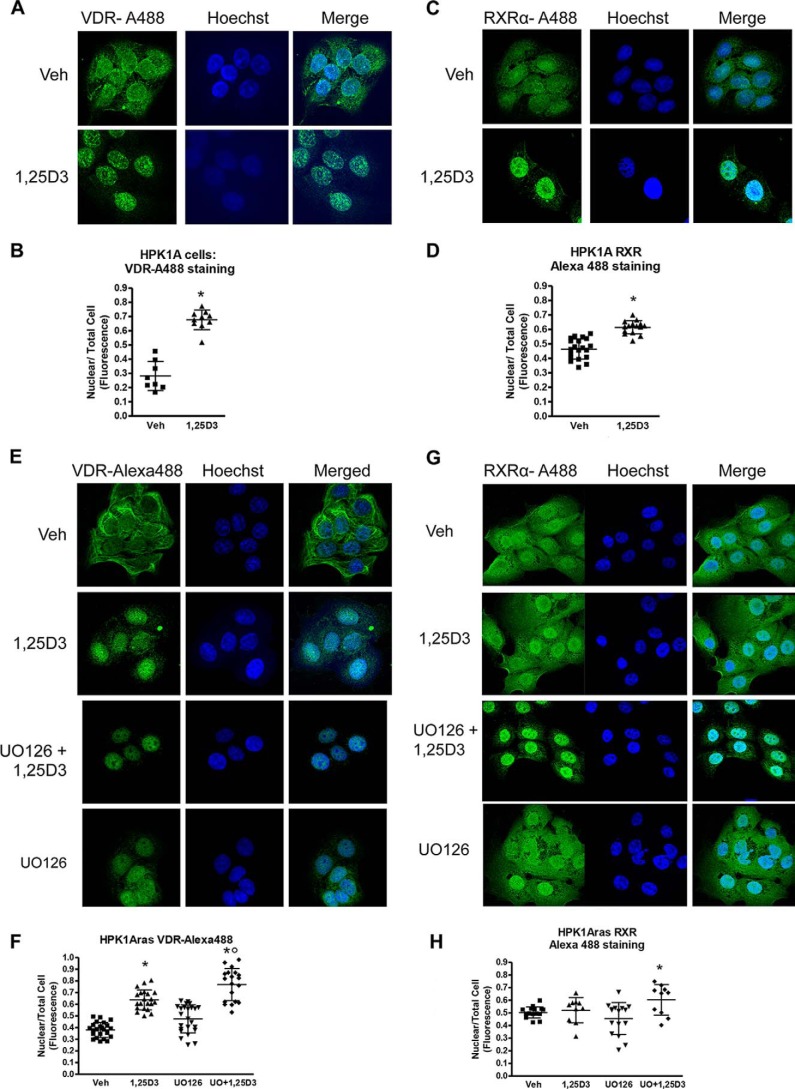
To control for possible effects of the fluorescent protein tag, we repeated the experiment in non-transfected cells using antibodies directed against endogenous hVDR and hRXRα. We incubated cells with conjugated Alexa-488 secondary antibody to detect endogenous VDR or RXR and measured the fluorescence by confocal microscopy. In HPK1A cells, hVDR distribution was both cytoplasmic and nuclear in the absence of the ligand (0.28 ± 0.01 nuclear). However, ligand addition significantly increased the nuclear localization of hVDR (0.68 ± 0.07 nuclear) (Fig. 3, A and B, p < 0.001). Similarly in the absence of the ligand, hRXRα was both cytoplasmic and nuclear (0.46 ± 0.07), but nuclear translocation increased after ligand addition (0.61 ± 0.06) (Fig. 3, C and D, p < 0.05).
FIGURE 3.
Effects of 1,25(OH)2D3 with or without UO126 treatment on nuclear localization of endogenous VDR and RXRα in HPK1A and HPK1ARas cells. HPK1A cells were grown and treated with either vehicle (Veh) or 1,25(OH)2D3 (1,25D3). The cells were also fixed and stained with VDR-Alexa-488 (A) or RXRα-Alexa-488 (C) antibodies to detect VDR or RXRα. Alexa-488 (C) followed by treatment with either vehicle (veh) or 1,25(OH)2D3. Hoechst dye was used as a DNA marker as discussed under “Experimental Procedures.” Similarly, HPK1ARas cells were grown and treated with either vehicle or 1,25(OH)2D3. The cells were then fixed and stained with VDR-Alexa-488 (E) or RXRα-Alexa-488 (G) antibodies to detect VDR or RXRα. Hoechst dye was also used as a DNA marker as discussed under “Experimental Procedures.” Nuclear localization was assessed as under “Experimental Procedures.” Scatter plots show the quantitation of fluorescence of nuclear receptors normalized to total cell fluorescence (B, D, F, and H). Values represent mean ± S.D. of at least 10 different cells. Asterisks indicate a significant difference in nuclear localization between 1,25(OH)2D3 treatment alone compared with vehicle-treated control. Open circles indicate a significant difference in nuclear localization of receptors in 1,25(OH)2D3-treated cells alone compared with combined UO126 and 1,25(OH)2D3 treatment. A p value of p < 0.05 was considered significant.
In HPK1ARas cells, VDR localization was found to be both cytoplasmic and nuclear in vehicle-treated cells (0.38 ± 0.06). Addition of 1α,25(OH)2D3 alone (0.64 ± 0.09) or a combination of the MEK inhibitor UO126 and 1α,25(OH)2D3 significantly increased VDR accumulation in the nucleus compared with control (0.77 ± 0.14) (Fig. 3, E and F, p < 0.05). However, addition of the MEK inhibitor UO126 alone did not increase the nuclear accumulation of VDR when compared with control (0.38 ± 0.06). The combination of UO126 and 1α,25(OH)2D3 increased nuclear accumulation of VDR by more than 1.5-fold compared with control and about 2-fold compared with 1α,25(OH)2D3 or UO126 treatment alone (0.48 ± 0.11) (Fig. 3, E and F, p < 0.005). Furthermore, treatment of HPK1ARas cells with 1α,25(OH)2D3 alone did not increase nuclear accumulation of endogenous RXRα (0.52 ± 0.10) compared with control (0.50 ± 0.04, Fig. 3, G and H, p > 0.05). However, pre-treatment with the MEK inhibitor UO126 followed by treatment with 1α,25(OH)2D3 significantly increased the nuclear accumulation of endogenous RXRα (0.60 ± 0.12, Fig. 3, G and H, p < 0.05) compared with vehicle or 1α,25(OH)2D3 alone treated cells (Fig. 3, G and H, p < 0.05).
Effects of 9-cis-Retinoic Acid Treatment on Subcellular Localization of RXRα in Non-transformed HPK1A and Ras-transformed HPK1ARas Cells
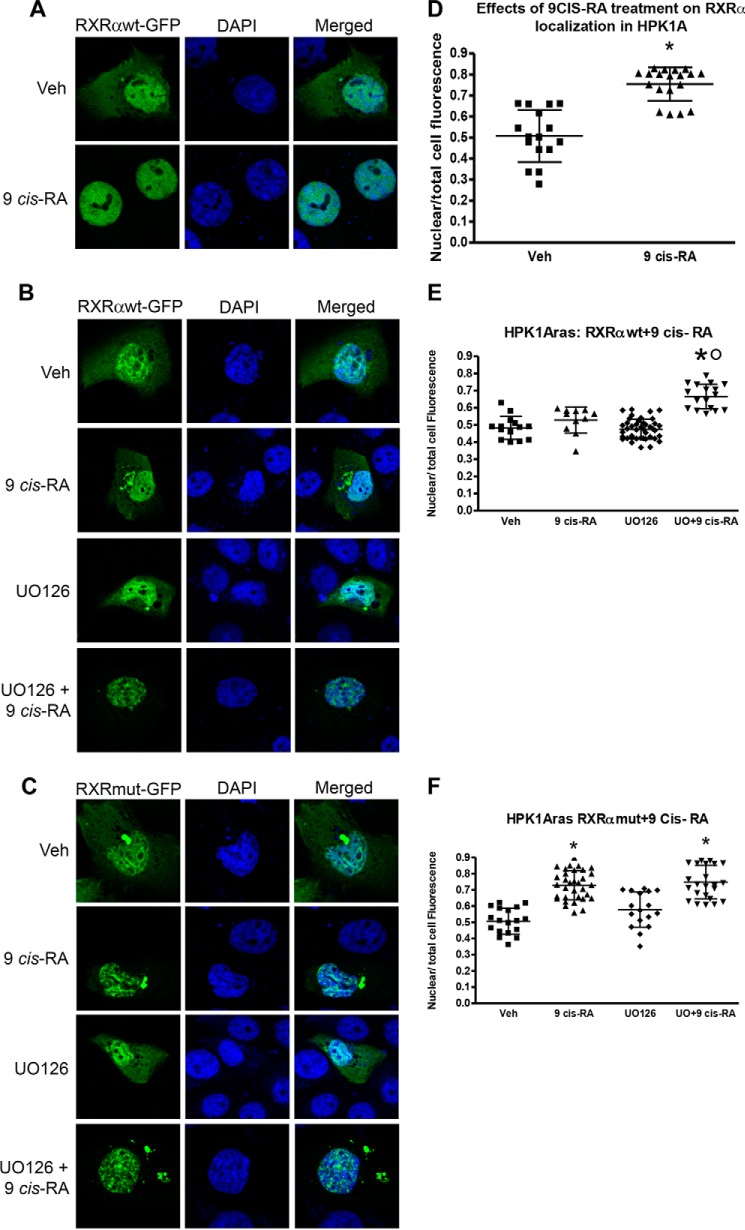
The intensity of RXRαWT-GFP was quantitated using confocal microscopy. In HPK1A cells, there was significant increase of RXRα in the nucleus following 9-cis-RA (0.75 ± 0.08) treatment when compared with control (0.51 ± 0.12, Figs. 4A and 3D, p < 0.05). In HPK1ARas cells transfected with RXRαWT-GFP plasmids, treatment with 9-cis-RA alone did not increase nuclear accumulation of RXRα (0.53 ± 0.08) compared with control (0.48 ± 0.07 Fig. 4, B and E, p > 0.05). However, pre-treatment with the MEK inhibitor UO126 followed by treatment with 9-cis-RA significantly increased the nuclear accumulation of RXRα compared with vehicle or 9-cis-RA alone treated cells (0.67 ± 0.07, Fig. 4, B and E, p < 0.05). In cells transfected with RXRαmut-GFP plasmids, 9-cis-RA treatment alone (0.73 ± 0.09) or a combination of UO126 and 9-cis-RA (0.75 ± 0.01) significantly increased nuclear accumulation of RXRα compared with vehicle (0.51 ± 0.08, Fig. 4, C and F, p < 0.05). Furthermore, treatment with the MEK inhibitor UO126 alone (0.58 ± 0.11) did not increase nuclear accumulation of RXRα compared with vehicle (0.51 ± 0.08) (Fig. 4, C and F, p > 0.05).
FIGURE 4.
Effects of 9-cis-retinoic acid on RXRα subcellular localization in HPK1A and HPK1ARas cells. HPK1A cells were transfected with RXRα-WT-GFP (A and D) followed by treatment with either vehicle (Veh) or 9-cis-RA for 4 h. Similarly, HPK1ARas cells were transfected with either RXRα-WT-GFP (B and E) or RXRα-mut-GFP (C and F) followed by treatment with either vehicle, 9-cis-RA, UO126 alone, or a combination of UO126 and 9-cis-RA. Nuclear localization was assessed as under “Experimental Procedures.” Scatter plots show the quantitation of fluorescence of nuclear receptors normalized to total cell fluorescence (D–F). Values represent mean ± S.D. of at least 10 different cells. Asterisks indicate a significant difference in nuclear localization between 9-cis-RA treatment alone compared with vehicle-treated control. Open circles indicate a significant difference in nuclear localization of receptors in 9-cis-RA-treated cells alone compared with combined UO126 and 9-cis-RA treatment. A p value of p < 0.05 was considered significant.
