Abstract
Objective:
To validate new mitochondrial myopathy serum biomarkers for diagnostic use.
Methods:
We analyzed serum FGF21 (S-FGF21) and GDF15 from patients with (1) mitochondrial diseases and (2) nonmitochondrial disorders partially overlapping with mitochondrial disorder phenotypes. We (3) did a meta-analysis of S-FGF21 in mitochondrial disease and (4) analyzed S-Fgf21 and skeletal muscle Fgf21 expression in 6 mouse models with different muscle-manifesting mitochondrial dysfunctions.
Results:
We report that S-FGF21 consistently increases in primary mitochondrial myopathy, especially in patients with mitochondrial translation defects or mitochondrial DNA (mtDNA) deletions (675 and 347 pg/mL, respectively; controls: 66 pg/mL, p < 0.0001 for both). This is corroborated in mice (mtDNA deletions 1,163 vs 379 pg/mL, p < 0.0001). However, patients and mice with structural respiratory chain subunit or assembly factor defects showed low induction (human 335 pg/mL, p < 0.05; mice 335 pg/mL, not significant). Overall specificities of FGF21 and GDF15 to find patients with mitochondrial myopathy were 89.3% vs 86.4%, and sensitivities 67.3% and 76.0%, respectively. However, GDF15 was increased also in a wide range of nonmitochondrial conditions.
Conclusions:
S-FGF21 is a specific biomarker for muscle-manifesting defects of mitochondrial translation, including mitochondrial transfer-RNA mutations and primary and secondary mtDNA deletions, the most common causes of mitochondrial disease. However, normal S-FGF21 does not exclude structural respiratory chain complex or assembly factor defects, important to acknowledge in diagnostics.
Classification of evidence:
This study provides Class III evidence that elevated S-FGF21 accurately distinguishes patients with mitochondrial myopathies from patients with other conditions, and FGF21 and GDF15 mitochondrial myopathy from other myopathies.
Mitochondrial diseases are the most common form of inherited metabolic disorders. The high variability in clinical manifestation, heterogeneity of genetic causes with >150 known disease genes,1 and scarcity of sensitive and specific biomarkers make their diagnosis challenging. Our original multicenter analysis identified fibroblast growth factor 21 (FGF21) induction in genetically confirmed muscle-manifesting mitochondrial disorders.2 Serum FGF21 (S-FGF21) was both sensitive and specific for mitochondrial myopathies (MMs), correlating with disease severity and respiratory chain (RC)-deficient muscle fibers, indicating high potential for a serum biomarker for mitochondrial diseases.2,3 The findings were thereafter replicated in 4 independent cohorts.4–7 Moderately increased S-FGF21 has also been reported in some nonmitochondrial genetically heterogeneous disease groups,8–15 leaving the specificity of FGF21 for MM partially open.
In all reported studies, single patients with MM showed low FGF21 values, raising the question of whether the FGF21 response depended on the type of RC deficiency. We therefore analyzed its induction in different types of RC dysfunction, and compared the results to those of a newly suggested mitochondrial disease serum biomarker, growth and differentiation factor 15 (GDF15).6,16–18 We examined S-FGF21 in a variety of nonmitochondrial disorders with some symptoms or signs common to mitochondrial diseases. To deepen understanding of the mechanism of induction, we analyzed samples from 6 genetically modified mouse models with mechanistically different RC defects. We present here evidence that FGF21 and GDF15 respond especially to MMs caused by mitochondrial translation defects or mitochondrial DNA (mtDNA) deletions, with both similarities and differences in the biomarker response and specificity.
METHODS
Study participants.
To study the disease spectrum with FGF21 and GDF15 induction, we analyzed previously collected (years 1993–2013) serum materials from individuals with a nonmitochondrial disorder/state that shared some phenotypic features with mitochondrial disease (muscle/cardiac involvement, late-stage disease, poor prognosis, catabolism, secondary mitochondrial dysfunction, liver involvement; summarized in table e-1 at Neurology.org). Details of these populations were published previously: amyotrophic lateral sclerosis (ALS),e1 cardiomyopathy,e2,e3 lymphoma,e4 metastasized colorectal cancer (mCRC),e5 nonagenarians,e6 primary biliary cirrhosis (PBC), primary sclerosing cholangitis (PSC),e7 and statin-induced myalgia.e8 The genetic background of patients with ALS was as follows: SOD1 n = 6; C9ORF72 n = 1; and unknown n = 2. Among cardiomyopathy patients, all had LMNA gene mutations, normal serum creatine kinase (CK) value, and no signs of skeletal muscle dysfunction. Statin-induced myalgia samples were drawn during an acute myalgia period and after clinical recovery. mCRC samples were drawn before initiation of palliative chemotherapy. PBC and PSC samples were taken prior to liver transplantation; clinical data were collected from the latest medical records or the national liver transplant registry. Fibromyalgia samples were collected from pain outpatient clinics excluding patients with a diagnosed muscle disorder and/or severe psychiatric conditions. Liver function tests were available only from the patients with liver disease (mCRC, PBC, PSC) and cardiomyopathy.
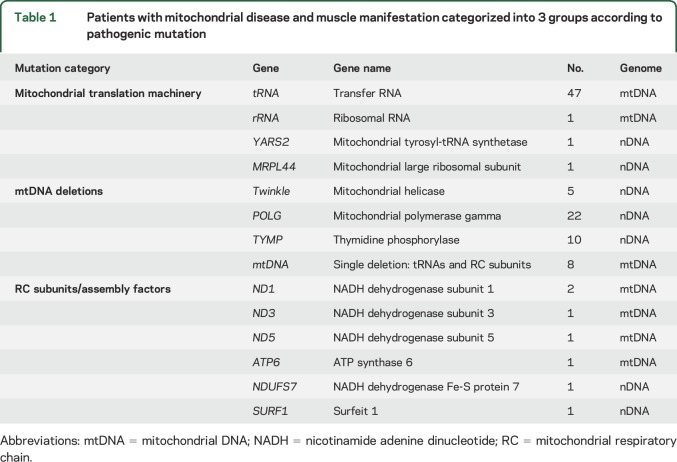
Figure e-1 summarizes the patient groups and inclusion criteria. Patients with MM included those with meta-analysis of muscle-manifesting mitochondrial disease2,4,6 and 17 new patients with a verified genetic diagnosis (table 1 summarizes their clinical and genetic findings). Serum was collected from 2009 to 2016 and stored at −80°C. Clinical data were collected from the latest medical records.
