Abstract
Purpose
Sentinel node biopsy (SNB) has been shown to accurately stage the regional lymphatics in oral carcinoma. However, intraoperative pathology is only moderately sensitive and final pathology takes several days to complete. The purpose of this study was to develop a rapid, automated, and quantitative real-time PCR (qRT-PCR) assay that can match final pathology in an intraoperative time frame.
Experimental Design
Four hundred forty-eight grossly tumor-negative lymph nodes were evaluated for expression of 3 markers [PVA (pemphigus vulgaris antigen), PTHrP (parathyroid hormone-related protein), and TACSTD1 (tumor-associated calcium signal transducer 1)]. Conformity of metastasis detection by qRT-PCR was determined using hematoxylin and eosin and immunohistochemistry staining as the gold standard. PVA and TACSTD1 were then multiplexed with β-glucuronidase to develop a rapid, automated single-tube qRT-PCR assay using the Cepheid GeneXpert system. This assay was used to analyze 103 lymph nodes in an intraoperative time frame.
Results
Four hundred forty-two nodes produced an informative result for both qRT-PCR and pathologic examination. Concordance of qRT-PCR for individual markers with final pathology ranged from 93% to 98%. The best marker combination was TACSTD1 and PVA. A rapid, multiplex assay for TACSTD1 and PVA was developed on the Cepheid GeneXpert and demonstrated an excellent reproducibility and linearity. Analysis of 103 lymph nodes demonstrated 94.2% accuracy of this assay for identifying positive and negative nodes. The average time for each assay to yield results was 35 minutes.
Conclusions
A rapid, automated qRT-PCR assay can detect lymph node metastasis in head and neck cancer with high accuracy compared to pathologic analysis and may be more accurate than intraoperative pathology. Combined, SNB and rapid qRT-PCR could more appropriately guide surgical treatment of patients with head and neck cancer.
Introduction
For patients with a diagnosis of head and neck squamous cell carcinoma (HNSCC), nodal involvement is one of the strongest prognostic indicators (1, 2), and guides use of adjuvant radiation and chemotherapy to reduce disease recurrence. HNSCC frequently metastasizes to the cervical nodal basins, yet clinical staging via physical exam and radiological modalities [PET-CT (positron emission tomography-computed tomography), CT scan, MRI, ultrasound] are inadequate, and usually cannot detect metastases less than 8 to 10 mm in size (3). Thus, the standard of care for the clinically node negative (N0) patient is an elective neck dissection (END), which leads to increased locoregional control and regional recurrence-free survival (4–6). However, END is a surgical procedure that represents overtreatment for approximately 70% of cN0 patients who are found to have a pathologically negative neck.
In a multicenter validation trial conducted by the American College of Surgeons Oncology Group (ACOSOGZ0360 trial), sentinel node biopsy (SNB) was demonstrated to be feasible and accurate as a means of avoiding unnecessary END by identifying pN0 patients (7). Although SNB holds great promise, widespread application is limited by the lack of rapid and accurate, intraoperative detection of metastatic disease in the sentinel node(s) (3, 8–14). Unfortunately, intraoperative frozen section has a sensitivity of approximately 60% in HNSCC (8, 15). The gold standard of H&E (hematoxylin and eosin) staining on formalin-fixed, paraffin embedded nodes with immunohistochemistry (IHC) to detect small tumor deposits is much more accurate, but takes several days to perform and thus cannot be performed intraoperatively. A return to the operating room for a completion procedure would lead to additional costs, discomfort, and increased difficulty in the subsequent surgery in addition to delaying adjuvant therapies.
Real-time quantitative reverse-transcription PCR (qRT-PCR) can be very sensitive for detection of cancer cells in a background of normal cells (16). qRT-PCR targeting tissue specific mRNA markers that are highly expressed by malignant cells with minimal expression in lymphoid tissue improves the specificity of this technique greatly, and qRT-PCR can detect histologically occult micrometastases in many cancer types (17–22). Despite the ability to assess large portions of a sentinel lymph node (SLN) for micrometastasis objectively, qRT-PCR also has some restrictions in its application to rapid, intraoperative analysis. It can be very labor and time intensive, requiring RNA isolation, reverse transcription, and PCR. These procedures are prone to contamination and RNA degradation, resulting in possible false positives and false negatives, respectively (23).
The goal of this study was to evaluate the best 3 tumor markers from our previous study (24) in a large number of lymph nodes with standard qRT-PCR and, based upon this evaluation, to develop and test an automated, rapid qRT-PCR assay that could be used intraoperatively to improve the sensitivity of SLN analysis.
Materials and Methods
Patients and lymph node collection
Institutional review board-approved, written informed consent was obtained from all patients donating specimens for this study. All cervical lymph nodes were collected from 92 patients representative of our patient population undergoing neck dissections for HNSCC, with various primary sites including the oral cavity, oropharynx, larynx, and hypopharynx (Table 1). A total of 448 lymph nodes were harvested at surgery, sectioned in alternating 1 mm sections, with half of the sections used for pathological diagnosis and the other half used for this study. In order to ensure no gross discrepancy between the 2 halves, the research halves were embedded in OCT (optimal cutting temperature) compound and 1 section was mounted on a slide, H&E stained, and read by a head and neck pathologist (R.S.). The OCT embedded nodes were stored at −80°C prior to processing and RNA isolation.
Table 1.
Clinical features and risk factors in 92 previously untreated SCCHN patients, from whom validation set lymph nodes (n = 5 ± 1 nodes per patient, 442 total nodes) were harvested
| Age | 60 y |
| Gender | 79M/13F |
| Smoking history | 78/92 (84.8%) |
| Alcohol history | 58/92 (63.0%) |
| pT status | |
| T1 | 28/92 (30.4%) |
| T2 | 25/92 (27.2%) |
| T3 | 20/92 (21.7%) |
| T4 | 19/92 (20.7%) |
| pN status | |
| N0 | 50/92 (54.3%) |
| N1 | 12/92 (13.1%) |
| N2 | 30/92 (32.6%) |
| N3 | 0/92 (0%) |
| Site | |
| Oral cavity | 57/92 (62%) |
| Oropharynx | 11/92 (12%) |
| Larynx | 20/92 (21.7%) |
| Hypopharynx | 2/92 (2.2%) |
| Other | 2/92 (2.2%) |
Pathologic analysis
Histologic analysis was performed at 3 separate times for each lymph node; H&E stained frozen sections of alternate 1 mm pieces were reviewed and entered into the medical record prior to release of nodes for research ("clinical frozen section"), these frozen pieces were also cut at a later date for both RNA isolation and histologic review by 2 pathologists and consensus was reached using both H&E and IHC ("research pathology"). Finally, alternate pieces of each node were fixed and embedded, and H&E stained sections were reviewed and entered into the medical record (permanent or "final" pathology). Nodes were classified as negative, positive, as isolated tumor cells (ITC), or if tissues were considered inadequate, as nondiagnostic. From these data, each node was also given an overall "consensus pathology" call based on all 3 evaluations, where any positive result trumped ITC and any ITC trumped negative results.