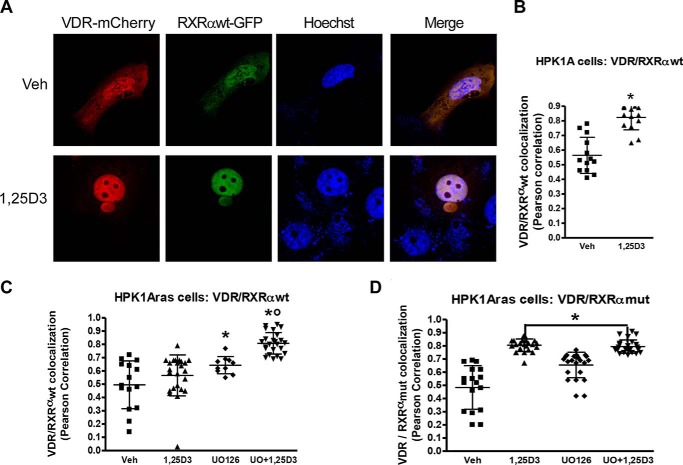
Effects of RXRα Phosphorylation on VDR/hRXRα Co-localization in Non-transformed HPK1A and Ras-transformed HPK1ARas Cells
We used Pearson correlation coefficient to assess VDR/RXRα co-localization in the nuclear compartment of the non-transformed and Ras-transformed cells. In HPK1A cells co-transfected with VDR-mCherry/RXRαWT-GFP, treatment with 1α,25(OH)2D3 (0.82 ± 0.09) significantly increased nuclear VDR/hRXRαWT co-localization when compared with vehicle (0.56 ± 0.82, Fig. 5, A and B, p < 0.0001). The effect observed is similar to the one seen when RXRα and VDR were transfected separately. In HPK1ARas cells co-transfected with VDR-mCherry/RXRαWT-GFP, treatment with 1α,25(OH)2D3 alone (0.50 ± 0.18) did not increase VDR/hRXRαWT co-localization when compared with vehicle (0.56 ± 0.10, Fig. 5C, p > 0.05). However, treatment with UO126 alone (0.64 ± 0.06) or a combination of UO126 and 1α,25(OH)2D3 (0.81 ± 0.08) significantly increased VDR/hRXRα nuclear co-localization when compared with control (Fig. 5C, p < 0.05). When the cells were co-transfected with VDR-mCherry/RXRαmut-GFP, treatment with 1α,25(OH)2D3 alone (0.81 ± 0.05) significantly increased VDR/hRXRαmut co-localization when compared with vehicle (0.49 ± 0.17, Fig. 5D, p < 0.05). Similarly, treatment with UO126 alone (0.66 ± 0.09) or a combination of UO126 and 1α,25(OH)2D3 (0.80 ± 0.05) significantly increased VDR/hRXRα nuclear co-localization when compared with control (Fig. 5D, p < 0.05).
FIGURE 5.
Effects of RXRα phosphorylation on VDR/hRXRα co-trafficking in HPK1A and HPK1ARas cells. A, cells were co-transfected with either VDR-mCherry/RXRαWT-GFP or VDR-mCherry/RXRαmut-GFP. Following transfection, cells were treated with either vehicle, 1,25(OH)2D3 (1,25D3), UO126 alone, or a combination of UO126 and 1,25(OH)2D3. Scatter plots show co-localization measurement using Pearson correlation coefficient of HPK1A cells co-transfected with VDR-mCherry/RXRαWT-GFP (B) or HPK1ARas cells co-transfected with either VDR-mCherry/RXRαWT-GFP (C) or with or VDR-mCherry/RXRαmut-GFP (D). Values are mean ± S.D. of at least 10 cells per treatment condition. Asterisks indicate a significant difference in interaction between 1,25(OH)2D3 treatment alone compared with vehicle-treated control. Open circles indicate a significant difference in interaction between 1,25(OH)2D3-treated cells alone compared with combined UO126 and 1,25(OH)2D3 treatment. A p value of p < 0.05 was considered significant.
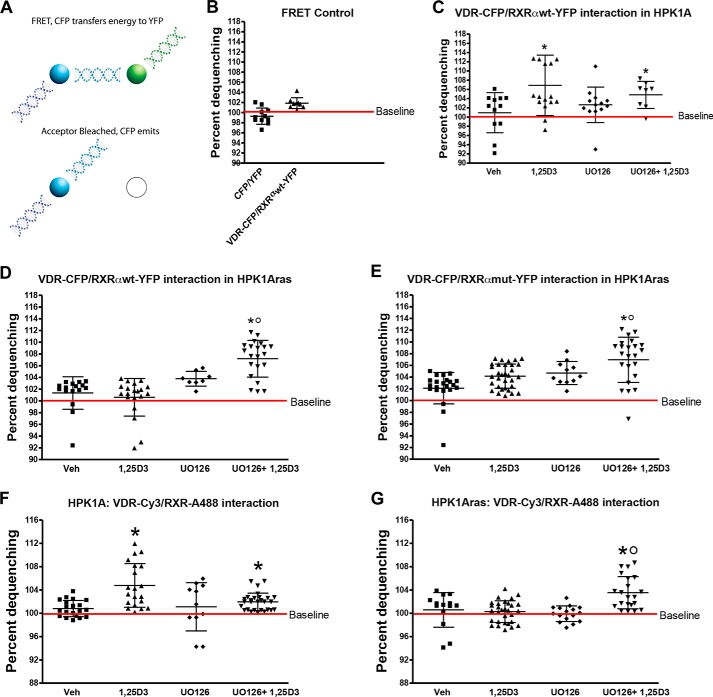
Effects of RXRα Phosphorylation on VDR and hRXRα Interaction in Living Cells Using FRET
We used FRET to investigate the interaction between VDR and RXRα after ligand binding in both cell lines. We assayed FRET between our YFP- and CFP-tagged proteins and compared it with co-transfected CFP and YFP probes that served as negative controls (Fig. 6A). As expected our results showed VDR/RXR heterodimeric interaction indicating that our FRET pairs were functional. No FRET signal was observed with the negative control CFP/YFP probes in both cell lines (Fig. 6B). Next, we tested for VDR and RXRα interaction in both cell lines using the VDR-CFP and RXRαWT-YFP or RXRαmut-YFP FRET pairs. Results show that VDR interacts with RXRα in the absence of ligand in both cell lines. In HPK1A cells co-transfected with VDR-CFP/RXRαWT-YFP, treatment with 1α,25(OH)2D3 alone (107 ± 6.56%) or a combination of UO126 and 1α,25(OH)2D3 (105.1 ± 2.95%) increased the interaction between VDR and RXRα compared with vehicle (100.91 ± 4.35%) or UO126 (103.04 ± 3.86%; Fig. 6C, p < 0.05). In HPK1A cells co-transfected with VDR-CFP/RXRαmut-YFP, treatment with 1α,25(OH)2D3 also increased FRET efficiency. However, treatment with UO126 or a combination of UO126 and 1α,25(OH)2D3 had similar effects and did not increase FRET efficiency when compared with vehicle alone (p > 0.05). Efficiency increased only with 1α,25(OH)2D3 treatment (data not shown).
FIGURE 6.
Effects of RXRα phosphorylation on VDR and RXR interaction in HPK1A and HPK1ARas cells. A, FRET was measured by acceptor photobleaching as described under “Experimental Procedures.” Cells were co-transfected with either VDR-CFP/RXRαWT-YFP or VDR-CFP/RXRαmut-YFP. Following transfection, cells were treated with either vehicle (Veh), 1,25(OH)2D3 (1,25D3), UO126 alone, or a combination of UO126 and 1,25(OH)2D3 (C–E). B, HPK1A cells were co-transfected with either CFP/YFP (negative control) or VDR-CFP/RXRαWT-YFP plasmids. Similarly, both HPK1A (F) and HPK1ARas (G) cells were grown and treated with either vehicle, 1,25(OH)2D3, UO126 alone, or a combination of UO126 and 1,25(OH)2D3. The cells were then fixed and co-stained with VDR-Cy3 and RXRα-Alexa-488 antibodies to detect both endogenous VDR and RXRα. FRET was measured by acceptor photobleaching as described under “Experimental Procedures.” Scatter plots show FRET measurement of HPK1A cells co-transfected with VDR-CFP/RXRαWT-YFP (C) or co-stained with VDR-cy3 and RXRα-Alexa-488 antibodies (F). Similarly, in HPK1ARas cells scatter plots show cells co-transfected with either VDR-CFP/RXRαWT-YFP (D), VDR-CFP/RXRαmut-YFP (E), or co-stained with VDR-cy3 and RXRα-Alexa-488 antibodies (G). Values are mean percentage dequenching ± S.D. of at least 10 cells per treatment condition. FRET baseline was set at 100%. Asterisks indicate a significant difference in interaction between 1,25(OH)2D3 treatment alone compared with vehicle-treated control. Open circles indicate a significant difference in interaction between 1,25(OH)2D3-treated cells alone compared with combined UO126 and 1,25(OH)2D3 treatment. A p value of p < 0.05 was considered significant.
In HPK1ARas cells, co-transfection with VDR-CFP/RXRαWT-GFP and treatment with either 1α,25(OH)2D3 (100.6 ± 3.20%) or UO126 (103.8 ± 1.26%) alone did not increase FRET efficiency compared with vehicle (101.3 ± 2.76) (Fig. 6D, p > 0.05). However, combined treatment of UO126 and 1α,25(OH)2D3 significantly increased VDR/RXRα interaction compared with vehicle or 1α,25(OH)2D3 treatment alone (107.2 ± 3.14%; Fig. 6D, p < 0.05). When the cells were co-transfected with VDR-CFP/RXRαmut-YFP, VDR, and RXRα, interaction significantly increased on treatment with 1α,25(OH)2D3 (104.17 ± 2.06%) when compared with vehicle (102.11 ± 2.69%; Fig. 6E, p < 0.05). Furthermore, combined treatment with UO126 and 1α,25(OH)2D3 significantly increased VDR/RXRα interaction (107.0 ± 3.86%) when compared with control (104.7 ± 1.97%; Fig. 6E, p < 0.0001). Taken together, these data suggest that VDR/RXRα interaction and heterodimerization are improved when RXRα phosphorylation is blocked or abolished in the Ras-transformed cells.
Effects of RXRα Phosphorylation on Endogenous VDR and hRXRα Interaction in Living Cells Using FRET
We used Alexa Fluor-488 and Cy3 fluorochrome-conjugated secondary antibodies to detect and measure interaction between endogenous VDR and RXRα in non-transfected cells in both cell lines. In HPK1A cells, treatment with 1α,25(OH)2D3 alone (104.8 ± 3.75% FRET) or a combination of UO126 and 1α,25(OH)2D3 (101.11 ± 4.14% FRET) increased the interaction between VDR and RXRα compared with vehicle (101.0 ± 1.40% FRET) or UO126 (101.3 ± 1.99% FRET; Fig. 6F, p < 0.05). In HPK1ARas cells, co-stained with VDR-Cy3/RXR-Alexa-488, treatment with either 1α,25(OH)2D3 (100.03 ± 1.68% FRET) or UO126 (99.93 ± 1.34% FRET) alone did not increase FRET efficiency compared with vehicle (100.58 ± 2.97% FRET; Fig. 6G, p > 0.05). However, the combined treatment of UO126 and 1α,25(OH)2D3 significantly increased VDR/RXRα interaction compared with vehicle or 1α,25(OH)2D3 treatment alone (103.53 ± 2.76% FRET; Fig. 6G, p < 0.05).
