ABSTRACT
RNA helicase Brr2 is implicated in multiple phases of pre-mRNA splicing and thus requires tight regulation. Brr2 can be auto-inhibited via a large N-terminal region folding back onto its helicase core and auto-activated by a catalytically inactive C-terminal helicase cassette. Furthermore, it can be regulated in trans by the Jab1 domain of the Prp8 protein, which can inhibit Brr2 by intermittently inserting a C-terminal tail in the enzyme's RNA-binding tunnel or activate the helicase after removal of this tail. Presently it is unclear, whether these regulatory mechanisms functionally interact and to which extent they are evolutionarily conserved. Here, we report crystal structures of Saccharomyces cerevisiae and Chaetomium thermophilum Brr2-Jab1 complexes, demonstrating that Jab1-based inhibition of Brr2 presumably takes effect in all eukaryotes but is implemented via organism-specific molecular contacts. Moreover, the structures show that Brr2 auto-inhibition can act in concert with Jab1-mediated inhibition, and suggest that the N-terminal region influences how the Jab1 C-terminal tail interacts at the RNA-binding tunnel. Systematic RNA binding and unwinding studies revealed that the N-terminal region and the Jab1 C-terminal tail specifically interfere with accommodation of double-stranded and single-stranded regions of an RNA substrate, respectively, mutually reinforcing each other. Additionally, such analyses show that regulation based on the N-terminal region requires the presence of the inactive C-terminal helicase cassette. Together, our results outline an intricate system of regulatory mechanisms, which control Brr2 activities during snRNP assembly and splicing.
KEYWORDS: Brr2, pre-mRNA splicing, remodeling of RNA-protein complexes, RNA helicase structure and function, spliceosome catalytic activation, X-ray crystallography
Introduction
Splicing entails the excision of non-coding sequences (introns) from eukaryotic pre-mRNAs and the ligation of neighboring coding sequences (exons) and is mediated by a large, molecular RNA-protein (RNP) machine, the spliceosome. The spliceosome is composed of 5 small nuclear (sn) RNPs (U1, U2, U4, U5 and U6 snRNP in the major spliceosome), each comprising a unique snRNA and different sets of proteins, as well as many non-snRNP proteins.1,2 For each splicing event, these subunits assemble anew and in a stepwise manner on a pre-mRNA substrate.1,2 Initial assembly steps lead to a catalytically inactive spliceosome, which is subsequently activated in several stages, before carrying out the 2 transesterification reactions (steps 1 and 2) of a splicing event. After splicing catalysis, the products are released and the spliceosomal subunits are recycled. Each transition in this splicing cycle is accompanied by conformational and compositional remodeling of the underlying spliceosomal protein-protein, RNA-RNA and protein-RNA interaction networks.1-3 These rearrangements are driven and controlled by the ATP/NTP-dependent RNP remodeling activities of at least 8 conserved superfamily 2 (SF2) RNA helicases.4
The most profound remodeling events occur during spliceosome activation. Initially, U4 and U6 snRNAs are extensively base-paired via 2 regions (stems I and II) and are bound by several proteins in a U4/U6 di-snRNP, which joins the spliceosome together with U5 snRNP as a pre-formed U4/U6•U5 tri-snRNP. During spliceosome activation, U4/U6 base pairing is disrupted by the Brr2 helicase5,6 and U4 snRNA as well as U4/U6-associated proteins are released,7,8 allowing U6 snRNA to engage in alternative interactions with U2 snRNA and the pre-mRNA substrate and to form an internal stem-loop structure required for splicing catalysis.9-11
Brr2 is the only member of the Ski2-like family of SF2 helicases in the spliceosome. It comprises a ∼450-residue N-terminal region (NTR) followed by a tandem array of similarly structured helicase units (cassettes; together referred to as the “helicase region”), only the first of which is active as an ATPase and RNA helicase.12,13 Each helicase cassette is composed of 2 RecA-like domains (RecA1 and RecA2), a winged-helix (WH) domain, a helical bundle (HB) domain, a helix-loop-helix (HLH) domain and an immunoglobulin-like (IG) domain, with the latter 3 domains forming a Sec 63 homology unit.13-15 Brr2 is a stable subunit of the U5 snRNP and thus encounters its U4/U6 di-snRNP substrate already outside the spliceosome in the U4/U6•U5 tri-snRNP, where mechanisms must exist to prevent pre-mature Brr2-mediated U4/U6 unwinding. While most other spliceosomal helicases only join the spliceosome during the phase in which their activities are required, Brr2 remains associated with the spliceosome after spliceosome activation throughout the remainder of the splicing cycle.7 Moreover, Brr2 has additionally been implicated in later phases of splicing, i.e. during the transition from the first to the second step of splicing16,17 and during spliceosome disassembly.18 However, it is presently not clear whether Brr2 ATPase and/or helicase activity is required during these later stages.16,19 Thus, Brr2 activity needs to be tightly regulated and possibly switched on and off repeatedly during each splicing event.
Several mechanisms have been elucidated, by which Brr2 can be regulated during a splicing event. The large NTR can fold back onto the 2 helicase cassettes and auto-inhibit the enzyme by substrate competition and conformational clamping.20 The NTR contains 4 functional regions (Fig. 1A): (i) A helical “plug” domain that can wedge between the RecA2 and HB domains of the NC and obstruct substrate RNA access; (ii) an inter-cassette (IC) clamp that connects the NC and the CC; (iii) a PWI-like domain that contacts the CC; and (iv) an NC-clamp that interconnects the RecA1, RecA2 and WH domains of the NC.20,21 Furthermore, the inactive C-terminal cassette (CC) has been shown to act as an intra-molecular activator of the active N-terminal cassette (NC).13 Additionally, Brr2 is regulated by the Jab1 domain of the Prp8 protein,22-25 which binds on top of the Sec 63 unit of the NC and can inhibit Brr2 by inserting an intrinsically disordered C-terminal tail into the Brr2 RNA-binding tunnel. Interestingly, upon removal of the C-terminal tail, the Jab1 domain is converted into a strong activator of Brr2 activity.22 Finally, an RNaseH-like domain preceding the Jab1 domain in Prp8 can transiently bind U4/U6 di-snRNAs and sequester them from Brr2.26
Figure 1.
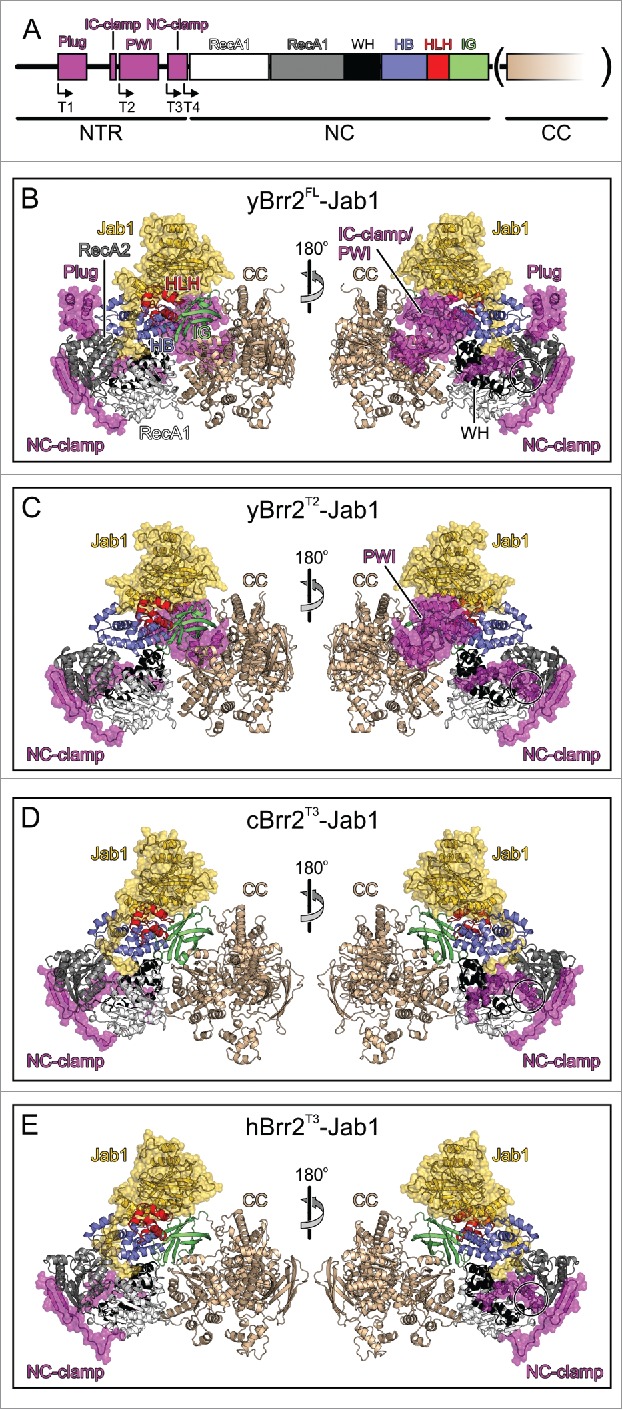
Structures of Brr2-Jab1 complexes. (A) Schematics of the Brr2 constructs used in the present and former studies. N-terminal truncations (T1, T2, T3 and T4; start sites indicated by angled arrows) were combined with deletion of the CC (in parentheses) in some cBrr2 variants investigated. (B) Diametric views of the yBrr2FL-Jab1 complex. (C) Diametric views of the yBrr2T2-Jab1 complex. (D) Diametric views of the cBrr2T3-Jab1 complex. (E) Diametric views of the hBrr2T3-Jab1 complex (PDB ID 4KIT).23 Domains and regions are colored identically in this and all following figures. NTR – magenta; RecA1 – light gray; RecA2 – dark gray; WH – black; HB – blue; HLH – red; IG – green; CC – beige; Jab1 – gold. Regions of the NTRs and Jab1 domains present in the various structures are highlighted by semi-transparent surfaces. Circles in (B-E) – residues of the NC-clamp (436–443 in yBrr2, 418–425 in hBrr2, 467–474 in cBrr2) not defined in the electron densities of the yBrr2FL-Jab1 (B) and yBrr2FL-Jab1ΔC 20 structures. Structures were aligned according to their Jab1 domains.
