ABSTRACT
Localized mRNA translation is a widespread mechanism for targeting protein synthesis, important for cell fate, motility and pathogenesis. In Drosophila, the spatiotemporal control of gurken/TGF-α mRNA translation is required for establishing the embryonic body axes. A number of recent studies have highlighted key aspects of the mechanism of gurken mRNA translational control at the dorsoanterior corner of the mid-stage oocyte. Orb/CPEB and Wispy/GLD-2 are required for polyadenylation of gurken mRNA, but unlocalized gurken mRNA in the oocyte is not fully polyadenylated.1 At the dorsoanterior corner, Orb and gurken mRNA have been shown to be enriched at the edge of Processing bodies, where translation occurs.2 Over-expression of Orb in the adjacent nurse cells, where gurken mRNA is transcribed, is sufficient to cause mis-expression of Gurken protein.3 In orb mutant egg chambers, reducing the activity of CK2, a Serine/Threonine protein kinase, enhances the ventralized phenotype, consistent with perturbation of gurken translation.4 Here we show that sites phosphorylated by CK2 overlap with active Orb and with Gurken protein expression. Together with our new findings we consolidate the literature into a working model for gurken mRNA translational control and review the role of kinases, cell cycle factors and polyadenylation machinery highlighting a multitude of conserved factors and mechanisms in the Drosophila egg chamber.
KEYWORDS: Drosophila, gurken, oogenesis, processing bodies, translation
Introduction
The ability to regulate localized mRNA translation allows a cell exquisite spatiotemporal control over protein synthesis. The localization, translation and degradation of mRNA requires the recruitment of trans-acting factors to the transcript, most commonly achieved through proteins binding to the 5′ and 3′ untranslated regions (UTRs). Once localized, initiation of translation involves scaffold factors and ribosomes being recruited to the mRNA by the 5′ cap and a sufficiently long 3′ poly(A) tail.5 Multiple mechanisms exist that can regulate when and where the recruitment of these factors required for translation takes place.6
One of the most common mechanisms is the packaging of transcripts into ribonucleoprotein complexes (RNPs) together with RNA-binding proteins that prevent the recruitment of the initiation machinery.6 RNPs serve to localize the mRNA through connection with the cytoskeleton. After localization is completed, translation is usually enabled through de-repression6 as exemplified by ASH1 mRNA in budding yeast and β-actin mRNA in chick fibroblasts.
During division of the budding yeast Saccharomyces cerevisiae, ASH1 mRNA is localized to the tip of the future daughter cell where the protein product acts to repress mating type switching.7 Translation of ASH1 mRNA is repressed in the mother cell through the binding of Puf6, inhibiting eIF5B function, thus blocking ribosome recruitment.8 In the daughter cell, ASH1 mRNA is de-repressed through CK2-dependent phosphorylation of Puf6, leading to localized translation.8
A similar mechanism functions at the leading edge of cultured migrating chick fibroblasts. Within the 3′ UTR of β-actin mRNA, the ‘zip-code’ sequence is bound by zip-code binding protein 1 (ZBP1), which acts to both localize and repress the translation of the mRNA.9 Once at the leading edge, the spatially restricted activity of Src kinase results in a change of ZBP1 binding affinity, allowing for localized β-actin translation.10
In the Drosophila egg chamber, localized translation of gurken/TGF-α (grk) mRNA is required for setting up the future body axes.11-13 During mid-oogenesis (stages 7-10a), grk mRNA is transcribed and packaged into RNPs in the nurse cells, support cells that are interconnected with each other and the developing oocyte.14 These grk mRNA containing RNPs are actively transported by the molecular motor dynein on microtubules to the dorsoanterior corner,15,16 where translation and subsequent secretion of Grk protein patterns the dorsoventral axis.13,17
Translational repression of grk mRNA in the nurse cells was recently shown by genetic analysis to not require any previously described repressors.3 Instead, the absence of the translational activator oo18-RNA binding protein (Orb), the Drosophila homolog of cytoplasmic polyadenylation element binding protein (CPEB) in the nurse cells prevents grk translation.3 Although there is no direct evidence of Orb binding grk mRNA, examination of the 3′ UTR shows that there are 6 weakly conserved cytoplasmic polyadenylation elements (CPEs) and in orb alleles that still allow development through mid-oogenesis, grk mRNA is not fully polyadenylated.1 The orb auto regulatory loop, dependent on an eIF4E binding protein (4E-BP) and dFMR1, the Drosophila homolog of Fragile X Mental Retardation Protein, functioning in the nurse cells to restrict Orb protein only to the oocyte.18-22 This absence of an activator supports a model for grk translational control where in the nurse cells, grk mRNA is synthesized and then exported to the oocyte with a short poly(A) tail maintaining translational repression until the mRNA is localized at the dorsoanterior corner. Sufficient levels of Orb at the dorsoanterior corner are able to direct cytoplasmic polyadenylation, resulting in localized activation of grk translation.1,3 This polyadenylation is thought to be achieved by the Drosophila GLD-2 homolog Wispy, as the loss of cytoplasmic polyadenylation in orb mutant egg chambers is enhanced further by mutations in wispy.1
At the same time as localized grk translation, bicoid (bcd) mRNA, the anterior determinant, is localized along the anterior margin of the oocyte and partially overlaps with grk mRNA23-25 However, bcd mRNA remains translationally silent during oogenesis. This differential control of translation is regulated through association with Processing bodies (P bodies),2 electron dense, non-membrane bound structures, conserved in eukaryotes, that do not support translation through a lack of ribosomes.
At the dorsoanterior corner, where both bcd and grk mRNA are localized, the core of P bodies are enriched in translational repressors, while the periphery or edge (70nm in width) is enriched for the translational activator Orb.2 bcd mRNA is concentrated 6-fold at the P body core, whereas at the outer edge, grk mRNA is 23-times more concentrated.2 This leads to a model whereby the edge of the P body (where grk mRNA is localized) is able to support translation through access to Orb and ribosomes, while the core (where bcd mRNA is localized) is not.2 Compared to the oocyte, nurse cell P bodies do not associate with grk mRNA and have significantly reduced levels of Orb.3 Overexpression of Orb in the nurse cells results in grk mRNA and Orb associating with the edge of P bodies and mis-expression of Grk protein in the nurse cells.3 This result further supports that it is the restricted access to Orb that maintains grk mRNA repression in the nurse cells.3
While access to Orb explains the lack of translational activation in the nurse cells, within the oocyte, Orb protein is not localized solely at the dorsoanterior corner.3 In addition to this, unlocalized grk is not polyadenylated suggests that there is a mechanism regulating the activity of Orb.1 Localizing the activity of Orb to the dorsoanterior corner could be achieved by phosphorylation, a conserved way of regulating CPEB function.26-28 Supporting this model is the presence of multiple phospho-isoforms of Orb in the egg chamber, which are altered or lost in mutants for either the catalytic or regulatory subunit of the Ser/Thr kinase CK2.4 In wild-type oocytes, hyper-phosphorylated Orb associates with Wispy, an homologous interaction that occurs in the oocytes of Xenopus laevis.4,28,29 These observations, together with data showing that partially reducing CK2 activity in orb egg chambers enhances dorsoventral polarity defects,4 suggests that CK2 is phosphorylating Orb, and that this is required for grk translation.