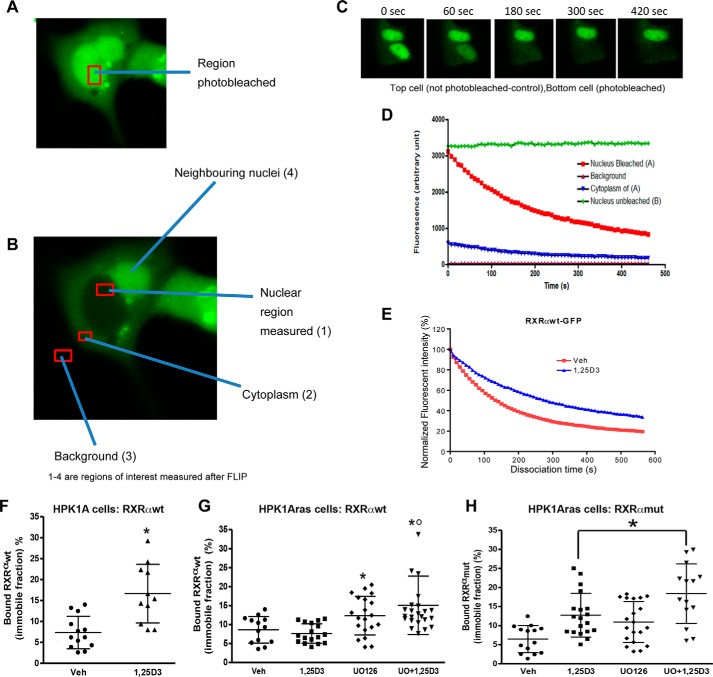
Effects of RXRα Phosphorylation at Serine 260 on Intra-nuclear Mobility of RXRα Using FLIP
We used FLIP to investigate the intra-nuclear movement of RXRα. In these experiments, a portion of the nucleus of the living cell is repeatedly bleached in the same spot using high laser power. The cell is imaged between each round of bleaching, and a spot elsewhere in the nucleus is monitored for loss of fluorescence. If the GFP-labeled molecules are shuttling between the bleaching and reporting points, then the fluorescence will decrease at both points. Relatively immobile proteins in contrast will be bleached effectively at the bleaching point but not at the reporting point. The rate of loss of fluorescence from the region of interest contains information on the rate of dissociation of the protein from the particular compartment (Fig. 7, A–E). Thus, by measuring the kinetics of loss of RXRα-GFP from the reporting point we could determine whether the protein is tightly bound within the nuclear compartment. HPK1A cells were transfected with either RXRαWT-GFP or RXRαmut-GFP and treated with or without 1α,25(OH)2D3. A nuclear area of 16 μm2 (4 μm width) was next selected as the region of interest and repeatedly photobleached at 1-s intervals for the duration of the experiments. Multiple data points gathered over time were fitted to a one-phase dissociation curve. Using this method, we found that more than 70% of the RXRα-GFP nuclear pool was mobile in both cell lines in the absence of treatment (Fig. 7F). Furthermore, we found that the nucleolus-associated pool of RXRα is continuously and rapidly exchanged with the nucleoplasmic pool. Repeated bleaching within the nucleus resulted in the complete loss of RXRα-GFP signal in the entire nucleus and also the cytoplasm. Thus, this experiment provided a direct demonstration of intra-nuclear mobility. The half-time for dissociation of the RXRαWT-GFP in the nuclear compartment after 1α,25(OH)2D3 treatment (112.02 ± 13.5 s) was longer than the vehicle control (98.11 ± 11.2 s, p > 0.05). The immobile fraction showed larger changes, indicating significant modulation of binding to nuclear structure. RXRα was found to be stably anchored (16.65 ± 7.01% bound) to other nuclear proteins within the nuclear compartment on treatment with 1α,25(OH)2D3. This immobile fraction was significantly larger when compared with vehicle-treated cells (7.35 ± 3.89% bound, Fig. 7F, p < 0.05). Taken together, the results indicate that ligand addition in the non-transformed cells increases the half-time of dissociation and bound fraction of the receptor in the unbleached portion of the nuclear compartment.
FIGURE 7.
Effects of RXRα phosphorylation on nucleocytoplasmic kinetics of RXRα in HPK1A and HPK1ARas cells. FLIP methodology was used to assess nucleocytoplasmic kinetics. HPK1A cells were transfected with RXRαWT-GFP (A–E). Following transfection, live cells were treated with either vehicle (Veh) or 1,25(OH)2D3 (1,25D3) and nucleocytoplasmic trafficking was measured using confocal microscopy (see “Experimental Procedures”). Nuclear area was selected and photobleached (A), and other regions of interests measured but not photobleached (B, 1–3). C, time course showing (i) an unbleached nucleus of a neighboring cell and (ii) a cell with a bleached nucleus. The normalized fluorescent intensity of the unbleached and bleached nuclei above is shown in D. The dissociation curve of vehicle and 1,25(OH)2D3-treated cells are shown in E. The bound fractions of HPK1A cells transfected with RXRαWT-GFP (F) or HPK1ARas cells transfected with either RXRαWT-GFP (G) or RXRαmut-GFP (H) are similarly shown. Values are mean ± S.D. of at least 10 cells per treatment. Asterisks indicate a significant difference in bound fraction between vehicle (Veh) and 1,25(OH)2D3 (1,25D3) cells. A p value < 0.05 was considered significant.
In the Ras-transformed cells transfected with RXRαWT-GFP, treatment with1α,25(OH)2D3 did not significantly increase the residence time of RXRα in the nucleus when compared with vehicle-treated cells (data not shown, p > 0.05). Also, the percentage of immobile fraction was comparable but not significantly different between 1α,25(OH)2D3 (8.61 ± 3.50%) and vehicle treatment (7.64 ± 2.55%, Fig. 7G, p > 0.05). Interestingly, treatment with UO126 alone (12.34 ± 5.10%) or pre-treatment with UO126 followed by 1α,25(OH)2D3 treatment (15.08 ± 7.81) significantly increased the percentage of immobile fractions compared with control (Fig. 7G, p < 0.05).
In HPK1ARas cells transfected with RXRα mutant GFP, treatment with 1α,25(OH)2D3 significantly increased the residence time of RXRα in the nuclear compartment when compared with vehicle control (data not shown, p < 0.005). Furthermore, the percentage of immobile fraction in 1α,25(OH)2D3-treated cells was significantly higher (12.72 ± 5.73%) compared with vehicle (6.47 ± 3.55%; Fig. 7H, p < 0.05). The effect observed was probably due to slowly exchanging receptor pools and binding to nucleoplasmic components. Furthermore, treatment with UO126 alone (10.94 ± 5.35%) or combined treatment with UO126 followed by 1α,25(OH)2D3 treatment (18.38 ± 7.81%) significantly increased the percentage of immobile fractions compared with control (Fig. 7H, p < 0.05). Taken together, the results show that blocking or abolishing the phosphorylation in the Ras-transformed cells will possibly increase binding of the receptors to their ligands and co-activator recruitment.
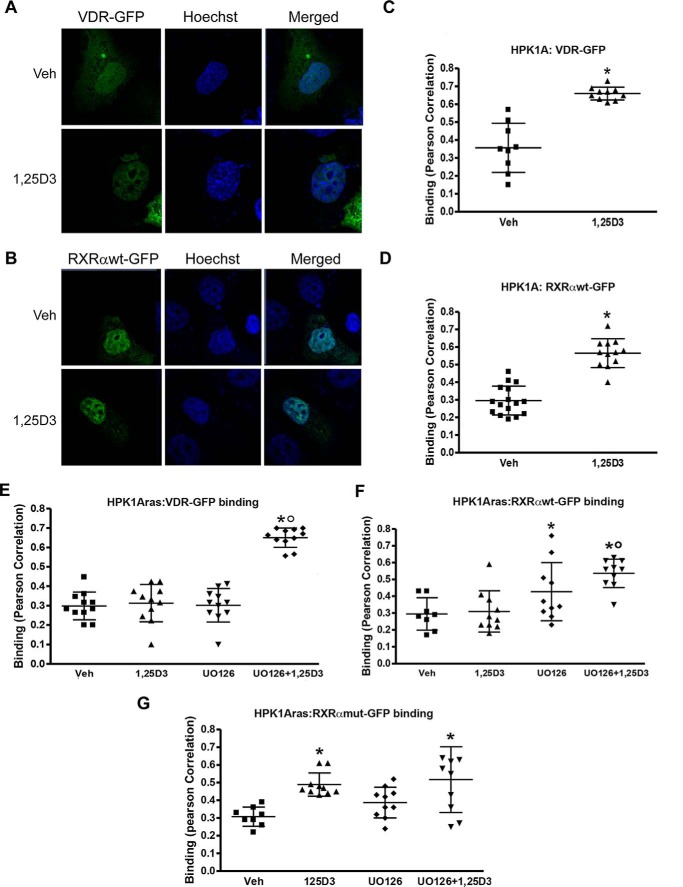
Effects of 1α,25(OH)2D3 on VDR-RXRα Complex Binding to DNA in Non-transformed HPK1A and Ras-transformed HPK1ARas Cells
We first assessed the effects of RXRα phosphorylation on VDR-RXR complex binding to DNA in HPK1A cells (Fig. 8, A and B). HPK1A cells transfected with VDR-GFP (Fig. 8A) or RXRαWT-GFP (Fig. 8B) and treated with vehicle or 1α,25(OH)2D3 were fixed and stained with Hoechst 33342, a widely used DNA-specific dye, which emits blue fluorescence under ultraviolet (UV) illumination when bound to DNA. Hoechst has a preference to bind to A/T-rich DNA sequences and to highlight a subset of the genome. Co-localization of RXRαWT-GFP or VDR-GFP and DNA (Hoechst 33342) using Pearson's coefficient correlation showed that the VDR-RXRα complex bound more to DNA in cells treated with 1,25(OH)2D3 compared with vehicle (Fig. 8, A and C, B and D, p < 0.001). We next compared VDR-RXR complex binding to DNA in HPK1ARas cells transfected with either VDR-GFP or RXRαWT-GFP. We found no significant difference in 1,25(OH)2D3 compared with vehicle-treated cells (Fig. 8, F and G, p > 0.05). However, when cells were pre-treated with UO126 followed by 1,25(OH)2D3 treatment, the VDR-RXRα complex binding to DNA significantly increased compared with vehicle or 1,25(OH)2D3 treatment alone (Fig. 8, F and G, p < 0.05). Similarly, in HPK1ARas cells transfected with RXRαmut-GFP and treated with 1,25(OH)2D3, a significant increase in the VDR-RXRα complex binding to DNA was observed when compared with vehicle (Fig. 8H, p < 0.05). Pre-treatment with UO126 followed by 1,25(OH)2D3 did not increase binding to DNA levels seen with 1,25(OH)2D3 treatment alone (Fig. 8H, p < 0.05).
FIGURE 8.
Determination of receptor/DNA interaction. HPK1A cells transfected with VDR-GFP (green) and treated with either vehicle or 1,25(OH)2D3 (1,25D3) post-transfection were stained with Hoechst dye (blue, A). Quantitation of binding between DNA (Hoechst) and VDR (GFP) was assessed using confocal microscopy and Pearson correlation (B). Similarly, cells were transfected with RXRαWT-GFP (C), and binding was assessed following treatment as above (D). Next, HPK1ARas cells were transfected with either VDR-GFP (E), RXRαWT-GFP (F), or RXRαmut-GFP (G), and binding was assessed following treatment with either vehicle (veh), 1,25(OH)2D3, UO126 alone, or a combination of UO126 and 1,25(OH)2D3. Values are mean ± S.D. of at least 10 cells per treatment. Asterisks indicate a significant increase in DNA/receptor interaction in vehicle compared with 1,25(OH)2D3-treated cells. Open circles indicate a significant difference in interaction between 1,25(OH)2D3-treated cells alone compared with combined UO126 and 1,25(OH)2D3 treatment. A p value of p < 0.05 was considered significant.
