ABSTRACT
The endoribonuclease toxins of the E. coli toxin-antitoxin systems arrest bacterial growth and protein synthesis by targeting cellular mRNAs. As an exception, E. coli MazF was reported to cleave also 16S rRNA at a single site and separate an anti-Shine-Dalgarno sequence-containing RNA fragment from the ribosome. We noticed extensive rRNA fragmentation in response to induction of the toxins MazF and MqsR, which suggested that these toxins can cleave rRNA at multiple sites. We adapted differential RNA-sequencing to map the toxin-cleaved 5′- and 3′-ends. Our results show that the MazF and MqsR cleavage sites are located within structured rRNA regions and, therefore, are not accessible in assembled ribosomes. Most of the rRNA fragments are located in the aberrant ribosomal subunits that accumulate in response to toxin induction and contain unprocessed rRNA precursors. We did not detect MazF- or MqsR-cleaved rRNA in stationary phase bacteria and in assembled ribosomes. Thus, we conclude that MazF and MqsR cleave rRNA precursors before the ribosomes are assembled and potentially facilitate the decay of surplus rRNA transcripts during stress.
Keywords: Differential RNA sequencing, MazF, MqsR, ribosome, rRNA precursors, Toxin-Antitoxin systems
Introduction
Bacterial toxin-antitoxin (TA) systems consist of a stable toxin and an unstable antitoxin that neutralizes its cognate toxin. If the levels of antitoxins drop, toxins become free from inhibition and target essential bacterial functions such as translation, DNA synthesis and energy production. In this way, TA systems are potentially able to regulate bacterial growth and proliferation.1 Despite their wide distribution among prokaryotes, the exact role of TA systems has remained elusive.2 According to a core concept in the field, antitoxins are degraded and toxins are liberated in response to stress and hunger.1,3 On the other hand, their benefit on recovery from the stress has not been demonstrated.4 During the last decade, TA systems have gained attention due to their role in the formation of persister bacteria, which are the minor subset of the bacterial population that is not killed by bactericidal antibiotics.5-7 Possibly, stochastic activation of TA toxins causes transient growth inhibition of a fraction of microbial cells that become persisters, whereas the bulk of proliferating bacteria are killed by antibiotics.8-10
The toxins of many well-studied TA systems are endoribonucleases. Some of them (e.g., RelE and HigB) are cotranslational endoribonucleases that cleave mRNA during translation. Others (e.g., MazF, MqsR, and HicA) cleave single-stranded RNA independently of the ribosome and translation. The cleavage of the latter group may be sequence-specific, as has been shown for MazF, which cuts single-stranded RNA at 5′↓ACA3′ sites,11 and MqsR, which cuts at 5′G↓CU3′, but also at 5′G↓CC3′, 5′G↓CA3′ and 5′G↓CG3′.12,13 Overproduction of ribonuclease toxins of either group leads to rapid inhibition of protein synthesis.14
mRNA has been considered the primary target of both co-translational and ribosome-independent TA-endoribonucleases.1 However, during past few years, several toxins of the MazF- and VapC families were demonstrated to target non-coding RNA. Certain VapC toxins of the enterobacterial, mycobacterial and spirochaetal vapBC systems cleave tRNA within the anticodon stem-loop,15-18 while VapC20 and VapC26 of M. tuberculosis cleave the sarcin–ricin loop (SRL) of 23S rRNA at the same position as α-Sarcin and other fungal ribotoxins.18,19 All these VapC family toxins are very specific endoribonucleases and cleave structured RNA: their inhibitory effect on translation and growth is based on cleavage of one unique target site within a stem-loop in tRNA or rRNA.17,19 SR loop is essential to the ribosome function20-22 and cleavage by VapC20 takes place in the context of assembled ribosomes: this toxin cleaves effectively 23S rRNA in vivo and within purified ribosomes, but does not cleave purified total RNA in vitro.19
Among the toxins of the MazF family, MazF-mt9 from Mycobacterium tuberculosis cleaves the tRNAPro14 D-loop and the tRNALys43 anticodon loop.23 Both MazF-mt3 and MazF-mt6 from the same organism cleave 23S rRNA within the helix/loop 70 in the ribosomal A site.24,25 MazF-mt3 also cleaves the anti-Shine-Dalgarno (aSD) sequence (5′U↓CCUU3′) within the unpaired 3′-end of 16S rRNA.24 Differently from the aforementioned VapC toxins, tRNA and rRNA are not unique targets of these MazF toxins: they all cleave mRNA and principally should target all single-stranded targets that bear their cognate nucleotide sequence. Although the cleavage site of MazF-mt3 and MazF-mt6 within 23S rRNA is essential to the ribosome function, this site is accessible only in purified total RNA and in dissociated 50S subunits but not in the 70S ribosomes.25 Cleavage of the aSD sequence by MazF-mt3 in vivo was first detected within 16S rRNA precursor, although this cleavage site is unpaired and potentially accessible within assembled ribosomes.24 Resolved structures of 2 MazF-RNA complexes (of Bacillus subtilis and Escherichia coli) convincingly explain why toxins of the MazF family can cleave only non-structured targets, which fit into the deep RNA-binding channel of MazF, whereas 5 adjacent bases of the target are facing toward the protein and the backbone phosphate moieties project outward.26,27
In Escherichia coli, MazF is the only toxin that, besides fragmenting mRNA, has been reported to cut rRNA. It was shown to cleave 16S rRNA at ↓1500ACA, close to the 3′-end.28 The cleaved-off 43 nt-fragment contains an anti-Shine-Dalgarno sequence and is separated from the ribosome. The ribosomes that have trimmed 16S rRNA are thought to preferentially translate MazF-processed mRNAs.28,29 According to the authors' initial hypothesis, these 70SΔ43 ribosomes selectively translate leaderless mRNAs (lmRNAs) that lack a Shine-Dalgarno (SD) sequence, which also has been cleaved off by MazF.28 That enables the synthesis of a specific set of proteins encoded by leaderless transcripts.28,30 Further study by the same authors showed that this hypothesis was exaggerated and that a diverse pool of mRNA is associated with the 70S ribosomes upon MazF induction.29 When MazF was incubated with purified ribosomes in vitro, it was shown to cleave 16S rRNA also at a second site, at ↓1396ACA in the ribosomal decoding center. Cleavage at this site is thought to be detrimental to ribosome activity.28 Considering the structure of the complex of E. coli MazF with its substrate,27 cleavage at both of these sites is surprising, because C1395, A1396 and C1501 are base-paired (E. coli 16S rRNA, http://rna.ucsc.edu/rnacenter/ribosome_images.html).
Upon checking the quality of the RNA from E. coli cells that over-express MazF, we noticed large rRNA fragments on gel electrophoresis,31 suggesting that MazF also cleaves rRNA elsewhere and/or initiates cleavage by other RNases. To further examine rRNA fragmentation by E. coli toxins, we induced MazF and MqsR, and mapped the cleavage sites adapting a version of differential RNA sequencing32 that was used in recent studies from the Woychik Lab, where the authors defined the cleavage specificities of MazF-mt3 and MazF-mt9 by overexpressing them in Escherichia coli.23,24 The method leverages the fact that E. coli MazF33 and several other toxins.23,24,34-36 yield 5′-OH and 2′,3′-cyclic phosphate (2′,3′-cP) ends, while the processing endoribonucleases of E. coli (e.g., RNases E, H, P and III) generate 5′-P and 3′-OH ends37 Thus, we mapped the 5′-OH and 2′,3′-cP termini at the toxin cognate target sites upon MazF and MqsR induction. Mapping the 5′-ends is sufficient to detect the toxin cleavage sites because E. coli does not have any 5′-to-3′ exoribonucleases, while mapping the 3′-ends would show what happens with the RNA after the cleavage. We imply that if rRNA is cleaved within assembled ribosomes, the 3′-ends must reside close to the cleavage sites, while cleavage of the rRNA precursors is probably followed by 3′-to-5′ exonucleolytic degradation.