Table 1.
Patients with mitochondrial disease and muscle manifestation categorized into 3 groups according to pathogenic mutation
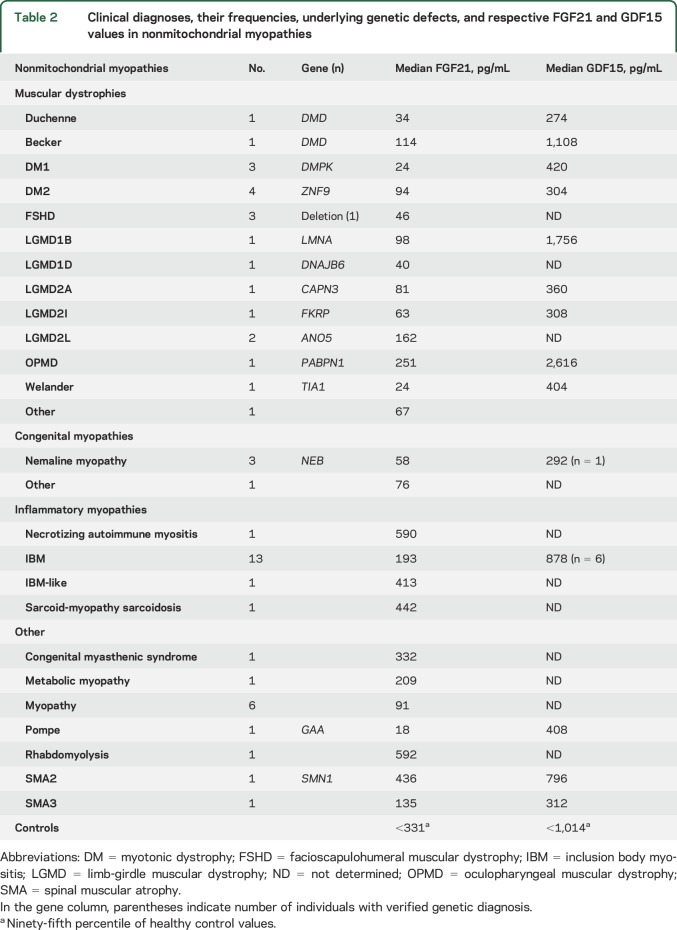
Non-MM included 39 patients from previous publications2,4 and 13 new non-MMs (figure e-1 and table 2 summarize clinical and genetic data). Controls (n = 87) were from 2 previous publications,2,6 with no significant difference between S-FGF21 concentrations. GDF15 was analyzed from all cohorts, except for lymphoma or statin myopathy samples (not available).
Table 2.
Clinical diagnoses, their frequencies, underlying genetic defects, and respective FGF21 and GDF15 values in nonmitochondrial myopathies
Mouse models and sampling.
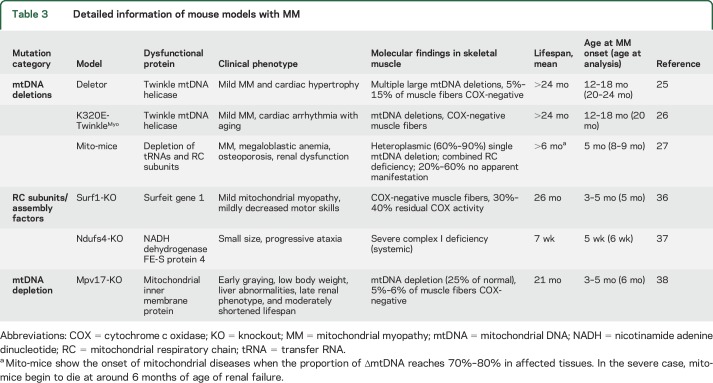
Samples were from previously characterized transgenic mouse models representing mechanistically different muscle-manifesting RC deficiencies (table 3). Blood, skeletal muscle, and/or liver samples were collected immediately after euthanization, snap-frozen in liquid nitrogen, and stored at −80°C. Serum was separated by letting the blood clot for 15 minutes at room temperature and centrifuged at 3,000g for 15 minutes at +4°C.
Table 3.
Detailed information of mouse models with MM
Measurement of serum/plasma FGF21 and GDF15 by ELISA.
We measured human serum/plasma FGF21 using 2 ELISA kits (primarily BioVendor, Brno, Czech Republic; results replicated with R&D Systems, Minneapolis, MN). GDF15 from human serum/plasma was analyzed with ELISA (R&D Systems). Mouse samples were measured with a Quantikine Mouse Fgf21 immunoassay (R&D Systems). Absorbance measurements were performed using a SpectraMax 190 absorbance microtiter plate reader (Molecular Devices, Sunnyvale, CA).
RNA extraction and analysis of Fgf21 messenger RNA amount in mice.
Fgf21 expression was analyzed using total extracted tissue RNA and quantitative PCR as previously described.e8 Primer sequences and the amplification protocol are summarized in appendix e-1.
mtDNA deletion analysis in mito-mice.
Quantification of mtDNA heteroplasmy in muscle samples of mito-mice was done with Southern blot, as previously described.e9 Briefly, XhoI digested total muscle DNA was separated in 0.6% agarose gel and transferred to a nylon membrane. Hybridization was performed with an mtDNA probe, nucleotide positions 1859–2762. Signal calculation was done with the NIH IMAGE program.
Statistical analysis.