Discussion
The effects of 1α,25(OH)2D3 on keratinocytes and other cancer cells have been reported to include growth inhibition, cell cycle arrest, induction of differentiation, and apoptosis (4, 8, 9, 20, 24, 28, 30, 33, 34). In our earlier studies (32–37), we found that the immortalized non-transformed (HPK1A) cells were more sensitive to the growth inhibitory action of 1α,25(OH)2D3 compared with the neoplastic Ras-transformed (HPK1ARas) keratinocytes. However, pre-treatment of HPK1ARas cells with MEK inhibitors partially restored the sensitivity of the Ras-transformed cells to 1α,25(OH)2D3 (33, 34, 36). Furthermore, Solomon and co-workers (33–35) showed that activation of MAPK using a constitutively active MAPKK expression vector transfected into HPK1A cells induces resistance to vitamin D-dependent signaling. This sensitivity was similarly restored following pre-treatment with an MAPK inhibitor (33–36). Here, these findings are further supported with cell cycle distribution data. Furthermore, we showed previously that genetic approaches using a non-phosphorylatable RXR mutant prevents phosphorylation of RXRα at Ser-260 and can reverse the sensitivity to 1,25(OH)2D3 using functional assays (33). The combination therapy appears to be additive in the HPK1ARas cells only but not in HPK1A cells. In both HPK1A and HPK1ARas cells, UO126 and 1,25-dihydroxyvitamin D appear to work independently. Our conclusion from these experiments is that UO126 and 1,25-dihydroxyvitamin D affect the cell cycle by an independent mechanism. However, UO126 by blocking phosphorylation indirectly potentiates the effects of 1,25 dihydroxyvitamin D independently of its direct effect on the cell cycle. In this study we found by FRET that in HPK1ARas cells 1α,25(OH)2D3-induced interaction of VDR with RXR is impaired and that this impairment is mediated through phosphorylation of RXR. Furthermore, we found that we can detect 1α,25(OH)2D3-induced immobilization of VDR-GFP and RXR-GFP onto nuclear structure in a photobleach-based binding assay and in a direct measure of co-localization of endogenous VDR and RXRα with chromatin fine structure. This immobilization is lost in HPK1ARas cells in a manner dependent on phosphorylation of RXRα. Our major conclusions are as follows: 1) failure of 1α,25(OH)2D3 to induce growth inhibition in Ras-transformed cells primarily results from Ras pathway-mediated phosphorylation of RXRα. 2) Phosphorylation of RXRα leads to non-responsiveness to 1α,25(OH)2D3 via impaired association of VDR with targets within the nucleus.
In this study we have gained insight into the mechanisms by which VDR/RXRα fail to mediate growth inhibition in Ras-transformed cells. We conducted a series of experiments examining changes in subcellular localization, VDR/RXRα interaction, and DNA binding. Investigating the subcellular localization of both VDR and RXRα in non-transformed and Ras-transformed cell lines, we found that although VDR can partition from the cytoplasm to the nucleus, its nuclear accumulation is increased when it forms a complex with RXRα. This nuclear accumulation appears to be inhibited by phosphorylation at Ser-260 because transfection of the Ras-transformed cells with a non-phosphorylatable mutant or pre-treatment with the MEK inhibitor UO126 increased the nuclear accumulation of RXRα.
Our findings on both VDR and RXRα distributions in the non-transformed cells are not only supported by other reports (12, 50, 52, 53) but additionally our data suggest that phosphorylation retains a fraction of RXRα in the cytoplasm of transformed cells (54). The FRET experiments carried out with fluorescent chimeras of VDR, RXRαWT, and a non-phosphorylatable RXRα mutant confirmed that VDR/RXRα heterodimer interaction in the Ras-transformed cells was compromised by phosphorylation, as blocking phosphorylation either by pre-treatment with the MEK inhibitor UO126 or the non-phosphorylatable RXRα mutant increased VDR/RXRα interaction. We obtained similar results from cells transfected with GFP-tagged VDR and RXRα and from FRET assays employing immunofluorescence staining of endogenous proteins, suggesting that the GFP tag is not interfering with aspects of VDR or RXRα function relevant to this study.
Our results further support the view that heterodimerization may influence the subcellular localization of VDR and RXRα. In our confocal microscopy experiments, we showed that dimerizing RXRα with VDR facilitates nuclear accumulation of VDR. Our FRET data gave mechanistic support to these subcellular localization studies. Although our observation supports a novel mechanism by which the RXR heterodimerization partner dominates the activity of the heterodimers, it differs from reports from Yasmin et al. (55), who suggested that nuclear accumulation of RXR-VDR heterodimers is mediated predominantly by the VDR. Previous research demonstrated that steroid receptors are in constant rapid motion within the nucleus and that they accumulate in discrete nuclear foci after hormone binding (56–58). Furthermore, recent FRET studies carried out in living cells using GFP fusion proteins and co-localization results using RXRα and vitamin D receptor (VDR) fluorescent chimeras have led to the conclusion that RXRα does not only dynamically shuttle between the nucleus and cytoplasm but it also heterodimerizes with VDR in the cytoplasm regardless of the calcitriol (1α,25(OH)2D3) binding status (59, 60). Previous and recent studies examined VDR-RXR (FRET) interaction in normal African green monkey kidney COS-7, rat osteosarcoma ROS17/2.8 (61), and human embryonic kidney (HEK) 293 cells (54). However, VDR-RXR interaction was never examined nor compared in normal and transformed cells.
Pruffer et al. (54, 61) reported that VDR/RXR heterodimerization can occur in the cytoplasm without ligand addition. We did not measure RXR/VDR heterodimerization in the cytoplasm. However, we have shown by co-localization studies and FRET that VDR/RXR heterodimerize in the nucleus even without ligand addition and that ligand addition increases heterodimerization and interaction. FRET has the advantage of examining directly the interaction between VDR and RXRα in an intact cellular environment as opposed to our previous studies, which used nuclear extracts and electromobility gel shift assays (34, 36). In these earlier studies we were only able to determine that VDR-RXR complex formation occurred in Ras-transformed keratinocytes, but this experimental approach did not permit us to measure VDR/RXR interaction. By using FRET in this study, we could measure directly the interaction and show that it was significantly affected by phosphorylation at serine 260.
We assayed binding of RXRα to intra-nuclear structures by FLIP and also by the use of Pearson correlation coefficient to assess co-localization with DNA fine structure. These distinct methods gave consistent results, showing that ligand addition induced binding of hRXRα to chromatin, that this binding was blunted in the nuclear compartment of Ras-transformed cells, and that phosphorylation of hRXRα mediated this insensitivity to ligand because blocking phosphorylation with a non-phosphorylatable RXRα mutant increased RXRα binding and restored vitamin D function in the Ras-transformed cells.
In the FLIP experiments, we recorded two distinct kinetic pools of RXRα-GFP in the nucleus as follows: a large mobile pool, which represents the continuously exchanging molecules within the nucleoplasmic compartment responsible for the fluorescence signal loss, and a smaller less mobile (bound) fraction that does not contribute to the fluorescence loss over the time scale of the experiment. We hypothesize that this immobile fraction could represent receptor bound to DNA or to chromatin. We tested this hypothesis using the Pearson correlation coefficient to quantify co-localization of the GFP-tagged receptor with Hoechst dye on chromatin structure at the level of individual pixels. Our results are in accordance with the FLIP results showing that binding to DNA is affected in the Ras-transformed keratinocytes. Because 1α,25(OH)2D3 does not bind to RXRα, it is possible that the non-phosphorylatable RXRαmut-GFP induced a conformational change allowing VDR to bind DNA upon ligand addition and resulting in docking of new factors or the recruitment of cofactors to the VDR-RXRα receptor complex. The increase in residence time after ligand addition as shown by FLIP might also reflect the strengthening of the interaction with slowly or non-diffusing nuclear components such as chromatin. Furthermore, the increased interaction could result from either an increased affinity or from stabilization of interaction leading to longer binding events (63, 64). In the Ras-transformed cells, protein phosphorylation could inhibit immobilization of receptors on chromatin. Failure to bind chromatin could explain inhibition of vitamin D signaling in the cancer cells (65, 66). Solomon and co-workers (33–35) previously reported that in Ras-transformed cells, RXRα phosphorylation at serine 260 would create conformational changes within the ligand binding domain, disrupting interactions with co-regulators and therefore decreasing transcriptional activities resulting in resistance to the growth inhibitory action of vitamin D. More recently, it was shown that phosphorylation at serine 260 impairs recruitment of DRIP205 and other co-activators to the VDR-RXRα heterodimer (36), and it also delays nuclear export and RXRα degradation in hepatocellular carcinoma (67). Quack and Carlberg (68) using limited protease digestion and gel shift clipping experiments demonstrated that binding of RXR to VDR not only induces conformational changes to VDR but also the conformational changes induced by ligand binding stabilized VDR/RXR dimers (69). Feige and co-workers (64) using FRET to study the PPAR/RXR interaction indicated that PPAR-RXR dimerization occurred prior to ligand binding or DNA binding; however, heterodimer binding to DNA was only observed to be stable in vivo after ligand had bound. Increasing evidence indicates that RXR does not play a passive role as a heterodimeric partner but impacts the responses of its nuclear receptor partner, regardless of its permissive, nonpermissive, or conditionally permissive status (11). Taken together, these results, including ours, demonstrate an important role of RXR in VDR-RXR heterodimer binding to target DNA sequences in living cells.
The physiological relevance of this study and the consequences of RXRα phosphorylation cannot be understated because RXR additionally functions as a homodimer and also heterodimerizes with multiple members of the nuclear receptor superfamily. RXR thus plays a central role in regulating a number of signaling pathways (11, 35, 36, 70–75). The clinical relevance of RXRα phosphorylation has been discussed in other cancer model systems, including cancers of the liver, lung, breast, colon, pancreas, and leukemia (75–82). Matsushima-Nishiwaki et al. (67, 80) and Moriwaki et al. (81) reported that RXRα protein was phosphorylated anomalously in human hepatocellular carcinoma (HCC) tissues as well as HCC cell lines. Furthermore, RXRα phosphorylation at serine 260 also resulted in impairment of its function and resistance to the growth inhibitory effects of all-trans-retinoic acid. Furthermore, Lu et al. (82) demonstrated that phosphorylation of hRXRα at serine 260 interferes with its function and delays its degradation in cultured human HCC, leading to enhanced cellular proliferation. Yamazaki et al. (78) showed that 75% of colorectal cancer tissues expressed phosphorylated RXRα protein when compared with corresponding normal colon epithelial tissues. Kanemura et al. (79) reported that abnormal phosphorylation of RXRα protein played a role in the enhancement of cell proliferation, while producing an antiapoptotic effect and also acquiring RA resistance in HL-60R human leukemia cells. Thus, phosphorylation of RXRα plays an important role in insensitivity to growth inhibition of a wide range of human cancers. Although phosphorylation at serine 260 appears to play the predominant role in these models through ERK1/2, it should be noted that other MAPK downstream pathways such as c-Jun N-terminal kinase (JNK) have been reported to alter vitamin D action through phosphorylation at alternative amino acid sequences (70, 84).
Proposed Model for Restoration of Vitamin D Sensitivity in Ras-transformed Keratinocytes
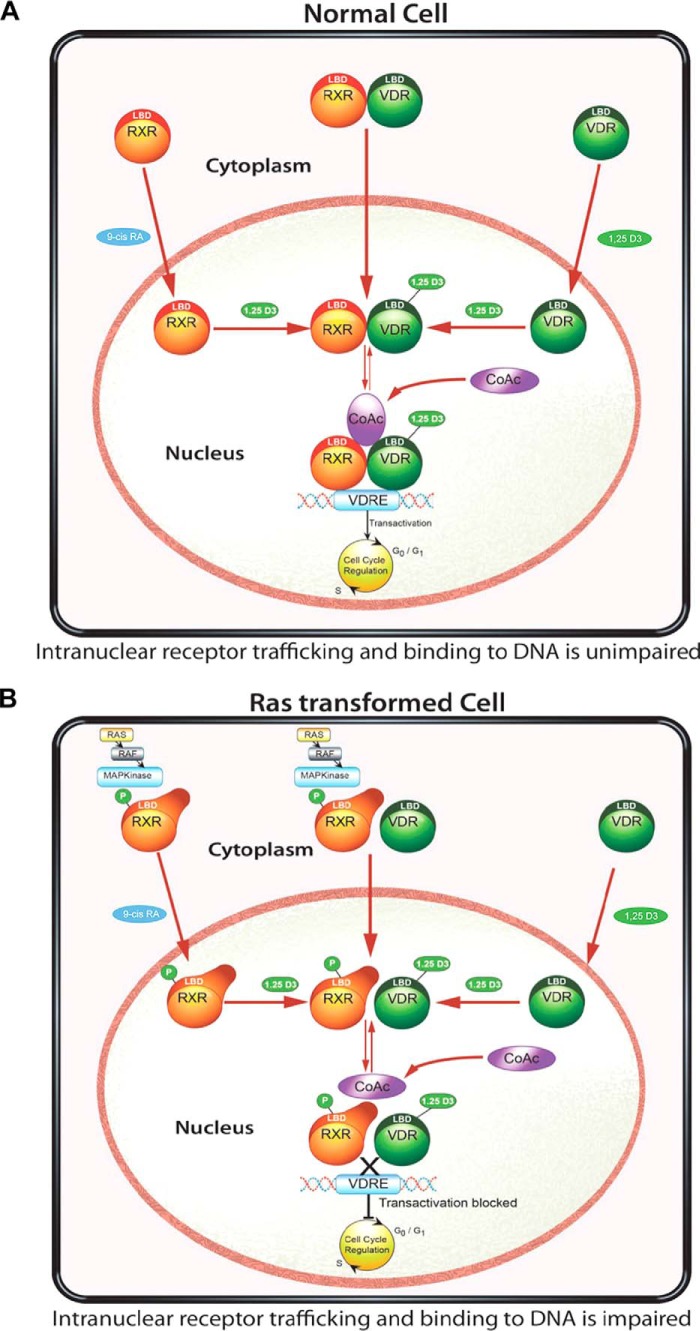
We propose a model (Fig. 9) explaining how RXRα phosphorylation at serine 260 could affect VDR and RXR interaction and nucleoplasmic trafficking (Fig. 9A). In normal cells, it is subject to debate whether VDR and RXR move independently or as a complex from the cytoplasm to the nucleus. However, the nuclear import of their mutual heterodimer is controlled predominantly by RXR and regulated by 1α,25(OH)2D3, and in the presence of 1α,25(OH)2D3, the VDR controls this process. Once in the nucleus, the VDR-RXR heterodimer can bind to chromatin and carry out gene transcription (Fig. 9B). In Ras-transformed cells, VDR and RXR could still move into the nucleus independently or as a complex. Phosphorylation of RXR alters the conformation of the RXR, which indirectly prevents the mobilization of a large proportion of the mutual heterodimer from entering the nuclear compartment leading to a decrease in localization and binding to chromatin.