Currently it is unclear, at which stage of a splicing cycle the various Brr2 regulatory mechanisms take effect and whether there is any functional cross-talk between them. Furthermore, as Jab1-mediated inhibition of Brr2 was so far only seen in the structure of a human (h) Brr2-Jab1 complex23 but not in a similar structure from yeast,24 it is presently not clear to which extent the regulatory mechanisms are evolutionarily conserved. Here, we report crystal structures of yeast (y) Brr2-Jab1 and Chaetomium thermophilum (c) Brr2-Jab1 complexes, in which the Brr2 subunits encompass different portions of the NTR. Our structures reveal that Brr2 inhibition via the Jab1 C-terminal tail is conserved but exhibits species-specific differences in the underlying molecular contacts. In addition, the structures in combination with systematic RNA binding and unwinding studies show that the Brr2 NTR influences the mode of interaction of the Jab1 C-terminal tail at the RNA-binding tunnel, that the NTR and the Jab1 C-terminal tail can cooperatively inhibit Brr2 by competing with the accommodation of different portions of an RNA substrate, and that NTR-mediated regulation requires the presence of the inactive CC.
Results
Structural organization of yeast, C. thermophilum and human Brr2-Jab1 complexes
Besides yeast and human Brr2 and Prp8 proteins, we included Brr2 and Prp8 orthologs from C. thermophilum in the present analyses, as this organism thrives at temperatures up to 60°C. Tolerance of high temperature might indicate that C. thermophilum proteins are conformationally more rigid at room temperature or below than proteins from mesophilic organisms and thus possibly more amenable to crystallization. The NTR (yBrr2 – residues 1–478, cBrr2 – 1–510, hBrr2 – 1–462), NC (yBrr2 – 479–1310, cBrr2 – 511–1340, hBrr2 – 463–1288) and CC (yBrr2 – 1311–2163, cBrr2 – 1341–2205, hBrr2 – 1289–2136) of yeast, C. thermophilum and human Brr2 exhibit 25–35%, 50–64% and 39–49% sequence identity, respectively. A high level of conservation (43–55%) is also observed for the Prp8 Jab1 domains (yJab1 – 2147–2335, cJab1 – 2112–2385, hJab1 – 2064–2335). Human Brr2 and Prp8 share a higher degree of sequence identity with the corresponding C. thermophilum than with the more distantly related yeast proteins. Despite this relatively high level of similarity, the insertion of the C-terminal tail of the Prp8 Jab1 domain into the RNA-binding tunnel of Brr2 was observed in the crystal structure of a hBrr2-Jab1 complex,23 in which the hBrr2 subunit lacked all parts of the NTR except the NC-clamp (hBrr2 truncation 3; hBrr2T3; see Fig. 1A for a scheme illustrating the various Brr2 N-terminal truncation variants used in this and previous studies), but not in the structure of a yBrr2-Jab1 complex,24 in which the yBrr2 subunit additionally lacked a portion of the NC-clamp (yBrr2T4). These observations raised the question, to which extent the binding of the Jab1 C-terminal tail to the RNA-binding tunnel of Brr2 is conserved among species. Furthermore, in a recent crystal structure of a full-length (FL) yBrr2-Jab1ΔC complex,20 which contained a Jab1 fragment lacking the C-terminal 15 residues, the NTR was observed to fold back onto the helicase region, with the plug domain bound between the RecA2 and HB domains of the Brr2 NC, in close proximity to the Jab1 domain, thus auto-inhibiting Brr2 by substrate competition and conformational clamping. However, the analysis left the question unresolved, if the NTR and the Jab1 C-terminal tail can act in concert to inhibit Brr2.
To start to address the above questions, we determined crystal structures of yBrr2-Jab1 complexes bearing yBrr2FL or a N-terminally truncated yBrr2 variant starting at the PWI-like domain (yBrr2T2), as well as of a cBrr2T3-Jab1 complex, in which the cBrr2 variant lacked elements N-terminal of the NC-clamp (Fig. 1B–D). yBrr2FL-Jab1 and yBrr2T2-Jab1 crystals each contained 2 complexes in an asymmetric unit (root-mean-square deviation [rmsd] 1.9 Å for 2,125 common Cα atoms and 1.1 Å for 1,937 common Cα atoms, respectively) and diffracted to 3.4 Å and 4.2 Å resolution, respectively. cBrr2T3-Jab1 crystals contained 4 copies of the complex per asymmetric unit (rmsd 0.5–0.9 Å for 1,520–1,768 common Cα atoms) and diffracted to 3.2 Å resolution (Table 1; Fig. S1). Although the yBrr2T2-Jab1 crystals diffracted only to comparatively low resolution, the structure of the yBrr2T2-Jab1 complex could be solved and refined unequivocally based on the related higher-resolution structures. While structural details, such as the exact conformations of all side chains, cannot be reliably modeled at this resolution, the overall features of the structure were clearly defined in the electron density.
Table 1.
Crystallographic data.
| Data collection | |||
|---|---|---|---|
| Structure | yBrr2FL-Jab1 | yBrr2T2-Jab1 | cBrr2T3-Jab1 |
| Wavelength [Å] | 0.9184 | 0.9184 | 0.9999 |
| Space group | P212121 | P21212 | P21 |
| Unit cell parameters | |||
| a [Å] | 179.3 | 186.0 | 92.5 |
| b [Å] | 181.2 | 196.9 | 269.6 |
| c [Å] | 210.3 | 191.0 | 231.7 |
| β [°] | 90.1 | ||
| Resolution [Å]a | 50.0 – 3.4 | 95.5 – 4.2 | 50.0 – 3.2 |
| (3.6 – 3.4) | (4.4 – 4.2) | (3.4 – 3.2) | |
| Reflections | |||
| Unique | 94451 (14794) | 51802 (5113) | 184926 (29309) |
| Multiplicity | 6.8 (6.4) | 10.8 (10.9) | 7.0 (6.8) |
| Completeness [%] | 99.6 (98.2) | 99.9 (99.8) | 98.8 (97.6) |
| Mean I/σ(I) | 7.1 (1.3) | 8.6 (1.4) | 8.1 (0.9) |
| Rsym [%] | 32.7 (156.0) | 31.5 (202.4) | 29.5 (222.1) |
| CC1/2 | 0.985 (0.451) | 0.995 (0.609) | 0.991 (0.266) |
| Wilson B-factor [Å2] | 78.1 | 147.0 | 75.7 |
| Refinement | |||
| Resolution (Å) | 48.7−3.4 | 95.5–4.2 | 49.1–3.2 |
| (3.5–3.4) | (4.4–4.2) | (3.3–3.2) | |
| Reflections | |||
| Unique | 92351 (9078) | 51752 (5112) | 184414 (18425) |
| Test set [%] | 5 | 5 | 5 |
| Rwork [%] | 25.3 (37.3) | 30.7 (38.4) | 24.3 (36.0) |
| Rfree [%] | 29.6 (38.0) | 33.5 (40.6) | 28.6 (38.0) |
| Protein residues | 4306 | 4196 | 7956 |
| Number of non-hydrogen atoms | 34522 | 33653 | 63709 |
| Average B-factor [Å2] | 94.3 | 157.9 | 88.30 |
| Rmsd from ideality | |||
| Bond lengths [Å] | 0.007 | 0.03 | 0.002 |
| Bond angles [°] | 0.94 | 0.76 | 0.55 |
| Molprobity | |||
| Overall score | 1.11 | 1.43 | 2.41 |
| Clash score | 0.59 | 1.77 | 9.45 |
| Ramachandran favored [%] | 93.8 | 92.0 | 92.0 |
| Ramachandran outliers [%] | 0.4 | 0.26 | 1.4 |
| PDB entry | 5M52 | 5M5P | 5M59 |
Values for the highest resolution shell in parentheses.
The helicase regions and the globular parts of the Jab1 domains of the present structures closely resemble the corresponding elements of the previously determined yBrr2FL-Jab1ΔC,20 hBrr2T3-Jab113 and yBrr2T4-Jab124 structures, with the Jab1 domains bound on top of the Sec 63 homology units of the NCs (Fig. 1B–E; Fig S1). Furthermore, the NTR of the yBrr2FL-Jab1 complex matches the NTR of the previous yBrr2FL-Jab1ΔC structure, with residues 1–112 not defined in the electron density, the helical plug domain positioned between the RecA2 and HB domains of the NC, the IC-clamp running alongside NC and CC, the PWI domain neighboring the CC, and the NC-clamp encircling the NC (Fig. 1B). In the yBrr2T2-Jab1 structure, the PWI-domain resides next to the CC, as in the yBrr2FL-Jab1 and yBrr2FL-Jab1ΔC structures (Fig 1C). Visible portions of the NC-clamps of the yBrr2T2-Jab1 and cBrr2T3-Jab1 complexes are positioned very similarly to the corresponding portions of the NC-clamps in the yBrr2FL-Jab1, yBrr2FL-Jab1ΔC and hBrr2T3-Jab1 complexes (Fig 1C–E). However, in the yBrr2FL-Jab1 and yBrr2FL-Jab1ΔC structures, residues 436–443 of the NC-clamp (corresponding to hBrr2 residues 418–425 and cBrr2 residues 467–474), are not defined in the electron densities (Fig. 1B, circled region), while these residues aid in interconnecting the RecA1, RecA2 and WH domains in the yBrr2T2-Jab1, cBrr2T3-Jab1 and hBrr2T3-Jab1 complexes (Fig. 1C–E, circled regions). These latter observations indicate that release of the plug domain from the cleft between the N-terminal RecA2 and HB domains is accompanied by more intimate binding of the NC-clamp around the NC.