Tissue processing for RNA isolation and research pathology review
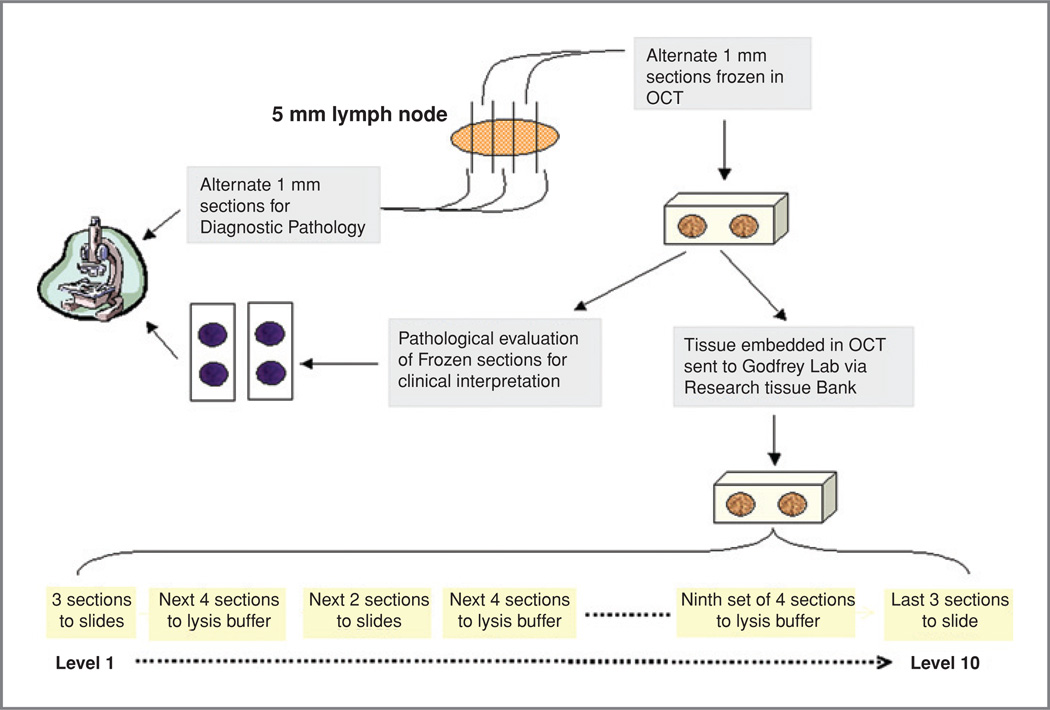
Five-micron serial sections were cut from each OCT embedded lymph node half, with the initial and final tissue sections being mounted on slides for histologic analysis with H&E staining (Fig. 1). The next adjacent tissue section was mounted on slides and stained with pancytokeratin IHC for histopathology. Immunohistochemical evaluation was done using the antibody pan keratin (AE1/AE3) mixture (DAKO), and Vector Elite ABC kit and Vector AEC Chromagen (Vector Laboratories). The intervening sections were distributed 4:1:4:1:4, etc., such that 4 sections were immediately placed in chaotropic lysis buffer for RNA isolation (total of 50–60 sections) and every fifth section was mounted on a slide for histologic review (every 5 sections are considered a level Fig. 1). All unstained slides from levels 2 to 9, were fixed in acetone and stored at −20°C. All H&E and IHC slides from levels 1 and 10 of the lymph nodes were reviewed by 2 specialized head and neck pathologists (R. S. and S.C.) to confirm the presence of tumor, percentage of tumor, and identify the presence of any contaminating tissues. Furthermore, the slides from the GeneXpert validation set were read by 2 independent head and neck pathologists (R.S and B.H).
Figure 1.
Pathologic and molecular tissue handling and sectioning protocol. Nodes were harvested at surgery and subjected to the sectioning process shown, for either histologic (H&E or IHC) or qRT-PCR analysis.
RNA isolation and cDNA synthesis
RNA was isolated using the Stratagene RNA isolation mini-kit (Stratagene) using the manufacturers described protocol. Reverse transcription was done in 100-µL reaction volumes with random hexamer priming and SuperScript II (Invitrogen) reverse transcriptase as per our previous studies (24).
Standard quantitative PCR
All quantitative PCR (qPCR) was performed on the Stratagene MX3000P QPCR System (Stratagene). Relative expression of the marker genes [PVA (pemphigus vulgaris antigen), TACSTD1 (tumor-associated calcium signal transducer 1), and PTHrP (parathyroid hormone-related protein)] was calculated using the ΔCT methods that were previously described (25), and with β-glucuronidase (GUSB) as the endogenous control gene. Primer and probe sequences for each gene were as published previously (24).
GeneXpert multiplex assay development
Multiplex qPCR assays can suffer from limited linear dynamic range due to common reagent utilization. The SmartCycler and the GeneXpert both employ a method called temperature controlled primer limiting to inhibit the assay that reaches threshold first in order to decrease competition between the simultaneous assays (26). The PVA, TACSTD1, GUSB triplex assay used in this study is essentially identical to our previously published GeneXpert assay for breast cancer (27) except that PVA is used in place of prolactin inducible protein (PIP). Duplex and then triplex assays were first developed on the SmartCycler before transitioning to the GeneXpert. Briefly, PCR efficiency and dynamic range were assessed for duplex assays (TACSTD1/GUSB and PVA/GUSB) using serial dilutions of cDNA with high expression of the respective target genes (normal esophagus RNA for PVA, normal colon RNA for TACSTD1) in a background of total spleen cDNA. Linear dynamic range for the triplex assay was then tested to ensure all 3 assays functioned well in the ranges needed for lymph node classification. At this point, the triplex qPCR assay was incorporated into the Cepheid GeneXpert system to facilitate automated RNA isolation, reverse transcription, and qPCR. Reproducibility of the quantification of each marker gene was then tested on the GeneXpert using serial dilutions of total RNA known to have high expression of PVA and TACSTD1. RNAs were diluted in a background of lysate from lymph nodes negative for disease in order to mimic a true test scenario. Six serial dilutions were performed and each assay was repeated 4 times on the GeneXpert in order to assess reproducibility across a range of target gene expression. Primer and probe sequences used in the GeneXpert for each gene are listed in Supplementary Table SA.