Median of S-FGF21 (interquartile range in parentheses) was used in intergroup comparisons, and 2-tailed p values <0.05 were considered significant. For mouse data, mean and SD were used. Mann–Whitney U test was used to compare S-FGF21 levels between 2 groups, and Kruskal–Wallis or 1-way analysis of variance for comparing multiple groups, followed by the Dunn multiple comparisons test. Comparison of S-FGF21 and CK values during and after acute statin-induced myalgia was performed with the Wilcoxon matched-pairs signed rank test. Spearman rank correlation analysis was used for associations of S-FGF21 levels to continuous numeric variables. Nonparametric association was considered significant if r value was ≥0.5 and p < 0.05. In case r exceeded 0.5, linear regression was performed, and R2 and P (capital p) value for goodness of fit are shown. To determine sensitivity and specificity, we plotted the receiver operating characteristic curves and calculated the area under the curve. Outlier analysis was performed for homogeneous mouse data, and cleaned data were used for analysis. All statistical analyses were performed and graphs generated using GraphPad PRISM 6 (GraphPad Software, La Jolla, CA).
Standard protocol approvals, registrations, and patient consents.
Institutional ethics review boards of all centers approved the study, participants gave written informed consents, and the Declaration of Helsinki was followed. Animal procedures were performed according to protocols approved by the ethical boards for animal experimentation for each collaborating center. All experiments were done in accordance with good practice of handling laboratory animals and of genetically modified organisms.
Primary research questions.
Are FGF21 and GDF15 induced in all muscle-manifesting mitochondrial disorders or specific subgroups? This study demonstrates that S-FGF21 and GDF15 responses are induced in MMs caused by mitochondrial translation defects and mtDNA deletions, but less in diseases with RC subunit or assembly mutations. The FGF21 finding is corroborated in comprehensive mouse model material.
Are FGF21 and GDF15 specific for MM? This study provides Class III evidence that S-FGF21 distinguishes MM from other diseases with phenotypic similarities.
RESULTS
S-FGF21 and GDF15 are induced in patients with muscle-manifesting diseases caused by mitochondrial translation defects or mtDNA deletions.
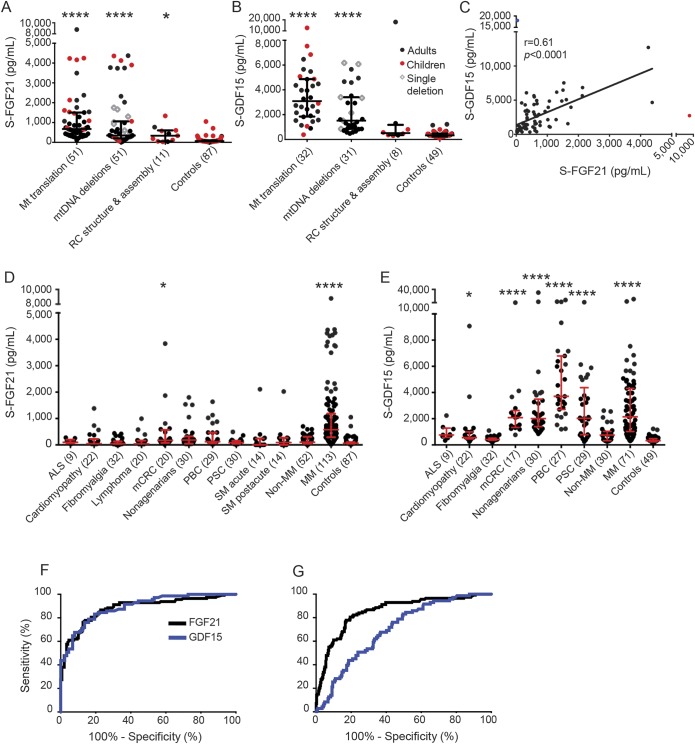
We categorized patients with MM into 3 groups according to their genetic defects: (1) directly affecting mitochondrial translation, (2) primary or secondary mtDNA deletions, and (3) defects affecting RC subunit or assembly factors (table 1). FGF21 was remarkably elevated in groups 1 and 2, whereas RC protein structure or assembly factor mutations elevated S-FGF21 levels only modestly (figure 1A).
Figure 1. S-FGF21 is increased in MMs caused by defective mitochondrial translation or mtDNA deletions.
(A) Median (IQR) of S-FGF21 in different MM groups: Mt translation 675 pg/mL (437–1,504), mtDNA deletions 347 pg/mL (206–1,062), RC structure and assembly 335 pg/mL (54–604), and controls 66 pg/mL (48–104). (B) Median S-GDF15 (IQR) in different MM groups: Mt translation 3,092 pg/mL (1,844–4,868), mtDNA deletions 1,520 pg/mL (852–3,403), RC structure and assembly 512 pg/mL (348–1,178), and controls 328 pg/mL (235–474). (C) Correlation between S-FGF21 and GDF15 in MMs. Blue and red dots indicate patients considered outliers and were not included in the linear regression. Blue patient: mtATP6 mutation; red patient: tRNALeu-UUR. (D) Median (IQR) of S-FGF21 for the nonmitochondrial diseases and traits, MM, and controls: ALS 98 pg/mL (62–196), cardiomyopathy 47 pg/mL (45–223), fibromyalgia 45 pg/mL (39–135), lymphoma 39 pg/mL (38–162), mCRC 170 pg/mL (75–566), nonagenarians 181 pg/mL (45–325), PBC 121 pg/mL (62–466), PSC 84 pg/mL (42–157), SM acute phase 4 pg/mL (1–241), SM post acute phase 90 pg/mL (1–269), non-MM 101 pg/mL (46–309), MM 576 pg/mL (289–1,187), and controls 66 pg/mL (48–104). (E) Median (IQR) of S-GDF15 for the nonmitochondrial diseases and traits, MM, and controls: ALS 760 pg/mL (582–1,284), cardiomyopathy 558 pg/mL (456–993), fibromyalgia 407 pg/mL (359–518), mCRC 2,082 pg/mL (1,332–2,625), nonagenarians 2,007 pg/mL (1,397–3,489), PBC 3,706 pg/mL (2,706–6,794), PSC 2,026 pg/mL (774–4,371), non-MM 796 pg/mL (344–1,056), MM 2,146 pg/mL (1,024–4,312), and controls 328 pg/mL (235–474). (F) ROC curves for FGF21 and GDF15: MM patients compared to nonmitochondrial muscle diseases and controls. AUC for FGF21 is 0.88 (95% CI 0.84–0.93) and for GDF15 0.89 (95% CI 0.84–0.94). (G) ROC curves with MM against non-MM, other diseases and traits, and controls. AUC for FGF21 is 0.86 (95% CI 0.81–0.90) and for GDF15 0.70 (95% CI 0.64–0.76). Note the discontinuous y-axes in A–E. Number of participants is in parentheses. Statistical significance: *p < 0.05, ****p < 0.0001. ALS = amyotrophic lateral sclerosis; AUC = area under the curve; CI = confidence interval; IQR = interquartile range; mCRC = metastasized colorectal cancer; MM = mitochondrial myopathy; Mt = mitochondrial; mtDNA = mitochondrial DNA; PBC = primary biliary cirrhosis; PSC = primary sclerosing cholangitis; RC = mitochondrial respiratory chain; ROC = receiver operating characteristic; SM = statin-induced myalgia.