Binding of the Prp8 Jab1 C-terminal tail at the Brr2 RNA-binding tunnel is evolutionarily conserved
Similar to the yBrr2T4-Jab1 complex, the RNA-binding tunnel in the yBrr2T2-Jab1 structure is unoccupied. In contrast, the C-terminal tails of the respective Jab1 domains are bound at the RNA-binding tunnels in the yBrr2FL-Jab1 and cBrr2T3-Jab1 complexes, resembling the hBrr2T3-Jab1 structure (Fig. 1B–E). Superpositioning of yBrr2FL-Jab1, cBrr2T3-Jab1 and hBrr2T3-Jab1 complexes according to the Jab1 domains revealed that the proximal parts of the Jab1 tails (yPrp8 residues 2388–2393, cPrp8 residues 2363–2368, hPrp8 residues 2310–2314; Fig. 2A) run in a similar fashion between the HLH and HB domains of the respective Brr2 NCs. However, the central (yPrp8 residues 2394–2399, cPrp8 residues 2369–2374, hPrp8 residues 2315–2321; Fig. 2A) and distal parts (yPrp8 residues 2400–2413, cPrp8 residues 2375–2385, hPrp8 residues 2322–2335; Fig. 2A) of the tails contact Brr2 differently in the different species. In yeast, the central Jab1 tail interacts with the HLH domain, whereas in C. thermophilum and human the corresponding portions solely contact the HB domains (Fig. 2B–D, top panels; Table 2). The distal portions of the Jab1 tails show strikingly different positions and engage in diverse interactions in the 3 organisms (Fig. 2B–D, bottom panels; Table 2). The yJab1 distal tail contacts mainly the RecA2 domain and has only few contacts with the HB and the RecA1 domains (Fig. 2B, bottom panel). The cJab1 distal tail interacts predominantly with the HB and the RecA1 domains and engages in additional contacts to the RecA2 domain (Fig. 2C, bottom panel). The hJab1 distal tail interacts mainly with the HB and RecA1 domains, displaying only few contacts to the RecA2 domain (Fig. 2D, bottom panel). Thus, while the general ability of the C-terminal Jab1 tail to occupy the RNA-binding tunnel of Brr2 is universally conserved, this interaction is based on different molecular contacts in different species.
Figure 2.
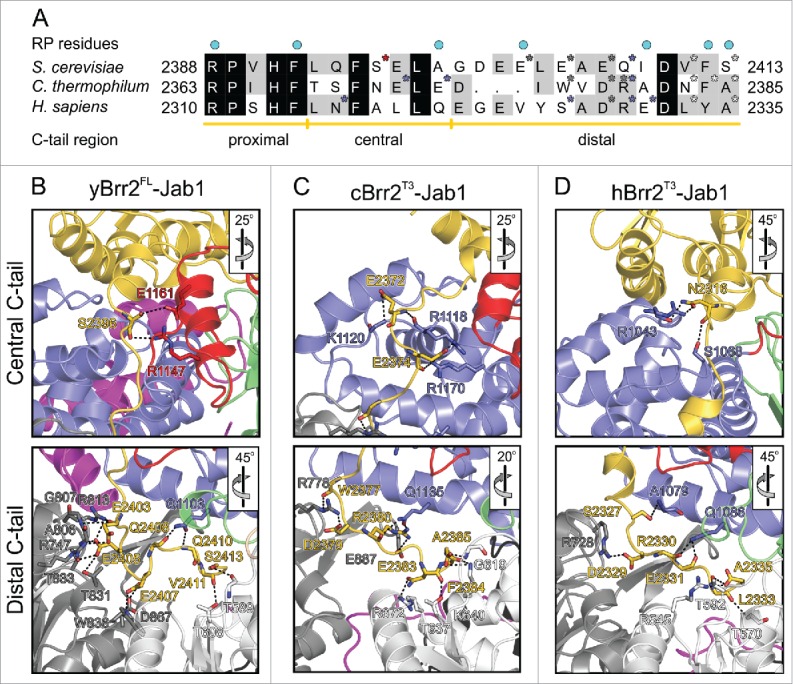
Interactions of the Jab1 C-terminal tail with the RNA-binding tunnel of Brr2. (A) Sequence alignment of the Jab1 tails of S. cerevisiae, C. thermophilum and H. sapiens. Residues interacting at the RNA-binding tunnel of the respective Brr2 protein are indicated by asterisks, colored according to the respective interacting Brr2 domain (RecA1 – light gray; RecA2 – dark gray; HB – blue; HLH – red). Residues affected by prp8 mutations that cause retinitis pigmentosa in humans are indicated by cyan dots above the alignment. Regions corresponding to the proximal, central and distal portions of the tails are indicated below the alignment. (B-D) Details of the interactions of the Jab1 C-terminal tails at the Brr2 RNA-binding tunnels in the yBrr2FL-Jab1 complex (B), cBrr2T3-Jab1 complex (C) and hBrr2T3-Jab1 complex (PDB ID 4KIT)23 (D). Top panels – interactions of the central regions of the Jab1 tails. Bottom panels – interactions of the distal regions of the Jab1 tails. Interacting residues are shown as sticks and colored by atom type (carbon – as the respective domain/region; nitrogen – blue; oxygen – red). For interactions involving only protein backbone atoms, side chains are not shown for clarity. Dashed lines indicate hydrogen bonds or salt bridges. Rotation symbols indicate orientations relative to Fig. 1B, D and E, respectively.
Table 2.
Contacts between the Jab1 C-terminal tail and the Brr2 N-terminal cassette.
| Organism | Jab1 residue | Tail region | Brr2 residue | Brr2 NC domain | Distance [Å] |
|---|---|---|---|---|---|
| Yeast | S2396 | central | E1161 | HLH | 3.9 |
| S2396 (BB)a | central | R1147 | HLH | 3.4 | |
| E2403 (BB) | distal | R747 | RecA2 | 3.1 | |
| E2403 | distal | G807 (BB) | RecA2 | 4.0 | |
| E2403 | distal | R813 | RecA2 | 3.3 | |
| E2405 | distal | R747 | RecA2 | 3.3 | |
| E2405 (BB) | distal | A806 (BB) | RecA2 | 3.4 | |
| E2405 | distal | T831 | RecA2 | 2.7 | |
| E2405 | distal | T833 | RecA2 | 3.7 | |
| E2407 | distal | W836 | RecA2 | 3.2 | |
| E2407 | distal | D867 | RecA2 | 3.0 | |
| Q2408 (BB) | distal | Q1103 | HB | 2.7 | |
| D2410 (BB) | distal | Q1103 | HB | 3.6 | |
| V2411 (BB) | distal | T608 | RecA1 | 2.9 | |
| S2413 | distal | T589 (BB) | RecA1 | 3.1 | |
| C. thermophilum | E2372 (BB) | central | R1118 (BB) | HB | 3.2 |
| E2372 | central | K1120 (BB) | HB | 3.6 | |
| E2374 | central | R1118 | HB | 3.0 | |
| E2374 (BB) | central | R1170 | HB | 3.0 | |
| E2374 | central | R1170 | HB | 4.0 | |
| W2377 (BB) | distal | R778 | RecA2 | 3.3 | |
| D2379 | distal | R778 | RecA2 | 2.4 | |
| R2380 | distal | Q1135 | HB | 2.3 | |
| R2380 (BB) | distal | Q1135 | HB | 3.4 | |
| R2380 | distal | E887 | RecA2 | 3.5 | |
| N2383 | distal | R672 | RecA1 | 3.1 | |
| N2383 (BB) | distal | T637 | RecA1 | 2.6 | |
| F2384 (BB) | distal | K640 | RecA1 | 2.3 | |
| A2385 (BB) | distal | G619 (BB) | RecA1 | 2.4 | |
| A2385 (BB) | distal | K640 | RecA1 | 3.4 | |
| Human | N2316 | central | R1043 (BB) | HB | 3.6 |
| N2316 | central | R1043 | HB | 2.6 | |
| N2316 (BB) | central | S1068 | HB | 3.3 | |
| S2327 | distal | A1079 (BB) | HB | 4.0 | |
| D2329 | distal | R728 | RecA2 | 2.6 | |
| R2330 (BB) | distal | Q1086 | HB | 2.7 | |
| E2331 (BB) | distal | Q1086 | HB | 2.9 | |
| L2333 (BB) | distal | K592 (BB) | RecA1 | 3.4 | |
| A2335 (BB) | distal | R545 | RecA1 | 3.3 | |
| A2335 (BB) | distal | T570 | RecA1 | 3.2 |
BB – backbone.
Structural comparisons suggest an inter-dependence of NTR-based auto-inhibition and Jab1 tail binding
While our structure of the yBrr2FL-Jab1 complex indicates that Brr2 auto-inhibition via the NTR folding back on the helicase region and inhibition in trans by the Jab1 C-terminal tail can manifest at the same time, it leaves open the question of any functional cross-talk between the 2 inhibitory mechanisms. Consistent with a lack of stable Jab1 tail binding to the RNA-binding tunnel of yBrr2 in the yBrr2T2-Jab1 and the yBrr2T4-Jab1 complex, the RecA2 domain in these 2 complexes are slightly repositioned compare with the yBrr2FL-Jab1 complex. These results suggest that the RecA2 domain is flexibly hinged to the remainder of the NC and that its position is sensitive to the presence of the NTR, possibly explaining the loss of stable Jab1 tail binding upon NTR deletion (Fig. 3A and Fig. S2). Likewise, conformational rearrangements in the yBrr2T2-Jab1 and the yBrr2T4-Jab1 complex would be expected to lead to loss of interactions of the Jab1 tail to the N-terminal Brr2 HLH domain, which might be involved in positioning the Jab1 tail for insertion into the RNA-binding tunnel (Fig. 3B and Fig. S2). Besides rationalizing the lack of stable binding of the Jab1 tail in the latter 2 scenarios, this analysis suggests that wedging of the plug between the RecA2 and HB domains influences the relative positions of other domains of the NC to support binding of the Jab1 C-terminal tail at the RNA-binding tunnel. However in contrast to the yeast system, the Jab1 tail was stably bound at the Brr2 RNA-binding tunnels in the human and C. thermophilum Brr2T3-Jab1 structures in the absence of most of the respective NTRs, indicating that different relative affinities underlie this structural interplay in different species.