Analysis of lymph nodes using the GeneXpert
One hundred three (n = 103) of the original 448 lymph nodes were analyzed on the GeneXpert. Frozen tissue pieces, which were recut for RNA and tissue sections, were evaluated blindly by 2 fellowship trained head and neck pathologists (GeneXpert Research Pathology). Twenty-four, 5-µmol/L sections of OCT-embedded tissue were sectioned into 800 µL of GeneXpert lysis buffer (Cepheid) for each node, with 2 initial and final tissue sections mounted on slides for H&E and IHC histopathology. The lysate was then filtered through a 0.22-µmol/L syringe filter (Osmonics Inc.) and loaded into a GeneXpert cartridge. The exact details of the RNA isolation, reverse transcription, and qRT-PCR on the automated GeneXpert system are described in our previous work (26).
Diagnostic accuracy of standard qRT-PCR and the GeneXpert assays
Two reference standards were established: (i) research pathology, a consensus of 2 academic pathologists applying H&E and IHC; (ii) a consensus pathology which combined research pathology with clinical frozen section pathology. To explore the diagnostic potential of individual markers with both standard qRT-PCR and the GeneXpert, receiver operating characteristic (ROC) curves were constructed and the cutoff value of each marker that produced the highest overall classification accuracy was selected. The diagnostic parameters sensitivity, specificity, positive predictive value, negative predictive value, and overall accuracy were calculated for each cutoff. In addition the area under the ROC curve (AUC) was estimated. For the purpose of developing a diagnostic model, and identifying which maker or combination of markers are most important a different strategy was used. In this approach, the sample of 442 lymph nodes were collected from 92 patients and therefore were not considered to be independently distributed. For this reason we applied a generalized linear model with random effects. This model used a logit link to estimate the binomial variable, positive or negative by pathology, as a function of marker RNA expression level controlling for the random effect of patient (28). The best fitting model was selected based on the likelihood ratio test and the number of model parameters (Akaike’s Information Criteria; ref. 29). A "GeneXpert" classification model was developed with conventional (fixed effects) logistic regression. The model was used to predict the probability that a node was positive and the range of predicted probabilities served as a list of potential cutoff values in an ROC analysis. In addition to logistic regression models, recursive portioning and nearest neighbor models were fit but neither method compared favorably to logistic regression. All diagnostic operating characteristics were cross-validated by either bootstrap resampling (200 reps) or leave-10-out cross-validation.
Results
Pathology analysis
A total of 448 lymph nodes were analyzed at 3 times; clinical frozen section, research frozen section (H&E plus IHC), and final pathology of fixed sections. Overall (consensus pathology) results classified 378 nodes as negative, 2 nodes as ITC, and 62 nodes as positive. Six nodes were determined to harbor incidental, metastatic papillary thyroid carcinoma (PTC) and these were excluded from further analyses leaving a total of 442 nodes. Comparison of each separate pathology review with the overall consensus pathology result (excluding ITC) indicates that conformity was >97% in all cases (Supplementary Tables SB–SE). However, consensus pathology identified a total of 61 positive nodes, of which research pathology identified 53 (86.9%), permanent pathology identified 56 (91.8%) but frozen section pathology only identified 49 (80.3%).
Evaluation of qRT-PCR markers
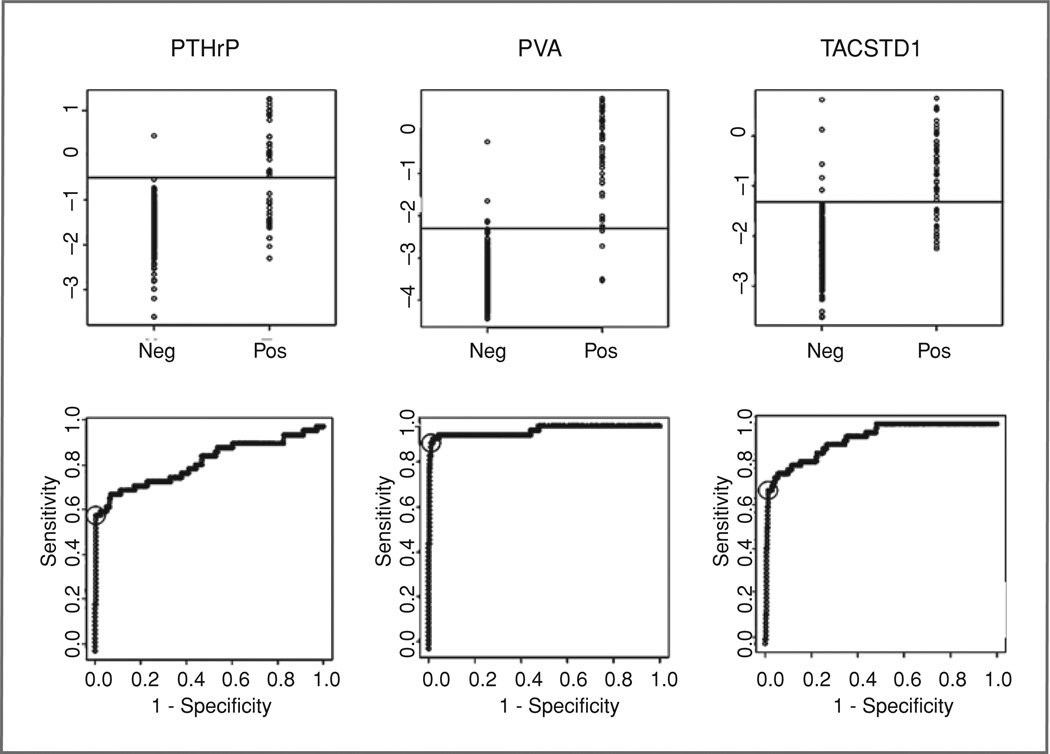
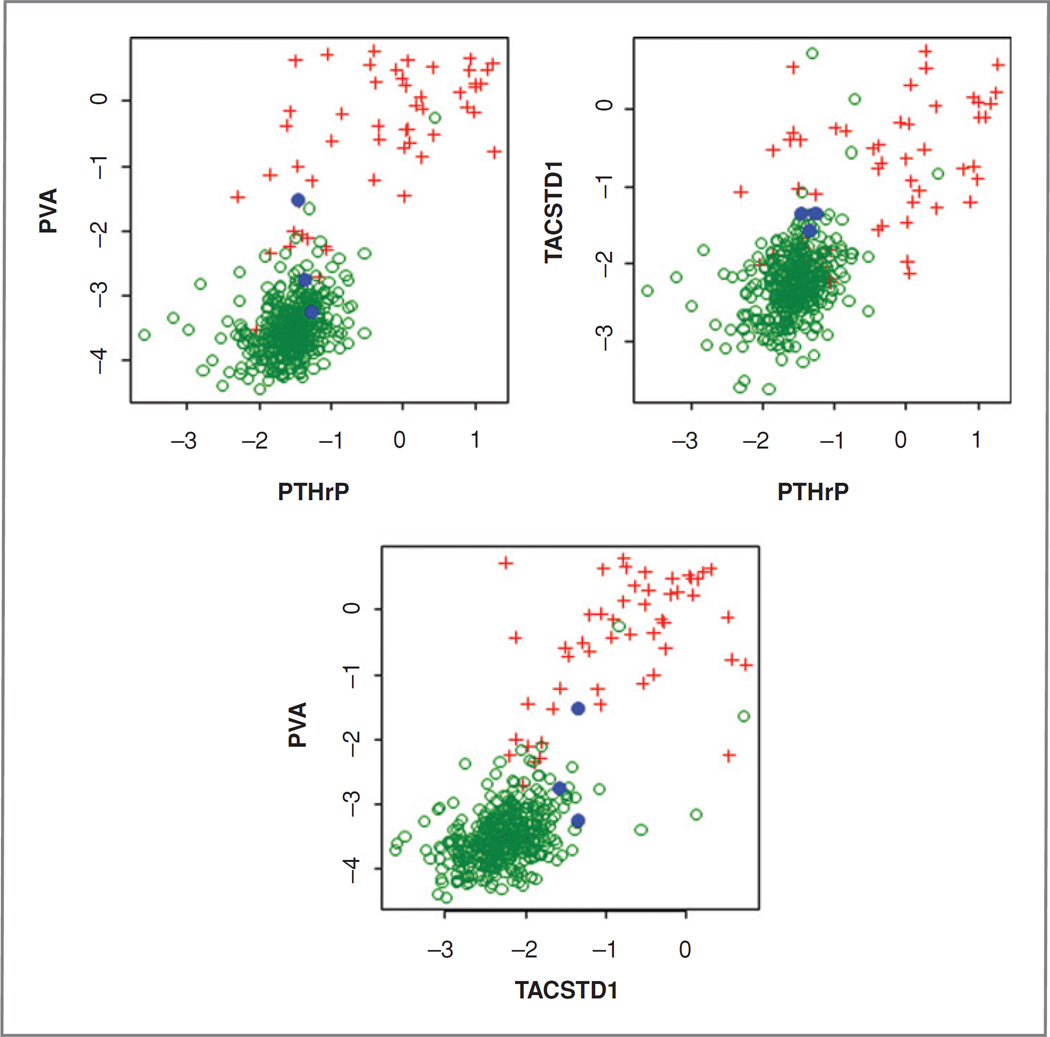