S-FGF21 was significantly higher in children with mitochondrial translation defects than in adults with disorders of that group (1,572 pg/mL [1,000–4,179] and 624 pg/mL [373–983], respectively, p < 0.001) (figure 1A). S-FGF21 showed no general correlation to age, as reported previously.3 FGF21 was similarly induced in patients with sporadic, single, large-scale mtDNA deletions and or multiple mtDNA deletions, associated with dominant or recessive disorders of mtDNA maintenance (531 pg/mL [300–1,231] and 317 pg/mL [173–1,062], respectively, p = 0.32).
Similar to S-FGF21, GDF15 was increased in mitochondrial translation and mtDNA deletion disorders, and less in RC complex and assembly defects (figure 1, A and B). S-FGF21 and GDF15 correlated significantly among MM groups (figure 1C). Of primary translation defects, both FGF21 and GDF15 showed the highest induction in patients with different mitochondrial transfer RNA (tRNA) mutations (Leu [UUR], Glu, Ile, Arg). Of mtDNA deletion disorders, FGF21 was especially increased in Alpers-Huttenlocher disease and mitochondrial neurogastrointestinal encephalomyopathy (secondary multiple mtDNA deletions caused by POLG1 and TYMP mutations), whereas GDF15 was highest in primary MMs of adult age, caused by multiple and single mtDNA deletions. GDF15 showed suggestive positive correlation, but FGF21 did not, with mutant mtDNA load in patients with single mtDNA deletions, all of whom had pure MM of similar clinical severity (figure e-2). These findings indicate only partial overlap in the pathogenic stimuli for the 2 biomarkers.
Nonmitochondrial diseases/traits do not induce S-FGF21 response.
In individuals with nonmitochondrial diseases and traits, S-FGF21 never reached 578 pg/mL, the median in the MM group. Liver-metastasized colon cancer showed a significant moderate increase (figure 1D). S-GDF15, analyzed from the same samples, showed increased concentrations in most diseases and conditions, except for ALS, fibromyalgia, and non-MM (figure 1E). The receiver operating characteristic curves of GDF15 and FGF21 were similar, when MM was compared to differential diagnostic disease controls (non-MM, ALS) and healthy participants (figure 1F). However, GDF15 was considerably less sensitive to distinguish MMs from a wider group of nonmitochondrial severe disorders (listed in figure 1, D and E) with some manifestations common to mitochondrial diseases (figure 1G).
Among conditions typically examined in muscle clinics (mitochondrial and non-MMs, ALS, and healthy people who are eventually deemed not to have a muscle disease), FGF21 sensitivity was 67.3% (95% confidence interval [CI] 57.8%–75.8%) and specificity 89.3% (95% CI 83.2%–93.8%). GDF15 sensitivity was 76.0% (95% CI 64.5%–85.4%) and specificity 86.4% (95% CI 77.4%–92.8%). We considered a value abnormal if the serum concentration exceeded >331 pg/mL for FGF21 and >1,014 pg/mL for GDF15 (95th percentile of healthy control values).
Liver is a source of circulating FGF21 during starvation19 and hepatic steatosis is reported to induce FGF21.12,15 We found no correlation of S-FGF21 and liver dysfunction measures (r > 0.5, p < 0.05) (table e-2). S-FGF21 did not associate with α-fetoprotein, Ca19-9, and carcinoembryonic antigen in hepatic metastases/cancer predisposing disorders (mCRC, PBC, and PSC) (data not shown). The degree of atypia in PSC patient biliary tract brush cytology samples (1 > 5: no atypia > cancer)20 showed no association with S-FGF21 (data not shown).
To study the prognostic value of S-FGF21 in late-stage disorders, we correlated S-FGF21 to survival (in years) in patients who had died within the follow-up period (n = 46) and to the MELD (model for end-stage liver disease) index (in PSC and PBC), but no association was found (figure e-3, E and F). In nonagenarians, frailty index, a rough measure of mortality and disability,21 did not mirror S-FGF21 (figure e-3G).
Discontinuation of statin treatment and clinical remission of myalgia did not alter S-FGF21 in statin-induced myalgia group (figure e-3, A and B). However, in cardiomyopathy patients, S-FGF21 showed correlation with plasma B-type natriuretic peptide (R2 = 0.73, p < 0.0001) and heart ejection fraction (%) (R2 = 0.56, p < 0.0001) (figure e-3, C and D), suggesting that decreasing heart dysfunction can contribute to S-FGF21. However, as a group, cardiomyopathies did not show significant S-FGF21 increase.
S-Fgf21 is induced in mouse models with muscle-manifesting RC deficiency caused by mtDNA deletions.