Here we present new data to suggest that the localized translation of grk mRNA at the dorsoanterior corner is achieved through the spatial restriction of CK2 activity. This leads to phosphorylated Orb associating with grk mRNA at the P body edge where it directs cytoplasmic polyadenylation leading to Grk protein synthesis. We also take this opportunity to review the literature and present a working model for grk mRNA translation, highlighting components conserved with other systems.
Results
CK2 activity is restricted to the oocyte and co-localizes with active Orb
CK2 is a highly promiscuous enzyme, with a consensus site that has been hard to define. Various studies have shown that phosphorylation by CK2 is dictated by multiple acidic residues mostly downstream of the acceptor amino acid where n+3 is the most crucial.30 A minimum consensus of S/T-X-X-E/D has been suggested, with the second most important residue (n+1), also normally acidic.30 To investigate whether CK2 phosphorylates Orb, an antibody which recognizes sites phosphorylated by CK2: pS/pT-D-X-E (where p represents a phosphorylated residue) was used in combination with an Orb antibody (see Methods).
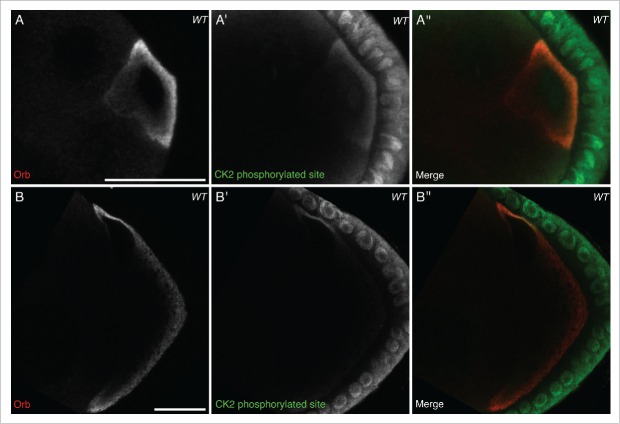
At stage 6, when grk mRNA is known to be localized at the posterior of the oocyte, Orb is present in the same pattern as grk mRNA and sites phosphorylated by CK2 (Fig. 1A-A″). Grk, secreted at the posterior of the oocyte, binds Torpedo (Top) receptors on the overlying follicle cells. This Grk-Top signaling event polarises the egg chamber23,31 and leads to the oocyte nucleus being pushed to the future dorsoanterior corner.32 grk mRNA becomes re-localized in a characteristic crescent at the dorsoanterior corner and then translated.11 At stage 8, while Orb is present throughout the oocyte (Fig. 1B) CK2 activity appears to be largely restricted to the dorsoanterior corner (Fig. 1B″), where grk mRNA is known to be translated.12 During both grk mRNA translation events, CK2 activity seems to be restricted only to the oocyte, with very little signal in the nurse cells (Fig. 1A′ and B′). Together with genetic data where ck2 mutant alleles in combination with orb mutants results in dorsoventral defects in the oocyte,4 our data suggests that CK2 could be acting to phosphorylate Orb at the dorsoanterior corner.
Figure 1.
CK2 phosphorylation is restricted to the oocyte and overlaps with active Orb. Double immunofluorescent staining of wild type (OregonR) egg chambers for Orb protein (red) and phosphorylated CK2 consensus sites (pS/pT-D-X-E, green) at stage 6 (A-A″) and stage 8 (B-B″) of oogenesis. (A), (B) Orb protein is localized throughout the developing oocyte. (A′) In stage 6 oocytes, sites phosphorylated by CK2 are present mainly at the posterior of the oocyte, between the nucleus and membrane, where localized grk translation is known to occur. There is minimal signal in the nurse cells, but all follicle cells show intense staining (n = 20/20). (B′) In stage 8 oocytes, sites phosphorylated by CK2 are present almost exclusively at the dorsal-anterior corner, where grk mRNA is locally translated. Again there is little or no signal in the nurse cells and all follicle cells show intense staining (n = 20/20). (A″), (B″) Merge of the two stains showing that CK2 phosphorylated sites overlap with Orb protein at sites of localized grk translation. Single slices, scale bar 20 μm.
CK2 activity overlaps with Grk protein
The functional CK2 enzyme is a hetero-tetramer composed of 2 catalytic subunits (α) and 2 regulatory subunits (β).33 When either of these subunits is mutated to eliminate function (even when heterozygous) there is a change in the phosphorylation state of Orb.4 There is also an increase in the percentage of dorsalised chorions in mature oocytes,4 characteristic of a grk mutant phenotype.
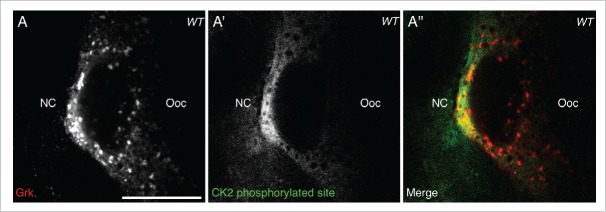
Having shown that CK2 activity overlaps with active Orb at the dorsoanterior corner (Fig. 1B-B″), it would be expected that if CK2-dependent phosphorylation of Orb is directing localized grk translation, Grk protein would also overlap with CK2 phosphorylation. Double staining for CK2 phosphorylated targets and Grk protein in stage 8 egg chambers shows that secreted Grk protein overlaps with the localized activity of CK2 (Fig. 2), further implicating a role for CK2 in directing localized grk translation.
Figure 2.
CK2 phosphorylation overlaps with Grk protein. Double immunofluorescent staining of wild-type (OregonR) egg chambers for Grk protein (red) and phosphorylated CK2 consensus sites (pS/pT-D-X-E, green) at stage 8 of oogenesis nurse cells (NC), oocyte (Ooc). (A) Grk protein is present at the dorsal-anterior corner of stage 8 oocytes (n = 15/15). (A′) CK2 phosphorylated sites are present at the dorsal-anterior corner and in the surrounding follicle cells, but absent from nurse cell cytoplasm and other regions of the oocyte (n = 15/15). (A″) Merge of the two stains showing that phosphorylated CK2 sites overlap with Grk protein secretion (n = 15/15). Single slices, scale bar 20 μm.
CK2 activity co-localizes with ectopically expressed Orb in nurse cells
The access to Orb in the nurse cells adjacent to the developing oocyte is restricted through the function of the orb auto-regulatory loop which confines the majority of Orb protein to the oocyte through the action of Cup and dFMR1 in the nurse cells.18,19,22 This repression is thought to be saturated when orb is over-expressed in the nurse cells since UAS-Orb egg chambers have ectopic Orb present in the nurse cells in a similar manner to that of cup mutants.3 The overexpressed Orb is present in puncta at the edge of P bodies, along with grk mRNA which undergoes ectopic translation.3 If CK2 activity is required to cause localized activation of Orb at the dorsoanterior corner in wild-type egg chambers, we hypothesized that in UAS-Orb egg chambers, the over-expressed Orb in the nurse cells would be phosphorylated by CK2.