FIGURE 9.
Proposed model for nuclear import of VDR, RXR, and VDR/RXR interaction and DNA binding in non-transformed and Ras-transformed cells. In normal cells (A), the nuclear import of VDR and RXR is mediated by their respective ligands. Once in the nucleus, 1,25(OH)2D3 binding to VDR is critical for the VDR-RXR heterodimer interaction and binding to the hormone-response elements, recruitment of co-factors (CoAc), and effect on 1,25(OH)2D3 signaling. In the Ras-transformed keratinocyte (B), phosphorylation of RXR prevents the nuclear translocation of RXR and binding of the VDR-RXR complex to the hormone-response element. The recruitment of co-factors was impaired thus preventing 1,25(OH)2D3 signaling. Using either the MEK inhibitor UO126 or a non-phosphorylable RXR mutant, we can restore the cells nuclear import of RXR, VDR/RXR, as well as interaction with DNA and 1,25(OH)2D3 and VDR signaling.
These studies suggest that blocking phosphorylation either by the use of a MEK inhibitor or using a non-phosphorylatable RXRα mutant in combination with active vitamin D compounds might provide novel strategies in the treatment of cancers with a Ras signature.
Experimental Procedures
Reagents and Antibodies
1α,25(OH)2D3 and 9-cis-RA were purchased from Sigma, and stock solutions were prepared in ethanol. The MEK1/2 inhibitor UO126 (1,4-diamino-2,3-dicyano-1,4-bis[2-aminophenylthio] butadiene) was purchased from Promega (Madison, WI), and stock solutions were prepared in DMSO. The following antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA): rabbit anti-hVDR (C-20, sc-5574); mouse anti-hVDR (D-6, sc-1313); rabbit anti-hRXR (D-20; sc-553); and mouse anti-hRXRα (F-1; sc-46659). Both mouse anti-Cy3 (A10521) and rabbit anti-Alexa Fluor-488 (A11034) secondary antibodies were purchased from Invitrogen-Molecular Probes.
Cell Lines and Culture
The HPK1A cell line was previously established by stably transfecting normal human keratinocytes with human papillomavirus type 16 (32). In culture, these cells have an indefinite life span but retain differentiation properties characteristic of normal keratinocytes and are non-tumorigenic when injected into nude mice. These immortalized cells were then transformed into the malignant HPK1ARas cell line after transfection with a plasmid carrying an activated Ha-ras oncogene (32). HPK1ARas cells are malignant cells that form colonies in soft agar and also produce invasive tumors when transplanted into nude mice. The cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 2 mm glutamine, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 10% fetal bovine serum (FBS) and passaged twice weekly in 6-, 24-, or 96-well Falcon plates (Corning, NY).
Proliferation Assay
For assessment of cell proliferation, HPK1A and HPK1ARas cells were seeded in 24-well plates at a density of 1 × 104 cells/well and were grown in DMEM supplemented with 2 mm glutamine, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 10% FBS. After 24 h, the medium was replaced with serum-free DMEM overnight to synchronize the cells. At time 0, the medium was replaced with DMEM containing 5% charcoal-stripped FBS in the presence of increasing concentrations of 1α,25(OH)2D3 (i.e. 10−9 to 10−7 m concentration) as a single agent or in combination with the MEK1/2 inhibitor UO126 (10−6 m concentration). Control cells were treated with vehicle (0.1% ethanol and 0.1% DMSO, v/v) or MEK1/2 inhibitor UO126 (10−6 m concentration) alone. Treatment was continued for 72 h. At the end of the experiment, cells were washed with PBS, trypsinized, and counted with a Coulter counter.
Alamar Blue Cell Viability Assay
Cells were seeded in 96-well plates at a density of 5 × 103 cells/well, and the experiment was carried out as above in the proliferation assay. At the end of each experiment, 50 μl of Alamar blue (Invitrogen) was added to each well, and the plate was further incubated at 37 °C for 4 h and then transferred to a plate reader, and absorbance at 550 nm was determined as per the manufacturer's instruction.
Cell Cycle Analysis with Propidium Iodide Staining
HPK1A and HPK1ARas cells were seeded into 6-well plates at a density of 5.0 × 105 cells/plate for 24 h, grown as described above in the proliferation assay, and treated with 10−7 m 1,25(OH)2D3 alone or in combination with 10−6 m UO126. The cells were trypsinized, washed in PBS, and fixed in 70% ethanol at 4 °C overnight. 1 × 105 cells were then resuspended in 40 μg/ml propidium iodide solution with 1 mg/ml RNase and incubated in the dark at 37 °C for 30 min. DNA content and cell cycle analyses were carried out using a FACScan flow cytometer (BD Biosciences). Different phases of the cell cycle were assessed by collecting the signal at channel FL2-A. The percentage of the cell population at a particular phase was estimated by the BD CellQuest software.
Cloning of Fluorescent (Plasmids)-tagged Constructs
Subcloning of hVDR Plasmids
VDR/pSG5 plasmid was a kind gift from Dr. John White's laboratory (McGill University, Montreal, Canada). The expression vector was originally constructed by inserting a 2.1-kb EcoRI fragment containing the entire coding region of the human VDR into the EcoRI site of pSG5.VDR-CFP, VDR-GFP, VDR-YFP, and VDR-mCherry plasmids were constructed by PCR amplification of hVDR sequence using hVDRpSG5 as a template, and forward GGTTAC CTCGAG ATG GAG GCA ATG GCG GCC AGC ACT TCC CTG and reverse GTTAC CCG CGG AGA GGA GAT CTC ATT GCC AAA CAC TTC G primers were designed with XhoI and SacII restriction sites. The hVDR PCR product was ligated to the GFP variants, a generous gift from Dr. Stephane Laporte (McGill University, Montreal, Canada), and mCherry (Clontech).
Subcloning of hRXRα Plasmids
The hRXRα wild type (WT) plasmid was a kind gift from Dr. Ronald Evans' laboratory (Salk Institute, CA). The hRXRα S260A mutant plasmid was previously constructed in our laboratory (33–36). The hRXRαWT and the hRXRα S260A mutant fluorescent GFP variants (i.e. GFP, CFP, YFP, and mCherry) were constructed by PCR amplification of hRXRWT and the hRXRα S260A mutant sequences using hRXRα WT and the hRXRα S260A mutant as templates, and forward GGTTAC CTCGAG ATG GAC ACC AAA CAT TTC CTG C and reverse GTTAC CCG CGG AGA AGT CAT TTG GTG CGG CGC CTC CAG C primers were designed to create new XhoI and SacII restriction sites. The resulting amplified PCR products were ligated to mCherry and the GFP variants, respectively.
Transfection
HPK1A and HPK1ARas cells were maintained in DMEM containing 10% FBS. For experimentation, cells were plated overnight in 6-well plates on No. 1 coverslips (Fisher) for fixed cells or 35-mm MatTek glass bottom dishes (MatTek Corp., Ashland, MA) for live cell experiments. Cells were plated at 8 × 104 cells/well (HPK1A) and 6 × 104cells/well (HPK1ARas) in DMEM containing 10% FBS. The next day the medium was changed to serum-free DMEM for an hour prior to initiating the experiment. Transfection was carried out in serum-free DMEM FuGENE HD at a FuGENE HD/DNA transfection ratio of 6 μl/2 μg DNA (Roche Applied Science). The cells were transfected with vectors encoding constructs of hRXRα-YFP, hRXRα-GFP (2.0 μg), or hVDR-CFP and hVDR-GFP (3.0 μg). In co-transfection studies, a total of 5 μg of the co-transfected vectors was used per well. After 4 h of incubation, the medium was supplemented with 10% FBS (by adding 200 μl of FBS/well). Following a 30-h incubation, the medium was changed to DMEM containing 5% FBS and incubated overnight. The next day, cells were treated with vehicle (ethanol + DMSO, 0.1% v/v) or 1,25(OH)2D3 (10−7 m), 9-cis-RA (10−7 m), and UO126 (10−6 m) alone or a combination of UO126 and 1,25(OH)2D3 for 4 h. For real time live cell microscopy, the transfected cells were first transferred onto a heated stage at 37 °C for drug treatments and data acquisition. For fixed cell experiments, the cells were washed with PBS after the treatment and fixed for 15 min in 4% paraformaldehyde at 37 °C. Following fixation, cells were re-washed in PBS and mounted using Shandon immuno-Mount mounting medium (Fisher). For subcellular localization studies, following fixation and re-washing, the cells were stained with either DAPI or Hoechst 33342 dye (Invitrogen) for 10 min and then mounted using Shandon immuno-Mount mounting medium (Fisher). Imaging was carried out the next day using a Zeiss LSM 510 or LSM 780 confocal microscope (Jena, Germany).
Immunofluorescence Staining
Cells were grown in 6-well plates on 22-mm No. 1 glass slides (Fisher). At experimental endpoints the coverslips were fixed in 4% formaldehyde at room temperature for 10 min. After rinsing with PBS, cells were permeabilized for 10 min with 0.25% Triton X-100 in PBS, washed in PBS three times for 5 min, and blocked with 1% BSA in PBS/Tween 20 for 30 min. Cells were incubated with anti-VDR (rabbit at 1:50 dilution), anti-RXR (mouse at 1:50 dilution), or a combination of rabbit-anti VDR and mouse-anti RXR antibodies (at 2:100 dilution) overnight at 4 °C. Samples were then incubated with the corresponding Alexa Fluor-488 and Cy3-conjugated secondary antibodies (Invitrogen-Molecular Probes, 1:500). Following re-washing with PBS, slides were counter-stained with either DAPI or Hoechst 33342 dye (Invitrogen) for 10 min and then mounted using Shandon immuno-Mount mounting medium (Fisher). Samples were visualized the next day using a Zeiss LSM 510 or LSM 780 confocal microscope (Jena, Germany) using a ×63 N.A.1.4 oil immersion objective and appropriate filter sets.
Fluorescence Microscopy, Time-Lapse Imaging, and Image Processing
HPK1A and HPK1ARas cells were grown on 35-mm glass-bottom dishes (MatTek Corp., Ashland, MA) (live cells) or 22-mm No. 1 glass slides (fixed cells). Time lapse imaging was performed using a confocal laser scanning microscope (model LSM 510 or LSM 780, Carl Zeiss, Inc., Jena, Germany) equipped with a motorized triple line argon laser, a 100 × 1.4 NA PlanApochromat oil immersion objective, a 63 × 1.3 NA PlanApochromat oil immersion objective, a 40 × 1.3 NA Neofluar oil immersion objective, a 25 × 0.8 NA Neofluar immersion corrected objective, and a temperature and CO2 controlled stage. Time-lapse sequences were recorded using the time series function of the Zeiss LSM software.