Figure 3.
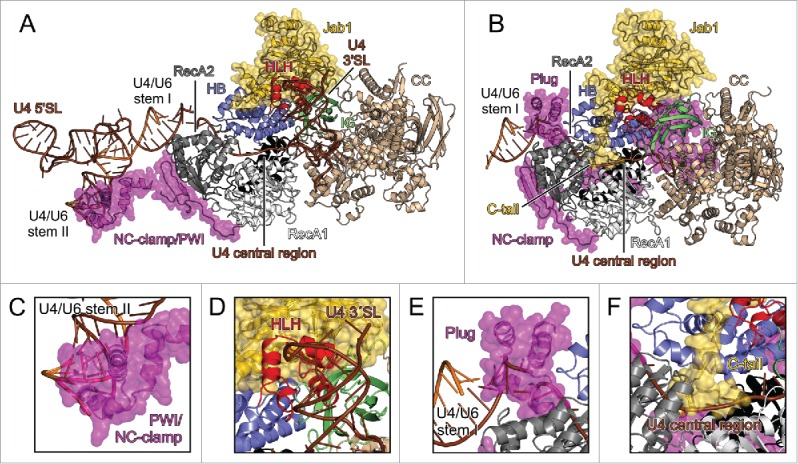
U4/U6 binding by Brr2 and possible inhibitory mechanisms. (A) yBrr2-Jab1 bound to U4/U6 as seen in the cryo-EM structure of a yeast U4/U6•U5 tri-snRNP (PDB ID 5GAO).27 (B) Structure of the isolated yBrr2FL-Jab1 complex with portions of U4/U6, obtained by superimposition with the structure in (A) according to the NCs of the Brr2 subunits. (C) The NC-clamp and part of the PWI domain of the NTR bind U4/U6 stem II in the yeast U4/U6•U5 tri-snRNP. (D) The HLH domain of the NC contacts the U4 3′-SL in the yeast U4/U6•U5 tri-snRNP. (E) In isolated yBrr2FL-Jab1, the plug domain of the NTR sterically hinders accommodation of the stem I portion of U4/U6 between the RecA2 and HB domains of the NC. (F) In isolated yBrr2FL-Jab1, the Jab1 C-terminal tail binds along and hinders opening of the HB and RecA2 domains to accommodate RNA and occupies part of the tunnel that accommodates the ss region of U4 snRNA neighboring U4/U6 stem I during U4/U6 unwinding. U4 snRNA – brown; U6 snRNA – orange.
The NTR and the Jab1 tail cooperate in inhibiting RNA binding by Brr2
A recent electron-cryomicroscopic (cryo-EM) structure of a yeast U4/U6•U5 tri-snRNP,27 in which Brr2 is loaded onto U4 snRNA and ready to unwind U4/U6 stem I, disclosed various Brr2-RNA interfaces (Fig. 3A). In that structure, the single-stranded (ss) U4 central domain runs through the RNA-binding tunnel of the NC (Fig. 3A), U4/U6 stem I is positioned between the HB and RecA2 domains (Fig. 3A), regions of the NTR corresponding to the NC-clamp and parts of the PWI domain are detached from the helicase core and instead interact with U4/U6 stem II (Fig. 3A,C) and a U4 3′-stem loop (SL) contacts the HLH domain of the Brr2 NC (Fig. 3A,D). Superpositioning of our yBrr2FL-Jab1 structure on the yBrr2-U4/U6 sub-complex of the yeast tri-snRNP (Fig. 3B) showed that the plug domain of the NTR, positioned between the N-terminal Brr2 RecA2 and HB domains, interferes with accommodation of U4/U6 stem I (Fig. 3B,E). The proximal and central parts of the Jab1 C-terminal tail bind along and thus prevent opening between the N-terminal HB and RecA2 domains of Brr2, which is required for Brr2 to engage the ss U4 central domain. Furthermore, the distal part of the tail partially occludes the RNA-binding tunnel and clashes with the ss U4 central domain (Fig. 3B,F). Consistently, the NTR and the Jab1 C-terminal tail were previously seen to inhibit Brr2-RNA interactions.20,23
To further delineate how regulatory mechanisms based on the NTR and the Jab1 tail might interact, we performed systematic binding studies with hBrr2 and hBrr2-Jab1 complexes and different RNA ligands. For single-stranded RNA binding assays, we used a single-stranded, 26-nt RNA (referred to as “ssRNA”), whose sequence corresponded to the yU4 snRNA central domain, where Brr2 loads onto the U4/U6 duplex. To address the effect of a duplex equivalent to U4/U6 stem I, we appended a 12-base pair double-stranded region to this ssRNA (dsRNA). Additionally, we monitored binding to full-length yU4/U6. ssRNA and dsRNA were labeled with 5-carboxyfluorescein (5-FAM) at the 5′-end and at the 3′-end of the complementary strand, respectively, and binding was monitored by fluorescence polarization (FP) assays (Fig. 4A,B, left and middle panels). Due to its large size, binding to U4/U6 was assessed by electrophoretic mobility shift assays (EMSAs; Fig. 4A,B, right panels).
Figure 4.
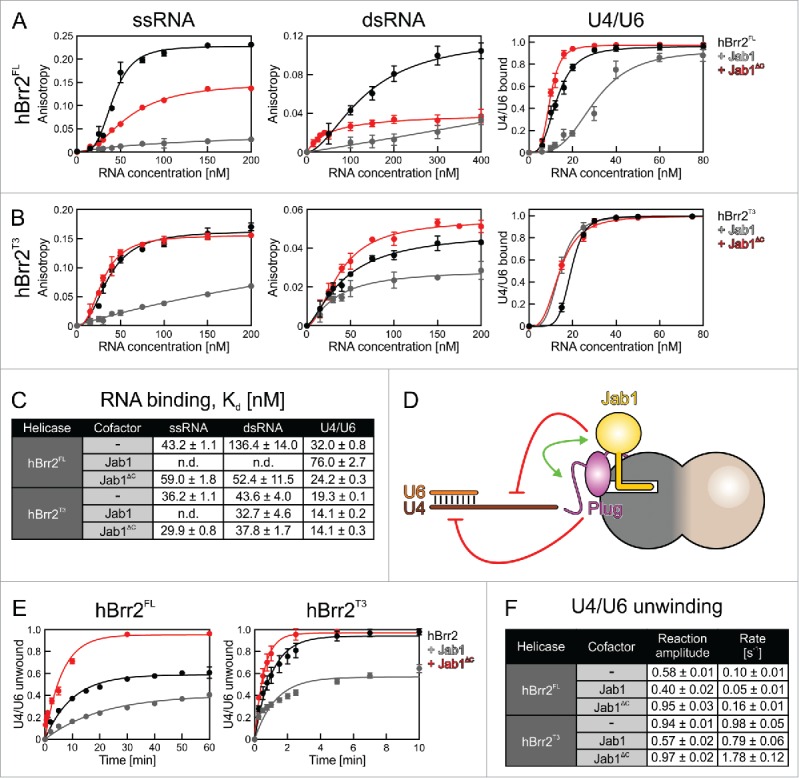
Effects of the NTR and the Prp8 Jab1 domain on RNA binding and unwinding by Brr2. (A,B) Assays monitoring RNA binding by hBrr2FL (A) or hBrr2T3 (B). Black curves – hBrr2FL or hBrr2T3 alone; gray curves – hBrr2FL or hBrr2T3 in complex with Jab1; red curves – hBrr2FL or hBrr2T3 in complex with JabΔC. Left panels – binding to ssRNA; central panels – binding to dsRNA; right panels – binding to yU4/U6 snRNA. Binding of ssRNA and dsRNA was monitored by FP using 5-FAM-labeled RNAs. Binding of yU4/U6 was monitored via EMSA. Data points and error bars represent means +/− SEMs of at least 2 independent experiments. (C) RNA affinities. Values represent means +/− SEMs of at least 2 independent experiments. Kd values were obtained by fitting the data from (A) and (B) to a single exponential Hill function (fraction bound = A[protein]n/([protein]n+Kdn); A – fitted maximum of RNA bound; n – Hill coefficient).50 (D) Scheme summarizing the effects of the NTR and the Jab1 domain on different portions of a substrate. Red symbols – inhibition; green double-arrow – reinforcement. (E) U4/U6 unwinding by Brr2 variants in the absence or presence of Jab1 variants. Black curves – Brr2FL or Brr2T3 alone; gray curves – Brr2FL or Brr2T3 in complex with Jab1; red curves – Brr2FL or Brr2T3 in complex with JabΔC. (F) Unwinding parameters. Amplitudes and unwinding rates were obtained by fitting the data to a first-order reaction (fraction unwound = A[1-exp{-ku t}]; A – amplitude of the reaction; ku – apparent first-order rate constant of unwinding; t – time). Values represent means +/− SEMs of at least 4 independent experiments.