Orb over-expression, as previously described,3 results in Orb present in nurse cells in large foci and at a significantly higher level compared to wild-type egg chambers (compare Fig. 3A to Fig. 3B).3 We find that CK2 phosphorylation is present in the nurse cells of UAS-Orb egg chambers, whereas in wild-type nurse cells, CK2 activity does not appear to be present (compare Fig. 3B′ to Fig. 3A′). Moreover, the CK2 phosphorylation is detected overlapping with foci of Orb (Fig. 3B″, white arrows). Together with Davidson et al., these data are consistent with a model where grk mRNA translation is directed through the localized activity of Orb at the edge of P bodies, achieved by localized CK2 activity.
Figure 3.
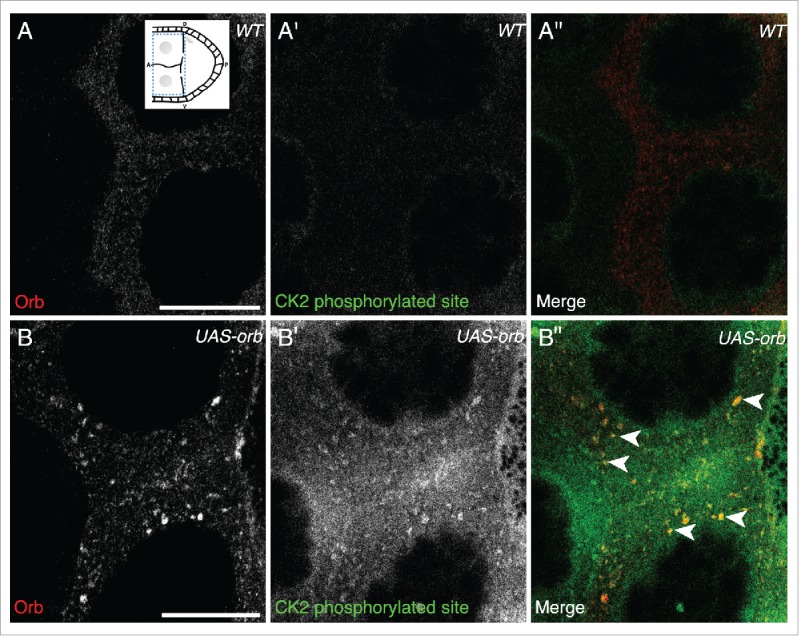
In UAS-Orb chamber, ectopically expressed Orb is phosphorylated by CK2. Double immunofluorescent staining of wild type (A-A″) and tubGal4-VP16; UAS-Orb (B-B″) egg chambers for Orb protein (red) and phosphorylated CK2 consensus sites (pS/pT-D-X-E, green). Inset: schematic illustrating the relative position of the oocyte and nurse cells for a stage 8 egg chamber, dashed blue box highlights area focused on in both (A-A″) and (B-B″). (A-A″) In the nurse cells of wild-type egg chambers, Orb protein is at a low level and CK2 activity is below the level of detection (n = 12/12). (B) tubGal4-VP16; UAS-Orb egg chambers, have ectopically expressed Orb in foci of varying sizes throughout the nurse cells (n=12/12). (B′) Phosphorylated CK2 consensus sites are visible in tubGal4-VP16; UAS-Orb egg chambers (n = 12/12). (B″) Merge of 2 stains showing that ectopically expressed Orb protein in the nurse cells is phosphorylated by CK2 (white arrowheads). Single slices, scale bar 20 μm.
Working model for grk mRNA translational control
Together with previous biochemical data and genetic interaction studies,4 our data suggests that CK2 is phosphorylating Orb protein at the dorsoanterior corner during mid-oogenesis. It appears that phosphorylation acts to direct Orb to the edge of P bodies, where grk mRNA is localized, resulting in translation. Coupled with previous work in this field, we suggest an updated model for the translational regulation of grk mRNA (Fig. 4).
Figure 4.
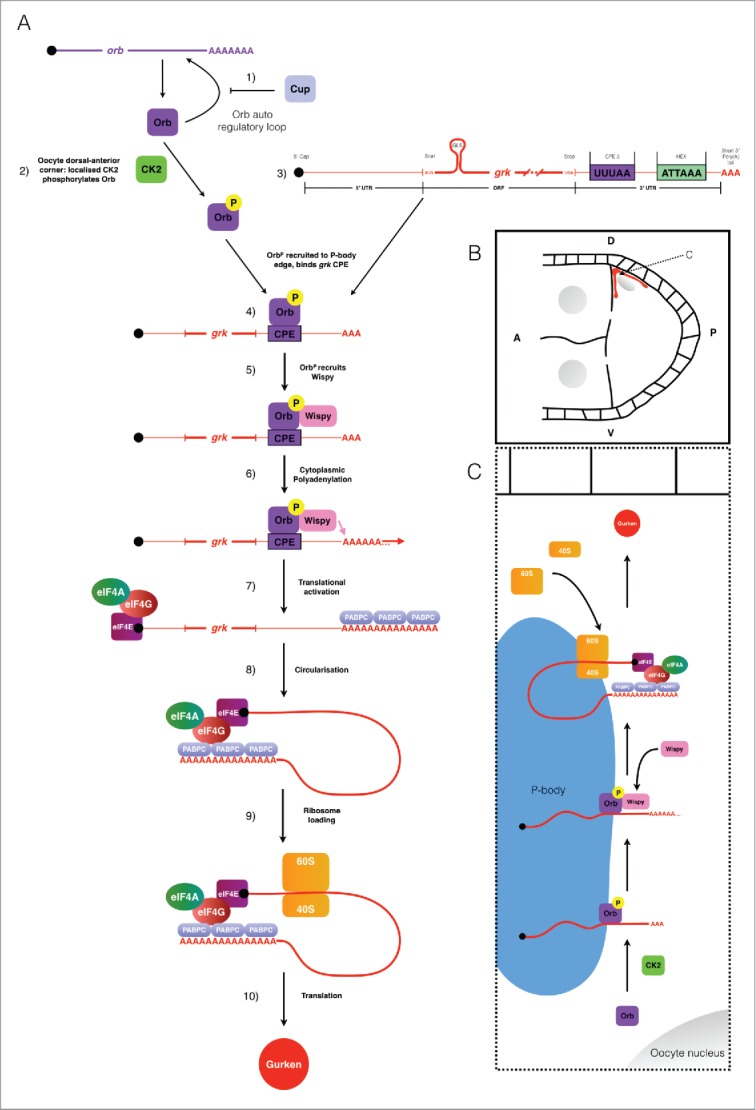
Schematic collating data from this and other studies (references refer to the text) to present a model for the translational control of grk mRNA during stage 8 of oogenesis. (A) 1) orb mRNA translation is repressed in the nurse cells though the action of Cup.3,18,19 Orb is present throughout the oocyte, but is locally phosphorylated by CK2 at the dorsal-anterior corner. 3) Also present at the dorsal-anterior corner is grk mRNA, localized by the gurken localization signal (GLS). 4) Phosphorylated Orb binds grk mRNA, presumably at one or many of the 6 weak CPE consensus sites present in the mRNAs 3′ UTR (only the strongest CPE is shown: CPE3).1 5) Phosphorylated Orb is able to recruit the Drosophila homolog of the GLD-2 cytoplasmic polymerase Wispy1,4,29. 6) Wispy catalyses the addition of around 60As to the 3′ tail of grk mRNA extending it's length from 30 to 90As.1 7) Cytoplasmic polyadenylation of grk mRNA results in translational activation: canonical translation initiation machinery binds the mRNA. 8) The mRNA is circularized through interactions between eIF4G and PABPC for efficient translation. 9) The ribosome is loaded onto the mRNA. 10) Translation of grk mRNA generates Grk protein. (B) Stage 8 egg chamber with Anterior (A) left and Dorsal (D) up. Grk protein (red) is synthesized only at the dorsal-anterior corner, in a characteristic crescent between the oocyte nucleus (grey) and oocyte membrane. (C) Model for the mechanism of grk translational control at the dorsal-anterior corner. grk translational activation occurs at the edge of P-bodies2 at the dorsal-anterior corner of the oocyte. Phosphorylation of Orb by CK2 directs the CPEB homolog to the edge of P-bodies where grk mRNA is localised.2 Orb then directs cytoplasmic polyadenylation resulting in translational activation of grk mRNA (see (A) for mechanistic detail).