To test for the rRNA cleavage under natural stress conditions, we sequenced RNA from stationary phase bacteria. It is noteworthy that the same conditions of stress and hunger that liberate the TA system toxins,1,3 trigger also ribosome degradation. According to current understanding, ribosomes are stable in proliferating bacteria, but become subject to degradation while cultures are approaching stationary phase.38-40 An endonucleolytic cleavage within rRNA by unknown RNase(s) is believed to initiate ribosome degradation.41 Thus, the TA toxins may be culprits for triggering ribosome decay and it is worth looking for the marks of toxin cleavage in the stationary phase rRNA. The cleaved RNA from decayed ribosomes is rapidly recycled and may go unnoticed. Therefore, we also sequenced the stationary phase RNA of an exoribonuclease mutant strain (exo-) where rRNA cleavage products accumulate.38
Results
Induction of MazF and MqsR causes fragmentation of E. coli rRNA
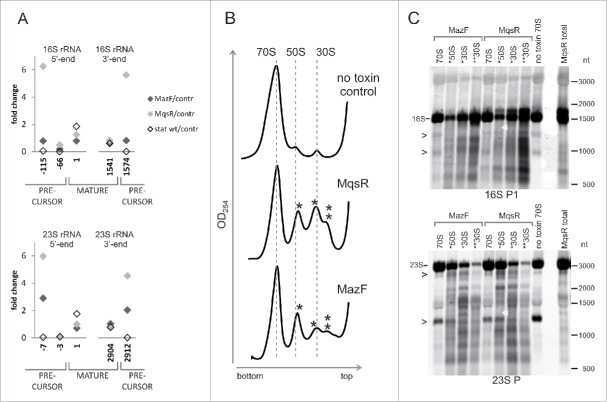
As with our former observations,31 we saw extra RNA bands on electrophoresis upon induction of MazF and MqsR that indicate rRNA cleavage and/or incomplete rRNA processing (Fig. 1A). Northern hybridization revealed considerable fragmentation of both 16S rRNA and 23S rRNA in response to both toxins (Fig. 1B, C). MazF and MqsR produce different patterns of rRNA fragments that suggest either sequence-specific cleavage by the toxin or toxin-specific induction of secondary rRNA fragmentation. We noted extra bands indicating rRNA fragmentation also in the stationary phase, both in the wt and exo− strains (Fig. 1A). Besides MazF and MqsR, we overexpressed also HicA of E. coli but did not detect any rRNA fragments (data not shown).
Figure 1.
Expression of MazF and MqsR leads to appearance of rRNA fragments. (A) Analysis of RNA samples by capillary electrophoresis. Total RNA was extracted from a log phase culture without toxin induction (no toxin control), the cultures after 2 hours of induction of either MqsR or MazF, and from the 24 h stationary phase cultures of the wt and exo− strains. The same RNA samples were used for preparation of the cDNA libraries for sequencing. (B) and (C). Northern blot analysis of rRNA fragments. Cultures of E. coli BW25113 contained plasmids for MazF and MqsR expression. Toxins were induced and RNA was extracted prior to induction (−1 min) and 15, 60, and 120 min after induction; RNA was separated on a 1.5% agarose (B) or a 6% PAA gel (C), transferred to a membrane, and hybridized with the oligoprobes provided below each panel. The graph at the top shows the location of the probes on the rRNA.
RNA-seq experimental setup
To identify the rRNA cleavage sites, we mapped the 5′- and 3′-ends of the RNA by differential RNA-sequencing. The 5′-end mapping is based on ligation of RNA adaptors to the cellular RNA molecules, which requires 5′-monophosphates (Fig. S1A). The 3′-end mapping is based on poly(A) tailing of cellular RNA, which requires 3′-OH groups (Fig. S1B). Transcription initiation, RNA processing, and toxin cleavage yield different types of RNA ends (Table 1). Most of the cellular RNases that take part in RNA processing and degradation produce 5′-P and 3′-OH,37 which do not require modification before the adaptor ligation or polyadenylation. E. coli MazF,33 several other toxins,24,34-36 and RNase I37 produce 5′-OH ends that must be phosphorylated before the adaptor ligation and 2′,3′-cyclic phosphates that must be dephosphorylated before polyadenylation. While planning experiments, we supposed that MqsR also yields 5′-OH and 2′, 3′-cP termini, although this had not been formally demonstrated. Both phosphorylation and dephosphorylation can be performed by T4 Polynucleotide Kinase (PNK)42 and may occur to some extent also within bacterial cells as the MazF-cleaved 43-nt 16S rRNA fragment was detected by 5′ RNA-seq without in vitro phosphorylation of RNA.43
Table 1.
Distinct phosphorylation status enables identification of toxin-cleaved RNA termini using RNA sequencing.
| 5′-end | Treatment before 5′-adaptor ligation | 3′-end | Treatment before 3′-polyadenylation | |
|---|---|---|---|---|
| Typical RNase cut site | P | — | OH | — |
| Toxin and RNaseI cut site | OH | T4 PNK + ATP | 2′,3′-cP | T4 PNK |
| Native transcript | P-P-P | TAP | OH | — |
We isolated RNA from a log phase culture without toxin induction (no toxin control), cultures of MazF or MqsR induction, and stationary phase cultures of wt and exo− strains. As our aim was rRNA analysis, we did not deplete rRNA before cDNA synthesis. From each RNA sample we prepared T4 PNK-treated (PNK+) and no treatment (PNK-) 5′-cDNA libraries (Fig. S1A) and 3′-cDNA libraries (Fig. S1B). Besides that, Tobacco Acid Pyrophosphatase (TAP)-treated 5′-end libraries were made as controls. TAP converts the 5′-triphosphate ends of primary transcripts to monophosphates; if the cDNA preparation and sequencing are technically sound, the TAP-treated and no treatment libraries must yield similar 5′-end counts because of a lack of internal transcription start sites within the rRNA genes.
Illumina sequencing and data analysis were performed as described in Materials and Methods. Overview of the sequence read mapping statistics is provided in Supplement (RNA-seq Mapping Statistics and Fig. S2-S4). The first base of each mapped 5′-read corresponds to the 5′-end of an RNA molecule and the last base of each mapped 3′-read corresponds to the 3′-end of an RNA molecule. For every position in a genome, we determined 5′-end count (the total number of 5′-reads whose first base aligns to this position) and 3′-end count (the total number of 3′-reads whose last base aligns to this position). We compiled all the sequence reads mapped on seven E. coli rRNA operons and aligned them to composite 16S rRNA and 23S rRNA sequences. We then identified sites for which the 5′-end count in the toxin induction PNK+ library was at least 30-fold higher than in the no toxin control PNK+ library. All the sites that passed this threshold and were located at the toxin recognition sequences (5′↓ACA3′ for MazF and 5′G↓CN3′ for MqsR) were considered primary toxin cleavage sites (Fig. 2, Fig. S5). For further analysis, different experimental conditions or treatments were compared between each other using MA scatter plots where the M values on y-axis show the fold change and the A values on x-axis show amount of the cleaved RNA termini at different rRNA positions (Fig. S6–S9 and interactive plotter). Such comparison of the TAP-treated and no treatment 5′-end libraries demonstrates that cDNA preparation and sequencing have been reliable (Fig. S9). Finally, to make the cleavage data comparable between several different libraries, we normalized the 5′- and 3′-end counts at every rRNA position to the end counts of mature full-length rRNAs (Fig. S10). RNA-seq results (the 5′- and 3′-end counts at each rRNA position) are available in Supplement and can be visualized using an interactive plotter (http://www.tuit.ut.ee/en/research/downloads).
Figure 2.
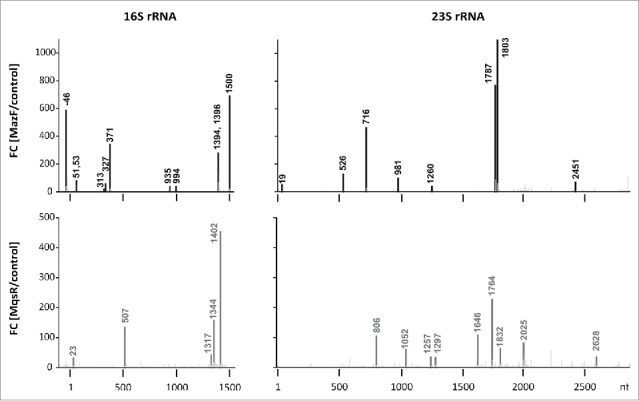
Detected MazF- and MqsR-cleavage sites. Histograms show the ratios of the 5′-end counts observed at each rRNA position in the PNK+ libraries of the toxin induction (MazF, MqsR) and the no toxin control.