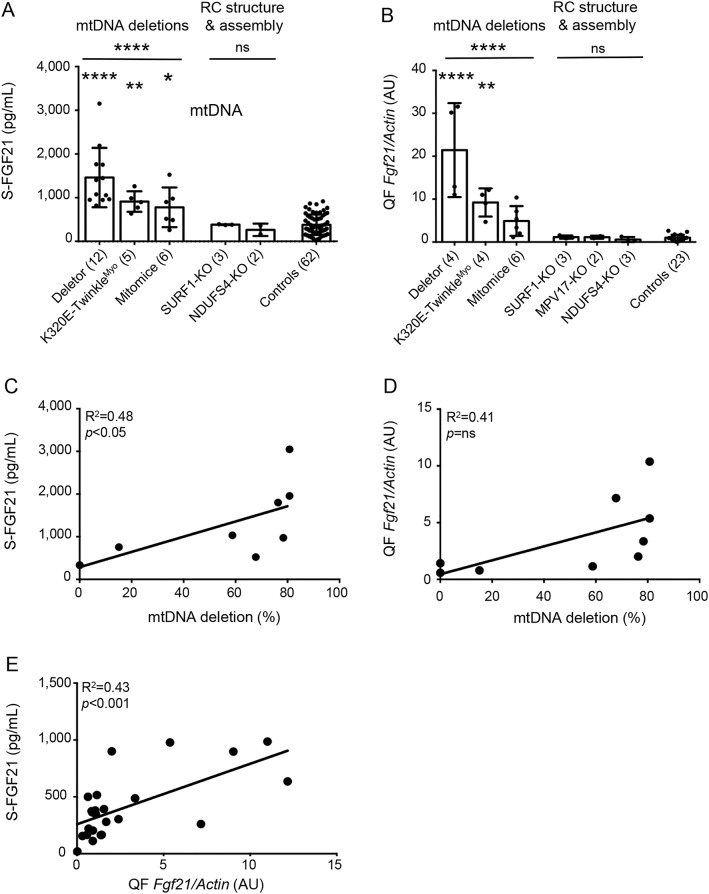
As human patients showed FGF21 induction in specific mitochondrial disease groups, we analyzed samples from 6 different congenic mouse models, which all manifested RC deficiency in the muscle, but had different types of underlying defect (table 2). Deletor, K320E-TwinkleMyo mice (secondary multiple mtDNA deletions), and mito-mice (single mtDNA deletion) showed a significant induction of S-Fgf21 (figure 2A), whereas Surf1-KO and Ndufs4-KO (RC complex structural or assembly defects) showed no increases (figure 2A). Together, the deletion disorders showed more than 3-fold-higher S-Fgf21 (1,163 pg/mL [±625]) than controls (379 pg/mL [±233]) or assembly/structural RC mutations (335 pg/mL [±96]). These results indicated a similar mechanistic induction of S-Fgf21 in mice as in human patients.
Figure 2. MM in mice induces secretion of Fgf21 by skeletal muscle.
(A) Mean (±SD) S-Fgf21 levels in mouse models: Deletor 1,460 pg/mL (±679), K320E-TwinkleMyo 911 pg/mL (±236), mito-mice 778 pg/mL (±454), Surf1-KO 382 pg/mL (±9), Ndufs4-KO 264 pg/mL (±141), and controls 379 pg/mL (±233). (B) Skeletal muscle (QF) expression of Fgf21 gene normalized to β-actin in MM mouse models. AU mean expression (±SD) compared to controls normalized as 1 (±0.71): Deletor 21.4X (±11), K320E-TwinkleMyo 9.23X (±3.28), mito-mice 4.90X (±3.47), Surf1-KO 1.15X (±0.37), Mpv17-KO 0.58X (±0.59), and Ndufs4-KO 1.13X (±0.35). (C) Association of S-Fgf21 with mtDNA deletion load in muscle of mito-mice. (D) Association of QF expression of Fgf21 with mtDNA deletion load in muscle of mito-mice. (E) S-Fgf21 association with corresponding QF expression of the Fgf21 gene in all mouse models. Number of mice is in parentheses. *p < 0.05, **p < 0.01, ****p < 0.0001. AU = artificial unit; MM = mitochondrial myopathy; mtDNA = mitochondrial DNA; ns = not significant; QF = quadriceps femoris; RC = mitochondrial respiratory chain.
Skeletal muscle is the source of circulating Fgf21 in MM mice.
To investigate the role of skeletal muscle as the source of circulating Fgf21 in MM, we analyzed muscle Fgf21 messenger RNA levels in all 6 mouse models and their controls (figure 2B). Consistent with serum results (figure 2A), we found a strong induction of Fgf21 expression in Deletor mice and a milder induction in K320E-TwinkleMyo mice compared to controls. In mito-mice, the expression of Fgf21 in the skeletal muscle trended upward. No Fgf21 expression was detected in the muscle of Surf1-KO, Ndufs4-KO, or Mpv17-KO mice, consistent with their low S-Fgf21. In mito-mice with a single mtDNA deletion, heteroplasmy level of mutant mtDNA associated significantly (r > 0.5, p < 0.05) with both S-Fgf21 level (R2 = 0.48, p < 0.05) and muscle Fgf21 expression (R2 = 0.41, not significant) (figure 2, C and D). In addition, we correlated S-FGF21 values with corresponding muscle expression of Fgf21 in mice and detected a significant correlation (R2 = 0.43, p < 0.001) (figure 2E). Since liver is another known source of circulating Fgf21, we analyzed liver expression of Fgf21 in Deletor, Surf1, and Mpv17 mice, which showed no induction compared to controls (data not shown).
Together, these data from human and mice indicate that FGF21 expression responds to defects with single large-scale/multiple mtDNA deletions or impaired mitochondrial translation machinery, and that the likely source of S-FGF21 is the skeletal muscle.
DISCUSSION
Our results define S-FGF21 as a specific tool in mitochondrial disease diagnostics, especially raised in MMs with affected mitochondrial translation—the most common cause of MM—but not in structural RC complex or assembly deficiencies. The newly reported GDF15 identified MMs well from non-MMs, with higher sensitivity and lower specificity, but was also induced in a wide variety of different kinds of severe diseases of nonmitochondrial origin. These data support the value of both FGF21 and GDF15 in first-line diagnosis of muscle disorders.