Four hundred forty-two lymph nodes from 92 patients were analyzed by qRT-PCR for 3 markers, PVA, PTHrP, and TACSTD1, and results were compared to both research pathology (Fig. 2 and Table 2) and overall consensus pathology (Supplementary Fig. SA and Table 2). In general, qRT-PCR results were slightly more concordant with research pathology than with consensus pathology (mean accuracy 96% vs. 94%); probably reflecting the increased sampling error inherent in the comparison with consensus pathology. Individually, all markers in combination with their most accurate cutoff demonstrated high (>93%) overall accuracy compared with both pathology endpoints but PVA clearly provided superior sensitivity (92% vs. research pathology) compared to PTHrP and TACSTD1 and hence, slightly higher negative predictive value (99%, Table 2). ROC curve analysis also identifies PVA as the best marker with an estimated AUC of 98% although the AUC confidence intervals for PVA and TACSTD1 overlap. PTHrP was clearly the least accurate of the 3 markers tested. Paired combinations of markers were also evaluated (Fig. 3) with TACSTD1 and PVA providing the best correlation with pathology although the combination was not significantly better than PVA alone.
Figure 2.
Distribution of relative expression (top) of PTHrP, PVA, and TACSTD1 in histologically positive and negative lymph nodes as determined by review of adjacent tissue sections (research pathology). The Y-axis shows relative expression as ΔCT and the horizontal line indicates the most accurate cutoff determined from ROC curve analysis (bottom).
Table 2.
Accuracy characteristics of individual markers compared with research pathology and overall consensus pathology.
| Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) | AUC | AUC-CI | |
|---|---|---|---|---|---|---|---|
| Research pathology | |||||||
| PTHrP | 60 | 98 | 97 | 95 | 95 | 0.84 | 0.77–0.91 |
| PVA | 92 | 98 | 92 | 99 | 98 | 0.98 | 0.95–1.00 |
| TACSTD1 | 70 | 99 | 88 | 96 | 95 | 0.88 | 0.88–0.98 |
| Overall pathology | |||||||
| PTHrP | 53 | 100 | 94 | 93 | 93 | 0.78 | 0.71–0.85 |
| PVA | 84 | 98 | 90 | 97 | 96 | 0.92 | 0.87–0.97 |
| TACSTD1 | 61 | 99 | 88 | 94 | 93 | 0.88 | 0.82–0.93 |
Abbreviations: PPV, positive predictive value; AUC, area under the ROC curve; CI, confidence interval.
Figure 3.
Marker combinations and correlation with lymph node classification according to research pathology. Green circles: negative nodes; red plus: positive node; blue circle: node with ITCs. Axes show ΔCT.
GeneXpert assay development and reproducibility
For automated, rapid analysis of lymph nodes, a triplex assay was developed for TACSTD1, PVA, and GUSB. PCR efficiencies for multiplex assays ranged from 96.3% to 98.8% and the triplex assay showed a good linearity in the range needed to discriminate positive from negative nodes (Supplementary Fig. SB). Replicate experiments on the GeneXpert (Supplementary Fig. SC) resulted in an intraclass correlation coefficient of 0.90 for PVA and 0.97 for TACSTD1. The coefficient of variation was 18.2% for PVA and 3.9% for TACSTD1.
GeneXpert data
A total of 103 lymph nodes were evaluated using the triplex assay on the GeneXpert. Results were compared to GeneXpert research pathology (examination of frozen sections taken adjacent to the tissue used for GeneXpert qPCR) and consensus pathology (a consensus based on all histologic analyses performed on the node). Of the 103 nodes, 43 were tumor positive by consensus pathology and 60 were tumor negative. For the positive nodes, estimated tumor representation ranged from less than 5% to grossly positive (mean 55%). Seven of the positive nodes were negative by research pathology, likely indicating the effect of sampling error, i.e., the positive result was found on either the frozen clinical section or the permanent, formalin-fixed, paraffin embedded (FFPE) section but not on the research pathology section. PCR assays were performed from sections sandwiched by those used for research pathology and therefore the comparison with research pathology most accurately reflects the accuracy of the qPCR assay.