Both hBrr2FL (Kd = 43.2 ± 1.1 nM) and hBrr2T3 (Kd = 36.2 ± 1.1 nM) bound ssRNA with high affinity, indicating that the presence of the plug domain had no major effect on ssRNA binding (Fig. 4A,B, left panels; Fig. 4C). In addition, hBrr2T3 bound with a comparable Kd to dsRNA (43.6 ± 4.0 nM) and ssRNA (36.2 ± 1.1 nM; Fig. 4B, middle panel; Fig. 4C). In contrast, hBrr2FL exhibited a more than 3-fold lower affinity for dsRNA (Kd = 136.4 ± 14.0 nM) than ssRNA (Kd = 43.2 ± 1.1 nM), suggesting that the plug domain inhibits binding to a substrate that contains a duplex equivalent to U4/U6 stem I (Fig. 4A, middle panel; Fig. 4C). Our FP data are supported by EMSAs performed with full-length U4/U6. hBrr2T3 bound with high affinity to U4/U6 (Kd = 19.3 ± 0.1 nM) (Fig. 4B, right panel; Fig. 4C), whereas hBrr2FL bound U4/U6 1.7 times more weakly (Kd = 32 ± 0.8 nM) (Fig. 4A, right panel; Fig. 4C). These data corroborate that the plug domain interferes with accommodation of a duplex portion of the substrate between the N-terminal RecA2 and HB domains. Most likely, the plug domain had a smaller effect on U4/U6 binding than on dsRNA binding because U4/U6 can additionally contact Brr2 via the U4 3′-SL and U4/U6 stem II, and interaction of the PWI/NC-clamp region with stem II might stimulate release of the plug domain from its inhibitory position between the N-terminal RecA2 and HB domains.
The Jab1 domain encompassing the C-terminal tail almost completely abrogated ssRNA binding to hBrr2FL (Fig. 4A, left panel; Fig. 4C) and hBrr2T3 (Fig. 4B, left panel; Fig. 4C; no reliable Kd could be extracted from the FP data), consistent with its RNA-competitive binding to Brr2 revealed in the structural analyses. Likewise, Jab1 further inhibited binding of hBrr2FL to dsRNA (no reliable Kd could be extracted from the FP data) or U4/U6 (Kd = 76.0 ± 2.7 nM; Fig 4A, middle and right panels; Fig. 4C). However, Jab1 did not inhibit, and even slightly increased, binding of hBrr2T3 to dsRNA (Kd = 32.7 ± 4.6 nM) or U4/U6 snRNA (Kd = 14.1 ± 0.2 nM; Fig. 4B, middle and right panels; Fig. 4C), indicating that in the absence of the plug domain the double-stranded portion of the RNA can be accommodated at the NC irrespective of the presence of the Jab1 tail. However, the binding data do not reveal whether under these conditions the ss portion of the RNA displaces the Jab1 tail from the RNA-binding tunnel.
In the presence of Jab1ΔC, hBrr2FL showed enhanced binding to dsRNA and U4/U6 and binding of hBrr2T3 to all substrates was upregulated (Fig. 4A–C). The strongest stimulation was seen for hBrr2FL binding to dsRNA (approximately 2.6-fold; Fig 4A, middle panel; Fig. 4C), suggesting that the globular part of Jab1 counteracts inhibition of RNA binding by the plug domain. Together, these analyses corroborate a functional interplay between NTR-based auto-inhibition and Brr2 inhibition in trans by the Jab1 C-terminal tail, in which the plug domain of the NTR is required for the Jab1 tail to elicit an effect on binding of a substrate that bears a duplex region preceding the ss loading region. The Jab1 domain in the absence of the C-terminal tail, in turn, favors displacement of the plug and accommodation of a duplex portion of the substrate (Fig. 4D).
The NTR and the Jab1 tail cooperate in inhibiting RNA unwinding by Brr2 in vitro
We next asked whether the cooperative effects of the Brr2 NTR and the Jab1 tail on RNA binding translate into a cooperative inhibition of U4/U6 unwinding by Brr2. To this end, we monitored unwinding of yU4/U6 by hBrr2 variants in the absence or presence of Jab1 variants by a multiple-round, gel-based unwinding assay. In this assay, differences in the maximum fraction of unwound duplex (unwinding amplitude) indicate differences in the equilibrium distribution of free Brr2 or Brr2 bound non-productively to U4/U6 and Brr2 bound productively to U4/U6. Differences in the unwinding rates indicate an effect on one or more unwinding steps. hBrr2FL exhibited a lower amplitude (A = 0.59 ± 0.01) and unwinding rate (ku = 0.1 ± 0.01) compare with hBrr2T3 (A = 0.94 ± 0.01, ku = 0.98 ± 0.05; Fig. 4E,F). The 1.6-fold reduced amplitude is consistent with inhibition of U4/U6 binding by the NTR in hBrr2FL. The about 10-fold reduction in unwinding rate of hBrr2FL compare with hBrr2T3 might indicate that in our reductionist in vitro system, regions of the NTR other than the plug might still attach to the helicase region after RNA loading and inhibit the helicase by restricting conformational rearrangements.
Upon addition of Jab1 to hBrr2FL, the unwinding amplitude further decreased 1.5-fold (A = 0.40 ± 0.01; Fig. 4E,F), most likely reflecting a shift of RNA-bound to unbound hBrr2FL by the action of the Jab1 C-terminal tail. A similar reduction of the amplitude was observed upon addition of Jab1 to hBrr2T3 (A = 0.57 ± 0.02; Fig. 4E,F). Comparison of the unwinding amplitude of hBrr2FL in the presence of Jab1 (A = 0.40 ± 0.01) to that of isolated hBrr2T3 (A = 0.94 ± 0.01; Fig. 4E,F) indicated that inhibition of U4/U6 binding by the joint action of the NTR and the Jab1 tail leads to a 2.4-fold inhibition of Brr2-mediated U4/U6 unwinding. Additionally, Jab1 led to an about 2-fold decrease in the unwinding rate of hBrr2FL (ku = 0.05 ± 0.01) and an about 1.2-fold decrease in the unwinding rates of hBrr2T3 (ku = 0.79 ± 0.06; Fig. 4E,F). Likewise, addition of Jab1 to hBrr2FL doubled the decrease in the unwinding rate compare with that of isolated hBrr2T3 (about 20-fold reduced rate for hBrr2FL-Jab1 compare with hBrr2T3 alone vs. about 10-fold reduced rate for isolated hBrr2FL compare with isolated hBrr2T3; Fig. 4E,F). The about 1.5-fold reduction of the amplitude of both, hBrr2FL and hBrr2T3, upon addition of Jab1 (Fig. 4E,F) suggests that the Jab1 C-terminal tail inhibits productive accommodation of the single-stranded U4 loading region at the RNA-binding tunnel of the helicase. This effect might be hidden in the RNA binding assays with hBrr2T3, as U4/U6 exhibits additional attachment sites (U4 3′-SL, U4/U6 stem II) for Brr2. Alternatively or in addition, the Brr2 conformation might change upon ATP addition, enabling the Jab1 tail to inhibit U4/U6 binding even in the absence of the NTR. The 2-fold decrease of the hBrr2FL unwinding rate vs. the almost unchanged unwinding rate of hBrr2T3 upon addition of Jab1 (Fig. 4E,F) indicates that the NTR supports Jab1-mediated inhibition of a subsequent unwinding step in our in vitro system, in agreement with the effect of the NTR on U4/U6 unwinding by isolated Brr2 (see above).
Upon addition of Jab1ΔC, hBrr2FL (A = 0.95 ± 0.03) reached a similar amplitude as the isolated hBrr2T3 (A = 0.94 ± 0.01) (Fig. 4E,F). These data are in full agreement with our EMSA results (Fig. 4A,B), suggesting that Jab1ΔC helps to circumvent the NTR-based U4/U6 binding inhibition, possibly by inducing conformational changes within Brr2. Both, hBrr2FL (ku = 0.16 ± 0.01) and hBrr2T3 (ku = 1.78 ± 0.12) showed a similar increase in unwinding rates (about 1.6–1.8-fold) upon Jab1ΔC addition compare with the isolated helicases (ku = 0.10 ± 0.01 and ku = 0.98 ± 0.05, respectively) (Fig. 4E,F). Thus, although Jab1ΔC counteracts inhibition of RNA binding via the Brr2 NTR (see above), Jab1ΔC does not entirely reverse the NTR-mediated inhibition of Brr2-dependent U4/U6 unwinding, in agreement with the above finding that the NTR to some extent inhibits the helicase independently of Jab1.
