ABSTRACT
The mRNA cap structure, which is added to nascent RNA pol II transcripts, recruits the protein complexes required for pre-mRNA transcript processing, mRNA export and translation initiation. The enzymes which catalyze mRNA cap synthesis are regulated by cellular signaling pathways which impact on their expression, localization and activity. Here we discuss the recent observation that the mRNA cap methyltransferase, RNMT, is phosphorylated on Thr-77 by CDK1-cyclin B1, which regulates its activity and the proteins with which it interacts. RNMT Thr-77 phosphorylation provides a burst of mRNA cap methyltransferase activity during early G1 phase at a time when transcription is reactivated following completion of the cell cycle. This co-ordination of transcription and mRNA capping makes an important contribution to gene expression in the cell; preventing RNMT Thr-77 phosphorylation inhibits cell proliferation. Here we discuss these findings and how mRNA cap synthesis may be regulated in other scenarios.
KEYWORDS: Cell cycle, gene expression, mRNA, mRNA cap, phosphorylation, RNMT, transcription, translation
Introduction
Eukaryotic cells contain a variety of RNA molecules, the vast majority of which assist with mRNA processing and translation including rRNA and tRNA. Transcripts which mature to become protein-encoding mRNA only account for about 5% of all cellular transcripts. The mRNA cap, a modification added to the first transcribed nucleotide of pre-mRNA and other RNA pol II transcripts, flags these RNAs for appropriate processing, nuclear export and translation into protein.1-3 The structure of the mRNA cap also protects transcripts from exonucleases. The importance of the mRNA cap for gene expression was recognized shortly after its discovery by a series of experiments in vitro and in yeast. More recently formation and degradation of the mRNA cap has been observed to be regulated by cellular signaling pathways in mammalian cells.4,5
The mRNA cap
The mRNA cap consists of a 7-methylguanosine group linked via a 5′ to 5′ triphosphate bridge to the first transcribed nucleotide of RNA pol II transcripts (Fig. 1).2,3 The first few nucleotides can also be methylated, but the position and extent of methylation varies between species. Transcripts are synthesized with a 5′ triphosphate on the first transcribed nucleotide to which the mRNA cap is added by the action of a series of enzymes.1,4 In mammals the basic mRNA cap structure, cap 0, is formed by the sequential action of CE/RNGTT and RNMT-RAM, 2 enzymes recruited to phosphorylated RNA pol II at the initiation of transcription. CE/RNGTT has triphosphatase and guanylyltransferase activities which cleave the transcript terminal phosphate and catalyze addition of guanosine monophosphate, to form the cap structure, G(5′)ppp(5′)N (N is the first transcribed nucleotide). Subsequently, RNMT-RAM catalyzes N-7 methylation of the guanosine cap to create, m7G(5′)ppp(5′)N. This cap 0 structure is functional in recruiting the Cap-binding complex (CBC) and the eukaryotic initiation factor 4E, eIF4E, and other factors which promote transcript splicing, export and translation initiation.5,6 Furthermore, in mammals the first 2 nucleotides are methylated on the O-2 position by CMTR1 and CMTR2.7,8 O-2 methylation has roles in translation initiation and identifying transcripts as “self” in innate immunity.4
Figure 1.
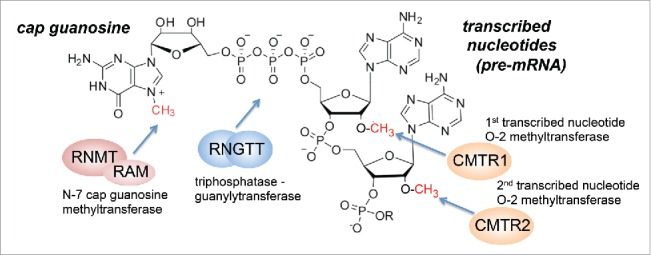
mRNA cap structure and mRNA capping enzymes. Structure of mRNA cap and action of mRNA capping enzymes.
Regulation of mRNA cap methylation
The mammalian mRNA cap methyltransferase, RNMT-RAM, consists of the enzymic subunit, RNMT (RNA guanine-7 methyltransferase) and the activator subunit, RAM (RNMT-activating miniprotein).9,10 RNMT has a catalytic core, which is homologous in structure and function to other eukaryotic cap methyltransferases.11 In addition, RNMT contains an N-terminal non-catalytic domain, which is conserved in mammals but not in other eukaryotes (Fig. 2).9 S.cerevisiae null for the cap methyltransferase are inviable and can be rescued by the expression of murine RNMT catalytic domain.12 RAM activates RNMT by altering the position of key active site residues, increasing methyl donor binding.11 RAM also has a high-affinity RNA binding domain which may increase the rate of recruitment of transcripts in general or may have specificity for particular transcript motifs.10,13
Figure 2.