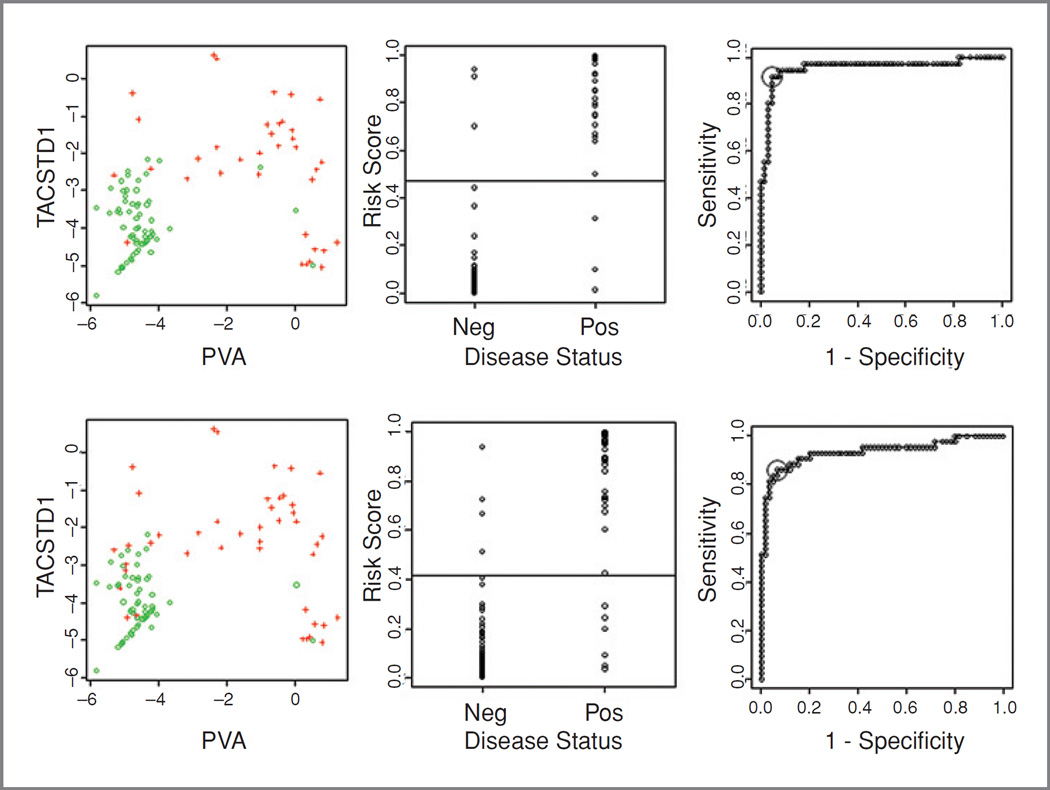
For each lymph node, TACSTD1 and PVA expression data from the triplex GeneXpert assay was used in a logistic model of pathologic status. From this model, the probability of being positive was estimated and the probability value leading to the most accurate classification was found (Fig. 4). Using this approach, the overall accuracy of qPCR compared with research pathology was 94.2% and compared with consensus pathology was 90.3%. The sensitivity and specificity compared with research pathology were 91.7% and 95.5%, respectively, and compared with consensus pathology were 86% and 93.3%. Ten-fold cross validation indicated that these accuracy levels are likely to be quite robust as the estimated accuracy only dropped by 4% to 5%.
Figure 4.
Results of the triplex GeneXpert assay compared to research pathology (top) and overall consensus pathology (bottom). First, expression of TACSTD1 and PVA in positive (+) and negative (○) nodes; second, the risk score (probability of being positive calculated based on the TACSTD1 and PVA expression) for each node; and third, the horizontal line indicates the most accurate cutoff determined by ROC curve analysis.
Discussion
We have developed a completely automated multiplex qRT-PCR assay that can accurately detect lymph node metastasis in head and neck cancer. This assay can be performed in 35 minutes or less and thus could be performed intraoperatively to detect metastasis to lymph nodes, even those with as little as 5% nodal involvement. To the best of our knowledge, this is the first study to demonstrate the use of a rapid, automated qRT-PCR assay that can be used in an intraoperative setting to detect head and neck cancer metastasis. The 2 distinct mRNA markers have been previously identified (24) and now independently validated as promising markers of tumor metastasis in both single and multiplex assays in a large sample set of lymph nodes.
This rapid, 2 marker qRT-PCR assay is a significant advance because of the inherent problem with intraoperative analysis via frozen section, i.e., sampling error and subjectivity. Less than 1% of a node is analyzed and may miss metastasis, with a published sensitivity of roughly 60% to 80% for micrometastasis (30–34). Improvement in results can be obtained if multisectioned frozen analysis is performed, but this limits material available for permanent H&E section and IHC analyses, as well as generates greater costs and expense of intraoperative time. Frozen section histology is also subject to both technical difficulties in sample and section preparation, including interpretive challenges, all of which are heavily dependent on the skill and experience of the pathologist and support staff (35). Further difficulty exists with frozen section since tissue that has been frozen, then thawed, and then processed for routine light microscopic examination may be compromised, and may not represent the true status of the entire specimen. Imprint cytology provides worse results compared to frozen section (34). Even final assessment of step sectioned FFPE sections, which samples a larger percent of the node and is the gold standard, may not find micrometastasis (tumor deposits <2 mm and >0.2 mm), as well as ITCs (tumor clusters <0.2 mm), as suggested by the fact that 7% to 15% of patients with no pathologic evidence of cervical lymph node involvement after END suffer recurrent disease in the neck (36–39). Hence, a method of evaluating a larger portion of SLN for metastasis that is both rapid and objective would be of great clinical benefit, to complement the sentinel lymph node biopsy (SLNB) procedure which accurately stages the clinically negative neck in approximately 95% of oropharyngeal squamous cell carcinoma (OSCC) patients.
In this regard, ideally our assay should be as accurate as "the gold standard," but the most important goal for an intraoperative analysis is to be more accurate than frozen section in a comparable time frame. After processing and embedding of a SLN, the face that is cut for staining may be completely different and far away from the face that was cut for the frozen section. Therefore, it is entirely possible for the frozen section to be positive while the permanent section is negative. It may be argued whether that scenario would constitute a "false positive" on the frozen section or a "false negative" for the permanent section (as observed in 8%–9% of cases; refs. 36, 40), warranting a re-evaluation of "gold standard" pathologic analysis in the molecular pathology era. We argue that any positive finding by pathology, that is agreed upon by 2 or more pathologists is a true positive regardless of whether it is determined by H&E or IHC on a frozen section, a research pathology section or a permanent section and we do not believe that this is changing the gold standard, it is simply accepting the possibility of sampling error. Expert pathologic review is the true "gold standard" regardless of when the section was taken and we believe that the pathology community would agree with this notion.
The potential role of SNB in OSCC may have greater applicability than in melanoma and breast cancer because nearly all OSCC patients undergo routine END. Thus, the ability to accurately and rapidly stage the cN0 neck would avoid morbidity, cost(s), and other health system resources utilized in open neck dissection, for the 70% of patients without metastatic disease. Because no reliable gene prediction profile has been developed, currently the primary tumor mRNA profile is not useful in classifying presence of lymphatic disease in these patients. Thus, direct analysis of the draining SLN is necessary, a procedure which has been recently validated in a large prospective multicenter trial (ACOSOG Z0360, ref. 7). Based on these trial results, we considered that positive and negative predictive values are influenced by disease prevalence. Since our GeneXpert cohort used a 35% sample of positive nodes, we estimated that what the negative predicted value (NPV) of our qRT-PCR assay would have been if we used the prevalence of positive nodes in the ACOSOG trial (29%). We assumed a 1,000 patient cohort in which the prevalence of pathologically positive sentinel lymph nodes was 30% (7). Applying the same 83% sensitivity and 92% specificity as the cross-validated GeneXpert classifier to this 1,000 patient cohort of head and neck cancer patients with neck dissection would slightly improve the NPV from the observed value of 0.912 versus research pathology to 0.930.