In nurse cells the levels of the Orb protein are very low due to the action of the orb auto-regulatory loop (1, Fig. 4A).18,19 This lack of Orb prevents grk mRNA translation in the nurse cells.3 In the oocyte, Orb protein is present at a significantly higher level (2, Fig. 4A).3 Orb is only targeted to the edge of P bodies at the dorsoanterior corner due to localized CK2-mediated phosphorylation (2, Fig. 4A & C).2,3 Phosphorylated Orb binds to localized grk mRNA, also present at the P body edge (3, 4, Fig. 4A), likely through one or more of the 6 weak CPE consensus sites in the 3′ UTR (3, 4, Fig. 4A & C, only the strongest consensus site, CPE3 is shown).1-3 Phosphorylated Orb interacts with Wispy1,4,29 (5, Fig. 4A & C) which polyadenylates grk mRNA (6, Fig. 4A & C).1 The grk mRNA poly(A) tail increases in length from 30As to 90As1 (7, Fig. 4A). The sufficiently long poly(A) tail of grk mRNA is then bound by the canonical translation initiation machinery leading to ribosome recruitment (8, 9, Fig. 4A) thus resulting in localized Grk protein production at the dorsoanterior corner (10, Fig. 4A, B and C).
Discussion
CPE-mediated translational control
Here we have shown similarities to other systems that regulate CPEB homologs to direct the cytoplasmic polyadenylation of specific mRNAs. Cytoplasmic polyadenylation allows for both spatial and temporal control of protein synthesis, thereby separating translation from nuclear transport.
This method of translational control is critical in oocytes as these specialized cells are biologically “paused” with no transcription or translation for extended periods of time, in extreme examples, years. The silenced state of many maternal mRNAs with a short poly(A) tail is reversed by cytoplasmic polyadenylation at one of 2 major developmental events: oocyte maturation or the oocyte-to-embryo transition.
Oocyte maturation occurs following the main phase of growth and differentiation of the oocyte. Maturation allows for the co-ordination of the completion of meiosis with fertilisation. During this event, the cell cycle machinery is re-activated by the translation of a subset of mRNAs. This results in an active CDK-CyclinB complex which initiates and maintains a second pause in the cell cycle at metaphase until egg activation.34
Typically coupled with fertilisation, egg activation releases the egg from the pause in metaphase established at maturation and is associated with a transient rise in the concentration of intracellular calcium (Ca2+) levels.35 Egg activation is part of the oocyte-to-embryo transition, when the mature, quiescent oocyte receives the necessary cues to form the totipotent, rapidly dividing early embryo.
While the role for cytoplasmic polyadenylation was first described at egg activation in sea urchin eggs,36 the majority of research on how cytoplasmic polyadenylation is regulated has been undertaken in Xenopus oocytes. This work has focused on the steps required to re-activate the translation of 2 different subsets of mRNAs at oocyte maturation, the passage from meiotic prophase I to metaphase II.
RNA injection experiments in maturing Xenopus oocytes revealed a cis-element within mRNAs known to undergo cytoplasmic polyadenylation, termed the cytoplasmic polyadenylation element (CPE). The CPE is U-rich and roughly octameric, with a consensus of U4-5A1-3U.37,38 To determine the potential CPE binding protein, radio-labeled CPE-containing mRNA was incubated in Xenopus extracts and then irradiated to cross-link bound proteins for purification.38 This identified cytoplasmic polyadenylation element binding protein (CPEB), an RNA-binding protein essential for the cytoplasmic polyadenylation of CPE containing mRNAs.38 In vertebrates there are 4 CPEB proteins that share the conserved 2 RNA Recognition Motifs followed by a zinc finger domain in the C-terminus. For CPEB1, the originally isolated isoform, all 3 of these domains are required for RNA binding.38 While the N-terminus is widely divergent between species, CPEB is evolutionary conserved, and has been identified in the worm, fly, fish, mouse, frog, a clam, a sea slug and human.39
The regulation of CPEB binding to CPE and subsequent recruitment of the poly(A) machinery must be regulated since CPEB is present in the cytosol along with CPE-containing mRNAs. This is most commonly achieved through post-translational modification, a wide-spread mechanism in post-transcriptional control.
Xenopus CPEB1 has been shown to undergo a number of phosphorylation events during oocyte maturation. Progesterone signaling from the surrounding follicle at maturation activates Eg2 (related to Aurora A Kinase), part of the events to co-ordinate further oocyte development.40,41 Phosphorylation of CPEB1 at S174 by Eg2 strengthens the interaction between CPEB1 and xGLD-2 (the Xenopus homolog of the cytoplasmic poly(A) polymerase),28 leading to poly(A) tail extension of a subset of mRNAs and subsequent translation. Later during maturation, a second phosphorylation event, which is dependent on CDK-CyclinB, occurs at S210 of CPEB1.42 This phosphorylation targets most but not all of CPEB1 for proteolytic degradation and drastically reduces the ratio of CPEB1 to CPEs in the cytoplasm. This is necessary to allow a second subset of mRNAs to undergo translational activation in the final steps of oocyte maturation.43-45
Drosophila oocyte maturation reveals some conservation of mechanism with Xenopus. Cortex (Cort) is a distant member of the Cdc20 family of APC/C (Anaphase Promoting Complex/Cyclosome) specificity factors and is required for female meiosis.46,47 Cort functions through binding APC/C (forming APC/CCort) to direct the proteolytic activity ensuring that the cell cycle progresses correctly.48 Cort is not detectable during early oogenesis, but is present at the beginning of maturation (stage 13 of oogenesis) and remains active until egg activation.34,49 The appearance of Cort and the activity of APC/CCort at maturation coincides with both the cytoplasmic polyadenylation of cort mRNA and the degradation of CyclinA, which is necessary for entry into metaphase.49,50
The cytoplasmic polyadenylation of cyclinB (cycB) mRNA occurs concurrently with cort mRNA.49 APC/CCort at maturation targets CyclinA. At this time, CycB and Securin are not targeted and can maintain the mature Drosophila oocyte in metaphase I49. For both cycB mRNA and cort mRNA, cytoplasmic polyadenylation is dependent on Wispy.29 Altogether, this demonstrates the high level of conservation involved in cytoplasmic polyadenylation at oocyte maturation for Drosophila and Xenopus. However, it has not been shown if Drosophila cort and cycB mRNA possess CPEs.