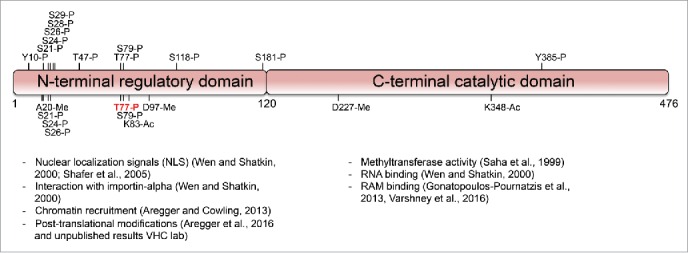
Post-translational modifications of RNMT. Human RNMT 1-120 is the N-terminal domain and RNMT 121-476 is the catalytic domain and interacts with RAM. Above depiction of RNMT are phosphorylation sites (P) obtained from PhosphositePlus (sites with only 1 mass spectrometry/high-throughput proteomics reference excluded). Below depiction of RNMT are phosphorylation, acetylation (Ac) and methylation (Me) sites identified from analysis in Aregger et al., 2016.
In human cells, RNMT-RAM is recruited via the N-terminal domain of RNMT to RNA pol II proximal to transcription initiation sites.14,15 The RNMT N-terminal domain also impacts on cap methyltransferase activity. Its removal increases catalytic activity, suggesting that this domain may have some regulatory function. We questioned whether RNMT could be regulated by N-terminal domain post-translational modifications or interacting proteins, which could potentially change the conformation or action of the N-terminus. To address this question we performed proteomic analyses to map post-translational modifications of RNMT.16
Our mass spectrometry analysis performed in HEK293 cells identified several phosphorylated, methylated and acetylated residues (Fig. 2)16 (VHC/MA unpublished observations). All of the phosphorylation sites, along with others, were also indicated in the PhosphositePlus database, which contains data from high-throughput phospho-proteomic experiments from many research groups (Fig. 2).17 Interestingly, the RNMT post-translational modifications were predominantly found in the N-terminus of RNMT and not in the catalytic domain. Sites of post-translational modification that arose in the catalytic domain may have been detrimental to activity and suppressed during evolution. The mammalian RNMT N-terminal domain may have evolved in order to allow regulation of cap methylation by signaling pathways. The phosphorylation sites found in the N-terminus cover a range of different amino acid motifs suggesting that several different kinases act on RNMT. Furthermore, the different methylation and acetylation events identified in our study demonstrate that several types of enzyme can modify RNMT. Therefore, the RNMT N-terminus is receiving a range of signaling information from different pathways, some of which may be regulating RNMT and thus formation of the mRNA cap.
Regulation and function of RNMT T77 phosphorylation
In HeLa cells, Thr-77 is the predominant site of phosphorylation. We have also observed RNMT Thr-77 phosphorylation in embryonic stem cells, mammary epithelial cells, T lymphocytes and HEK293 cells16 (VHC lab unpublished observations). RNMT Thr-77 is present in a CDK (cyclin-dependent kinase) recognition site adjacent to a cyclin binding site. The cell cycle is governed by the sequential activation of different CDK-cyclin complexes. RNMT Thr-77 phosphorylation levels oscillate throughout the cell cycle with highest phosphorylation levels during late S phase, through G2/M phase and into early G1 phase. The major kinase operational during these phases is CDK1-cyclin B1, although other CDK-cyclins are active at this time. We provided evidence in cells and in vitro that CDK1-cyclin B1 can phosphorylate RNMT directly. However, other CDK-cyclins may also phosphorylate RNMT Thr-77 and other proline-directed kinases may phosphorylate RNMT in different biological contexts.