MazF and MqsR cleave rRNA at multiple sites
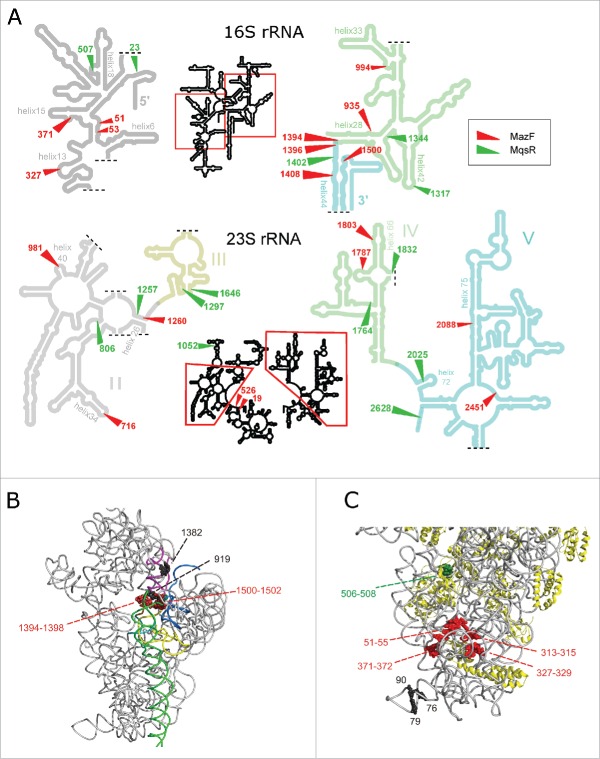
RNA-seq revealed that both MazF and MqsR cut 16S rRNA and 23S rRNA at several sites (Fig. 2, Fig. S5, S6). These cleavage sites became detectable only after the T4 PNK treatment, verifying that the cleaved ends were indeed 5′-OH ends (Fig. S8). Mapping of these cleavages sites onto rRNA secondary and tertiary structures shows that RNA is cleaved within regions that are at least partially base-paired in the assembled ribosomes (Fig. 3A) and/or buried within the ribosomal subunits and likely inaccessible in mature ribosomes, as exemplified in Fig. 3C. Therefore, almost certainly, rRNA at these sites was cleaved within rRNA precursors, before secondary structure formation and ribosome assembly. A clear example of detection of a cutting site within rRNA precursor by 5′-RNA-seq is the cleavage of 16S rRNA by MazF at ↓1500ACA, which is characterized by a big fold change (but low 5′-end count). As the length of our sequencing reads was 100 nt, the 43-nt distance between the cutting site and the mature 16S rRNA 3′-end indicates that RNA-seq has identified cleavage of the rRNA precursor before its processing into mature form. Also, a MazF cleavage site was identified within the 5′-precursor sequence of 16S rRNA (at ↓−46ACA; Fig. 2, Fig. S5).
Figure 3.
Locations of the cleavage sites on rRNA secondary structures and in the 30S ribosomal subunit. (A) MazF and MqsR cleavage sites on rRNA secondary structures (according http://rna.ucsc.edu/rnacenter/ribosome_images.html). (B,C) The major cleavage sites near the ribosome decoding center (B) and in the 5′-domain of 16S rRNA (C) in the 30S ribosomal subunit (PDB 4U1U).68 RNA is colored in gray and proteins in yellow. The helices that make up the decoding center (A) are colored in yellow (h1), teal (h2), purple (h28), green (h44), and blue (h45). The cleaved MazF ↓ACA cutting sites are highlighted in red and the MqsR G↓CU cutting site in green. The black spheres depict nucleotides at the 3′-ends (G76, G79, C1382) or 5′- ends (C90, A919) of the stationary phase cleavage fragments.
At most of the cleavage sites, we detected only the 5′-ends, which indicates 3′-to-5′ exonucleolytic trimming of the 3′-ends and agrees well with the notion that the toxins cleave rRNA precursors. However, at several MazF-cleaved sites, for example at U1393 and C1395 within 16S rRNA 3′-region, we observed also 2′,3′-cP ends (Fig. S7). We did not detect a 3′-end at A1499 in 16S rRNA (at the MazF ↓1500ACA cutting site) neither in response to MazF induction nor in the stationary phase and thus could not verify the presence of the stress ribosomes.28 Also, we did not identify any other potentially toxin-cleaved 5′-OH and 2′,3′-cP termini at the MazF and MqsR target sites in the stationary phase. The cleaved 5′-ends at C806 and A981 within 23S rRNA in the stationary phase cultures of exo- strain (Fig. S5) were 5′-P ends and the 3′-ends at U1393 and C1395 within 16S rRNA were 3′-OH ends (Fig. S10). Thus, most probably, the rRNA that originates from decaying ribosomes is often cleaved at these sites but MazF and MqsR play no role in these cleavages.
In conclusion, RNA-seq allowed us to map the toxin-cleaved 5′- and 3′-ends and identify the toxin cleavage sites within rRNA (Fig. 1). In spite of the presence of several 3′-to-5′ exoribonucleases in E. coli, 3′-RNA-seq identified several MazF-cleaved 3′-ends. We also verified that MqsR produces 5′-OH ends, as we had hypothesized.
MazF and MqsR induce secondary rRNA cleavages
In addition to the sequence-specific primary cleavages, expression of MazF and MqsR induced secondary cleavages at sites that lack the toxin-specific sequence motives. The products of these secondary cleavages have 5′-P and 3′-OH ends which are detectable independently of the PNK treatment (Fig. S10). Several of these secondary cleavage sites are the same sites where rRNA is cleaved during ribosome degradation in the stationary phase. For example, 16S rRNA is extensively cleaved 5′ of A919 in response to MazF production and in the stationary phase. Cleavage at this site was readily detected in the exo- strain, where the rRNA fragments from degraded ribosomes accumulate, and was formerly identified in studies of ribosome degradation.38,44 In secondary and tertiary structure, this cleavage site is located near the ribosome decoding center (Fig. 3B), as well as the cleaved or trimmed 3′-OH ends at C1382, U1393 and C1395, which were abundant in the stationary phase RNA of the exo- strain (Fig. S10).
MazF- and MqsR-cleaved rRNA precursors build up in aberrant ribosome subunits
During production of new ribosomes, the rRNA gene transcripts are at first cleaved by RNase III to produce precursors that are longer than the rRNA in mature ribosomes.41 After that, mature ends of rRNA are formed in the translating ribosomes.45,46 Our RNA sequencing data show clearly that induction of MqsR causes accumulation of the 5′- and 3′-precursors of 16S rRNA and 23S rRNA (Fig. 4A). Accumulation of the rRNA precursors was confirmed by Northern analysis and primer extension mapping (Fig. S11).
Figure 4.
Accumulation of the rRNA precursors and aberrant ribosomal subunits. (A) The ratios of the 5′-end counts and 3′-end counts of the mature- and precursor rRNA ends. Comparison of the PNK+ toxin induction (MazF, MqsR) and wt stationary phase libraries to the no toxin control. (B) Sucrose density gradient centrifugation profiles of the no toxin control and the cultures after 2 hours of induction of either MqsR or MazF. Peaks of irregular subunits (*50S, *30S, and **30S) are marked by asterisks. (C) The irregular ribosomal subunits contain fragmented rRNA. RNA from the sucrose gradient fractions was analyzed by Northern hybridization. RNA (2 µg per lane) was separated on a 1.5% agarose gel, transferred to a membrane, and hybridized with the oligoprobes 16S P1 and 23S P. Total RNA of the MqsR induction culture was used as a control. Bands that occur in all 70S lanes due to the RNA cleavages during ribosome preparation are marked by arrowheads (>).
As MqsR and MazF repress the synthesis of ribosomal proteins, the precursor rRNAs are produced in excess and may end up in aberrant ribosomal subunits. Indeed, sucrose density gradient centrifugation showed that irregular subribosomal particles accumulate in response to both MqsR and MazF induction (Fig. 4B). We isolated 70S ribosomes and the smaller particles from a sucrose density gradient and analyzed their RNA by Northern hybridization. The aberrant subribosomal particles contained much more fragmented RNA than the 70S ribosomes (Fig. 4C). Also, the MqsR-induced 16S rRNA precursors accumulated in the aberrant particles (Fig. S11A). This indicates that most of the rRNA cleavages take place before the ribosomes are fully assembled and is in good agreement with the observation that MqsR and MazF cleave rRNA at the sites that are accessible only prior to the ribosome assembly.