In our previous work, we identified 4 novel mRNA markers (SCCA1/2, PVA, TACSTD1, and PTHrP) for the detection of cervical lymph node metastasis in squamous cell carcinoma of the head and neck (SCCHN), and showed the feasibility of an automated multiplex assay in the detection of PVA, one of the most promising markers from the initial screen (24). In the present study, we have not only validated 3 of the 4 mentioned markers with a large sample set of histologically negative and positive lymph nodes, but also developed a rapid triplex qRT-PCR assay that combines the 2 best markers (PVA and TACSTD1) with an endogenous control (GUSB). Furthermore, this assay is fully automated (RNA isolation, reverse transcription, and qPCR) using the Cepheid GeneXpert system and can be completed in less than 35 minutes.
The GeneXpert assay has several advantages over prior methods of molecular diagnosis, as well as conventional pathologic analysis, in detecting occult metastasis in SLNs of head and neck cancer. First of all, this is the first instrument that can perform a fully automated RNA isolation and qRT-PCR in less than 35 minutes, which could easily fit into an intraoperative time frame. Multiple SLNs can be assessed in parallel since each assay is independently controlled by the GeneXpert. Excision of the primary oral cancer could be undertaken after SLN removal and during pathologic and qRT-PCR analysis of the SLNs. Secondly, the objective nature of the assay eliminates the uncertainty and discordance rates with frozen section pathology analysis, which is a clearly documented issue (35). Finally, with further work, this wet assay can become automated into a dry assay which will remove any doubt of potential human error with the sample processing (i.e., pipetting error). This technology has already been developed by our group with breast cancer sentinel lymph node analysis (41) and was recently evaluated in a multicenter test (27). The development of the TACSTD1 assay for application to breast cancer explains its currently better reproducibility over the PVA assay, which will improve as greater developmental effort is devoted to preparing rapid PVA amplification for clinical evaluation.
After detailed analysis of the 3 positive nodes that were missed by the GeneXpert (rapid) assay for TACSTD1 and PVA, we note that all 3were originally foundtocontain5%or less tumor cells on only 1 section. When accounting for sectioning for H&E, IHC, and additional sections for RNA isolation and qRT-PCR, the potential for significant sampling error develops. Four sections at each of 10 levels were analyzed using the rapid PCR assay, leading to approximately 50 to 60, 5-µm sections (roughly 0.3 mm) between the first and last slides examined histologically. In each of the 3 missed cases, only the last section was determined to be positive histologically. While this may occur in the clinical scenario of intraoperative SLN analysis, the positive result by either frozen section or PCR would lead the surgeon to perform a completion neck dissection. This scenario must be formally evaluated in prospective trial(s) utilizing the assays developed here, in a clinical situation and time frame.
The clinical importance of ITC and micrometastatic disease in dictating disease progression in SLNs of HNSCC is currently unclear. Sobin and Wittekind have shown that according to the tumor classification proposed by Hermanek and colleagues, ITC do not typically show evidence of metastatic activity (e.g., proliferative or stromal reaction) or penetration of vascular or lymphatic sinus walls (42, 43). Furthermore, studies from van Den Brekel and colleagues and Woolgar showed that ITC and micrometastatic disease have the same short-term significance as the clinically N0 neck (44, 45). Currently, this area of clinical controversy will likely remain a topic for future debate and research in head and neck cancer.
While other lymph node qRT-PCR studies report homogenizing whole portions of, or even complete lymph nodes for RNA isolation, we believe that lymph nodes can be processed in such a way that both qRT-PCR and routine pathologic evaluation can be performed in parallel on immediately adjacent tissue sections (46). At the Third International Conference on Sentinel Node Biopsy in mucosal head and neck cancer (Miami, FL, March 2007), it was established that all SLNs should be step-sectioned at 150 µmol/L in 3 levels from 2 mm blocks of each node (33). A similar protocol of alternate parallel sections throughout the entire SLN to be cut into a lysate buffer for qRT-PCR could be clinically feasible. The rest of the lymph node would be paraffin-embedded and sent for permanent IHC analysis, as per normal histological protocol for SLN biopsy. However, it is important to note that the GeneXpert assay can be used in the common clinical scenario in which the pathologist suspects metastatic disease in the frozen section room, however, cannot confirm the diagnosis solely based on H&E analysis. Also, this assay may be useful to rule out micrometastasis in a grossly negative SLN. Instead of running the risk of having the patient potentially return back for surgery in the case that IHC staining picks up metastatic disease, one can run the GeneXpert assay in less than 35 minutes intraoperatively, providing over 94% accuracy that the SLN is free of tumor. The high negative predictive value is of greatest clinical importance, and was the primary clinical endpoint of the recently published ACOSOG SLN trial (7), because it saves those patients free of disease from having a more extensive, END. Future work prior to commercialization includes the conversion to a dry assay on the GeneXpert system, as well as the implementation of this technology into a multicenter sentinel lymph node trial.
Supplementary Material
Translational Relevance.