RNMT Thr-77 phosphorylation directly increases cap methyltransferase activity.16 Presumably phosphorylation reverses some of the inhibitory effects of the N-terminal domain. The structure of the RNMT N-terminus has not been solved limiting molecular insight into Thr-77 function. Following reformation of the nuclear envelope after mitosis, factors involved in transcription are reassembled on chromatin and transcription initiates. Thus CDK1-dependent Thr-77 phosphorylation co-ordinates the peak of RNMT cap methyltransferase activity with transcription initiation. Furthermore, high levels of transcripts carrying the mRNA cap may also contribute to the increase in cap-dependent translation that is observed during the initiation of G1 phase.18
The mRNA cap binds to factors involved in transcription, mRNA processing and translation, and also protects the transcript from exonucleases.4 However, the precise role of the mRNA cap on these different gene expression processes on a genome-wide level is only beginning to be documented in mammalian cells. Mutation of RNMT Thr-77 to alanine (T77A), which blocks phosphorylation did, not have a detectable effect on transcript level, despite significant depth of RNA sequencing analysis.16 However, the T77A mutation did inhibit the expression of a subset of proteins, and inhibited cell proliferation. Although RNMT Thr-77 phosphorylation is cell cycle-dependent, the proteins whose expression is dependent on this modification were not found to be associated with synthesis at a specific stage of the cell cycle. It is important to highlight that this gene expression analysis was performed in unsynchronized cells and therefore a small effect on transcript levels during a specific phase of the cell cycle may have been undetected.
RNMT has a relatively long half-life in cells investigated to date (over 12 hours) (VHC lab unpublished observations). Therefore, the observation that RNMT Thr-77 phosphorylation is lost rapidly during G1 phase is likely to be the result of a rapidly acting phosphatase. The identity of the phosphatase or phosphatases involved is not known but a change in the activity or expression of these enzymes has the power to alter the pattern of gene expression.
Does RNMT Thr-77 have other functions?
Although RNMT Thr-77-dependent phosphorylation has a role in co-ordinating transcription and mRNA cap methylation during early G1, it is also highly phosphorylated at the end of S phase and through G2/M.16 Although the majority of gene expression occurs during G1 phase of the cell cycle, translation has been described to occur at other phases including during mitosis.19,20 It may be that in certain circumstances, in particular cell lineages, the increase in mRNA cap methyltransferase activity induced by RNMT Thr-77 phosphorylation is needed to drive the expression of a subset of transcripts during late S or G2/M phase.
RNMT is imported into the nucleus following synthesis or after mitosis by interaction with importin-α.21,22 Three classical lysine-rich nuclear localization signals lie in the RNMT N-terminus. Although we observed that phosphorylation of RNMT Thr-77 suppresses RNMT interaction with importin-α, in HeLa cells nuclear import of the RNMT T77A mutant or T77D mutant (phosphor mimic mutation) was not altered.16 RAM carries a nuclear localization signal and thus in the presence of Thr-77 phosphorylation RNMT may rely on its interaction with RAM for nuclear import.12 However, it may be that in certain situations Thr-77 phosphorylation regulates RNMT nuclear import, for example if RAM is limiting.
We observed that importin-α inhibits RNMT activity. If RNMT has a role in methylating mature cytoplasmic transcripts, Thr-77 phosphorylation-dependent inhibition of importin-α binding would increase cytoplasmic cap methyltransferase activity, which may have a positive effect on gene expression.23,24 This role could be particularly relevant when the nuclear envelope breaks down during mitosis and RNMT is dispersed into the cytoplasm. Increased RNMT activity and delocalisation from importin-α may be required for RNMT to function when it is dissociated from chromatin.
RNMT Thr-77 phosphorylation specificity
In HeLa cells, blocking RNMT Thr-77 phosphorylation did not affect the expression of all genes equally.16 What is the nature of the specificity? Why are only certain genes dependent on RNMT Thr-77 phosphorylation? As with all enzymes, specificity comes from either the enzyme itself or from the availability of the substrate. RNMT Thr-77 is in the N-terminus, the domain that interacts with RNA pol II (probably indirectly).15 However, no differential recruitment of RNMT wild type and the T77A mutant was observed by chromatin immunoprecipitation performed on candidate genes. It remains a possibility that RNMT Thr-77 phosphorylation influences transcript recruitment in a gene-specific manner. Sequence motifs and structures in the 5′ untranslated region of mRNAs may contribute to sensitivity to RNMT phosphorylation. Thus, analysis of the transcripts bound by RNMT/RAM using CLIP-Seq, or a more thorough analysis of the changes in translation rates of specific mRNAs using ribosome profiling/Ribo-Seq during specific stages of the cell cycle may provide further insights into the specificity of RNMT Thr-77 phosphorylation.