The usefulness of FGF21 as a serum marker for mitochondrial disease has been validated in 4 studies.2–7 However, single MM outliers without induction led us to ask whether the mechanism of response was related to specific kinds of dysfunction. Our patient and mouse data, supplemented by literature review, pointed to the highest induction of FGF21 response in disorders that primarily or secondarily affect mitochondrial translation—direct mutations of translation machinery or mtDNA deletions leading to imbalance of mtDNA-encoded tRNAs and rRNAs—but not mutations in structural RC complexes or their assembly factors. mtDNA maintenance disorders, such as those caused by nuclear gene mutations in POLG, C10orf2, or TYMP, also cause mtDNA deletions or point mutations,22–24 and induced FGF21. Consistent with human data, mouse models accumulating multiple mtDNA deletions in skeletal muscle,25,26 or mice with a single large heteroplasmic mtDNA deletion,27 induced Fgf21, clearly linking mtDNA deletions to the cytokine response. Mpv17 KO mice did not elicit Fgf21 response, which may be linked with mtDNA depletion or the unknown Mpv17 function. Recently, heart- and muscle-specific DARS2-KO (mitochondrial aspartyl-tRNA synthetase) mice, with impaired mitochondrial translation machinery and mosaic RC deficiency, were reported to show a robust increase of Fgf21 in their heart.28 These data strongly support the conclusion that impaired mitochondrial translation is a primary trigger for FGF21 response.
FGF21 concentration has been suggested to reflect the severity of disease.2,7 Previously, S-FGF213 or FGF21 messenger RNA2 were found to correlate to the amount of RC-deficient muscle fibers in human patients and Deletor mice. Here, we show mutant mtDNA amount correlating with S-Fgf21 and muscle Fgf21-RNA in mito-mice, and S-FGF21 in patients with single mtDNA deletions. These data suggest that FGF21 reflects muscle disease severity, indicating potential for follow-up of MM progression.
High S-FGF21 values were reported in patients with end-stage mitochondrial disease,2 suggesting that FGF21 could be induced by tissue degeneration or multiple organ failure. Furthermore, GDF15 concentrations were high in a variety of severe disorders, including pulmonary hypertension and end-stage heart failure.29,30 We found that S-FGF21 correlated with prohormone brain natriuretic peptide and ejection fraction %, suggesting that heart failure could induce FGF21. Despite reports of increased expression of FGF21 in the liver in hepatic stress and hepatocarcinogenesis in mice and humans,31 we found no S-FGF21 increase in liver pathology (mCRC, PBC, PSC), nor did it associate with liver dysfunction tests or degree of atypia in biliary tracts, indicating that FGF21 is not a general consequence of liver disease. Poor disease prognosis or general frailty did not stimulate S-FGF21. All these conditions did increase GDF15, even to similar levels as in MM. However, within the group of muscle disorders, including ALS and inclusion body myositis with secondary mitochondrial abnormalities, both GDF15 and FGF21 sensitively identified primary MM patients, suggesting that both biomarkers could be useful in serum-diagnostic work of patients in a muscle clinic.
To elucidate the role of secondary muscle mitochondrial pathology in FGF21 induction, we studied patients with statin-induced myalgia as well as elderly patients. Statins are inhibitors of HMG-CoA (3-hydroxy-3-methyl-glutaryl-coenzyme A) reductase, which participates in mitochondrial ubiquinone synthesis.32 The mechanisms of statin myopathy are complex, also involving mitochondrial RC complex III deficiency by direct binding of the drug to the enzyme.33 S-FGF21 did not respond to statin-induced muscle events, even in the acute phase. Furthermore, despite low amounts of mtDNA deletions in skeletal muscle of normal aging individuals,34 S-FGF21 was not increased even in people in their 10th decade. However, single individuals with nonmitochondrial diseases showed high S-FGF21 values and without a genetic diagnosis, the possibility of an undiagnosed mitochondrial disease remains. Overall, the findings support the specificity of FGF21 induction in primary mitochondrial disease.
Our study corroborates the role of S-FGF21 as a valuable biomarker for MMs. Increased values in patients' sera point to defects in mitochondrial translation machinery or mtDNA maintenance, the most common causes of mitochondrial disease. We propose this biomarker for first-line diagnosis of muscle clinic patients. However, in clinical manifestations consistent with Leigh syndrome or Leber hereditary optic atrophy, typically associated with structural or assembly defects of RC complexes, S-FGF21 can remain low. Our evidence indicates that GDF15 has a related mechanism of induction in skeletal muscle as FGF21, but is also increased in a spectrum of nonmitochondrial pathologic conditions and upon high age. Both biomarkers outperform the sensitivity of conventional biomarkers CK and lactate,3,35 but induction of GDF15 in many severe nonmitochondrial disorders is important to acknowledge and may limit its use in diagnostics in relation to FGF21. Of note, our data show that the physiologic consequences of RC deficiency in skeletal muscle are dictated by the primary molecular disease mechanism, and not by its downstream endpoint, RC deficiency.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Antti Hervonen and Marja Jylhä from University of Tampere, Eija Kalso, Kirsi Pietiläinen, Sini Weckström, Anna Hakonen, Ksenia Sevastianova, Hannele Yki-Järvinen, and Minna Taskinen from the University of Helsinki, Ria de Haas and Saskia Koene from Radboud UMC, and Costanza Lamperti from Carlo Besta Neurological Institute, Milan, for their contributions in collecting materials. Anu Harju, Markus Innilä, and Tuula Manninen from the University of Helsinki are thanked for technical assistance.