Sentinel node biopsy (SNB) has been shown to accurately stage the regional lymphatics in oral carcinoma. However, intraoperative pathology is only moderately sensitive and final pathology takes several days to complete. We are developing a rapid, automated and quantitative real-time PCR (qRT-PCR) assay that can match final pathology in an intraoperative time frame. Concordance of qRT-PCR for individual markers with final pathology in 442 nodes was 93% to 98%. The best marker combination was TACSTD1 (tumor-associated calcium signal transducer 1) and PVA (pemphigus vulgaris antigen). Analysis of 103 lymph nodes demonstrated 94.2% accuracy of this assay for identifying positive and negative nodes in an average time for each assay of 35 minutes. A rapid, automated qRT-PCR assay, combined with SNB or fine needle aspiration biopsy, could more appropriately guide surgical treatment of patients with oral cancer.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clinicalcancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Thekdi AA, Ferris RL. Diagnostic assessment of laryngeal cancer. Otolaryngol Clin North Am. 2002;35:953–969. doi: 10.1016/s0030-6665(02)00041-5. [DOI] [PubMed] [Google Scholar]
- 2.Takes RP. Staging of the neck in patients with head and neck squamous cell cancer: imaging techniques and biomarkers. Oral Oncol. 2004;40:656–667. doi: 10.1016/j.oraloncology.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Shoaib T, Soutar DS, MacDonald DG, Camilleri IG, Dunaway DJ, Gray HW, et al. The accuracy of head and neck carcinoma sentinel lymph node biopsy in the clinically N0 neck. Cancer. 2001;91:2077–2083. doi: 10.1002/1097-0142(20010601)91:11<2077::aid-cncr1235>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 4.Kligerman J, Lima RA, Soares JR, Prado L, Dias FL, Freitas EQ, et al. Supraomohyoid neck dissection in the treatment of T1/T2 squamous cell carcinoma of oral cavity. Am J Surg. 1994;168:391–394. doi: 10.1016/s0002-9610(05)80082-0. [DOI] [PubMed] [Google Scholar]
- 5.McGuirt WF, Jr, Johnson JT, Myers EN, Rothfield R, Wagner R. Floor of mouth carcinoma. The management of the clinically negative neck. Arch Otolaryngol Head Neck Surg. 1995;121:278–282. doi: 10.1001/archotol.1995.01890030020004. [DOI] [PubMed] [Google Scholar]
- 6.Taylor RJ, Wahl RL, Sharma PK, Bradford CR, Terrell JE, Teknos TN, et al. Sentinel node localization in oral cavity and oropharynx squamous cell cancer. Arch Otolaryngol Head Neck Surg. 2001;127:970–974. doi: 10.1001/archotol.127.8.970. [DOI] [PubMed] [Google Scholar]
- 7.Civantos FJ, Zitsch RP, Schuller DE, Agrawal A, Smith RB, Nason R, et al. Sentinel lymph node biopsy accurately stages the regional lymph nodes for T1–T2 oral squamous cell carcinomas: results of a prospective multi-institutional trial. J Clin Oncol. 28:1395–1400. doi: 10.1200/JCO.2008.20.8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rassekh CH, Johnson JT, Myers EN. Accuracy of intraoperative staging of the NO neck in squamous cell carcinoma. Laryngoscope. 1995;105:1334–1336. doi: 10.1288/00005537-199512000-00013. [DOI] [PubMed] [Google Scholar]
- 9.Pitman KT, Johnson JT, Myers EN. Effectiveness of selective neck dissection for management of the clinically negative neck. Arch Otolaryngol Head Neck Surg. 1997;123:917–922. doi: 10.1001/archotol.1997.01900090023004. [DOI] [PubMed] [Google Scholar]
- 10.Yuen AP, Wei WI, Wong YM, Tang KC. Elective neck dissection versus observation in the treatment of early oral tongue carcinoma. Head Neck. 1997;19:583–588. doi: 10.1002/(sici)1097-0347(199710)19:7<583::aid-hed4>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 11.Koch WM, Choti MA, Civelek AC, Eisele DW, Saunders JR. Gamma probe-directed biopsy of the sentinel node in oral squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 1998;124:455–459. doi: 10.1001/archotol.124.4.455. [DOI] [PubMed] [Google Scholar]
- 12.Zitsch RP, 3rd, Todd DW, Renner GJ, Singh A. Intraoperative radiolymphoscintigraphy for detection of occult nodal metastasis in patients with head and neck squamous cell carcinoma. Otolaryngol Head Neck Surg. 2000;122:662–666. doi: 10.1016/S0194-5998(00)70192-6. [DOI] [PubMed] [Google Scholar]
- 13.Hyde NC, Prvulovich E, Newman L, Waddington WA, Visvikis D, Ell P. A new approach to pre-treatment assessment of the N0 neck in oral squamous cell carcinoma: the role of sentinel node biopsy and positron emission tomography. Oral Oncol. 2003;39:350–360. doi: 10.1016/s1368-8375(02)00121-5. [DOI] [PubMed] [Google Scholar]
- 14.Pitman KT, Johnson JT, Brown ML, Myers EN. Sentinel lymph node biopsy in head and neck squamous cell carcinoma. Laryngoscope. 2002;112:2101–2113. doi: 10.1097/00005537-200212000-00001. [DOI] [PubMed] [Google Scholar]
- 15.Civantos FJ, Gomez C, Duque C, Pedroso F, Goodwin WJ, Weed DT, et al. Sentinel node biopsy in oral cavity cancer: correlation with PET scan and immunohistochemistry. Head Neck. 2003;25:1–9. doi: 10.1002/hed.10213. [DOI] [PubMed] [Google Scholar]
- 16.Mori M, Mimori K, Ueo H, Karimine N, Barnard GF, Sugimachi K, et al. Molecular detection of circulating solid carcinoma cells in the peripheral blood: the concept of early systemic disease. Int J Cancer. 1996;68:739–743. doi: 10.1002/(SICI)1097-0215(19961211)68:6<739::AID-IJC8>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 17.Liefers GJ, Cleton-Jansen AM, van de Velde CJ, Hermans J, van Krieken JH, Cornelisse CJ, et al. Micrometastases and survival in stage II colorectal cancer. N Engl J Med. 1998;339:223–228. doi: 10.1056/NEJM199807233390403. [DOI] [PubMed] [Google Scholar]
- 18.Mitas M, Cole DJ, Hoover L, Fraig MM, Mikhitarian K, Block MI, et al. Real-time reverse transcription-PCR detects KS1/4 mRNA in mediastinal lymph nodes from patients with non-small cell lung cancer. Clin Chem. 2003;49:312–315. doi: 10.1373/49.2.312. [DOI] [PubMed] [Google Scholar]
- 19.Godfrey TE, Raja S, Finkelstein SD, Gooding WE, Kelly LA, Luketich JD. Prognostic value of quantitative reverse transcription-polymerase chain reaction in lymph node-negative esophageal cancer patients. Clin Cancer Res. 2001;7:4041–4048. [PubMed] [Google Scholar]
- 20.Shivers SC, Wang X, Li W, Joseph E, Messina J, Glass LF, et al. Molecular staging of malignant melanoma: correlation with clinical outcome. JAMA. 1998;280:1410–1415. doi: 10.1001/jama.280.16.1410. [DOI] [PubMed] [Google Scholar]