The RNA fragmentation patterns in the aberrant ribosomal particles match the fragmentation patterns of total RNA, implying that this fragmentation has occurred in vivo and is not an artifact, while several bands that are common to all 70S preparations but missing in total RNA lanes (Fig. 4C) indicate non-physiological RNA fragmentation during ribosome preparation and/or sucrose gradient runs, which has been observed also by other researchers.19
Validation of the RNA-seq results
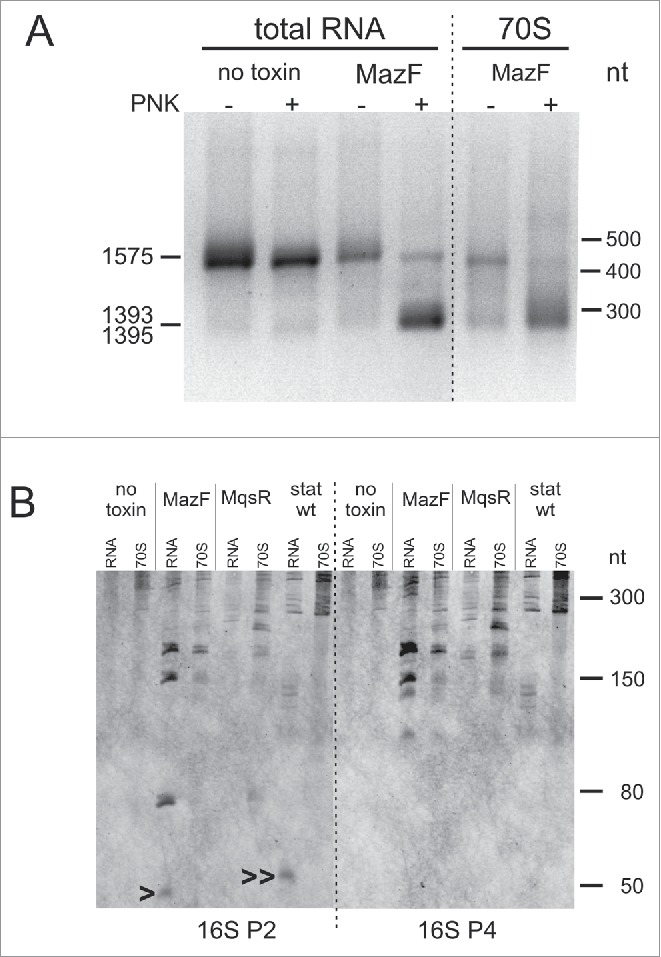
To validate the findings of RNA-seq, we mapped the toxin cleavage sites by primer extension. Because we knew the expected locations of the 5′-ends beforehand, we used the labeled oligonucleotides (Table S2) as size markers instead of the sequence ladder (explained in Fig. S12). We verified the MazF-cleaved 5′-ends at ↓981ACA, ↓1803ACA, ↓2451ACA in 23S rRNA, and at ↓51ACA, ↓53ACA, ↓371ACA, ↓1394ACA, ↓1396ACA, ↓1500ACA in 16S rRNA (Fig. S12). In 16S rRNA, we also verified the MazF-cleavage sites at ↓1225ACA and ↓1227ACA, which were identified by 3′ RNA-seq (Fig. S7) but remained below the 30-fold change threshold using 5′ RNA-seq. The MqsR-cleaved 5′-ends were verified at C507 in 16S rRNA and at C2628 in 23S rRNA (Fig. S13). All these cleavage sites were detectable in total RNA but not in the RNA extracted from the 70S fraction (Fig. S12, S13).
To identify the 16S rRNA 3′-ends which are formed in response to MazF induction, we adapted 3′-RACE (Rapid Amplification of cDNA Ends). We performed poly(A) tailing of total RNA and the RNA from 70S fractions before and after the T4 PNK treatment, synthesized cDNA, PCR-amplified the 3′-end fragments, separated them by electrophoresis and sequenced. The PCR products fall into two size categories. The longer products captured the 16S rRNA 3′-precursor, which was detected in all the total RNA samples and also in the 70S fraction of the MazF-expressing bacteria (Fig. 5A, Fig. S14A). Presence of the unprocessed rRNA precursors in the 70S fraction is uncommon; however, this is in agreement with the results of primer extension analysis that revealed the 23S rRNA 5′-precursor in the 70S fractions of the MazF and MqsR expressing bacteria (Fig. S11B). Ribosomes that contain unprocessed rRNA are not unique as the last steps in rRNA processing that occur in polysomes,47 are not obligate for the formation of 70S ribosomes.45,48
Figure 5.
Fragmentation of the 16S rRNA 3′-region. (A) 3′-RACE of 16S rRNA 3′-ends. Total RNA was isolated from the no toxin control and after the MazF induction. Ribosomes were isolated after the MazF induction and RNA was extracted from the 70S sucrose gradient fraction (Fig. 4B). Poly(A) tail was synthesized either before or after the T4 PNK treatment. 0.5 μg-aliquots of the amplified 3′-end fragments were subjected to electrophoresis in a 1.5% agarose gel. DNA was visualized by EtBr staining, purified from the gel, and sequenced. The longer fragments correspond to the 3′-end of 16S rRNA precursor at 1575. The shorter fragments correspond to the MazF-cleaved 3′-ends at 1393 and 1395, upstream of the MazF cleavage sites at ↓1394ACA and ↓1396ACA. (B) Northern blot analysis of the total RNA and the RNA from 70S fractions. Total RNA and ribosomes were isolated two hours after induction of either MazF or MqsR, from the no toxin control and from the 24h-stationary phase cultures. Ribosomal particles were separated by sucrose density gradient centrifugation (Fig. 4B) and RNA was isolated from the 70S fraction. RNA was separated on a 6% PAA gel, transferred to a membrane, and hybridized with the oligoprobes 16S P2 (complementary to nucleotides 1520…1542) and 16S P4 (1473…1494). The 43-nt fragment produced by MazF cleavage at ↓1500ACA is marked by an arrowhead (>) and the 48-nt fragment produced by the cleavage between G1494 and U1495 in the stationary phase is marked by 2 arrowheads (>>).
The shorter PCR products captured the MazF-cleaved 3′-ends at U1393 and C1395 from the MazF induction samples. These 3′-ends were amplified only from the T4 PNK-treated samples, verifying that we captured 2′,3′-cP ends, and were detected in both total RNA and 70S fraction (Fig. 5A, Fig. S14B). We suggest that, in the latter case, the rRNA is cleaved in unprocessed 70S ribosomes, but not in regular mature ribosomes. As 3′-RNA-seq, 3′-RACE did not detect the anticipated 16S rRNA 3′-end of the 70SΔ43 stress ribosomes at A1499. Here we must consider that both 3′-RACE and 3′-RNA-seq favored detection of precursors and trimmed 3′-ends over mature 16S rRNA 3′-end because the methylated nucleotides downstream of the ↓1394ACA and ↓1396ACA sites (m4Cm1402, m5C1407, m3U1498, m2G1516, m62A1518, and m62A1519) block reverse transcription and detection of the mature 3′-end.
The results of RNA sequencing, primer extension mapping, and 3′-RACE were validated with Northern hybridization focusing on the 16S RNA 3′-region. MazF induction produced fragments of approximate sizes 150-nt and 180-nt, which were present in both total RNA and the 70S fraction and were formed by cutting the mature 16S rRNA and the 16S rRNA precursor, respectively, at ↓1394ACA and/or ↓1396ACA site (Fig. 5B). Two fragments with approximate sizes of 50 and 80 nt that occurred in total RNA but were missing in the 70S fraction (Fig. 5B) correspond to the 43-nt and 76-nt products of MazF cleavage at ↓1500ACA within the mature 16S rRNA and the precursor, respectively. Lack of these fragments in the 70S ribosomes is expected, because the 43-nt fragment dissociates from the ribosome.28 Thus, the Northern blot data agree with our 3′-RACE results and demonstrate one more time that the 70S ribosome fraction of toxin-expressing bacteria contains unprocessed rRNA precursors.
In conclusion, we confirmed several MazF and MqsR cleavages that were identified by RNA-seq with conventional methods. As much as we can corroborate, rRNA at these sites is cleaved prior to ribosome assembly and is intact in mature ribosomes. The 70S sucrose gradient fractions of toxin-expressing bacteria contain unprocessed rRNA precursors and even if we spot MazF-cleaved rRNA in this fraction, it may originate from these unprocessed or immature 70S ribosomes.
rRNA fragmentation in stressed bacteria
Electrophoresis (Fig. 1A) and RNA-seq (Fig. S10) indicated rRNA fragmentation in the stationary phase cultures of wild type and exo- E. coli. RNA-seq did not identify any potentially toxin-cleaved sites in the stationary phase rRNA but nevertheless detected two cleavage sites in wt E. coli, where 5′-and 3′-ends are in close proximity suggesting that the cleavages may potentially occur within intact ribosomes. One of these sites is within the hypervariable region V1, a.k.a. “spur” in helix 6 of 16S rRNA (3′-OH ends at G76 and 79 (A or G); 5′-P end at 90 (C or U) (Fig. 3C) and the other is located within helix 45 of 23S rRNA (3′-OH end at 1170 (A or G); 5′-P end at C1171) (Fig. S10). Cutting at these sites did not depend on the primary sequence and the transcripts of all rRNA operons were cleaved to some extent (data not shown). Both sites are exposed on the surface of the ribosome and the cleavage within helix 6 of 16S rRNA in the stationary phase ribosomes was recently reported.49
As activation of MazF,50,51 and the cleavage of 16S rRNA at ↓1500ACA28 were reported in stress conditions, we expected to identify the truncated 3′-end of 16S rRNA at A1499 and verify the presence of the stress ribosomes upon stress. Therefore, we applied 3′-RACE and Northern hybridization to analyze the RNA from stationary phase cells, CAM treated cultures and cultures where amino acid starvation was induced by mupirocin (MUP). 3′-RACE did not identify 16S rRNA 3′-end at A1499 in these conditions, but detected 3′-termini at several locations between C1397 and G1405 (data not shown). Truncated 3′-ends at these positions were also identified in the Δ10 strain where all TA endonuclease genes are deleted,7 thus showing that TA systems are not required for such truncation.
Northern blots of the MUP- and CAM-treated samples show rRNA fragmentation but no MazF-specific cleavage products of the 16S rRNA 3′-end, even when we used a sensitive ECL (enhanced chemiluminescence) detection method and performed these treatments in the exo- strain, where cleavage products accumulate (Fig. S16).