GLOSSARY
- ALS
amyotrophic lateral sclerosis
- CI
confidence interval
- CK
creatine kinase
- FGF21
fibroblast growth factor 21
- GDF15
growth and differentiation factor 15
- mCRC
metastasized colorectal cancer
- MM
mitochondrial myopathy
- mtDNA
mitochondrial DNA
- PBC
primary biliary cirrhosis
- PSC
primary sclerosing cholangitis
- RC
respiratory chain
- S-FGF21
serum FGF21
- tRNA
transfer RNA
Footnotes
Supplemental data at Neurology.org
Editorial, page 2286
AUTHOR CONTRIBUTIONS
J.M. Lehtonen: study design, acquisition of data, performed experiments, analysis and interpretation of the data, writing of the manuscript. S. Forsström: study design, acquisition of data, performed experiments, analysis and interpretation of the data, writing of the manuscript. E. Bottani: acquisition of data, revision of the manuscript for important intellectual content. C. Viscomi: acquisition of data, revision of the manuscript for important intellectual content. O.R. Baris: acquisition of data, revision of the manuscript for important intellectual content. H. Isoniemi: acquisition of data, revision of the manuscript for important intellectual content. K. Höckerstedt: acquisition of data, revision of the manuscript for important intellectual content. P. Österlund: acquisition of data, revision of the manuscript for important intellectual content. M. Hurme: acquisition of data, revision of the manuscript for important intellectual content. J. Jylhävä: acquisition of data, revision of the manuscript for important intellectual content. S. Leppä: acquisition of data, revision of the manuscript for important intellectual content. R. Markkula: acquisition of data, revision of the manuscript for important intellectual content. T. Heliö: acquisition of data, revision of the manuscript for important intellectual content. G. Mombelli: acquisition of data, revision of the manuscript for important intellectual content. J. Uusimaa: acquisition of data, revision of the manuscript for important intellectual content. R. Laaksonen: acquisition of data, revision of the manuscript for important intellectual content. H. Laaksovirta: acquisition of data, revision of the manuscript for important intellectual content. M. Auranen: clinical evaluation of patients, acquisition of data, revision of the manuscript for important intellectual content. M. Zeviani: acquisition of data, revision of the manuscript for important intellectual content. J. Smeitink: acquisition of data, revision of the manuscript for important intellectual content. R.J. Wiesner: acquisition of data, revision of the manuscript for important intellectual content. K. Nakada: acquisition of data, revision of the manuscript for important intellectual content. P. Isohanni: study design, acquisition of data, clinical evaluation of patients, analysis and interpretation of the data, revision of the manuscript for important intellectual content, supervision of the study. A. Suomalainen: study design, analysis and interpretation of the data, writing of the manuscript, supervision of the study.
STUDY FUNDING
No targeted funding reported.
DISCLOSURE
J. Lehtonen has received personal study funding from the Finnish Medical Association, Biomedicum Helsinki Foundation, and Helsinki Doctoral Programme in Biomedicine. S. Forsström, E. Bottani, C. Viscomi, O. Baris, H. Isoniemi, K. Höckerstedt, P. Österlund, M. Hurme, J. Jylhävä, S. Leppä, and R. Markkula report no disclosures relevant to the manuscript. T. Heliö has received financial support from the Finnish Foundation of Cardiovascular Research, Finnish Medical Foundation, Aarno Koskelo Foundation, and the special governmental subsidy for health sciences research of the University Hospital of Helsinki. G. Mombelli, J. Uusimaa, R. Laaksonen, H. Laaksovirta, M. Auranen, and M. Zeviani report no disclosures relevant to the manuscript. J. Smeitink is the founder CRO of Khondrion, and has received financial support from the Netherlands Organization for Scientific Research (Centres for Systems Biology Research) initiative from the Netherlands Organisation for Scientific Research (NOW; NO; CSBR09/013V). R. Wiesner has received financial support from DFG Wi 889/6-2, Cluster of Excellence CECAD & SFB 728/C2. K. Nakada and P. Isohanni report no disclosures relevant to the manuscript. A. Suomalainen has received funding from the Jane and Aatos Erkko Foundation, Sigrid Jusélius Foundation, Academy of Finland, European Research Council, University of Helsinki, and Helsinki University Central Hospital. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Wong LJ. Molecular genetics of mitochondrial disorders. Dev Disabil Res Rev 2010;16:154–162. [DOI] [PubMed] [Google Scholar]
- 2.Suomalainen A, Elo JM, Pietilainen KH, et al. FGF-21 as a biomarker for muscle-manifesting mitochondrial respiratory chain deficiencies: a diagnostic study. Lancet Neurol 2011;10:806–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tyynismaa H, Carroll CJ, Raimundo N, et al. Mitochondrial myopathy induces a starvation-like response. Hum Mol Genet 2010;19:3948–3958. [DOI] [PubMed] [Google Scholar]
- 4.Davis RL, Liang C, Edema-Hildebrand F, Riley C, Needham M, Sue CM. Fibroblast growth factor 21 is a sensitive biomarker of mitochondrial disease. Neurology 2013;81:1819–1826. [DOI] [PubMed] [Google Scholar]
- 5.Salehi MH, Kamalidehghan B, Houshmand M, et al. Association of fibroblast growth factor (FGF-21) as a biomarker with primary mitochondrial disorders, but not with secondary mitochondrial disorders (Friedreich ataxia). Mol Biol Rep 2013;40:6495–6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita Y, Ito M, Kojima T, et al. GDF15 is a novel biomarker to evaluate efficacy of pyruvate therapy for mitochondrial diseases. Mitochondrion 2015;20:34–42. [DOI] [PubMed] [Google Scholar]
- 7.Koene S, de Laat P, van Tienoven DH, et al. Serum FGF21 levels in adult m.3243A>G carriers: clinical implications. Neurology 2014;83:125–133. [DOI] [PubMed] [Google Scholar]