Cytoplasmic polyadenylation is not only limited to the germ-line. In mouse neurons, Calmodulin-dependent Kinase II (CaMKII) dependent phosphorylation of CPEB at T171 (a site highly similar to Xenopus CPEB1 S174) is able to regulate translational activation.26 This co-ordination of translation of CPE containing mRNAs in response to increased intracellular calcium facilitates synaptic plasticity.26 CaMKII mRNA is also known to undergo cytoplasmic polyadenylation mediated by Aurora kinase-catalyzed CPEB phosphorylation at synapses in response to external cues.27
CPEB homologs have also been implicated in disease through their role in directing localized translation. CPEB1 in mammary glands is required to correctly localize mRNAs encoding polarity proteins that are required for the construction of tight junctions. CPEB1 absence in these cells results in the loss of apico-basal polarity,51 which can lead to the epithelial-to-mesenchymal transition (EMT) resulting in metastasis.52 Depletion of CPEB1 can also lead to cancer progression by directly altering poly(A) tail lengths of EMT/metastasis-related mRNAs.53 One example, Matrix metalloproteinase 9 (MMP9) mRNA, encodes a metastasis-promoting factor54 and in CPEB1 depleted cells, there is an increase in the poly(A) tail length of MMP9 that results in translational activation.53
grk mRNA as a model for CPE-mediated translational control
The regulation of Orb in grk translational control appears to be very distinct from that of CPEB1 regulation in Xenopus oocytes. In Xenopus, CPEB1 is believed to be weakly associated with mRNA prior to maturation and Eg2 phosphorylation strengthens this and other interactions, with factors required for cytoplasmic polyadenylation such as xGLD-228. However, we have demonstrated that phosphorylation of the CPEB homolog Orb at the dorsoanterior corner is important to localize Orb with grk mRNA to direct cytoplasmic polyadenylation at the edge of P bodies. Therefore for grk translational control, phosphorylation of Orb appears to regulate the activity of the protein spatially, rather than temporally. This reflects a key difference in the mRNAs undergoing translational control in these systems, Grk protein must be secreted in the correct place for patterning, however for mRNAs in maturing Xenopus oocytes, the polyadenylation must occur at the right time to ensure cell cycle reactivation.
This localized translation of grk mRNA does resemble the translational control of ASH1 mRNA in budding yeast and β-actin mRNA in migrating chick fibroblasts. In both these examples, the activity of a kinase is localized and required to initiate the translation of these mRNAs in the correct sub-cellular region.8,10 Contrastingly to mRNAs undergoing translational activation in the maturing Xenopus oocyte, their spatial control is much more important than their temporal control.
What is particularly interesting is the conserved use of localized CK2 activity between Drosophila and Saccharomyces cerevisiae, despite the two systems being separated by millions of years of evolution. CK2-GFP in yeast is present throughout the dividing cell, but localizes strongly in the cortex of the future daughter cell, prior to the expression of ASH1 mRNA.8
However, this presents a further set of questions: how is CK2 activity localized first at the posterior and then relocalized at the dorsoanterior corner for dorsoventral axis establishment? How is CK2 activity restricted to the oocyte only, given that in UAS-Orb egg chambers CK2 phosphorylates the ectopically expressed Orb? One simple explanation is that CK2 is active in the whole egg chamber but due to the low levels of Orb in the nurse cells, the signal is not visible and ultimately no grk translation occurs. Thus the spatial restriction of Orb to the oocyte is key to regulating grk mRNA translational control.
The role of the orb auto-regulatory loop in grk translational control
During the early stages of oogenesis, Orb is critical for the specification of the oocyte.55 Orb preferentially accumulates in the oocyte due to an auto-regulatory loop where Orb is required to localize its own mRNA.18 Concurrently, factors act in the nurse cells to prevent incorrect activation of the auto-regulatory loop. One key repressor is Cup, a 4E-BP.19 In cup mutants the orb auto-regulatory loop is not confined to the oocyte, resulting in high levels of Orb protein in the nurse cells.3,19 Our previous work has shown that this loss of Orb restriction solely to the oocyte is sufficient to result in the ectopic translation of grk mRNA in nurse cells that leading to dorsoventral defects.3 Together our data suggest an indirect role for Cup in the translational control of grk mRNA.3 This is in contrast to previous work, prior to the full elucidation of the orb auto-regulatory loop, which suggested a direct role for Cup in bind grk mRNA.56 Therefore, with our UAS-Orb data presented here, and based on previous work, a complete understanding of the orb auto-regulatory loop is important to understand how grk translational control is achieved in the developing oocyte.
In cup mutants, orb mRNAs have longer poly(A) tails and the majority of Orb protein is hyper-phosphorylated.19 However, the role of Cup as a 4E-BP does not seem important for regulating the translation of orb, as specific loss of the eIF4E binding domain in cupΔ212 does not have a strong effect on dorsoventral polarity as would be expected if this were the sole role of Cup (Davidson, unpublished). This would suggest that there are functions for Cup other than eIF4E sequestration that restrict the orb auto-regulatory loop to the oocyte. One potential mechanism by which Cup could be acting to repress orb translation in the nurse cells is through promoting the recruitment of the deadenylation machinery to the mRNA.57
In both cup and UAS-Orb egg chambers, Grk is expressed along the margins of nurse cells although less severely in UAS-Orb.3 In UAS-Orb nurse cells, we have shown that Orb co-localizes with CK2 phosphorylated sites (Fig. 3) and previously shown that this ectopic Orb is present at the edge of nurse cell P bodies resulting in grk translation.3 The phosphorylation of Orb by CK2 in the nurse cells when Orb is overexpressed, would fit with the increased amount of hyper-phosphorylated Orb in cup mutants.19 Together this would suggest that Cup-mediated repression of orb translation in the nurse cells is not only directly important for oocyte specification, but also indirectly for the translational control of grk mRNA.
Loss of Orb repression does not fully explain the cup phenotype as in a particularly strong allele, cup5, grk mRNA is less efficiently localized.56 Another component shown to be involved in the control of grk translation is the heterogeneous nuclear ribonucleoprotein (hnRNP) Squid (Sqd).58-60 Sqd and Cup have been shown to interact through both biochemical and genetic analysis where cup mutants enhance the dorsalised phenotype of sqd homozygotes.56 However, whether this enhancement of sqd by cup is specifically due to loss of function of Cup independent of the role in the orb auto-regulatory loop remains to be fully investigated.
sqd1 mutants show Grk protein expressed along the entire anterior margin of the developing oocyte.61 How specifically Sqd functions remains unclear, particularly as this loss of sqd does not lead to ectopic expression in the nurse cells.3 Loss of sqd does result in a loss of grk mRNA enrichment to the dorsoanterior corner and leads to ectopic translation along the anterior margin.3,58,61 This suggests that there is a mechanism involved in controlling the translational activation of grk mRNA in the oocyte that is dependent on Sqd. In light of the data on the role of Cup in the orb auto-regulatory loop, it seems that the roles of Sqd and Cup in grk translational control in the oocyte need to be re-considered.