Northern analysis of the stationary phase RNA revealed a small fragment of the 16S rRNA 3′-end that was slightly larger than the 43-nt MazF-cleaved fragment and was present in total RNA but not in the 70S fraction (Fig. 5B). The 5′-end of this fragment was mapped to U1495 (Fig. S15C,D), indicating a cleavage site between G1494 and U1495 in helix 44 of 16S rRNA, next to the cutting site of colicin E3 between A1493 and G1494.52 To our knowledge, this stationary phase-specific cleavage site has not been previously described. The 48-nt cleavage fragment was abundant in the exo- strain as well as in the ΔmazEF and ΔmqsRA deletion strains constructed in the exo- background (Fig. S15A,B). Thus, it is generated independently of MazF and MqsR.
Discussion
The vast majority of the ribosome studies are conducted on ribosomes from growing cultures and it is silently assumed that ribosomes are structurally and functionally similar under all physiological conditions. In contrast with this assumption, a study from Isabella Moll's Lab reported that MazF cleaves off the anti-Shine-Dalgarno (aSD) sequence from the 3′-end of 16S rRNA in stressed E. coli cells. The authors concluded that these cells contain a distinct subpopulation of 70SΔ43 stress ribosomes.28,30
Our work shows that the toxins MazF and MqsR cleave rRNA also in many other sites. Considering the structure of MazF in complex with its target,27 it is clear that this toxin can cleave only single-stranded and unstructured RNA. As the newly identified cleavage sites are buried inside the ribosomal subunits (Fig. 3C) and/or are partially base-paired (Fig. 3A), these sites must have been cleaved before the ribosome assembly. Analysis of the sucrose gradient fractions is supporting this notion because we found fragmented rRNA in the aberrant ribosomal subunits, not in mature 70S ribosomes (Fig. 4C, Fig S12–13). Formation of these aberrant subunits may be caused by the deficit of ribosomal proteins, as it happens in the presence of protein synthesis inhibiting antibiotics53-55 or by direct cleavage of the rRNA precursors by MazF and MqsR. The inhibition of protein synthesis is not always accompanied by cleavage of rRNA precursors; while the toxins RelE and HicA shut down protein synthesis as well, their overexpression did not cause any rRNA fragmentation (data not shown). Toxin GraT of Pseudomonas was recently shown to cause a cold-sensitive ribosome biogenesis defect. GraT induced accumulation of free 50S and 30S subunits that contained unprocessed and unmodified rRNA but did not cause rRNA fragmentation.56
Why have the fragmentation of rRNA precursors and formation of aberrant ribosomal subunits in response to MazF gone unreported earlier11,28,57? We suggest that this can be explained by differences in the experimental conditions, first of all the growth phase of the culture where toxin was induced. We induced toxins in fast-growing and dilute cultures, where the synthesis of new rRNA is most active. Details of the experimental setup may also have an effect on the extent of RNA cleavage by MazF during stationary phase, amino acid starvation and antibiotic treatments, where we did not find MazF-cleaved rRNA. We do not think that our current observations principally contradict the results of other labs and hope that additional studies under diverse culture conditions will explain these discrepancies.
We identified the toxin cleavage sites using differential RNA sequencing.32 This method enables to observe toxin activity directly, if the toxin-cleaved RNA fragments are stable enough and the cleaved ends are not rapidly converted into 5′-P and 3′-OH ends. Therefore, it would be a suitable method to detect for toxin liberation during different stresses, which so far has been detected indirectly, using transcriptional activation of the TA operons as a marker.5,50,58 Differently from what was done in the studies, which used RNA-seq to define the cleavage specificities of mycobacterial MazF-mt3 and MazF-mt9,23,24 we sequenced also the 3′-ends and did not degrade the 5′-monophosphate-bearing transcripts. The 5′-P ends allowed us to identify the non-toxin rRNA cleavage sites (secondary and stationary phase cleavage sites). Dephosphorylation of the 2′,3′-cP ends by T4 PNK allowed us to specifically detect the toxin-cleaved 3′-ends by 3′-end sequencing and 3′-RACE (Fig. 5A, Fig. S8E,F). The 3′-ends of the endonucleolytically cleaved RNA are not necessarily located at the cleavage sites but may be subject to 3′-to-5′ trimming by exonucleases and, therefore, must be mapped separately. For example, we readily identified the MazF-cleaved 5′-end at ↓1500ACA in 16S rRNA (Fig. 2, Fig. S12). We also detected the respective MazF-cleaved fragment of 16S rRNA precursor and the 43-nt cleavage fragment of mature 16S rRNA, as well as 48-nt stationary phase-specific cleavage fragment of 16S rRNA (Fig. 5B). However, in both cases, we did not find the newly formed 3′-ends at respective cleavage sites. Thus, further studies that apply 3′-end mapping under various relevant conditions are needed to clarify the issue of MazF-generated 70SΔ43 stress ribosomes.
In contrast to the toxin-cleaved sites, the rRNA cleavage sites within helix 45 of 23S rRNA and helix 6 of 16S rRNA in the stationary phase wt E. coli are exposed on the ribosome surface and are very probably cleaved within mature ribosomes (Fig. 3C). It was recently shown that cleavages within the helix 6 of 16S rRNA reduce the performance of stationary phase ribosomes.49 At the same time, in many bacterial species, rRNA is normally discontinuous and is formed through excision of intervening sequences (IVS) which are located in the same variable helices.59-61 Sequence tags have been inserted into these helices with no deleterious effects,62 63 and when a Salmonella IVS was inserted into the helix 45 of E. coli 23S rRNA, it was excised by RNase III while the ribosomes retained functionality.64 Therefore, it is possible that the stationary phase-specific cleavages at these sites do not disrupt ribosome function.
The most important conclusion of this study is that detection of the rRNA fragments does not necessarily indicate fragmentation of the RNA in mature ribosomes and a set of different experiments is needed to characterize the state of rRNA. Summarizing our findings, we suggest that the principal outcome of the cleavage of rRNA precursors by toxins is the clean-up of unnecessary rRNA in stressed bacteria. Toxins help to decay surplus rRNA precursors that have been synthesized in growth-promoting conditions when a large amount of the resources are dedicated to the making of new ribosomes. New ribosomes are not required under unfavorable conditions and it is beneficial to first recycle the excessive building components before starting to demolish complete ribosomes. In this way, TA systems would help to scale down the translational machinery and work in concert with the stringent response which reduces the synthesis of the ribosomal components.65
Materials and methods
Bacterial cultures
Lysogeny broth (LB) and LB agar plates were used for growth. Liquid cultures were grown on Infors HT Multitron shaker, at 190 RPM, at 37°C. Antibiotics were used at the following concentrations: ampicillin, 30 μg/ml or 100 μg/ml; chloramphenicol, 50 μg/ml, and kanamycin, 25 μg/ml.
Expression of MazF and MqsR was induced in BW25113 and MG1655exo-, harboring plasmids pSC228 and pSC3326 for the induction of MazF and MazE or pTX3 and pAT3 for the induction of MqsR and MqsA. The overnight medium was supplemented with 0.2% glucose to avoid the toxin leakage and 1 mM IPTG to induce the antitoxin. The overnight cultures were started from a fresh colony and diluted 1000-fold into 200 ml of LB in 1 l flasks. At OD600nm ≈ 0.2, 1 mM L-arabinose was added to induce the toxin and incubation was continued for 2h. Under these conditions, MazF and MqsR do not kill E. coli and all bacterial cells restart growth, although some do this with a delay.31
Total RNA of the log phase control-, stationary phase-, CAM-treated and MUP-treated samples was isolated from the wild type strain MG1655 and the mutant strains listed in Table S1. For growing the logarithmic phase and stationary phase samples, the 16–17h overnight cultures were diluted 1000-fold into 100 ml of LB in 1 l flasks. At OD600nm ≈ 0.2, 25 ml of the culture was used for the log phase RNA extraction. The remaining culture was grown for 24 h and used for the stationary phase RNA extraction. For the chloramphenicol (CAM) and mupirocin (MUP) treatments, the overnight cultures were diluted 1000-fold into 200 ml of LB in 1 l flasks. At OD600nm ≈0.2, 25 µg/ml CAM or 30 µg/ml MUP was added, and incubation was continued for 2h.
Primer extension analysis
Primer extension was performed according to a modification of the protocol used by Kasari, et al.31 To verify the 5′-ends identified by RNA-seq, labeled marker oligonucleotides that correspond to the anticipated extension products were loaded as the size markers instead of the sequencing reactions. Primers and markers (Supplementary Table S2) were labeled with [γ32P]ATP by T4 PNK (Thermo Scientific) and purified using Oligo Clean & Concentrator Kit (Zymo Research). RNA samples (5 μg) were mixed with labeled primer, incubated for 2 min at 75 °C and slowly cooled down to 47 °C. Extension reactions with primers 16S:1530→1504_ext and 16S:943→924_ext were performed in HY buffer (45 mM Tris-HCl pH 8.4, 30 mM KCl), while all the other extension reactions were performed in HB buffer (45 mM Hepes-KOH pH 7.0, 90 mM KCl) at 42°C for 30 min, using 20 units of AMV reverse transcriptase (Promega). cDNA was precipitated with ethanol overnight, resuspended in 12 µl of formamide loading buffer and subjected to electrophoresis in a 12% PAA, 7M urea gel.