- 8.Han SH, Choi SH, Cho BJ, et al. Serum fibroblast growth factor-21 concentration is associated with residual renal function and insulin resistance in end-stage renal disease patients receiving long-term peritoneal dialysis. Metabolism 2010;59:1656–1662. [DOI] [PubMed] [Google Scholar]
- 9.Lin Z, Zhou Z, Liu Y, et al. Circulating FGF21 levels are progressively increased from the early to end stages of chronic kidney diseases and are associated with renal function in Chinese. PLoS One 2011;6:e18398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Payne BA, Price DA, Chinnery PF. Elevated serum fibroblast growth factor 21 levels correlate with immune recovery but not mitochondrial dysfunction in HIV infection. AIDS Res Ther 2013;10:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin Z, Wu Z, Yin X, et al. Serum levels of FGF-21 are increased in coronary heart disease patients and are independently associated with adverse lipid profile. PLoS One 2010;5:e15534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yilmaz Y, Eren F, Yonal O, et al. Increased serum FGF21 levels in patients with nonalcoholic fatty liver disease. Eur J Clin Invest 2010;40:887–892. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, Yeung DC, Karpisek M, et al. Serum FGF21 levels are increased in obesity and are independently associated with the metabolic syndrome in humans. Diabetes 2008;57:1246–1253. [DOI] [PubMed] [Google Scholar]
- 14.Mraz M, Bartlova M, Lacinova Z, et al. Serum concentrations and tissue expression of a novel endocrine regulator fibroblast growth factor-21 in patients with type 2 diabetes and obesity. Clin Endocrinol 2009;71:369–375. [DOI] [PubMed] [Google Scholar]
- 15.Tyynismaa H, Raivio T, Hakkarainen A, et al. Liver fat but not other adiposity measures influence circulating FGF21 levels in healthy young adult twins. J Clin Endocrinol Metab 2011;96:E351–E355. [DOI] [PubMed] [Google Scholar]
- 16.Yatsuga S, Fujita Y, Ishii A, et al. Growth differentiation factor 15 as a useful biomarker for mitochondrial disorders. Ann Neurol 2015;78:814–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montero R, Yubero D, Villarroya J, et al. GDF-15 is elevated in children with mitochondrial diseases and is induced by mitochondrial dysfunction. PLoS One 2016;11:e0148709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalko SG, Paco S, Jou C, et al. Transcriptomic profiling of TK2 deficient human skeletal muscle suggests a role for the p53 signalling pathway and identifies growth and differentiation factor-15 as a potential novel biomarker for mitochondrial myopathies. BMC Genomics 2014;15:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gälman C, Lundåsen T, Kharitonenkov A, et al. The circulating metabolic regulator FGF21 is induced by prolonged fasting and PPARalpha activation in man. Cell Metab 2008;8:169–174. [DOI] [PubMed] [Google Scholar]
- 20.Layfield LJ, Wax TD, Lee JG, Cotton PB. Accuracy and morphologic aspects of pancreatic and biliary duct brushings. Acta Cytol 1995;39:11–18. [PubMed] [Google Scholar]
- 21.Fried LP, Tangen CM, Walston J, et al. Frailty in older adults: evidence for a phenotype. J Gerontol A Biol Sci Med Sci 2001;56:M146–M156. [DOI] [PubMed] [Google Scholar]
- 22.Hudson G, Chinnery PF. Mitochondrial DNA polymerase-gamma and human disease. Hum Mol Genet 2006;15:R244–R252. [DOI] [PubMed] [Google Scholar]
- 23.Spelbrink JN, Li FY, Tiranti V, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet 2001;28:223–231. [DOI] [PubMed] [Google Scholar]
- 24.Nishino I, Spinazzola A, Hirano M. Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science 1999;283:689–692. [DOI] [PubMed] [Google Scholar]
- 25.Tyynismaa H, Mjosund KP, Wanrooij S, et al. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc Natl Acad Sci USA 2005;102:17687–17692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baris OR, Ederer S, Neuhaus JF, et al. Mosaic deficiency in mitochondrial oxidative metabolism promotes cardiac arrhythmia during aging. Cell Metab 2015;21:667–677. [DOI] [PubMed] [Google Scholar]
- 27.Inoue K, Nakada K, Ogura A, et al. Generation of mice with mitochondrial dysfunction by introducing mouse mtDNA carrying a deletion into zygotes. Nat Genet 2000;26:176–181. [DOI] [PubMed] [Google Scholar]
- 28.Dogan SA, Pujol C, Maiti P, et al. Tissue-specific loss of DARS2 activates stress responses independently of respiratory chain deficiency in the heart. Cell Metab 2014;19:458–469. [DOI] [PubMed] [Google Scholar]
- 29.Rhodes CJ, Wharton J, Wilkins MR. Pulmonary hypertension: biomarkers. Handb Exp Pharmacol 2013;218:77–103. [DOI] [PubMed] [Google Scholar]
- 30.Kramer F, Milting H. Novel biomarkers in human terminal heart failure and under mechanical circulatory support. Biomarkers 2011;16(suppl 1):S31–S41. [DOI] [PubMed] [Google Scholar]
- 31.Yang C, Lu W, Lin T, et al. Activation of liver FGF21 in hepatocarcinogenesis and during hepatic stress. BMC Gastroenterol 2013;13:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nawarskas JJ. HMG-CoA reductase inhibitors and coenzyme Q10. Cardiol Rev 2005;13:76–79. [DOI] [PubMed] [Google Scholar]
- 33.Schirris TJ, Renkema GH, Ritschel T, et al. Statin-induced myopathy is associated with mitochondrial complex III inhibition. Cell Metab 2015;22:399–407. [DOI] [PubMed] [Google Scholar]
- 34.Bratic A, Larsson NG. The role of mitochondria in aging. J Clin Invest 2013;123:951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis RL, Liang C, Sue C. A comparison of current serum biomarkers as diagnostic indicators of mitochondrial disease. Neurology 2016;86:2010–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dell'agnello C, Leo S, Agostino A, et al. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Hum Mol Genet 2007;16:431–444. [DOI] [PubMed] [Google Scholar]
- 37.Kruse SE, Watt WC, Marcinek DJ, Kapur RP, Schenkman KA, Palmiter RD. Mice with mitochondrial complex I deficiency develop a fatal encephalomyopathy. Cell Metab 2008;7:312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viscomi C, Spinazzola A, Maggioni M, et al. Early-onset liver mtDNA depletion and late-onset proteinuric nephropathy in Mpv17 knockout mice. Hum Mol Genet 2009;18:12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.