Future perspectives
mRNA capping is now established as a gene expression mechanism which integrates cellular signaling pathways, resulting in reshaping of the gene expression landscape and alterations in cell physiology and fate. Transcription factors upregulate RNA pol II phosphorylation and methionine metabolism resulting in gene-specific regulation of mRNA cap methylation.25-28 Furthermore, embryonic stem cell differentiation requires ERK1/2-dependent phosphorylation of RAM, which triggers its degradation and loss of pluripotency associated gene expression.29 It seems likely that many other signaling pathways will be regulating mRNA capping enzymes. The challenge going forward is to understand the role of mRNA capping regulation on a gene-specific level and how it impacts on mammalian development and disease.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Michael Aregger is funded by a postdoctoral fellowship from the Swiss National Science Foundation. Victoria Cowling is funded by a Medical Research Council UK Senior Fellowship (MR/K024213/1) and Lister Institute Fellowship at the Center for Gene Regulation and Expression, University of Dundee (Wellcome Trust Center Award 097945/Z/11/Z).
References
- 1.Shuman S. RNA capping: progress and prospects. Rna 2015; 21:735-7; PMID:25780214; http://dx.doi.org/ 10.1261/rna.049973.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shatkin AJ. Capping of eucaryotic mRNAs. Cell 1976; 9:645-53; PMID:1017010; http://dx.doi.org/ 10.1016/0092-8674(76)90128-8 [DOI] [PubMed] [Google Scholar]
- 3.Furuichi Y. Discovery of m(7)G-cap in eukaryotic mRNAs. Proc Jpn Acad Ser B Phys Biol Sci 2015; 91:394-409; PMID:26460318; http://dx.doi.org/ 10.2183/pjab.91.394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramanathan A, Robb GB, Chan SH. mRNA capping: biological functions and applications. Nucleic Acids Res 2016; 44(16):7511-26; PMID:27317694; http://dx.doi.org/21957010 10.1093/nar/gkw551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA 2011; 2:277-98; PMID:21957010; http://dx.doi.org/ 10.1002/wrna.52 [DOI] [PubMed] [Google Scholar]
- 6.Gonatopoulos-Pournatzis T, Cowling VH. The cap binding complex. Biochem J 2014; 457:231-42; PMID:24354960; http://dx.doi.org/ 10.1042/BJ20131214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belanger F, Stepinski J, Darzynkiewicz E, Pelletier J. Characterization of hMTr1, a human Cap1 2′-O-ribose methyltransferase. J Biol Chem 2010; 285:33037-44; PMID:20713356; http://dx.doi.org/ 10.1074/jbc.M110.155283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Werner M, Purta E, Kaminska KH, Cymerman IA, Campbell DA, Mittra B, Zamudio JR, Sturm NR, Jaworski J, Bujnicki JM. 2′-O-ribose methylation of cap2 in human: function and evolution in a horizontally mobile family. Nucleic Acids Res 2011; 39:4756-68; PMID:21310715; http://dx.doi.org/ 10.1093/nar/gkr038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowling VH. Regulation of mRNA cap methylation. Biochem J 2009; 425:295-302; PMID:20025612; http://dx.doi.org/ 10.1042/BJ20091352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonatopoulos-Pournatzis T, Dunn S, Bounds R, Cowling VH. RAM/Fam103a1 is required for mRNA cap methylation. Mol Cell 2011; 44:585-96; PMID:22099306; http://dx.doi.org/ 10.1016/j.molcel.2011.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varshney D, Petit AP, Bueren-Calabuig JA, Jansen C, Fletcher DA, Peggie M, Weidlich S, Scullion P, Pisliakov AV, Cowling VH. Molecular basis of RNA guanine-7 methyltransferase (RNMT) activation by RAM. Nucleic Acids Res 2016; PMID: 27422871; http://dx.doi.org/10347220 10.1093/nar/gkw637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saha N, Schwer B, Shuman S. Characterization of human, Schizosaccharomyces pombe, and Candida albicans mRNA cap methyltransferases and complete replacement of the yeast capping apparatus by mammalian enzymes. J Biol Chem 1999; 274:16553-62; PMID:10347220; http://dx.doi.org/ 10.1074/jbc.274.23.16553 [DOI] [PubMed] [Google Scholar]