NTR-mediated inhibition depends on the inactive CC of Brr2
Previously, we have shown that Jab1-mediated regulation of Brr2 depends on the presence of the inactive CC.23 Folding back of the NTR onto the NC and CC in crystal structures of Brr2FL-Jab1 complexes (ref. 20 and Fig 1B) suggests that the CC is also important for the function of the NTR as an auto-inhibitory element. To address this question, we performed in vitro U4/U6 binding and unwinding analyses using C. thermophilum Brr2 constructs that lack the CC and exhibit stepwise truncations of the NTR (Brr2FL-NC, Brr2T1-NC, Brr2T2-NC, Brr2T3-NC, Brr2T4-NC; Fig. 1A). In contrast to the corresponding dual-cassette constructs,20 Brr2T2-NC and Brr2T3-NC variants did not lead to strongly altered RNA binding (Brr2FL-NC: Kd = 48.51 ± 1.58 nM; Brr2T2-NC: Kd = 31.70 ± 1.31 nM; Brr2T3-NC: Kd = 30.59 ± 5.37 nM) or U4/U6 unwinding (Brr2FL-NC: A = 0.72 ± 0.03, ku = 0.08 ± 0.01; Brr2T2-NC: A = 0.75 ± 0.03, ku = 0.07 ± 0.01; Brr2T3-NC: A = 0.70 ± 0.02, ku = 0.08 ± 0.01; Fig 5A–C). The Brr2T1-NC variant, missing the first 118 residues and starting at the plug domain, showed aberrant behavior, antagonistic to the effects seen with the corresponding dual-cassette construct20; compare with Brr2FL-NC, Brr2T2-NC and Brr2T3-NC, it exhibited a strongly decreased RNA affinity (Kd = 301.40 ± 70.38 nM; Fig. 5A,C) and a drastically reduced U4/U6 unwinding amplitude (A = 0.28 ± 0.01), but showed a similar unwinding rate (ku = 0.08 ± 0.01; Fig. 5B,C). These observations indicate that the Brr2T1-NC preparations contained a higher fraction of inactive protein that is unable to bind or productively accommodate RNA, possibly due to misfolding. Irrespectively, the lack of effects in the Brr2T2-NC and Brr2T3-NC variants compare with the stepwise activation seen with the corresponding constructs in the presence of the CC, shows that substrate competition via the plug domain depends on the CC. Most likely, the CC allows fastening of the IC-clamp and PWI-like domain, which in turn stabilizes the plug domain in its inhibitory position. Additionally, linking NC and CC in dual-cassette Brr2 constructs, the IC-clamp and PWI-like domain may further inhibit Brr2 by restricting the ability of the 2 cassettes to adopt different relative orientations, as may be required during RNA unwinding.
Figure 5.
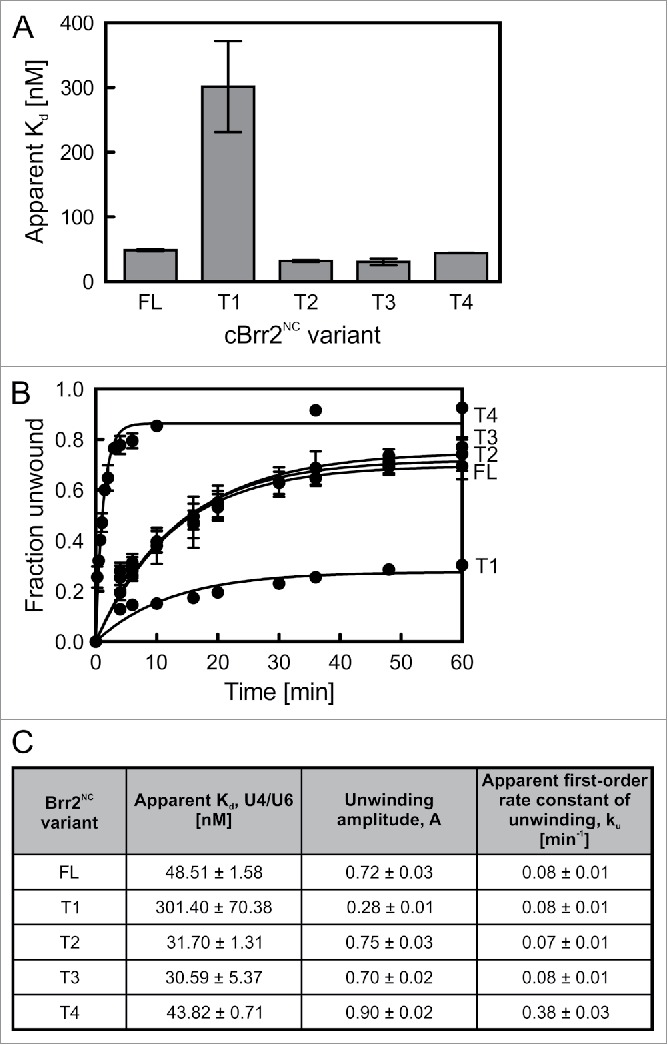
Effects of NTR truncations on C. thermophilum Brr2NC variants. (A) Binding of N-terminal Brr2NC truncations to U4/U6 di-snRNA monitored by electrophoretic mobility shift assays. Values represent means ± SEMs of at least 2 independent experiments. Kd values were obtained by fitting quantified binding data to a single exponential Hill function (fraction bound = A[protein]n/([protein]n+Kdn); A – fitted maximum of RNA bound; n – Hill coefficient). (B) Time courses of U4/U6 unwinding by Brr2NC variants. Data represent means ± SEM of at least 3 independent experiments. Amplitudes and unwinding rates were obtained by fitting the quantified data to a first-order reaction (fraction unwound = A[1-exp{-ku t}]; A – amplitude of the reaction; ku – apparent first-order rate constant of unwinding; t – time). (C) U4/U6 di-snRNA affinities and unwinding parameters of Brr2NC variants. Values represent means ± SEM of at least 2 independent experiments.
In contrast, Brr2T4-NC, which is lacking part of the NC-clamp (Fig. 1A), exhibited a similar RNA affinity (Kd = 43.82 ± 0.71 nM) yet an increased unwinding activity (A = 0.90 ± 0.02, ku = 0.38 ± 0.03) compared with the other Brr2NC variants (Fig. 5A–C), recapitulating the trend seen with the corresponding double-cassette construct.20 These findings are consistent with the NC-clamp only contacting elements of the NC (Fig. 1B–E) and its effect thus not depending on the presence of the CC. Tight binding of the NC-clamp to the NC might additionally support proper positioning of the preceding PWI-like domain on the CC and of the IC-clamp across both cassettes. Consistent with this notion, in structures of Brr2 and Brr2-Jab1 complexes lacking N-terminal parts of the NTR, the truncated NTR folds back onto the helicase cassettes in the same fashion as in Brr2FL-Jab1 (Fig. 1B,C). The above results clearly show that the CC not only directly influences the activity of the NC13 and Jab123 but is also required for NTR-mediated auto-inhibition to take full effect. Furthermore, the data demonstrate an inter-dependence of the various NTR elements that mutually stabilize each other's auto-inhibitory positions.
Discussion
In addition to their core motor domains, RNA helicases often contain accessory domains or regions that regulate helicase activities and specificities. Furthermore, helicases often cooperate with other proteins that further modulate their functions. Some protein co-factors directly bind and influence accessory domains of helicases. E.g., the spliceosomal helicase Prp43 contains an accessory OB-fold domain, which aids in RNA binding and stimulates the Prp43 ATPase activity in the presence of RNA.28 The G-patch protein Ntr1 binds to the Prp43 OB-fold domain and enhances the helicase activity of Prp43.29,30 Likewise, the G-patch protein SPP2 binds to an accessory OB-fold domain in the spliceosomal RNA helicase Prp231 and helps to couple Prp2 ATPase activity to spliceosome remodeling.32 Brr2 constitutes a particularly complex RNA helicase. The large NTR of Brr2 is composed of several domains and regions that can auto-inhibit the enzyme.20 WH, HB, HLH and IG accessory domains are appended to the dual RecA-like motor domains in the active NC of Brr2 and support RecA-dependent ATP and RNA transactions.13,14 Additionally, HB, HLH and IG domains of the NC serve as a landing pad for the Prp8 Jab1 domain that regulates Brr2 either negatively or positively in trans.22-24 Finally, the catalytically inactive CC features the same domain composition as the NC, acts as an intra-molecular activator of the active NC,13 and also constitutes a protein-protein interaction platform.27,33 Before our work, it was not clear to which extent the various Brr2 cis- and trans-acting regulatory mechanisms mutually influence each other.
By combining structural and biochemical analyses, we demonstrated a functional cross-talk between an intra-molecular and an intermolecular regulatory mechanism of Brr2. Our results indicate that the Brr2 NTR and the Prp8 Jab1 domain reinforce each other in inhibiting substrate RNA binding by Brr2, and that this binding inhibition translates into a corresponding inhibition of Brr2-mediated U4/U6 unwinding in vitro. Our crystal structure of a yBrr2FL-Jab1 complex and comparison with the corresponding subunits of a yeast tri-snRNP, in which Brr2 is loaded on the U4/U6 duplex, showed that NTR-based auto-inhibition and Prp8 Jab1-based intermolecular inhibition of RNA binding by Brr2 can take effect at the same time, with the plug domain of the NTR and the C-terminal tail of the Jab1 domain interacting with spatially separated RNA-binding surfaces of Brr2. While the plug domain can reside in a cleft between the RecA2 and HB domains to obstruct accommodation of a portion of the RNA duplex to be unwound, the Jab1 C-terminal tail can inhibit RNA loading by cross-strutting the N-terminal HB and RecA2 domains and can invade the RNA-binding tunnel to compete binding or accommodation of ssRNA regions. Structural comparisons of yBrr2-Jab1 complexes bearing different NTR truncations revealed that the plug domain, via its direct contacts to the N-terminal RecA2 and HB domains, influences the arrangement of Brr2 elements forming the RNA-binding tunnel, thus improving accommodation of the Jab1 tail.
In contrast to the yeast system, binding of the Jab1 tail to the Brr2 RNA-binding tunnel was observed even in the absence of the plug domain for the human and C. thermophilum factors, suggesting that the inhibitory Jab1 tail interaction at the Brr2 RNA-binding tunnel is more stable in the latter 2 organisms. Consistently, the corresponding interaction networks differ in the 3 cases investigated. However, our RNA binding and unwinding studies on the human system clearly showed that also in this organism the intra- and intermolecular inhibition mechanisms reinforce each other. Jab1 alone inhibited ssRNA binding by Brr2 but the plug domain was required for the Jab1 tail to significantly contribute to inhibition of dsRNA and U4/U6 binding. In contrast, addition of Jab1 led to a similar decrease in the amplitude of U4/U6 unwinding by hBrr2FL and hBrr2T3. Thus, Jab1 might interfere with the accommodation of single-stranded regions of these RNAs in the RNA-binding tunnel and/or the addition of ATP changes the Brr2 conformation such that the Jab1 tail can inhibit U4/U6 binding. Effects on the rates of U4/U6 unwinding revealed that the Jab1 tail depends on the NTR to inhibit one or more steps during the unwinding process. Additionally, our unwinding data showed that Jab1ΔC stimulates both, hBrr2FL and hBrr2T3, to a similar extent and did not fully counteract NTR-dependent inhibition.