The role of Bruno in grk mRNA translational control
Over expression of the grk locus further suggests that there is not a role for a translational repressor or grk mRNA in the nurse cells.2,3 In UAS-grk egg chambers, there is no ectopic grk translation in the nurse cells, suggesting that there is not a saturatable binding of repressors that occurs in the nurse cells to repress grk translation.3 In these grk over-expression egg chambers, previous works shows that some excess grk mRNA is localized to the repressive P body core.2 There is, however, some Grk overexpression as dorsoventral patterning defects, characteristic of a failure in grk translational control are observed.2,3
Previously a number of studies had implicated Bruno (Aret) in the translational repression of grk mRNA in the oocyte.56,61-63 Over-expression of Bruno does not affect the localization of grk mRNA, but does result in loss Grk protein.62 Further evidence in support of Bruno-mediated repression showed that Bruno bound to the 3′ UTR of grk mRNA.62 This is at odds with recent imaging data, where in different aret mutants, grk mRNA is not ectopically translated in the nurse cells or the oocyte.3 Interestingly, Bruno has been shown to associate with presumed non-active hypo-phosphorylated Orb,4 which we suggest is only present at low levels at the dorsoanterior corner, while previous work has shown Bruno is enriched at the dorsoanterior corner.64 One potential explanation is that Bruno may function as a redundant repressor for the translational control of grk mRNA in the oocyte. Bruno would not be required in the nurse cells, due to the low levels of Orb. Complementing direct, in vivo vizualization and genetic approaches should begin to unravel these conflicting observations.
Future directions
Combining our work with others, we present a model of grk mRNA translational control that is dependent upon the localized activity of CK2 at the dorsoanterior corner of the mid-stage oocyte (Fig. 4). The observation of the localization of CK2 itself during the two periods of grk translation would further validate our own imaging and other genetic and biochemical data. Generation of a GFP-tagged CK2 would allow for vizualization of the location of the enzyme. This could also be used to follow how CK2::GFP is localized over the course of oogenesis to direct grk translation. Examining CK2-GFP localization in nurse cells would also confirm our suggestion that CK2 is present and active in the nurse cells, but there is little Orb protein for the kinase to phosphorylate, due to the activity of Cup in the orb auto-regulatory loop.
The model also draws a number of similarities between the regulation of the poly(A) machinery in maturing Xenopus oocyte. Particularly, the phosphorylation of Orb by CK2 recruits Wispy to grk mRNA at the edge of P bodies.4,29 Direct vizualization of the interactions between Wispy and grk mRNA in wild-type and ck2 backgrounds would begin to uncover the mechanism by which Wispy is recruited to grk mRNA.
This work continues to highlight the Drosophila egg chamber as a powerful model for elucidating mechanisms of mRNA translational control. The proposed model demonstrates that post-translational modification of CPEB homologs is a conserved mechanism, not only for temporal, but also spatial control of cytoplasmic polyadenylation.
Methods
Fly strains
Stocks were raised on standard cornmeal-agar medium at 25°C. The wild type was Oregon R (OrR). Transgenic lines: maternal tubulin driver TubulinGal4-VP16; and UASp-orb (Li et al., 2014).
| Short name | X | 2 | 3 | Reference/Bloomington Number |
|---|---|---|---|---|
| UAS-Orb | + | UASp-Orb/CyO | + | Siegfried Roth |
| tubGAL4 | TubulinGAL4-VP16 | Sco/CyO | + | Siegfried Roth |
Fixation of oocytes
For all fixed oocyte immunofluorescence, the procedure was modified from Davidson et al. 2016.
Antibodies
Primary antibodies: Orb 4H8 (DSHB), mouse (1:40); Grk 1D120c (DSHB), mouse (1:300); Sites phosphorylated by CK2 (CST #8738), rabbit (1:200). Secondary antibodies: Donkey-647 (1:200); Goat-Cy3 (1:200).
Imaging
Images were acquired using a Leica SP5 inverted scanning confocal microscope with a 100x, 1.4NA oil immersion objective with sequential settings.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
Richard Parton, Paul Conduit and Tudor Fulga for their comments on the manuscript. Alex Davidson, Richard Parton, Ilan Davis and Catherine Rabouille for their initial input into the project. Nan Hu for assistance with drosophila stocks. Matt Wayland for microscopy assistance.
Funding
The University of Cambridge, Isaac Newton Trust/Wellcome Trust ISSF Research Grants (grant 097814 to T.T.W.).
References
- [1].Norvell A, Wong J, Randolph K, Thompson L. Wispy and Orb cooperate in the cytoplasmic polyadenylation of localized gurken mRNA. Dev Dyn Off Publ Am Assoc Anat 2015; 244:1276-1285. [DOI] [PubMed] [Google Scholar]
- [2].Weil TT, Parton RM, Herpers B, Soetaert J, Veenendaal T, Xanthakis D, Dobbie IM, Halstead JM, Hayashi R, Rabouille C, et al.. Drosophila patterning is established by differential association of mRNAs with P bodies. Nat Cell Biol 2012; 14:1305-1313; PMID:23178881; http://dx.doi.org/ 10.1038/ncb2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Davidson A, Parton RM, Rabouille C, Weil TT, Davis I. Localized translation of gurken/TGF-α mRNA during axis specification is controlled by access to Orb/CPEB on processing bodies. Cell Rep 2016; 14:2451-2462; PMID:26947065; http://dx.doi.org/ 10.1016/j.celrep.2016.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wong LC, Costa A, McLeod I, Sarkeshik A, Yates J 3rd, Kyin S, Perlman D, Schedl P, et al.. The functioning of the drosophila CPEB protein Orb is regulated by phosphorylation and requires casein kinase 2 activity. PLoS One 2011; 6:e24355; PMID:21949709; http://dx.doi.org/ 10.1371/journal.pone.0024355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jackson RJ, Hellen CU. T, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 2010; 11:113-127; PMID:20094052; http://dx.doi.org/ 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat Rev Mol Cell Biol 2008; 9:971-980; PMID:19023284; http://dx.doi.org/ 10.1038/nrm2548 [DOI] [PubMed] [Google Scholar]
- [7].Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen RP. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 1997; 277:383-387; PMID:9219698; http://dx.doi.org/ 10.1126/science.277.5324.383 [DOI] [PubMed] [Google Scholar]