3′ RACE
RNA (10 µg) was incubated with T4 PNK (Thermo Scientific) in a lack of ATP for 30 min at 37°C, to remove the 2′–3′-cyclic phosphate groups, and purified using RNA Clean & Concentrator-5 kit (Zymo Research). Poly(A) tailing of the T4 PNK-treated or untreated RNA (5 µg) was performed using Poly(A) Tailing Kit (Applied Biosystems). RNA was incubated with 0.01 U/µlof E. coli Poly(A) polymerase at 37°C for 1h, and purified using RNA Clean & Concentrator-5 kit (Zymo Research). During the course of purification, the RNA was treated with DNase according to the manufacturer's instructions. First-strand cDNA synthesis was primed with poly(T) primer (Supplementary Table S4) and performed by AMV reverse transcriptase (15 U) for 1h at 42 °C. Recombinant RNasin Ribonuclease Inhibitor (Promega) was added to all enzymatic reactions. The RNA was hydrolyzed and the single-stranded cDNA was purified as described in the manual of DNA Clean and Concentrator-5 kit (Zymo Research). cDNA was PCR amplified (30 cycles) using primers 16S_3prime and Arb2, and subjected to electrophoresis in a 1.5% agarose gel in 1xTBE buffer. EtBr-stained bands were cut out; DNA was purified with a Zymoclean Gel DNA Recovery kit (Zymo Research) and sequenced using primer 3race_SEQ1. The last nucleotide matching 16S rRNA and preceding a run of A-s indicates the position of a 3′-end.
Isolation and sucrose gradient analysis of ribosomes
Bacteria were grown, harvested, and stored as for RNA isolation. Bacterial pellets were suspended in TNM-10 buffer (10 mM MgCl2, 100 mM NH4Cl, 20mM Tris-HCl pH 8.0, and 6mM ß-mercaptoethanol). After addition of lysozyme (final concentration of 1 mg/ml) and DNase I (final concentration of 50 units/ml), the cells were disrupted by glass beads in Precellys 24 homogenizer (Bertin Technologies) (program: 6000 rpm, 4°C, 3×1 min, pause 1 min). S30 lysate was prepared by centrifugation at 12,000 g for 20 min. 30–50 A260 units of S30 lysate was loaded onto a 10–25% sucrose gradient in TNM-10 buffer, followed by centrifugation at 21,000 rpm for 16 h in a Beckman SW 28 rotor. The absorbance of the fractions at 260 nm was monitored with an UVis-920 UV monitor (GE), fractions containing ribosomal particles were collected and precipitated by addition of 2.5 volumes of ice-cold ethanol. Ribosomal pellets were suspended in TNM-10 buffer and stored at −80C.
cDNA library preparation and next-generation sequencing
cDNA libraries were constructed and sequenced by vertis Biotechnologie AG (Germany). To construct the 5′-fragment cDNA library, the total RNA samples were first treated with either T4 PNK (NEB), TAP (Epicentre), or left untreated. An RNA adaptor was ligated to the 5′-monophosphates of the RNA fragments. First-strand cDNA synthesis by M-MLV reverse transcriptase was primed with an N6 randomized primer. Then, Illumina adaptors containing barcode sequences were ligated to the 5′-ends of the antisense cDNA. The adaptors had 2 random nucleotides at the 3′-end to diminish the ligation bias. The cDNA was amplified with PCR (14 to 18 cycles) using a 5′-biotinylated primer and a proof reading enzyme. The cDNA preparations were degraded using NEBNext dsDNA fragmentase and the 5′-cDNA fragments were then bound to streptavidin magnetic beads. The bound cDNAs were blunted and the 3′ Illumina sequencing adaptor was ligated to the 3′-ends of the cDNA fragments. The bead-bound cDNAs were finally PCR-amplified (4–7 cycles).
To construct the 3′-fragment cDNA library, the total RNA samples were first treated either with T4 PNK or mock treated without the enzyme. This was followed by poly(A)-tailing using poly(A) polymerase. The samples were then fragmented with ultrasound (4 pulses of 30 sec at 4°C). The 3′-RNA fragments, which carry poly(A)-tails, were captured using oligo-dT chromatography. The newly generated 5′-ends were dephosphorylated using Antarctic Phosphatase (NEB) and rephosphorylated with PNK. Then, RNA adaptors containing barcode sequences were ligated to the 5′-phosphates of the 3′-RNA fragments. First-strand cDNA synthesis was performed using an oligo(dT)-adaptor primer and M-MLV reverse transcriptase. The resulting cDNA was PCR-amplified using a high fidelity DNA polymerase (13–15 cycles).
For Illumina sequencing, the 5′ and 3′ cDNAs were first pooled separately in approximately equimolar amounts. The 300–500 bp fragments of 5′ cDNA (carrying 123 bp flanking adaptor sequences) and the 250–350 bp fragments of 3′ cDNA (carrying 148 bp flanking sequences) were isolated for Illumina sequencing using the Agencourt AMPure XP kit (Beckman Coulter Genomics). This set the lower limit of detectable 5′-RNA fragments near 150 bases. The 5′ and 3′ cDNA pools were finally combined in equimolar amounts and single end-sequenced on an Illumina HiSeq2000 system using 100 bp read length. Flanking sequences were trimmed and sequence reads were sorted by the service provider.
RNA-seq data analysis
Before mapping, the sequence reads were processed as described in Supplementary Materials and Methods. The poly(A) tails of the 3′-reads were trimmed until the first non-A nucleotide. Processed 5′- and 3′- reads were aligned with the E. coli strain MG1655 genome (NC_000913.3) using bowtie1 (-v 1 -k 7 -p 2 -S –best –strata).66 5′-end positions were determined as the first positions of the mapped 5′-reads and 3′-end positions as the last positions of the 3′-reads. Total 5′-end counts and 3′-end counts were calculated for each genomic position using Samtools67 executed via Python scripts using pysam (https://github.com/pysam-developers/pysam). The 5′- and 3′-end counts of the 7 rRNA operons were compiled to composite 16S rRNA and 23S rRNA sequences. The sites where the 5′-end count in the toxin induction PNK+ library was at least 30-fold higher than in the no toxin control PNK+ library and which were located at the MazF and MqsR cognate recognition sequences were considered primary toxin cleavage sites.
Reproducibility of experiments
Northern blots and sucrose gradient centrifugation were repeated at least twice with similar results. RNA sequencing was performed once and the RNA from replicate bacterial cultures was used to verify the RNA-seq results by primer extension and 3′ RACE.
Data accession code
The RNA sequencing data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-4204.
Supplementary Material
Disclosure of potential conflicts of interests
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Ulvi Gerst Talas and Maido Remm for their help with the RNA sequencing data analysis, Murray Deutscher for strains and comments on the manuscript, Remy Loris for discussions, Kenn Gerdes for strains and plasmids, and Isabella Moll for discussion of experimental details and comments on the manuscript.
Funding
This work was supported by Estonian Science Foundation grants 8822 and 9040, by Estonian Research Council grant IUT2–22, and by the European Regional Development Fund through the Center of Excellence in Molecular Cell Engineering.