Receptor Expression and Subcellular Distribution Using Confocal Microscopy
GFP vector alone, hVDR-GFP, hRXRαWT-GFP, and hRXRαmut-GFP expression vectors were monitored by viewing and counting fluorescing cells using a Plan-Neofluar ×40/1.3 oil objective and 488-nm excitation and 515–565-nm emission filters (Carl Zeiss Inc.). To monitor subcellular distribution of the receptors, at least 10 healthy cells were observed at random from at least 10 fields. Repeated experiments were done using the same parameters. Z-stacks of double-labeled images were collected to account for total cellular fluorescence.
Morphometric Analysis of Subcellular Localization
For evaluation of nuclear/cytoplasmic signal distribution, confocal images were taken of each fluorescing cell. A single optical slice was taken of each cell with a focus set to maximize the circumference of the nucleus. At least 10 cells were evaluated for each experimental condition. Cells that showed clear morphological changes due to protein overexpression were excluded from statistical analysis. Image analysis was performed using the ImageJ version 1.41 software (National Institutes of Health, Bethesda) to determine the nuclear (Fn), cytoplasmic (Fc), and background (Fb) fluorescence. Briefly, a mean density measurement of pixel numbers was made on a non-saturated ROI consisting of the total nucleus, the whole cell (nucleus and cytoplasmic compartments combined), and a background region outside of the cell. The ratio of nuclear to cytoplasmic fluorescence (Fn/c) was then determined according to the following formula: Fn/c = (Fn − Fb)/(Fc − Fb). Data are presented as mean ± S.D. (85, 86).
Fixed Cell Imaging and FRET Microscopy in Transfected Cells
HPK1A and HPK1ARas cells were grown on 22-mm no. 1 coverslips and co-transfected with either VDR-CFP/hRXRWT-YFP or VDR-CFP/hRXRmut-YFP, respectively. Transfected cells were next treated with either vehicle control (ethanol or DMSO 0.01% v/v), 1,25(OH)2D3 (10−7 m) alone, or a combination of UO126 (10−6 m) and 1,25(OH)2D3. Treatments were carried out for 4 h before fixing, mounting, and FRET data acquisition using a Zeiss LSM 510 confocal microscope with a Zeiss 40 × NA 1.4 Neofluar oil objective and a chamber to maintain a temperature of 37 °C and 5% CO2. To assay dequenching of donor after photobleaching, a series of eight images of the CFP channel were taken. YFP within the nucleus was bleached after image 4 by scanning with the 514 nm laser line at maximum intensity. CFP intensities inside the nucleus were compared between the immediately pre-bleach image (image 4) and the post-bleach image (image 5). Dequenching was defined as nuclear CFP intensity in image 4 divided by nuclear CFP intensity in image 5. The remainder of the image sequence served as a control for focus stability. At least 10 cells per treatment were photobleached for each experiment. Experiments were repeated twice.
To calculate FRET percentage, the fluorescence intensities of three ROIs, a region in close proximity to the cell (background), a region in the nucleus that was photobleached (bleached region), and a region of the nucleus that was not bleached (unbleached region) was selected, and data were acquired for both YFP and CFP. The fluorescent intensities of CFP immediately before the bleach and immediately after the bleach were next background corrected by subtracting fluorescence intensity of the background region in the CFP channel of the same image. The pre-bleach and post-bleach CFP corrected intensities were then used to calculate the percent dequenching, which is a measure of FRET in this experimental design. At least 10 images were analyzed per experimental condition. Percent dequenching was calculated as follows: dequenching % = (CFP_corrected post-bleach/CFP_corrected pre-bleach) × 100. In this measure, 100% represents a baseline with no change in fluorescence, indicative of no significant FRET, although values greater than 100% are consistent with FRET prior to dequenching (21, 51, 62, 83).
Fixed Cell Imaging and FRET Microscopy in Antibody Co-stained Cells
HPK1A and HPK1ARas cells were grown on 22-mm no. 1 coverslips, treated, fixed, and permeabilized as discussed above under confocal immunofluorescence microscopy. Cells were incubated with combination of rabbit-anti VDR and mouse-anti RXR antibodies (at 2:100 dilution) overnight at 4 °C. Samples were then incubated with the corresponding Alexa Fluor-488 and Cy3-conjugated secondary antibodies (Invitrogen-Molecular Probes, 1:500). Following re-washing with PBS, slides were counter-stained with either DAPI or Hoechst 33342 dye (Invitrogen) for 10 min and then mounted using Shandon immuno-Mount mounting medium (Fisher). Samples were visualized the next day for FRET data acquisition on a Zeiss LSM 780 confocal microscope (Jena, Germany) with pinhole set at 1 airy unit using 405, 488, and 568 nm excitation and a ×63/1.4 oil objective lens. To assay dequenching of donor after photobleaching, a series of eight images of the Alexa-488 channel were taken. Cy3 within the nucleus was bleached after image 4 by scanning with the 561 nm laser line at maximum intensity. Alexa-488 intensities inside the nucleus were compared between the immediately pre-bleach image (image 4) and the post-bleach image (image 5). Dequenching was defined as nuclear Alexa-488 intensity in image 4 divided by nuclear Alexa-488 intensity in image 5 similarly to the calculation of CFP/YFP FRET. The remainder of the image sequence served as a control for focus stability. At least 10 cells per treatment were photobleached for each experiment. Experiments were repeated twice.
To calculate FRET percentage, the fluorescence intensities of three ROIs, a region in close proximity to the cell (background), a region in the nucleus that was photobleached (bleached region), and a region of the nucleus that was not bleached (unbleached region) were selected and data were acquired for both Cy3 and Alexa-488. The fluorescent intensities of Alexa-488 immediately before the bleaching and immediately after the bleaching were next background corrected by subtracting fluorescence intensity of the background region in the Alexa-488 channel of the same image. The pre-bleach and post-bleach Alexa-488 corrected intensities were then used to calculate the percent dequenching, which is a measure of FRET in this experimental design. A total of 10 images were analyzed per experimental condition. Percent dequenching was calculated as follows: dequenching % = (Alexa-488_corrected post-bleach/Alexa-488_corrected pre-bleach) × 100. In this measure, 100% represents a baseline with no change in fluorescence, indicative of no significant FRET, although values greater than 100% are consistent with FRET prior to dequenching. At least 10 cells were selected at random for each experimental condition. Statistical analysis using ANOVA and t test were carried out in GraphPad Prism.
Live Cell Imaging Using FLIP Microscopy
FLIP (79, 80) was used to assess real time intranuclear mobility of GFP-tagged proteins in the presence or absence of ligand. HPK1A and HPK1ARas cells were transfected with GFP tagged hRXRαWT or hRXRαmut respectively. After 30 h of transfection, the medium was changed to one containing 5% charcoal-stripped FBS, and the cells were treated with vehicle control (ethanol or DMSO 0.01%, v/v) or 1,25(OH)2D3 (10−7 m). All photobleach image series were obtained on a 37 °C heated stage using a ×40/1.3 NA oil immersion lens. Fluorescence in a narrow strip spanning one-fourth the width of the nucleus was bleached using repeated (50–200) scans of 488 nm illumination with 100% laser transmission. Bleaching alternated with image acquisition at 5% laser transmission. Cells were scanned 0.8 to 3 s per image with two-to-eight line averaging. Fluorescence intensity in an ROI on the opposite side of the nucleus and outside of the bleach ROI was quantitated at each time point and normalized to the fluorescence intensity before bleaching. A neighboring unbleached cell served as a control for focus drift and photobleaching during image acquisition. Normalization was done to the pre-bleach data point (39). At least 10 cells were collected per treatment condition.
The Zeiss LSM software package was used to define ROIs collecting mean fluorescent intensities of the background, whole nucleus, and the whole cell for each data set measured under the same experimental condition. Images were background subtracted and data normalized and exported into Microsoft Excel before quantitation and processing. The analyzed data were used to plot curves, calculate the mobile fraction, diffusion constants, and half-time of recovery. The mobile fraction (Mf) was calculated using the equation, Mf = (Fpre − Fend)/(Fpre) (21, 51), where Fpre is the average fluorescence in the ROI before bleaching, and Fend is the fluorescence immediately after the bleach. The immobile fraction (If) was calculated as If = 1 − Mf or If = 100 − Mf, where the normalized data were converted to percentages. Decay rates were calculated by fitting a one-phase exponential decay curve Y = span·exp(−K·X) + plateau from GraphPad Prism. The half-time of fluorescence loss (t½) is the time required for the fluorescent intensity in the bleached ROI to reduce by 50% (21, 51). The t½ was calculated as 0.69/K, which assumes pseudo-first order kinetics. To determine a model-independent half-life of fluorescence loss, the fluorescent intensity data were transformed using a 0–100% scale, and the time at which fluorescence intensity fell to 50% of full intensity was defined as the t½. At least 10 cells were selected at random for each experimental condition. Statistical analyses using ANOVA and t test were carried out in GraphPad Prism.
Binding of Receptor DNA to Hoechst Dye
HPK1A and HPK1ARas cells were seeded on 22-mm (MatTek Corp.) glass slides at a concentration of 1 × 105 cells/well for 24 h in DMEM containing 10% FBS. The next day, the cells were transfected with hVDR-GFP, hRXRαWT-GFP, or hRXRαmut-GFP using FuGENE HD at a DNA/FuGENE ratio of 2 μg /6 μl as described under “Transfection” above, and cells were treated with experimental reagents for 2 h prior to fixation as described in the text. The cells were next washed with PBS and fixed in 4% paraformaldehyde for 15 min. Cells were re-washed with PBS and stained with Hoechst 33342 dye (1 μg/ml) for 10 min at room temperature. Finally, cells were re-washed with PBS, and slides were mounted with Shandon immuno-Mount medium. Imaging was carried out the next day using both the 488- and 405-nm lasers to compare co-localization of the GFP and Hoechst dye in the cells. Image analysis was carried out by selecting the nuclei in the images using the regions of interest tool in the Zeiss LSM 780 Image Examiner. Pearson's coefficient was measured in the region of interest using the Image Examiner to quantitate pixel-by-pixel co-localization of Hoechst dye (a DNA marker) with GFP within the nucleus.
Statistical Analysis
We used analysis of variance (ANOVA) and t test in GraphPad Prism software. Results are presented as means ± S.D. of at least eight independent measurements. Data were analyzed statistically by one-way analysis of variance followed by a post hoc test and Student's t test. Means were considered significantly different when p values were at least below 0.05.
Author Contributions
R. K., S. J., and J. F. P. conceived and designed the work; S. J., J. F. P., and R. K. developed the methodology; S. J., J. F. P., and R. K. did the data analysis; S. J., J. F. P., and R. K. reviewed and wrote the manuscript.
Acknowledgments
We thank Dr. J. White for providing the human vitamin D receptor expression plasmid and Dr. Stephan Laporte for the GFP expression plasmids. We thank Dr. Benoit Ochietti for the work on subcloning the GFP-tagged plasmids and Dr. Min Fu for assistance with the LSM780 confocal microscopy. We also thank Patricia Hales for secretarial assistance.
This work was supported by Grant MOP10839 from Canadian Institutes of Health Research (to R. K.) and Grant RGPIN 262040 from Natural Sciences and Engineering Research Council of Canada (to J. F. P.). The authors declare that they have no conflicts of interest with the contents of this article.
- 1α,25(OH)2D3
- 1α,25-dihydroxyvitamin D3
- hRXRα
- human retinoid X receptor α
- hVDR
- human vitamin D receptor
- FLIP
- fluorescence loss in photobleaching
- VDR
- vitamin D receptor
- RXR
- retinoid X receptor
- PPAR
- peroxisome proliferator-activated receptor
- HCC
- hepatocellular carcinoma
- CFP
- cyan fluorescent protein
- ANOVA
- analysis of variance
- RA
- retinoic acid
- ROI
- region of interest.