- [8].Deng Y, Singer RH, Gu W. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev 2008; 22:1037-1050; PMID:18413716; http://dx.doi.org/ 10.1101/gad.1611308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Condeelis J, Singer RH. How and why does beta-actin mRNA target? Biol Cell Auspices Eur Cell Biol Organ 2005; 97:97-110. [DOI] [PubMed] [Google Scholar]
- [10].Hüttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature 2005; 438:512-515; PMID:16306994; http://dx.doi.org/ 10.1038/nature04115 [DOI] [PubMed] [Google Scholar]
- [11].Neuman-Silberberg FS, Schüpbach T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell 1993; 75:165-174; PMID:7691414; http://dx.doi.org/ 10.1016/S0092-8674(05)80093-5 [DOI] [PubMed] [Google Scholar]
- [12].Roth S. The origin of dorsoventral polarity in Drosophila. Philos Trans R Soc Lond B Biol Sci 2003; 358:1317-1329; PMID:14511478; http://dx.doi.org/ 10.1098/rstb.2003.1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Van Buskirk C, Schüpbach T. Versatility in signalling: multiple responses to EGF receptor activation during Drosophila oogenesis. Trends Cell Biol 1999; 9:1-4; PMID:10087609; http://dx.doi.org/ 10.1016/S0962-8924(98)01413-5 [DOI] [PubMed] [Google Scholar]
- [14].Cáceres L, Nilson LA. Production of gurken in the nurse cells is sufficient for axis determination in the Drosophila oocyte. Dev Camb Engl 2005; 132:2345-2353. [DOI] [PubMed] [Google Scholar]
- [15].MacDougall N, Clark A, MacDougall E, Davis I. Drosophila gurken (TGFalpha) mRNA localizes as particles that move within the oocyte in two dynein-dependent steps. Dev Cell 2003; 4:307-319; PMID:12636913; http://dx.doi.org/ 10.1016/S1534-5807(03)00058-3 [DOI] [PubMed] [Google Scholar]
- [16].Delanoue R, Herpers B, Soetaert J, Davis I, Rabouille C. Drosophila Squid/hnRNP helps Dynein switch from a gurken mRNA transport motor to an ultrastructural static anchor in sponge bodies. Dev Cell 2007; 13:523-538; PMID:17925228; http://dx.doi.org/ 10.1016/j.devcel.2007.08.022 [DOI] [PubMed] [Google Scholar]
- [17].Huynh J-R, St Johnston D. The origin of asymmetry: early polarisation of the Drosophila germline cyst and oocyte. Curr Biol CB 2004; 14:R438-449; PMID:15182695; http://dx.doi.org/ 10.1016/j.cub.2004.05.040 [DOI] [PubMed] [Google Scholar]
- [18].Tan L, Chang JS, Costa A, Schedl P. An autoregulatory feedback loop directs the localized expression of the Drosophila CPEB protein Orb in the developing oocyte. Dev Camb Engl 2001; 128:1159-1169. [DOI] [PubMed] [Google Scholar]
- [19].Wong LC, Schedl P. Cup Blocks the Precocious Activation of the Orb Autoregulatory Loop. PLOS One 2011; 6:e28261; PMID:22164257; http://dx.doi.org/ 10.1371/journal.pone.0028261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nelson MR, Leidal AM, Smibert CA. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J 2004; 23:150-159; PMID:14685270; http://dx.doi.org/ 10.1038/sj.emboj.7600026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell 2004; 6:69-78; PMID:14723848; http://dx.doi.org/ 10.1016/S1534-5807(03)00400-3 [DOI] [PubMed] [Google Scholar]
- [22].Costa A, Wang Y, Dockendorff TC, Erdjument-Bromage H, Tempst P, Schedl P, Jongens TA. The drosophila fragile X protein functions as a negative regulator in the orb autoregulatory pathway. Dev Cell 2005; 8:331-342; PMID:15737929; http://dx.doi.org/ 10.1016/j.devcel.2005.01.011 [DOI] [PubMed] [Google Scholar]
- [23].Roth S, Neuman-Silberberg FS, Barcelo G, Schüpbach T. cornichon and the EGF receptor signaling process are necessary for both anterior-posterior and dorsal-ventral pattern formation in Drosophila. Cell 1995; 81:967-978; PMID:7540118; http://dx.doi.org/ 10.1016/0092-8674(95)90016-0 [DOI] [PubMed] [Google Scholar]
- [24].Weil TT. mRNA localization in the Drosophila germline. RNA Biol 2014; 11:1010-1018; PMID:25482896; http://dx.doi.org/ 10.4161/rna.36097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Becalska AN, Gavis ER. Lighting up mRNA localization in Drosophila oogenesis. Dev Camb Engl 2009; 136:2493-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Atkins CM, Nozaki N, Shigeri Y, Soderling TR. Cytoplasmic polyadenylation element binding protein-dependent protein synthesis is regulated by calcium/calmodulin-dependent protein kinase II. J Neurosci 2004; 24:5193-5201; PMID:15175389; http://dx.doi.org/ 10.1523/JNEUROSCI.0854-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Huang Y-S, Jung M-Y, Sarkissian M, Richter JD. N-methyl-d-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and αCaMKII mRNA polyadenylation at synapses. EMBO J 2002; 21:2139-2148; PMID:11980711; http://dx.doi.org/ 10.1093/emboj/21.9.2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 2004; 119:641-651; PMID:15550246; http://dx.doi.org/ 10.1016/j.cell.2004.10.029 [DOI] [PubMed] [Google Scholar]
- [29].Benoit P, Papin C, Kwak JE, Wickens M, Simonelig M. PAP- and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Dev Camb Engl 2008; 135:1969-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB J 2003; 17:349-368; PMID:12631575; http://dx.doi.org/ 10.1096/fj.02-0473rev [DOI] [PubMed] [Google Scholar]
- [31].Price JV, Clifford RJ, Schüpbach T. The maternal ventralizing locus torpedo is allelic to faint little ball, an embryonic lethal, and encodes the Drosophila EGF receptor homolog. Cell 1989; 56:1085-1092; PMID:2493993; http://dx.doi.org/ 10.1016/0092-8674(89)90641-7 [DOI] [PubMed] [Google Scholar]
- [32].Zhao T, Graham OS, Raposo A, St Johnston D. Growing microtubules push the oocyte nucleus to polarize the Drosophila dorsal-ventral axis. Science 2012; 336:999-1003; PMID:22499806; http://dx.doi.org/ 10.1126/science.1219147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Litchfield DW. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem J 2003; 369:1-15; PMID:12396231; http://dx.doi.org/ 10.1042/bj20021469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Stetina JRV, Orr-Weaver TL. Developmental control of oocyte maturation and egg activation in metazoan models. Cold Spring Harb Perspect Biol 2011; 3:a005553; PMID:21709181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Stricker SA. Comparative biology of calcium signaling during fertilization and egg activation in animals. Dev Biol 1999; 211:157-176; PMID:10395780; http://dx.doi.org/ 10.1006/dbio.1999.9340 [DOI] [PubMed] [Google Scholar]
- [36].Slater DW, Slater I, Gillespie D. Post-fertilization synthesis of polyadenylic acid in sea urchin embryos. Nature 1972; 240:333-337; PMID:4570496; http://dx.doi.org/ 10.1038/240333a0 [DOI] [PubMed] [Google Scholar]
- [37].Bilger A, Fox CA, Wahle E, Wickens M. Nuclear polyadenylation factors recognize cytoplasmic polyadenylation elements. Genes Dev 1994; 8:1106-1116; PMID:7926790; http://dx.doi.org/ 10.1101/gad.8.9.1106 [DOI] [PubMed] [Google Scholar]