References
- 1.Yamaguchi Y, Park JH, Inouye M. Toxin-antitoxin systems in bacteria and archaea. Ann Rev Genet 2011; 45:61-79; PMID:22060041; http://dx.doi.org/ 10.1146/annurev-genet-110410-132412 [DOI] [PubMed] [Google Scholar]
- 2.Magnuson RD. Hypothetical functions of toxin-antitoxin systems. J Bacteriol 2007; 189:6089-92; PMID:17616596; http://dx.doi.org/ 10.1128/JB.00958-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol 2005; 3:371-82; PMID:15864262; http://dx.doi.org/ 10.1038/nrmicro1147 [DOI] [PubMed] [Google Scholar]
- 4.Tsilibaris V, Maenhaut-Michel G, Mine N, Van Melderen L. What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J Bacteriol 2007; 189:6101-8; PMID:17513477; http://dx.doi.org/ 10.1128/JB.00527-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol 2004; 186:8172-80; PMID:15576765; http://dx.doi.org/ 10.1128/JB.186.24.8172-8180.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maisonneuve E, Castro-Camargo M, Gerdes K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 2013; 154:1140-50; PMID:23993101; http://dx.doi.org/ 10.1016/j.cell.2013.07.048 [DOI] [PubMed] [Google Scholar]
- 7.Maisonneuve E, Shakespeare LJ, Jorgensen MG, Gerdes K. Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci U S A 2011; 108:13206-11; PMID:21788497; http://dx.doi.org/ 10.1073/pnas.1100186108 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Tuomanen E, Cozens R, Tosch W, Zak O, Tomasz A. The rate of killing of Escherichia coli by beta-lactam antibiotics is strictly proportional to the rate of bacterial growth. J Gen Microbiol 1986; 132:1297-304; PMID:3534137; http://dx.doi.org/ 10.1099/00221287-132-5-1297 [DOI] [PubMed] [Google Scholar]
- 9.Rotem E, Loinger A, Ronin I, Levin-Reisman I, Gabay C, Shoresh N, Biham O, Balaban NQ. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc Natl Acad Sci U S A 2010; 107:12541-6; PMID:20616060; http://dx.doi.org/ 10.1073/pnas.1004333107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Persistence Balaban NQ.: mechanisms for triggering and enhancing phenotypic variability. Curr Opin Genet Dev 2011; 21:768-75; PMID:22051606; http://dx.doi.org/ 10.1016/j.gde.2011.10.001 [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell 2003; 12:913-23; PMID:14580342; http://dx.doi.org/ 10.1016/S1097-2765(03)00402-7 [DOI] [PubMed] [Google Scholar]
- 12.Yamaguchi Y, Park JH, Inouye M. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J Biol Chem 2009; 284:28746-53; PMID:19690171; http://dx.doi.org/ 10.1074/jbc.M109.032904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen-Dalsgaard M, Jorgensen MG, Gerdes K. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol Microbiol 2010; 75:333-48; PMID:19943910; http://dx.doi.org/ 10.1111/j.1365-2958.2009.06969.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook GM, Robson JR, Frampton RA, McKenzie J, Przybilski R, Fineran PC, Arcus VL. Ribonucleases in bacterial toxin-antitoxin systems. Biochim Biophys Acta 2013; 1829:523-31; PMID:23454553; http://dx.doi.org/ 10.1016/j.bbagrm.2013.02.007 [DOI] [PubMed] [Google Scholar]
- 15.Cruz JW, Sharp JD, Hoffer ED, Maehigashi T, Vvedenskaya IO, Konkimalla A, Husson RN, Nickels BE, Dunham CM, Woychik NA. Growth-regulating Mycobacterium tuberculosis VapC-mt4 toxin is an isoacceptor-specific tRNase. Nat Commun 2015; 6:7480; PMID:26158745; http://dx.doi.org/ 10.1038/ncomms8480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopes AP, Lopes LM, Fraga TR, Chura-Chambi RM, Sanson AL, Cheng E, Nakajima E, Morganti L, Martins EA. VapC from the leptospiral VapBC toxin-antitoxin module displays ribonuclease activity on the initiator tRNA. PloS One 2014; 9:e101678; PMID:25047537; http://dx.doi.org/ 10.1371/journal.pone.0101678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winther KS, Gerdes K. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc Natl Acad Sci U S A 2011; 108:7403-7; PMID:21502523; http://dx.doi.org/ 10.1073/pnas.1019587108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winther K, Tree JJ, Tollervey D, Gerdes K. VapCs of Mycobacterium tuberculosis cleave RNAs essential for translation. Nucleic Acids Res 2016; 44:9860-71; PMID:27599842; http://dx.doi.org/24225902 10.1093/nar/gkw781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winther KS, Brodersen DE, Brown AK, Gerdes K. VapC20 of Mycobacterium tuberculosis cleaves the sarcin-ricin loop of 23S rRNA. Nat Commun 2013; 4:2796; PMID:24225902; http://dx.doi.org/ 10.1038/ncomms3796 [DOI] [PubMed] [Google Scholar]
- 20.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science 2010; 330:835-8; PMID:21051640; http://dx.doi.org/ 10.1126/science.1194460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallin G, Kamerlin SC, Aqvist J. Energetics of activation of GTP hydrolysis on the ribosome. Nat Commun 2013; 4:1733; PMID:23591900; http://dx.doi.org/ 10.1038/ncomms2741 [DOI] [PubMed] [Google Scholar]
- 22.Shi X, Khade PK, Sanbonmatsu KY, Joseph S. Functional role of the sarcin-ricin loop of the 23S rRNA in the elongation cycle of protein synthesis. J Mol Biol 2012; 419:125-38; PMID:22459262; http://dx.doi.org/ 10.1016/j.jmb.2012.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schifano JM, Cruz JW, Vvedenskaya IO, Edifor R, Ouyang M, Husson RN, Nickels BE, Woychik NA. tRNA is a new target for cleavage by a MazF toxin. Nucleic Acids Res 2016; 44:1256-70; PMID:26740583; http://dx.doi.org/ 10.1093/nar/gkv1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schifano JM, Vvedenskaya IO, Knoblauch JG, Ouyang M, Nickels BE, Woychik NA. An RNA-seq method for defining endoribonuclease cleavage specificity identifies dual rRNA substrates for toxin MazF-mt3. Nat Commun 2014; 5:3538; PMID:24709835; http://dx.doi.org/ 10.1038/ncomms4538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schifano JM, Edifor R, Sharp JD, Ouyang M, Konkimalla A, Husson RN, Woychik NA. Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal A site. Proc Natl Acad Sci U S A 2013; 110:8501-6; PMID:23650345; http://dx.doi.org/ 10.1073/pnas.1222031110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simanshu DK, Yamaguchi Y, Park JH, Inouye M, Patel DJ. Structural basis of mRNA recognition and cleavage by toxin MazF and its regulation by antitoxin MazE in Bacillus subtilis. Mol Cell 2013; 52:447-58; PMID:24120662; http://dx.doi.org/ 10.1016/j.molcel.2013.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zorzini V, Mernik A, Lah J, Sterckx YG, De Jonge N, Garcia-Pino A, De Greve H, Versées W, Loris R. Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF. J Biol Chem 2016; 291:10950-60; PMID:27026704; http://dx.doi.org/ 10.1074/jbc.M116.715912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, Moll I. Selective Translation of Leaderless mRNAs by Specialized Ribosomes Generated by MazF in Escherichia coli. Cell 2011; 147:147-57; PMID:21944167; http://dx.doi.org/ 10.1016/j.cell.2011.07.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sauert M, Wolfinger MT, Vesper O, Muller C, Byrgazov K, Moll I. The MazF-regulon: a toolbox for the post-transcriptional stress response in Escherichia coli. Nucleic Acids Res 2016; 44:6660-75; PMID:26908653; http://dx.doi.org/19282968 10.1093/nar/gkw115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amitai S, Kolodkin-Gal I, Hananya-Meltabashi M, Sacher A, Engelberg-Kulka H. Escherichia coli MazF leads to the simultaneous selective synthesis of both “death proteins” and “survival proteins.” PLoS Genet 2009; 5:e1000390; PMID:19282968; http://dx.doi.org/ 10.1371/journal.pgen.1000390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kasari V, Mets T, Tenson T, Kaldalu N. Transcriptional cross-activation between toxin-antitoxin systems of Escherichia coli. BMC Microbiol 2013; 13:45; PMID:23432955; http://dx.doi.org/ 10.1186/1471-2180-13-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermüller J, Reinhardt R, et al.. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 2010; 464:250-5; PMID:20164839; http://dx.doi.org/ 10.1038/nature08756 [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Zhang J, Hara H, Kato I, Inouye M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J Biol Chem 2005; 280:3143-50; PMID:15537630; http://dx.doi.org/ 10.1074/jbc.M411811200 [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Zhu L, Zhang J, Inouye M. Characterization of ChpBK, an mRNA interferase from Escherichia coli. J Biol Chem 2005; 280:26080-8; PMID:15901733; http://dx.doi.org/ 10.1074/jbc.M502050200 [DOI] [PubMed] [Google Scholar]
- 35.Pellegrini O, Mathy N, Gogos A, Shapiro L, Condon C. The Bacillus subtilis ydcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor. Mol Microbiol 2005; 56:1139-48; PMID:15882409; http://dx.doi.org/ 10.1111/j.1365-2958.2005.04606.x [DOI] [PubMed] [Google Scholar]