Our results indicate that NTR- and Jab1-mediated inhibition can reinforce each other but that they can also be alleviated independent of each other, in agreement with a recent yBrr2FL-Jab1ΔC structure20 and the yBrr2FL-Jab1 structure presented here, in which the NTR adopts an auto-inhibitory conformation without or with additional Jab1-based inhibition, respectively. These features would in principle enable differential adjustment in the strength of Brr2 inhibition during U5 snRNP or tri-snRNP assembly and during different stages of a splicing cycle. Comparing the unwinding rate of the least active hBrr2FL-Jab1 complex, in which both inhibitory mechanisms are at work, to that of the most active hBrr2T3-Jab1ΔC complex, which is almost completely uninhibited, our data show that Brr2 helicase activity can be up- or downregulated at least 35-fold in the human system.
In line with our results, NTR- and Jab1-based inhibitory mechanisms are apparently put to work in a differential fashion in the tri-snRNP or during different states of splicing. While in cryo-EM structures of yeast tri-snRNPs Brr2 is fully uninhibited and loaded on the U4 snRNA,27,34 the recent structure of a human U4/U6•U5 tri-snRNP35 revealed an inactive state, in which Brr2 is remote from its entry site on U4 snRNA and inhibited by the NTR folded back onto the helicase region; however, additional inhibition via the Jab1 C-terminal tail is not obvious in the latter structure, possibly due to the limited resolution. In the structure of a spliceosome immediately after branching,36 neither the NTR nor the Jab1 C-terminal tail were modeled, again possibly in part explained by the limited resolution in the corresponding portion of the cryo-EM map. In contrast, cryo-EM structures of yeast Bact complexes,37,38 showed Brr2 to be inhibited both via the Jab1 C-terminal tail as well as via its NTR. Brr2 could not be located in the recent cryo-EM maps of another post-step 1 spliceosome that lacked the Prp16 helicase39 or in a post-splicing intron-lariat spliceosome,40 presumably due to its flexible attachment to the core of the spliceosome during these stages of splicing. Thus, different strategies for structure probing as well as structures of the spliceosome at yet other stages of a splicing cycle will be required to fully assess how Brr2 is regulated throughout an entire splicing cycle.
Materials and methods
Cloning and expression
Codon-optimized DNA fragments encoding selected regions of hBrr2 (hBrr2FL – residues 1–2136, hBrr2T3 – 395–2129), cBrr2 (cBrr2T3 – 426–2193; cBrr2FL-NC – 1–1371, cBrr2T1-NC – 119–1371, cBrr2T2-NC – 287–1371, cBrr2T3-NC – 426–1371, cBrr2T4-NC – 473–1371) or yBrr2 (yBrr2FL – 1–2163; yBrr2T2 – 271–2163) were cloned into a modified pFL vector (EMBL, Grenoble) to produce proteins with a TEV-cleavable N-terminal His10-tag. Codon-optimized DNA fragments encoding human Jab1 (hJab1 – 2064–2335; hJab1ΔC – 2064–2310) were cloned with a PreScission-cleavable N-terminal GST-tag into the pFL vector. Virus production and expression were performed as described.13,20,23 Codon-optimized DNA fragments encoding yJab1 (residues 2147–2413) and cJab1 (residues 2112–2385) were cloned into the pETM-11 vector (EMBL, Heidelberg) under the control of a T7 promotor for production of the recombinant protein bearing a TEV-cleavable N-terminal His6-tag. Escherichia coli Rosetta2 DE3 cells were transformed with the vector and cultivated in auto-inducing medium.41
Protein purification
For all preparations, cell pellets were resuspended, supplemented with protease inhibitors (Roche), lyzed by sonication using a Sonoplus Ultrasonic Homogenizer HD 3100 (Bandelin) and the lysates were cleared by centrifugation. yBrr2FL, yBrr2T2, cBrr2T3, hBrr2FL, hBrr2T3, yJab1, hJab1 and hJab1ΔC were purified as described previously.13,20,23 cBrr2 variants lacking the CC were purified as described previously for other cBrr2 variants.20 Purification of cJab1 was identical to yJab1 purification. For purification of yNtr2, cells were lyzed in 100 mM Tris-HCl, pH 7.5, 200 mM NaCl, 2 mM DTT. GST-tagged tagged yNtr2 was captured on glutathione sepharose matrix (GE Healthcare) equilibrated in the same buffer, and the retained fraction treated overnight with PreScission protease. Elution fractions containing the target protein were further purified by Superdex 200 (GE Healthcare) size-exclusion chromatography in 10 mM TRIS, pH 7.5, 200 mM NaCl, 2 mM DTT.
Crystallographic procedures
yBrr2FL and yJab1 were mixed in a 1:2 molar ratio in 10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM DTT and separated by Superdex 200 (GE Healthcare) size-exclusion chromatography. Fractions containing the target complex were pooled and concentrated to 2 mg/ml. Crystals were grown in 48-well plates using the sitting-drop vapor diffusion technique at 18°C with drops containing 1.3 µl protein complex solution and 1 µl reservoir solution (0.1 M Tris-HCl, pH 7.5, 10.5% (w/v) PEG 3350, 0.2 M MgCl2). Crystals were cryo-protected by transfer into mother liquor containing 22.5% (v/v) ethylene glycol and flash-cooled in liquid nitrogen. Diffraction data were collected at 100 K on beamline 14.1 of the BESSY II storage ring (Berlin, Germany)42 and processed with XDS43 (Table 1). The structure was solved by molecular replacement with PHASER,44 using yBrr2FL-Jab1ΔC structure coordinates as the search model (PDB ID 5DCA).20 The C-terminal residues of yJab1 not contained in the model were appended by manual model-building in COOT.45 The model was refined by alternating rounds of manual model building with COOT and automated refinement with REFMAC546.
yBrr2T2, yJab1 and yNtr2 were mixed in a 1:5:5 molar ratio in 10 mM Tris-HCl, pH 7.5, 200 mM NaCl, 2 mM DTT and a ternary complex was purified by Superdex 200 size-exclusion chromatography. Fractions containing the target complex were pooled, concentrated to 4 mg/ml. Crystals were grown in 24-well plates using the hanging-drop vapor diffusion technique at 18°C with drops containing 0.5 µl protein complex solution, 0.25 µl reservoir solution (0.1 M MES-NaOH, pH 6.5, 9.2% (w/v) PEG 4000, 0.4 M MgCl2) and 0.25 µl 0.33% (w/v) 1,5-naphtalenedisulfonic acid. Crystals were cryo-protected by transfer into mother liquor containing 20% (v/v) glycerol and flash-cooled in liquid nitrogen. Diffraction data were collected on beamline 14.1 and 14.2 of the BESSY II storage ring, Berlin, Germany, at 100 K using a monochromated X-ray beam (λ = 0.9184 Å) and processed with XDS (Table 1). The structure was solved by molecular replacement with PHASER, using yBrr2T4-Jab1 structure coordinates as the search model (PDB ID 4BGD24). The initial model was refined with CNS applying deformable elastic network (DEN) restraints.47 After DEN refinement, additional density for a helicase-proximal N-terminal portion (modeled according to a similar extended N-terminus in the crystal structure of hBrr2T3; PDB ID 4F9113) and a PWI domain (modeled according to the PWI domain of cBrr2; PDB ID 4RVQ21) were clearly visible. Refinement was done by alternating rounds of manual model building in COOT and automated refinement with CNS and PHENIX,48 applying DEN restraints and non-crystallographic symmetry restraints between the 2 copies of the complex in an asymmetric unit. During refinement no additional density for Ntr2 appeared.
cBrr2T3 was mixed with cJab1 in a 1:3 molar ratio in 20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 2 mM DTT and separated by Superdex 200 size-exclusion chromatography. Fractions containing the target complex were pooled and concentrated to 5 mg/ml. Crystals were grown in 24 well plates using the sitting-drop vapor diffusion technique at 4°C with drops containing containing 1 µl protein complex solution, supplemented with 0.3 µl 2 mM spermine-tetrahydrochloride and 1 µl reservoir solution (3% (v/v) Tacsimate, pH 7.0, 2% (w/v) PEG MME5000, 0.1 M HEPES-NaOH, pH 7.1) and optimized by seeding. Crystals were cryo-protected by transfer into mother liquor containing 25% (v/v) propylene glycol and flash-cooled in liquid nitrogen. Diffraction data were collected at 100 K on the European Molecular Biology Laboratory beamline P14 of the PETRA III storage (DESY, Hamburg, Germany) and processed with XDS (Table 1). The structure was solved by molecular replacement with PHASER using hBrr2T3-Jab1 structure coordinates as a search model (PDB ID 4KIT23). After molecular replacement, sequences of the model were adjusted using SCULPTOR.49 The structure were refined by alternating rounds of manual model building with COOT and automated refinement with REFMAC5 and PHENIX.