- [38].Hake LE, Richter JD. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell 1994; 79:617-627; PMID:7954828; http://dx.doi.org/ 10.1016/0092-8674(94)90547-9 [DOI] [PubMed] [Google Scholar]
- [39].Fernández-Miranda G, Méndez R. The CPEB-family of proteins, translational control in senescence and cancer. Ageing Res Rev 2012; 11:460-472; http://dx.doi.org/ 10.1016/j.arr.2012.03.004 [DOI] [PubMed] [Google Scholar]
- [40].Andrésson T, Ruderman JV. The kinase Eg2 is a component of the Xenopus oocyte progesterone-activated signaling pathway. EMBO J 1998; 17:5627-5637; http://dx.doi.org/ 10.1093/emboj/17.19.5627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Frank-Vaillant M, Haccard O, Thibier C, Ozon R, Arlot-Bonnemains Y, Prigent C, Jessus C. Progesterone regulates the accumulation and the activation of Eg2 kinase in Xenopus oocytes. J Cell Sci 2000; 113 (Pt 7):1127-1138; PMID:10704364 [DOI] [PubMed] [Google Scholar]
- [42].Mendez R. Differential mRNA translation and meiotic progression require Cdc2-mediated CPEB destruction. EMBO J 2002; 21:1833-1844; PMID:11927567; http://dx.doi.org/ 10.1093/emboj/21.7.1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Barkoff AF, Dickson KS, Gray NK, Wickens M. Translational Control of Cyclin B1 mRNA during meiotic maturation: Coordinated repression and cytoplasmic polyadenylation. Dev Biol 2000; 220:97-109; PMID:10720434; http://dx.doi.org/ 10.1006/dbio.2000.9613 [DOI] [PubMed] [Google Scholar]
- [44].Richter JD. Breaking the code of polyadenylation-induced translation. Cell 2008; 132:335-337; PMID:18267064; http://dx.doi.org/ 10.1016/j.cell.2008.01.024 [DOI] [PubMed] [Google Scholar]
- [45].Piqué M, López JM, Foissac S, Guigó R, Méndez R. A combinatorial code for CPE-mediated translational control. Cell 2008; 132:434-448; http://dx.doi.org/ 10.1016/j.cell.2007.12.038 [DOI] [PubMed] [Google Scholar]
- [46].Chu T, Henrion G, Haegeli V, Strickland S. Cortex, a Drosophila gene required to complete oocyte meiosis, is a member of the Cdc20/fizzy protein family. Genes N Y N 2000 2001; 29:141-152. [DOI] [PubMed] [Google Scholar]
- [47].Swan A, Schüpbach T. The Cdc20 (Fzy)/Cdh1-related protein, Cort, cooperates with Fzy in cyclin destruction and anaphase progression in meiosis I and II in Drosophila. Development 2007; 134:891-899; PMID:17251266; http://dx.doi.org/ 10.1242/dev.02784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol 2011; 12:427-438; PMID:21633387; http://dx.doi.org/ 10.1038/nrm3132 [DOI] [PubMed] [Google Scholar]
- [49].Pesin JA, Orr-Weaver TL. Developmental role and regulation of cortex, a meiosis-specific anaphase-promoting complex/cyclosome activator. PLOS Genet 2007; 3:e202; PMID:18020708; http://dx.doi.org/ 10.1371/journal.pgen.0030202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Den Elzen N, Pines J. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J Cell Biol 2001; 153:121-136; PMID:11285279; http://dx.doi.org/ 10.1083/jcb.153.1.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Nagaoka K, Udagawa T, Richter JD. CPEB-mediated ZO-1 mRNA localization is required for epithelial tight-junction assembly and cell polarity. Nat Commun 2012; 3:675; PMID:22334078; http://dx.doi.org/ 10.1038/ncomms1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med 2013; 19:1438-1449; PMID:24202396; http://dx.doi.org/ 10.1038/nm.3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Nagaoka K, Fujii K, Zhang H, Usuda K, Watanabe G, Ivshina M, Richter JD. CPEB1 mediates epithelial-to-mesenchyme transition and breast cancer metastasis. Oncogene 2016; 35:2893-2901; PMID:26411364; http://dx.doi.org/ 10.1038/onc.2015.350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hiratsuka S, Nakamura K, Iwai S, Murakami M, Itoh T, Kijima H, Shipley JM, Senior RM, Shibuya M. MMP9 induction by vascular endothelial growth factor receptor-1 is involved in lung-specific metastasis. Cancer Cell 2002; 2:289-300; PMID:12398893; http://dx.doi.org/ 10.1016/S1535-6108(02)00153-8 [DOI] [PubMed] [Google Scholar]
- [55].Lantz V, Chang JS, Horabin JI, Bopp D, Schedl P. The Drosophila orb RNA-binding protein is required for the formation of the egg chamber and establishment of polarity. Genes Dev 1994; 8:598-613; PMID:7523244; http://dx.doi.org/ 10.1101/gad.8.5.598 [DOI] [PubMed] [Google Scholar]
- [56].Clouse KN, Ferguson SB, Schüpbach T. Squid, Cup, and PABP55B function together to regulate gurken translation in Drosophila. Dev Biol 2008; 313:713-724; PMID:18082158; http://dx.doi.org/ 10.1016/j.ydbio.2007.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Igreja C, Izaurralde E. CUP promotes deadenylation and inhibits decapping of mRNA targets. Genes Dev 2011; 25:1955-1967; PMID:21937713; http://dx.doi.org/ 10.1101/gad.17136311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Cáceres L, Nilson LA. Translational repression of gurken mRNA in the Drosophila oocyte requires the hnRNP Squid in the nurse cells. Dev Biol 2009; 326:327-334; http://dx.doi.org/ 10.1016/j.ydbio.2008.11.030 [DOI] [PubMed] [Google Scholar]
- [59].Kelley RL. Initial organization of the Drosophila dorsoventral axis depends on an RNA-binding protein encoded by the squid gene. Genes Dev 1993; 7:948-960; PMID:7684991; http://dx.doi.org/ 10.1101/gad.7.6.948 [DOI] [PubMed] [Google Scholar]
- [60].Li W, Klovstad M, Schüpbach T. Repression of Gurken translation by a meiotic checkpoint in Drosophila oogenesis is suppressed by a reduction in the dose of eIF1A. Development 2014; 141(20):3910-21; http://dx.doi.org/ 10.1242/dev.109306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Norvell A, Kelley RL, Wehr K, Schüpbach T. Specific isoforms of squid, a Drosophila hnRNP, perform distinct roles in Gurken localization during oogenesis. Genes Dev 1999; 13:864-876; PMID:10197986; http://dx.doi.org/ 10.1101/gad.13.7.864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Filardo P, Ephrussi A. Bruno regulates gurken during Drosophila oogenesis. Mech Dev 2003; 120:289-297; PMID:12591598; http://dx.doi.org/ 10.1016/S0925-4773(02)00454-9 [DOI] [PubMed] [Google Scholar]
- [63].Yan N, Macdonald PM. Genetic interactions of Drosophila melanogaster arrest reveal roles for translational repressor Bruno in accumulation of Gurken and activity of Delta. Genetics 2004; 168:1433-1442; PMID:15579696; http://dx.doi.org/ 10.1534/genetics.104.033985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Webster PJ, Liang L, Berg CA, Lasko P, Macdonald PM. Translational repressor bruno plays multiple roles in development and is widely conserved. Genes Dev 1997; 11:2510-2521; PMID:9334316; http://dx.doi.org/ 10.1101/gad.11.19.2510 [DOI] [PMC free article] [PubMed] [Google Scholar]