References
- 1. Bikle D. D. (2012) Vitamin and bone. Curr. Osteoporos. Rep. 10, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van Leeuwen J. P., van Driel M., van den Bemd G. J., and Pols H. A. (2001) Vitamin D control of osteoblast function and bone extracellular matrix mineralization. Crit. Rev. Eukaryot. Gene Expr. 11, 199–226 [PubMed] [Google Scholar]
- 3. Chen J., Dosier C. R., Park J. H., De S., Guldberg R. E., Boyan B. D., and Schwartz Z. (2016) Mineralization of three-dimensional osteoblast cultures is enhanced by the interaction of 1α,25-dihydroxyvitamin D3 and BMP2 via two specific vitamin D receptors. J. Tissue Eng. Regen. Med. 10, 40–51 [DOI] [PubMed] [Google Scholar]
- 4. Haussler M. R., Whitfield G. K., Kaneko I., Haussler C. A., Hsieh D., Hsieh J. C., and Jurutka P. W. (2013) Molecular mechanisms of vitamin D action. Calcif. Tissue Int. 92, 77–98 [DOI] [PubMed] [Google Scholar]
- 5. Shevde N. K., Plum L. A., Clagett-Dame M., Yamamoto H., Pike J. W., and DeLuca H. F. (2002) A novel potent analog of 1α,25-dihydroxyvitamin D3 selectively induces bone formation. Proc. Natl. Acad. Sci. U.S.A. 99, 13487–13491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deluca H. F., Cantorna M. T. (2001) Vitamin D: its role and uses in immunology. FASEB J. 15, 2579–2585 [DOI] [PubMed] [Google Scholar]
- 7. Adams J. S., and Hewison M. (2008) Unexpected actions of vitamin D: new perspectives on the regulation of innate and adaptive immunity. Nat. Clin. Pract. Endocrinol. Metab. 4, 80–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Suda T., Miyaura C., and Abe E. (1986) in Bone and Mineral Research (Peck W. A., ed) Vol. 4, pp. 1–48, Elsevier, New York [Google Scholar]
- 9. Colston K. W., and Hansen C. M. (2002) Mechanisms implicated in the growth regulatory effects of vitamin D in breast cancer. Endocr. Relat. Cancer 9, 45–59 [DOI] [PubMed] [Google Scholar]
- 10. Bikle D. D. (2012) Vitamin D and the skin. Physiology and pathophysiology. Rev. Endocr. Metab. Disord. 13, 3–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bettoun D. J., Burris T. P., Houck K. A., Buck D. W. 2nd., Stayrook K. R., Khalifa B., Lu J., Chin W. W., and Nagpal S. (2003) Retinoid X receptor is a non-silent major contributor to vitamin D receptor-mediated transcriptional activation. Mol. Endocrinol. 17, 2320–2328 [DOI] [PubMed] [Google Scholar]
- 12. Narayanan R., Sepulveda V. A., Falzon M., and Weigel N. L. (2004) The functional consequences of cross-talk between the vitamin D receptor and ERK signaling pathways are cell-specific. J. Biol. Chem. 279, 47298–47310 [DOI] [PubMed] [Google Scholar]
- 13. Dong D., and Noy N. (1998) Heterodimer formation by retinoid X receptor: regulation by ligands and by the receptor's self-association properties. Biochemistry 37, 10691–10700 [DOI] [PubMed] [Google Scholar]
- 14. Bourguet W., Vivat V., Wurtz J. M., Chambon P., Gronemeyer H., and Moras D. (2000) Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol. Cell 5, 289–298 [DOI] [PubMed] [Google Scholar]
- 15. Orlov I., Rochel N., Moras D., and Klaholz B. P. (2012) Structure of the full human RXR/VDR nuclear receptor heterodimer complex with its DR3 target DNA. EMBO J. 31, 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Olmos-Ortiz A., Avila E., Durand-Carbajal M., and Díaz L. (2015) Regulation of calcitriol biosynthesis and activity: focus on gestational vitamin D deficiency and adverse pregnancy outcomes. Nutrients 7, 443–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pike J. W., and Meyer M. B. (2010) The vitamin D receptor: new paradigms for the regulation of gene expression by 1,25-dihydroxyvitamin D(3). Endocrinol. Metab. Clin. North Am. 39, 255–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haussler M. R., Jurutka P. W., Mizwicki M., and Norman A. W. (2011) Vitamin D receptor (VDR)-mediated actions of 1α,25(OH)2 vitamin D3: genomic and non-genomic mechanisms. Best Pract. Res. Clin. Endocrinol. Metab. 25, 543–559 [DOI] [PubMed] [Google Scholar]
- 19. Bikle D. D. (2014) Vitamin D metabolism, mechanism of action, and clinical applications. Chem. Biol. 21, 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Abe E., Miyaura C., Sakagami H., Takeda M., Konno K., Yamazaki T., Yoshiki S., and Suda T. (1981) Differentiation of mouse myeloid leukemia cells induced by 1,25-dihydroxyvitamin D3. Proc. Natl. Acad. Sci. U.S.A. 78, 4990–4994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Truong K., and Ikura M. (2001) The use of FRET imaging microscopy to detect protein-protein interactions and conformational changes in vivo. Curr. Opin. Struct. Biol. 11, 573–578 [DOI] [PubMed] [Google Scholar]
- 22. Rossdeutscher L., Li J., Luco A. L., Fadhil I., Ochietti B., Camirand A., Huang D. C., Reinhardt T. A., Muller W., and Kremer R. (2015) Chemoprevention activity of 25-hydroxyvitamin D in the MMTV-PyMT mouse model of breast cancer. Cancer Prev. Res. 8, 120–128 [DOI] [PubMed] [Google Scholar]
- 23. El Abdaimi K., Dion N., Papavasiliou V., Cardinal P. E., Binderup L., Goltzman D., Ste-Marie L. G., and Kremer R. (2000) The vitamin D analogue EB 1089 prevents skeletal metastasis and prolongs survival time in nude mice transplanted with human breast cancer cells. Cancer Res. 60, 4412–4418 [PubMed] [Google Scholar]
- 24. Huang D. C., Papavasiliou V., Rhim J. S., Horst R. L., and Kremer R. (2002) Targeted disruption of the 25-hydroxyvitamin D3 1α-hydroxylase gene in ras-transformed keratinocytes demonstrates that locally produced 1α,25-dihydroxyvitamin D3 suppresses growth and induces differentiation in an autocrine fashion. Mol. Cancer Res. 1, 56–67 [PubMed] [Google Scholar]
- 25. Li J., Luco A. L., Ochietti B., Fadhil I., Camirand A., Reinhardt T. A., St-Arnaud R., Muller W., and Kremer R. (2016) Tumoral vitamin D synthesis by CYP27B1 1-α-hydroxylase delays mammary tumor progression in the PyMT-MMTV mouse model and its action involves NF-κB modulation. Endocrinology 157, 2204–2216 [DOI] [PubMed] [Google Scholar]
- 26. Kane K. F., Langman M. J., and Williams G. R. (1996) Antiproliferative responses of two human colon cancer cell lines to vitamin D3 are differentially modified by 9-cis retinoic acid. Cancer Res. 56, 623–632 [PubMed] [Google Scholar]
- 27. Polek T. C., and Weigel N. L. (2002) Vitamin D and prostate cancer. J. Androl. 23, 9–17 [DOI] [PubMed] [Google Scholar]
- 28. Getzenberg R. H., Light B. W., Lapco P. E., Konety B. R., Nangia A. K., Acierno J. S., Dhir R., Shurin Z., Day R. S., Trump D. L., and Johnson C. S. (1997) Vitamin D inhibition of prostate adenocarcinoma growth and metastasis in the Dunning rat prostate model system. Urology 50, 999–1006 [DOI] [PubMed] [Google Scholar]
- 29. Yu J., Papavasiliou V., Rhim J., Goltzman D., and Kremer R. (1995) Vitamin D analogs: new therapeutic agents for the treatment of squamous cancer and its associated hypercalcemia. Anticancer Drugs 6, 101–108 [PubMed] [Google Scholar]
- 30. Colston K., Colston M. J., and Feldman D. (1981) 1,25-Dihydroxyvitamin D3 and malignant melanoma: the presence of receptors and inhibition of cell growth in culture. Endocrinology 108, 1083–1086 [DOI] [PubMed] [Google Scholar]
- 31. Fujioka T., Hasegawa M., Ishikura K., Matsushita Y., Sato M., and Tanji S. (1998) Inhibition of tumor growth and angiogenesis by vitamin D3 agents in murine renal cell carcinoma. J. Urol. 160, 247–251 [PubMed] [Google Scholar]
- 32. Sebag M., Henderson J., Rhim J., and Kremer R. (1992) Relative resistance to 1,25-dihydroxyvitamin D3 in a keratinocyte model of tumor progression. J. Biol. Chem. 267, 12162–12167 [PubMed] [Google Scholar]
- 33. Solomon C., White J. H., and Kremer R. (1999) Mitogen-activated protein kinase inhibits 1,25-dihydroxyvitamin D3-dependent signal transduction by phosphorylating human retinoid X receptor α. J. Clin. Invest. 103, 1729–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Solomon C., Kremer R., White J. H., and Rhim J. S. (2001) Vitamin D resistance in RAS-transformed keratinocytes: mechanism and reversal strategies. Radiat. Res. 155, 156–162 [DOI] [PubMed] [Google Scholar]
- 35. Solomon C., Sebag M., White J. H., Rhim J., and Kremer R. (1998) Disruption of vitamin D receptor-retinoid X receptor heterodimer formation following ras transformation of human keratinocytes. J. Biol. Chem. 273, 17573–17578 [DOI] [PubMed] [Google Scholar]
- 36. Macoritto M., Nguyen-Yamamoto L., Huang D. C., Samuel S., Yang X. F., Wang T. T., White J. H., and Kremer R. (2008) Phosphorylation of the human retinoid X receptor α at serine 260 impairs coactivator(s) recruitment and induces hormone resistance to multiple ligands. J. Biol. Chem. 283, 4943–4956 [DOI] [PubMed] [Google Scholar]
- 37. Brosseau C., Colston K., Dalgleish A. G., Galustian C. (2012) The immunomodulatory drug lenalidomide restores a vitamin D sensitive phenotype to the vitamin D resistant breast cancer cell line MDA-MB-231 through inhibition of BCL-2: potential for breast cancer therapeutics. Apoptosis 17, 164–173 [DOI] [PubMed] [Google Scholar]
- 38. Rawson J. B., Sun Z., Dicks E., Daftary D., Parfrey P. S., Green R. C., Gallinger S., McLaughlin J. R., Wang P. P., Knight J. A., and Bapat B. (2012) Vitamin D intake is negatively associated with promoter methylation of the Wnt antagonist gene DKK1 in a large group of colorectal cancer patients. Nutr. Cancer 64, 919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chandler P. D., Giovannucci E. L., Scott J. B., Bennett G. G., Ng K., Chan A. T., Hollis B. W., Emmons K. M., Fuchs C. S., and Drake B. F. (2014) Null association between vitamin D and PSA levels among black men in a vitamin D supplementation trial. Cancer Epidemiol. Biomarkers Prev. 23, 1944–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hellström E., Robèrt K. H., Samuelsson J., Lindemalm C., Grimfors G., Kimby E., Oberg G., Winqvist I., Billström R., and Carneskog J. (1990) Treatment of myelodysplastic syndromes with retinoic acid and 1α-hydroxy-vitamin D3 in combination with low-dose ara-C is not superior to ara-C alone. Results from a randomized study. Eur. J. Haematol. 45, 255–261 [DOI] [PubMed] [Google Scholar]
- 41. Gulliford T., English J., Colston K. W., Menday P., Moller S., and Coombes R. C. (1998) A phase I study of the vitamin D analogue EB 1089 in patients with advanced breast and colorectal cancer. Br. J. Cancer 78, 6–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schwartz G. G., Hall M. C., Stindt D., Patton S., Lovato J., and Torti F. M. (2005) Phase I/II study of 19-nor-1α-25-dihydroxyvitamin D2 (paricalcitol) in advanced, androgen-insensitive prostate cancer. Clin. Cancer Res. 11, 8680–8685 [DOI] [PubMed] [Google Scholar]