- 36.Kamphuis MB, Bonvin AM, Monti MC, Lemonnier M, Munoz-Gomez A, van den Heuvel RH, Díaz-Orejas R, Boelens R. Model for RNA binding and the catalytic site of the RNase Kid of the bacterial parD toxin-antitoxin system. J Mol Biol 2006; 357:115-26; PMID:16413033; http://dx.doi.org/ 10.1016/j.jmb.2005.12.033 [DOI] [PubMed] [Google Scholar]
- 37.Cannistraro VJ, Kennell D. The 5′ ends of RNA oligonucleotides in Escherichia coli and mRNA degradation. Eur J Biochem 1993; 213:285-93; PMID:7682943; http://dx.doi.org/ 10.1111/j.1432-1033.1993.tb17761.x [DOI] [PubMed] [Google Scholar]
- 38.Basturea GN, Zundel MA, Deutscher MP. Degradation of ribosomal RNA during starvation: comparison to quality control during steady-state growth and a role for RNase PH. Rna 2011; 17:338-45; PMID:21135037; http://dx.doi.org/ 10.1261/rna.2448911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maivali U, Paier A, Tenson T. When stable RNA becomes unstable: the degradation of ribosomes in bacteria and beyond. Biol Chem 2013; 394:845-55; PMID:23612597; http://dx.doi.org/ 10.1515/hsz-2013-0133 [DOI] [PubMed] [Google Scholar]
- 40.Piir K, Paier A, Liiv A, Tenson T, Maivali U. Ribosome degradation in growing bacteria. EMBO Rep 2011; 12:458-62; PMID:21460796; http://dx.doi.org/ 10.1038/embor.2011.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deutscher MP. Maturation and degradation of ribosomal RNA in bacteria. Prog Mol Biol Transl Sci 2009; 85:369-91; PMID:19215777; http://dx.doi.org/ 10.1016/S0079-6603(08)00809-X [DOI] [PubMed] [Google Scholar]
- 42.Das U, Shuman S. Mechanism of RNA 2′,3′-cyclic phosphate end healing by T4 polynucleotide kinase-phosphatase. Nucleic Acids Res 2013; 41:355-65; PMID:23118482; http://dx.doi.org/ 10.1093/nar/gks977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romero DA, Hasan AH, Lin YF, Kime L, Ruiz-Larrabeiti O, Urem M, Bucca G, Mamanova L, Laing EE, van Wezel GP, et al.. A comparison of key aspects of gene regulation in Streptomyces coelicolor and Escherichia coli using nucleotide-resolution transcription maps produced in parallel by global and differential RNA sequencing. Mol Microbiol 2014; 94:963-87; PMID:25266672; http://dx.doi.org/27298395 10.1111/mmi.12810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sulthana S, Basturea GN, Deutscher MP. Elucidation of pathways of ribosomal RNA degradation: an essential role for RNase E. Rna; 22:1163-71; PMID:27298395; http://dx.doi.org/ 10.1261/rna.056275.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dahlberg AE, Dahlberg JE, Lund E, Tokimatsu H, Rabson AB, Calvert PC, Reynolds F, Zahalak M. Processing of the 5′ end of Escherichia coli 16S ribosomal RNA. Proc Natl Acad Sci U S A 1978; 75:3598-602; PMID:358190; http://dx.doi.org/ 10.1073/pnas.75.8.3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sirdeshmukh R, Schlessinger D. Ordered processing of Escherichia coli 23S rRNA in vitro. Nucleic Acids Res 1985; 13:5041-54; PMID:2991850; http://dx.doi.org/ 10.1093/nar/13.14.5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Srivastava AK, Schlessinger D. Coregulation of processing and translation: mature 5′ termini of Escherichia coli 23S ribosomal RNA form in polysomes. Proc Natl Acad Sci U S A 1988; 85:7144-8; PMID:3050989; http://dx.doi.org/ 10.1073/pnas.85.19.7144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sirdeshmukh R, Schlessinger D. Why is processing of 23 S ribosomal RNA in Escherichia coli not obligate for its function? J Mol Biol 1985; 186:669-72; PMID:3912511; http://dx.doi.org/ 10.1016/0022-2836(85)90139-1 [DOI] [PubMed] [Google Scholar]
- 49.Luidalepp H, Berger S, Joss O, Tenson T, Polacek N. Ribosome shut-down by 16S rRNA fragmentation in stationary phase E. coli. J Mol Biol 2016; 428:2237-47; PMID:27067112; http://dx.doi.org/12972253 10.1016/j.jmb.2016.01.033 [DOI] [PubMed] [Google Scholar]
- 50.Christensen SK, Pedersen K, Hansen FG, Gerdes K. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J Mol Biol 2003; 332:809-19; PMID:12972253; http://dx.doi.org/ 10.1016/S0022-2836(03)00922-7 [DOI] [PubMed] [Google Scholar]
- 51.Sat B, Hazan R, Fisher T, Khaner H, Glaser G, Engelberg-Kulka H. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J Bacteriol 2001; 183:2041-5; PMID:11222603; http://dx.doi.org/ 10.1128/JB.183.6.2041-2045.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lancaster LE, Savelsbergh A, Kleanthous C, Wintermeyer W, Rodnina MV. Colicin E3 cleavage of 16S rRNA impairs decoding and accelerates tRNA translocation on Escherichia coli ribosomes. Mol Microbiol 2008; 69:390-401; PMID:18485067; http://dx.doi.org/ 10.1111/j.1365-2958.2008.06283.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siibak T, Peil L, Donhofer A, Tats A, Remm M, Wilson DN, Tenson T, Remme J. Antibiotic-induced ribosomal assembly defects result from changes in the synthesis of ribosomal proteins. Mol Microbiol 2011; 80:54-67; PMID:21320180; http://dx.doi.org/ 10.1111/j.1365-2958.2011.07555.x [DOI] [PubMed] [Google Scholar]
- 54.Siibak T, Peil L, Xiong L, Mankin A, Remme J, Tenson T. Erythromycin- and chloramphenicol-induced ribosomal assembly defects are secondary effects of protein synthesis inhibition. Antimicrob Agents Chemother 2009; 53:563-71; PMID:19029332; http://dx.doi.org/ 10.1128/AAC.00870-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dodd J, Kolb JM, Nomura M. Lack of complete cooperativity of ribosome assembly in vitro and its possible relevance to in vivo ribosome assembly and the regulation of ribosomal gene expression. Biochimie 1991; 73:757-67; PMID:1764521; http://dx.doi.org/ 10.1016/0300-9084(91)90055-6 [DOI] [PubMed] [Google Scholar]
- 56.Ainelo A, Tamman H, Leppik M, Remme J, Horak R. The toxin GraT inhibits ribosome biogenesis. Mol Microbiol 2016; 100:719-34; PMID:26833678; http://dx.doi.org/ 10.1111/mmi.13344 [DOI] [PubMed] [Google Scholar]
- 57.Suzuki M, Zhang J, Liu M, Woychik NA, Inouye M. Single protein production in living cells facilitated by an mRNA interferase. Mol Cell 2005; 18:253-61; PMID:15837428; http://dx.doi.org/ 10.1016/j.molcel.2005.03.011 [DOI] [PubMed] [Google Scholar]
- 58.Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc Natl Acad Sci U S A 2001; 98:14328-33; PMID:11717402; http://dx.doi.org/ 10.1073/pnas.251327898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ray AE, Connon SA, Sheridan PP, Gilbreath J, Shields M, Newby DT, Fujita Y, Magnuson TS. Intragenomic heterogeneity of the 16S rRNA gene in strain UFO1 caused by a 100-bp insertion in helix 6. FEMS Microbiol Ecol 2010; 72:343-53; PMID:20557571; http://dx.doi.org/ 10.1111/j.1574-6941.2010.00868.x [DOI] [PubMed] [Google Scholar]
- 60.Evguenieva-Hackenberg E. Bacterial ribosomal RNA in pieces. Mol Microbiol 2005; 57:318-25; PMID:15978067; http://dx.doi.org/ 10.1111/j.1365-2958.2005.04662.x [DOI] [PubMed] [Google Scholar]
- 61.Winkler ME. Ribosomal ribonucleic acid isolated from Salmonella typhimurium: absence of the intact 23S species. J Bacteriol 1979; 139:842-9; PMID:383696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cochella L, Brunelle JL, Green R. Mutational analysis reveals two independent molecular requirements during transfer RNA selection on the ribosome. Nat Struct Mol Biol 2007; 14:30-6; PMID:17159993; http://dx.doi.org/ 10.1038/nsmb1183 [DOI] [PubMed] [Google Scholar]
- 63.Leonov AA, Sergiev PV, Bogdanov AA, Brimacombe R, Dontsova OA. Affinity purification of ribosomes with a lethal G2655C mutation in 23 S rRNA that affects the translocation. J Biol Chem 2003; 278:25664-70; PMID:12730236; http://dx.doi.org/ 10.1074/jbc.M302873200 [DOI] [PubMed] [Google Scholar]
- 64.Gregory ST, O'Connor M, Dahlberg AE. Functional Escherichia coli 23S rRNAs containing processed and unprocessed intervening sequences from Salmonella typhimurium. Nucleic Acids Res 1996; 24:4918-23; PMID:9016661; http://dx.doi.org/ 10.1093/nar/24.24.4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gourse RL, Gaal T, Bartlett MS, Appleman JA, Ross W. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Ann Rev Microbiol 1996; 50:645-77; PMID:8905094; http://dx.doi.org/ 10.1146/annurev.micro.50.1.645 [DOI] [PubMed] [Google Scholar]
- 66.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10:R25; PMID:19261174; http://dx.doi.org/ 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25:2078-9; PMID:19505943; http://dx.doi.org/ 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Noeske J, Huang J, Olivier NB, Giacobbe RA, Zambrowski M, Cate JH. Synergy of streptogramin antibiotics occurs independently of their effects on translation. Antimicrob Agents Chemother; 58:5269-79; PMID:24957822; http://dx.doi.org/ 10.1128/AAC.03389-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.