- 21.Bostick PJ, Morton DL, Turner RR, Huynh KT, Wang HJ, Elashoff R, et al. Prognostic significance of occult metastases detected by sentinel lymphadenectomy and reverse transcriptase-polymerase chain reaction in early-stage melanoma patients. J Clin Oncol. 1999;17:3238–3244. doi: 10.1200/JCO.1999.17.10.3238. [DOI] [PubMed] [Google Scholar]
- 22.Nieuwenhuis EJ, Jaspars LH, Castelijns JA, Bakker B, Wishaupt RG, Denkers F, et al. Quantitative molecular detection of minimal residual head and neck cancer in lymph node aspirates. Clin Cancer Res. 2003;9:755–761. [PubMed] [Google Scholar]
- 23.Keilholz U, Willhauck M, Rimoldi D, Brasseur F, Dummer W, Rass K, et al. Reliability of reverse transcription-polymerase chain reaction (RT-PCR)-based assays for the detection of circulating tumour cells: a quality-assurance initiative of the EORTC Melanoma Cooperative Group. Eur J Cancer. 1998;34:750–753. doi: 10.1016/s0959-8049(97)10105-8. [DOI] [PubMed] [Google Scholar]
- 24.Ferris RL, Xi L, Raja S, Hunt JL, Wang J, Gooding WE, et al. Molecular staging of cervical lymph nodes in squamous cell carcinoma of the head and neck. Cancer Res. 2005;65:2147–2156. doi: 10.1158/0008-5472.CAN-04-3717. [DOI] [PubMed] [Google Scholar]
- 25.Tassone F, Hagerman RJ, Taylor AK, Gane LW, Godfrey TE, Hagerman PJ. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am J Hum Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raja S, El-Hefnawy T, Kelly LA, Chestney ML, Luketich JD, Godfrey TE. Temperature-controlled primer limit for multiplexing of rapid, quantitative reverse transcription-PCR assays: application to intraoperative cancer diagnostics. Clin Chem. 2002;48:1329–1337. [PubMed] [Google Scholar]
- 27.Hughes SJ, Xi L, Gooding WE, Cole DJ, Mitas M, Metcalf J, et al. A quantitative reverse transcription-PCR assay for rapid, automated analysis of breast cancer sentinel lymph nodes. J Mol Diagn. 2009;11:576–582. doi: 10.2353/jmoldx.2009.090037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolfinger R, O’Connell M. Generalized linear models: A pseudo-likelihood approach. J Stat Comput Simul. 1993;48:233–243. [Google Scholar]
- 29.Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–723. [Google Scholar]
- 30.Goerkem M, Braun J, Stoeckli SJ. Evaluation of clinical and histomorphological parameters as potential predictors of occult metastases in sentinel lymph nodes of early squamous cell carcinoma of the oral cavity. Ann Surg Oncol. 17:527–535. doi: 10.1245/s10434-009-0755-3. [DOI] [PubMed] [Google Scholar]
- 31.Stoeckli SJ, Alkureishi LW, Ross GL. Sentinel node biopsy for early oral and oropharyngeal squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2009;266:787–793. doi: 10.1007/s00405-009-0955-2. [DOI] [PubMed] [Google Scholar]
- 32.Stoeckli SJ. Sentinel node biopsy for oral and oropharyngeal squamous cell carcinoma of the head and neck. Laryngoscope. 2007;117:1539–1551. doi: 10.1097/MLG.0b013e318093ee67. [DOI] [PubMed] [Google Scholar]
- 33.Stoeckli SJ, Pfaltz M, Ross GL, Steinert HC, MacDonald DG, Wittekind C, et al. The second international conference on sentinel node biopsy in mucosal head and neck cancer. Ann Surg Oncol. 2005;12:919–924. doi: 10.1245/ASO.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 34.Terada A, Hasegawa Y, Yatabe Y, Hyodo I, Ogawa T, Hanai N, et al. Intraoperative diagnosis of cancer metastasis in sentinel lymph node of oral cancer patients. Oral Oncol. 2008;44:838–843. doi: 10.1016/j.oraloncology.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Roberts CA, Beitsch PD, Litz CE, Hilton DS, Ewing GE, Clifford E, et al. Interpretive disparity among pathologists in breast sentinel lymph node evaluation. Am J Surg. 2003;186:324–329. doi: 10.1016/s0002-9610(03)00268-x. [DOI] [PubMed] [Google Scholar]
- 36.Greenberg JS, El Naggar AK, Mo V, Roberts D, Myers JN. Disparity in pathologic and clinical lymph node staging in oral tongue carcinoma. Implication for therapeutic decision making. Cancer. 2003;98:508–515. doi: 10.1002/cncr.11526. [DOI] [PubMed] [Google Scholar]
- 37.Ambrosch P, Brinck U. Detection of nodal micrometastases in head and neck cancer by serial sectioning and immunostaining. Oncology (Williston Park) 1996;10:1221–1226. discussion 6, 9. [PubMed] [Google Scholar]
- 38.Enepekides DJ, Sultanem K, Nguyen C, Shenouda G, Black MJ, Rochon L. Occult cervical metastases: immunoperoxidase analysis of the pathologically negative neck. Otolaryngol Head Neck Surg. 1999;120:713–717. doi: 10.1053/hn.1999.v120.a91761. [DOI] [PubMed] [Google Scholar]
- 39.Kocaturk S, Yilmazer D, Onal B, Erkam U, Urunal B. Do micrometastases detected with cytokeratin immunoperoxidase reactivity affect the treatment approach to neck in supraglottic cancers? Otolaryngol Head Neck Surg. 2003;128:407–411. doi: 10.1067/mhn.2003.103. [DOI] [PubMed] [Google Scholar]
- 40.Ambrosch P, Freudenberg L, Kron M, Steiner W. Selective neck dissection in the management of squamous cell carcinoma of the upper digestive tract. Eur Arch Otorhinolaryngol. 1996;253:329–335. doi: 10.1007/BF00178287. [DOI] [PubMed] [Google Scholar]
- 41.Hughes SJ, Xi L, Raja S, Gooding W, Cole DJ, Gillanders WE, et al. A rapid, fully automated, molecular-based assay accurately analyzes sentinel lymph nodes for the presence of metastatic breast cancer. Ann Surg. 2006;243:389–398. doi: 10.1097/01.sla.0000201541.68577.6a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sobin L, Wittekind CH, editors. TNM Classification of Malignant Tumours. 6th. New York: John Wiley & Sons; 2002. [Google Scholar]
- 43.Hermanek P, Hutter RV, Sobin LH, Wittekind C. International Union Against Cancer. Classification of isolated tumor cells and micrometastasis. Cancer. 1999;86:2668–2673. [PubMed] [Google Scholar]
- 44.Van Den Brekel MW, Stel HV, Van Der Valk P, Van Der Waal I, Meyer CJ, Snow GB. Micrometastases from squamous cell carcinoma in neck dissection specimens. Eur Arch Otorhinolaryngol. 1992;249:349–353. doi: 10.1007/BF00179388. [DOI] [PubMed] [Google Scholar]
- 45.Woolgar JA. Micrometastasis in oral/oropharyngeal squamous cell carcinoma: incidence, histopathological features and clinical implications. Br J Oral Maxillofac Surg. 1999;37:181–186. doi: 10.1054/bjom.1999.0037. [DOI] [PubMed] [Google Scholar]
- 46.Elsheikh MN, Rinaldo A, Hamakawa H, Mahfouz ME, Rodrigo JP, Brennan J, et al. Importance of molecular analysis in detecting cervical lymph node metastasis in head and neck squamous cell carcinoma. Head Neck. 2006;28:842–849. doi: 10.1002/hed.20368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.