- 43. Petrich A., Kahl B., Bailey H., Kim K., Turman N., and Juckett M. (2008) Phase II study of doxercalciferol for the treatment of myelodysplastic syndrome. Leuk. Lymphoma 49, 57–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Akiyama N., Miyazawa K., Kanda Y., Tohyama K., Omine M., Mitani K., and Ohyashiki K. (2010) Multicenter phase II trial of vitamin K2 monotherapy and vitamin K2 plus 1α-hydroxyvitamin D3 combination therapy for low-risk myelodysplastic syndromes. Leukemia Research 34, 1151–1157 [DOI] [PubMed] [Google Scholar]
- 45. Jain R. K., Trump D. L., Egorin M. J., Fernandez M., Johnson C. S., and Ramanathan R. K. (2011) A phase I study of the vitamin D3 analogue ILX23–7553 administered orally to patients with advanced solid tumors. Invest. New Drugs 29, 1420–1425 [DOI] [PubMed] [Google Scholar]
- 46. Giovannucci E. (2005) The epidemiology of vitamin D and cancer incidence and mortality: a review (United States). Cancer Causes Control 16, 83–95 [DOI] [PubMed] [Google Scholar]
- 47. Autier P., and Gandini S. (2007) Vitamin D supplementation and total mortality: a meta-analysis of randomized controlled trials. Arch. Intern. Med. 167, 1730–1737 [DOI] [PubMed] [Google Scholar]
- 48. Davis C. D. (2008) Vitamin D and cancer: current dilemmas and future research needs. Am. J. Clin. Nutr. 88, 565S–569S [DOI] [PubMed] [Google Scholar]
- 49. Bjelakovic G., Gluud L. L., Nikolova D., Whitfield K., Wetterslev J., Simonetti R. G., Bjelakovic M., and Gluud C. (2014) Vitamin D supplementation for prevention of mortality in adults. Cochrane Database Syst. Rev. 10.1002/14651858.CD007470.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Barsony J., Pike J. W., DeLuca H. F., and Marx S. J. (1990) Immunocytology with microwave-fixed fibroblasts shows 1α,25-dihydroxyvitamin D3-dependent rapid and estrogen-dependent slow reorganization of vitamin D receptors. J. Cell Biol. 111, 2385–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jares-Erijman E. A., and Jovin T. O. (2003) FRET Imaging. Nat. Biotechnol. 21, 1378–1395 [DOI] [PubMed] [Google Scholar]
- 52. Barsony J., Renyi I., and McKoy W. (1997) Subcellular distribution of normal and mutant vitamin D receptors in living cells. J. Biol. Chem. 272, 5774–5782 [DOI] [PubMed] [Google Scholar]
- 53. Colston K. W., Mackay A. G., Finlayson C., Wu J. C., and Maxwell J. D. (1994) Localisation of vitamin D receptor in normal human duodenum and in patients with coeliac disease. Gut 35, 1219–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Prüfer K., and Barsony J. (2002) Retinoid X receptor dominates the nuclear import and export of the unliganded vitamin D receptor. Mol. Endocrinol. 16, 1738–1751 [DOI] [PubMed] [Google Scholar]
- 55. Yasmin R., Williams R. M., Xu M., and Noy N. (2005) Nuclear import of the retinoid X receptor, the vitamin D receptor, and their mutual heterodimer. J. Biol. Chem. 280, 40152–40160 [DOI] [PubMed] [Google Scholar]
- 56. McNally J. G., Müller W. G., Walker D., Wolford R., and Hager G. L. (2000) The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287, 1262–1265 [DOI] [PubMed] [Google Scholar]
- 57. Stenoien D. L., Patel K., Mancini M. G., Dutertre M., Smith C. L., O'Malley B. W., and Mancini M. A. (2001) FRAP reveals that mobility of oestrogen receptor-α is ligand- and proteasome-dependent. Nat. Cell Biol. 3, 15–23 [DOI] [PubMed] [Google Scholar]
- 58. Tyagi R. K., Lavrovsky Y., Ahn S. C., Song C. S., Chatterjee B., and Roy A. K. (2000) Dynamics of intracellular movement and nucleocytoplasmic recycling of the ligand-activated androgen receptor in living cells. Mol. Endocrinol. 14, 1162–1174 [DOI] [PubMed] [Google Scholar]
- 59. Barsony J., Carroll J., McKoy W., Renyi I., Gould D. L., Htun H., and Hager G. L. (1997) in Microscopy and Microanalysis (Bailey G. W., Dimlich R. V., McCarthy J. J., and Pretlov T. P., eds) pp. 131–132, Springer-Verlag, New York [Google Scholar]
- 60. Dawson M. I., and Xia Z. (2012) The retinoid X receptors and their ligands. Biochim. Biophys. Acta 1821, 21–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Prüfer K., Racz A., Lin G. C., and Barsony J. (2000) Dimerization with retinoid X receptors promotes nuclear localization and subnuclear targeting of vitamin D receptors. J. Biol. Chem. 275, 41114–41123 [DOI] [PubMed] [Google Scholar]
- 62. Lippincott-Schwartz J., Snapp E., and Kenworthy A. (2001) Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2, 444–456 [DOI] [PubMed] [Google Scholar]
- 63. Misteli T., Gunjan A., Hock R., Bustin M., and Brown D. T. (2000) Dynamic binding of histone H1 to chromatin in living cells. Nature 408, 877–881 [DOI] [PubMed] [Google Scholar]
- 64. Feige J. N., Gelman L., Tudor C., Engelborghs Y., Wahli W., and Desvergne B. (2005) Fluorescence imaging reveals the nuclear behavior of peroxisome proliferator-activated receptor/retinoid X receptor heterodimers in the absence and presence of ligand. J. Biol. Chem. 280, 17880–17890 [DOI] [PubMed] [Google Scholar]
- 65. Lu M. J., Mpoke S. S., Dadd C. A., and Allis C. D. (1995) Phosphorylated and ephosphorylated linker histone H1 reside in distinct chromatin domains in Tetrahymena macronuclei. Mol. Biol. Cell 6, 1077–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dou Y., Mizzen C. A., Abrams M., Allis C. D., and Gorovsky M. A. (1999) Phosphorylation of linker histone H1 regulates gene expression in vivo by mimicking H1 removal. Mol. Cell 4, 641–647 [DOI] [PubMed] [Google Scholar]
- 67. Matsushima-Nishiwaki R., Okuno M., Adachi S., Sano T., Akita K., Moriwaki H., Friedman S. L., and Kojima S. (2001) Phosphorylation of retinoid X receptor α at serine 260 impairs its metabolism and function in human hepatocellular carcinoma. Cancer Res. 61, 7675–7682 [PubMed] [Google Scholar]
- 68. Quack M., and Carlberg C. (2000) The impact of functional vitamin D(3) receptor conformations on DNA-dependent vitamin D(3) signaling. Mol. Pharmacol. 57, 375–384 [PubMed] [Google Scholar]
- 69. Carlberg C., Quack M., Herdick M., Bury Y., Polly P., and Toell A. (2001) Central role of VDR conformations for understanding selective actions of vitamin D(3) analogues. Steroids 66, 213–221 [DOI] [PubMed] [Google Scholar]
- 70. Lee H. Y., Suh Y. A., Robinson M. J., Clifford J. L., Hong W. K., Woodgett J. R., Cobb M. H., Mangelsdorf D. J., and Kurie J. M. (2000) Stress pathway activation induces phosphorylation of retinoid X receptor. J. Biol. Chem. 275, 32193–32199 [DOI] [PubMed] [Google Scholar]
- 71. Li M., Indra A. K., Warot X., Brocard J., Messaddeq N., Kato S., Metzger D., and Chambon P. (2000) Skin abnormalities generated by temporally controlled RXRα mutations in mouse epidermis. Nature 407, 633–636 [DOI] [PubMed] [Google Scholar]
- 72. Li M., Chiba H., Warot X., Messaddeq N., Gérard C., Chambon P., and Metzger D. (2001) RXR-α ablation in skin keratinocytes results in alopecia and epidermal alterations. Development 128, 675–688 [DOI] [PubMed] [Google Scholar]
- 73. Li D., Li T., Wang F., Tian H., and Samuels H. H. (2002) Functional evidence for retinoid X receptor (RXR) as a nonsilent partner in the thyroid hormone receptor/RXR heterodimer. Mol. Cell. Biol. 22, 5782–5792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schneider Aguirre R., and Karpen S. J. (2013) Inflammatory mediators increase SUMOylation of retinoid X receptor α in a c-Jun N-terminal kinase-dependent manner in human hepatocellular carcinoma cells. Mol. Pharmacol. 84, 218–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Crowe D. L., and Chandraratna R. A. (2004) A retinoid X receptor (RXR)-selective retinoid reveals that RXR-α is potentially a therapeutic target in breast cancer cell lines, and that it potentiates antiproliferative and apoptotic responses to peroxisome proliferator-activated receptor ligands. Breast Cancer Res. 6, R546–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Konopleva M., Elstner E., McQueen T. J., Tsao T., Sudarikov A., Hu W., Schober W. D., Wang R. Y., Chism D., Kornblau S. M., Younes A., Collins S. J., Koeffler H. P., and Andreeff M. (2004) Peroxisome proliferator-activated receptor γ and retinoid X receptor ligands are potent inducers of differentiation and apoptosis in leukemias. Mol. Cancer Ther. 3, 1249–1262 [PubMed] [Google Scholar]
- 77. Avis I., Martínez A., Tauler J., Zudaire E., Mayburd A., Abu-Ghazaleh R., Ondrey F., and Mulshine J. L. (2005) Inhibitors of the arachidonic acid pathway and peroxisome proliferator-activated receptor ligands have superadditive effects on lung cancer growth inhibition. Cancer Res. 65, 4181–4190 [DOI] [PubMed] [Google Scholar]
- 78. Yamazaki K., Shimizu M., Okuno M., Matsushima-Nishiwaki R., Kanemura N., Araki H., Tsurumi H., Kojima S., Weinstein I. B., and Moriwaki H. (2007) Synergistic effects of RXRα and PPARγ ligands to inhibit growth in human colon cancer cells–phosphorylated RXRα is a critical target for colon cancer management. Gut 56, 1557–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kanemura N., Tsurumi H., Okuno M., Matsushima-Nishiwaki R., Shimizu M., and Moriwaki H. (2008) Retinoid X receptor α is highly phosphorylated in retinoic acid-resistant HL-60R cells and the combination of 9-cis retinoic acid plus MEK inhibitor induces apoptosis in the cells. Leuk. Res. 32, 884–892 [DOI] [PubMed] [Google Scholar]
- 80. Matsushima-Nishiwaki R., Okuno M., Takano Y., Kojima S., Friedman S. L., and Moriwaki H. (2003) Molecular mechanism for growth suppression of human hepatocellular carcinoma cells by acyclic retinoid. Carcinogenesis 24, 1353–1359 [DOI] [PubMed] [Google Scholar]
- 81. Moriwaki H., Shimizu M., Okuno M., and Nishiwaki-Matsushima R. (2007) Chemoprevention of liver carcinogenesis with retinoids: basic and clinical aspects. Hepatol. Res. 37, S299–S302 [DOI] [PubMed] [Google Scholar]
- 82. Lu N., Liu J., Liu J., Zhang C., Jiang F., Wu H., Chen L., Zeng W., Cao X., Yan T., Wang G., Zhou H., Lin B., Yan X., Zhang X. K., and Zeng J. Z. (2012) Antagonist effect of triptolide on AKT activation by truncated retinoid X receptor-α. PLoS ONE 7, e35722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Snapp E. L., Altan N., and Lippincott-Schwartz J. (2003) Measuring protein mobility by photobleaching GFP chimeras in living cells. Curr. Protoc. Cell Biol. 2003 Chapter 21, Unit 21.1 [DOI] [PubMed] [Google Scholar]
- 84. Zhang Z., Kovalenko P., Cui M., Desmet M., Clinton S. K., and Fleet J. C. (2010) Constitutive activation of the mitogen activated protein kinase pathway impairs vitamin D signaling in human prostate epithelial cells. J. Cell Physiol. 224, 433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Racz A., and Barsony J. (1999) Hormone-dependent translocation of vitamin D receptors is linked to transactivation. J. Biol. Chem. 274, 19352–19360 [DOI] [PubMed] [Google Scholar]
- 86. Kuusisto H. V., Wagstaff K. M., Alvisi G., Roth D. M., and Jans D. A. (2012) Global enhancement of nuclear localization-dependent nuclear transport in transformed cells. FASEB J. 26, 1181–1193 [DOI] [PubMed] [Google Scholar]