- 13.Gonatopoulos-Pournatzis T, Cowling VH. RAM function is dependent on Kapbeta2-mediated nuclear entry. Biochem J 2014; 457:473-84; PMID:24200467; http://dx.doi.org/ 10.1042/BJ20131359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol 2008; 15:71-8; PMID:18157150; http://dx.doi.org/ 10.1038/nsmb1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aregger M, Cowling VH. Human cap methyltransferase (RNMT) N-terminal non-catalytic domain mediates recruitment to transcription initiation sites. Biochem J 2013; 455:67-73; PMID:23863084; http://dx.doi.org/ 10.1042/BJ20130378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aregger M, Kaskar A, Varshney D, Fernandez-Sanchez ME, Inesta-Vaquera FA, Weidlich S, Cowling VH. CDK1-Cyclin B1 activates RNMT, coordinating mRNA cap methylation with G1 phase transcription. Mol Cell 2016; 61:734-46; PMID:26942677; http://dx.doi.org/ 10.1016/j.molcel.2016.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res 2012; 40:D261-270; PMID:22135298; http://dx.doi.org/ 10.1093/nar/gkr1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pyronnet S, Sonenberg N. Cell-cycle-dependent translational control. Curr Opin Genet Dev 2001; 11:13-8; PMID:11163145; http://dx.doi.org/ 10.1016/S0959-437X(00)00150-7 [DOI] [PubMed] [Google Scholar]
- 19.Stumpf CR, Moreno MV, Olshen AB, Taylor BS, Ruggero D. The translational landscape of the mammalian cell cycle. Mol Cell 2013; 52:574-82; PMID:24120665; http://dx.doi.org/ 10.1016/j.molcel.2013.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanenbaum ME, Stern-Ginossar N, Weissman JS, Vale RD. Regulation of mRNA translation during mitosis. Elife 2015; 4:e07957; PMID:26305499; http://dx.doi.org/ 10.7554/eLife.07957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shafer B, Chu C, Shatkin AJ. Human mRNA cap methyltransferase: alternative nuclear localization signal motifs ensure nuclear localization required for viability. Mol Cell Biol 2005; 25:2644-9; PMID:15767670; http://dx.doi.org/ 10.1128/MCB.25.7.2644-2649.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wen Y, Shatkin AJ. Cap methyltransferase selective binding and methylation of GpppG-RNA are stimulated by importin-α. Genes Dev 2000; 14:2944-9; PMID: 11114884; http://dx.doi.org/25541487 10.1101/gad.848200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiss DL, Oman K, Bundschuh R, Schoenberg DR. Uncapped 5′ ends of mRNAs targeted by cytoplasmic capping map to the vicinity of downstream CAGE tags. FEBS Lett 2015; 589:279-84; PMID:25541487; http://dx.doi.org/ 10.1016/j.febslet.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee C, Patil DP, Kennedy BA, Bakthavachalu B, Bundschuh R, Schoenberg DR. Identification of cytoplasmic capping targets reveals a role for cap homeostasis in translation and mRNA stability. Cell Rep 2012; 2:674-84; PMID:22921400; http://dx.doi.org/ 10.1016/j.celrep.2012.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aregger M, Cowling VH. E2F1-dependent methyl cap formation requires RNA pol II phosphorylation. Cell Cycle 2012; 11:2146-8; PMID:22592528; http://dx.doi.org/ 10.4161/cc.20620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandez-Sanchez ME, Gonatopoulos-Pournatzis T, Preston G, Lawlor MA, Cowling VH. S-adenosyl homocysteine hydrolase is required for Myc-induced mRNA cap methylation, protein synthesis, and cell proliferation. Mol Cell Biol 2009; 29:6182-91; PMID:19805518; http://dx.doi.org/ 10.1128/MCB.00973-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cole MD, Cowling VH. Specific regulation of mRNA cap methylation by the c-Myc and E2F1 transcription factors. Oncogene 2009; 28:1169-75; PMID:19137018; http://dx.doi.org/ 10.1038/onc.2008.463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cowling VH, Cole MD. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol Cell Biol 2007; 27:2059-73; PMID:17242204; http://dx.doi.org/ 10.1128/MCB.01828-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grasso L, Suska O, Davidson L, Gonatopoulos-Pournatzis T, Williamson R, Wasmus L, Wiedlich S, Peggie M, Stavridis MP, Cowling VH. mRNA cap methylation in pluripotency and differentiation. Cell Rep 2016; 16:1352-65; PMID:27452456; http://dx.doi.org/ 10.1016/j.celrep.2016.06.089 [DOI] [PMC free article] [PubMed] [Google Scholar]