RNA production
Fluorescently labeled ssRNA ([5-FAM]-5′-GAAAUUUAAUUAUAAACCAGACCGUC-3′) and dsRNA (5′-GACCAGCACGCG-3′-[5-FAM])/5′-CGCGUGCUGGUCGAAAUUUAAUUAUAAACCAGACCGUC-3′) were chemically synthesized (IBA). yU4 and yU6 snRNAs were produced by T7 RNA polymerase-based in vitro transcription, 5′-end labeled using [γ-32P]ATP and T4 polynucleotide kinase, annealed and gel-purified as described.13,20,23
RNA binding assays based on fluorescence anisotropy
To reconstitute hBrr2-Jab1 complexes for RNA binding studies, the respective components were mixed in a 1:2 (Brr2:Jab1) molar ratio in 40 mM Tris-HCl, pH 8.0, 200 mM NaCl, 2 mM DTT, 20% (v/v) glycerol, and the mixture was separated by Superdex 200 (GE Healthcare) size-exclusion chromatography. Fractions containing the target complex were pooled and concentrated by ultrafiltration. 5 nM fluorescently labeled ssRNA or 10 nM dsRNA were titrated with increasing concentrations of hBrr2 or pre-assembled hBrr2-Jab1 complex in 40 µl binding buffer (40 mM Tris-HCl, pH 7.5, 50 mM NaCl, 8% (v/v) glycerol, 0.5 mM MgCl2, 100 ng/µl acetylated BSA, 1.5 mM DTT). Binding was monitored via FP measurements in a 384-well plate in a Victor plate reader. To extract binding constants, changes in anisotropy were plotted against protein concentration and the data were fitted to a single exponential Hill function (fraction bound = A[protein]n/([protein]n+Kdn); A – fitted maximum of RNA bound; n – Hill coefficient)50 using GraphPad Prism (GraphPad Software, Inc.).
Electrophoretic gel mobility shift assays
For cBrr2 variants lacking the CC, 1 nM yU4/U6 di-snRNAs were titrated with increasing amounts of protein in 40 mM HEPES-NaOH, pH 7.9, 15 mM NaCl, 2.5 mM Mg(OAc)2, 1 mM DTT, 0.1 mg/ml acetylated BSA. For all other experiments, 5 nM yU4/U6 di-snRNAs were titrated with increasing amounts of protein or protein complex in 40 mM HEPES-NaOH, pH 7.9, 50 mM NaCl, 2.5 mM Mg(OAc)2, 1 mM DTT, 0.1 mg/ml acetylated BSA. Samples were separated using 4% (75:1) non-denaturing PAGE. Gels were scanned on a Storm phosphoimager (GE Healthcare), bands were quantified by densitometry. Apparent Kd values were obtained by fitting the resulting data points to a single exponential Hill function (fraction bound = A[protein]n/([protein]n+Kdn); A – fitted maximum of RNA bound; n – Hill coefficient)50 using GraphPad Prism.
U4/U6 unwinding assays
Unwinding assays were conducted and evaluated as described.13,20,23 Briefly, for cBrr2 variants lacking the CC, yU4/U6 complex (2 nM) and cBrr2 variants (100 nM) were mixed in 40 mM Tris-HCl, pH 7.5, 50 mM NaCl, 8% glycerol, 1.5 mM DTT, 0.1 mg/ml acetylated BSA. After incubation for 3 min at 30°C, reactions were started by the addition of 1 mM ATP/MgCl2. For all other experiments, yU4/U6 complex (0.5 nM) and Brr2 constructs (200 nM) were pre-incubated for 3 min at 30°C with or without Jab1 variants (2 µM) in 40 mM Tris-HCl, pH 7.5, 50 mM NaCl, 8% glycerol, 1.5 mM DTT, 0.1 mg/ml acetylated BSA. Reactions were started by the addition of 1 mM ATP/MgCl2. 10 µl samples were withdrawn at selected time points, mixed with 10 µl 40 mM Tris-HCl, pH 7.4, 50 mM NaCl, 25 mM EDTA, 1% (w/v) SDS, 10% (v/v) glycerol, 0.05% (w/v) xylene cyanol, 0.05% (w/v) bromophenol blue and separated by 6% native PAGE (19:1). Gels were scanned on a phosphoimager, bands were quantified by densitometry and data were fit to a first-order reaction (fraction unwound = A{1-exp[-ku t]}; A – amplitude of the reaction; ku – apparent first-order rate constant of unwinding; t – time).
Data deposition
Coordinates and structure factors have been deposited in the RCSB Protein Data Bank (www.pdb.org) with accession codes 5M52 (yBrr2FL-Jab1), 5M5P (yBrr2T2-Jab1) and 5M59 (cBrr2T3-Jab1) and will be released upon publication.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Reinhard Lührmann, Berthold Kastner and Holger Stark, Max-Planck-Institute for Biophysical Chemistry, Göttingen, Germany, for sharing data before publication. We acknowledge access to beamline BL14.2 of the BESSY II storage ring (Berlin, Germany) via the Joint Berlin MX-Laboratory sponsored by the Helmholtz Zentrum Berlin für Materialien und Energie, the Freie Universität Berlin, the Humboldt-Unversität zu Berlin, the Max-Delbrück Centrum and the Leibniz-Institut für Molekulare Pharmakologie, and to beamline P14 of the PETRA III storage ring at DESY (Hamburg, Germany), operated by the European Molecular Biology Laboratory.
Funding
This work was funded by the Deutsche Forschungsgemeinschaft (SFB 740 to MCW), the Bundesministerium für Bildung und Forschung (05K10KEC to MCW) and the Einstein Foundation Berlin (A-2012–140 to MCW). KFS was supported by a Dahlem International Network PostDoc Fellowship from Freie Universität Berlin.
References
- [1].Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell 2009; 136:701-18; PMID:19239890; http://dx.doi.org/ 10.1016/j.cell.2009.02.009 [DOI] [PubMed] [Google Scholar]
- [2].Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol 2011; 3:1-24; http://dx.doi.org/ 10.1101/cshperspect.a003707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Brow DA. Allosteric cascade of spliceosome activation. Annu Rev Genet 2002; 36:333-60; PMID:12429696; http://dx.doi.org/ 10.1146/annurev.genet.36.043002.091635 [DOI] [PubMed] [Google Scholar]
- [4].Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 1998; 92:315-26; PMID:9476892; http://dx.doi.org/ 10.1016/S0092-8674(00)80925-3 [DOI] [PubMed] [Google Scholar]
- [5].Raghunathan PL, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol 1998; 8:847-55; PMID:9705931; http://dx.doi.org/ 10.1016/S0960-9822(07)00345-4 [DOI] [PubMed] [Google Scholar]
- [6].Laggerbauer B, Achsel T, Lührmann R. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc Natl Acad Sci USA 1998; 95:4188-92; PMID:9539711; http://dx.doi.org/ 10.1073/pnas.95.8.4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Agafonov DE, Deckert J, Wolf E, Odenwalder P, Bessonov S, Will CL, Urlaub H, Lührmann R. Semiquantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method. Mol Cell Biol 2011; 31:2667-82; PMID:21536652; http://dx.doi.org/ 10.1128/MCB.05266-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Theuser M, Hobartner C, Wahl MC, Santos KF. Substrate-assisted mechanism of RNP disruption by the spliceosomal Brr2 RNA helicase. Proc Natl Acad Sci USA 2016; 113:7798-803; PMID:27354531; http://dx.doi.org/ 10.1073/pnas.1524616113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sashital DG, Cornilescu G, McManus CJ, Brow DA, Butcher SE. U2-U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat Struct Mol Biol 2004; 11:1237-42; PMID:15543154; http://dx.doi.org/ 10.1038/nsmb863 [DOI] [PubMed] [Google Scholar]
- [10].Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA. RNA catalyses nuclear pre-mRNA splicing. Nature 2013; 503:229-34; PMID:24196718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hang J, Wan R, Yan C, Shi Y. Structural basis of pre-mRNA splicing. Science 2015; 349:1191-8; PMID:26292705; http://dx.doi.org/ 10.1126/science.aac8159 [DOI] [PubMed] [Google Scholar]
- [12].Kim DH, Rossi JJ. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. RNA 1999; 5:959-71; PMID:10411139; http://dx.doi.org/ 10.1017/S135583829999012X [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Santos KF, Mozaffari-Jovin S, Weber G, Pena V, Lührmann R, Wahl MC. Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome. Proc Natl Acad Sci USA 2012; 109:17418-23; PMID:23045696; http://dx.doi.org/ 10.1073/pnas.1208098109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pena V, Mozaffari-Jovin S, Fabrizio P, Orlowski J, Bujnicki JM, Lührmann R, Wahl MC. Common design principles in the spliceosomal RNA helicase Brr2 and in the Hel308 DNA helicase. Mol Cell 2009; 35:454-66; PMID:19716790; http://dx.doi.org/ 10.1016/j.molcel.2009.08.006 [DOI] [PubMed] [Google Scholar]
- [15].Zhang L, Xu T, Maeder C, Bud LO, Shanks J, Nix J, Guthrie C, Pleiss JA, Zhao R. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat Struct Mol Biol 2009; 16:731-9; PMID:19525970; http://dx.doi.org/ 10.1038/nsmb.1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hahn D, Kudla G, Tollervey D, Beggs JD. Brr2p-mediated conformational rearrangements in the spliceosome during activation and substrate repositioning. Genes Dev 2012; 26:2408-21; PMID:23124065; http://dx.doi.org/ 10.1101/gad.199307.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mayerle M, Guthrie C. Prp8 retinitis pigmentosa mutants cause defects in the transition between the catalytic steps of splicing. RNA 2016; 22:793-809; PMID:26968627; http://dx.doi.org/ 10.1261/rna.055459.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Small EC, Leggett SR, Winans AA, Staley JP. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol Cell 2006; 23:389-99; PMID:16885028; http://dx.doi.org/ 10.1016/j.molcel.2006.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fourmann JB, Schmitzova J, Christian H, Urlaub H, Ficner R, Boon KL, Fabrizio P, Lührmann R. Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev 2013; 27:413-28; PMID:23431055; http://dx.doi.org/ 10.1101/gad.207779.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Absmeier E, Wollenhaupt J, Mozaffari-Jovin S, Becke C, Lee CT, Preussner M, Heyd F, Urlaub H, Lührmann R, Santos KF, et al.. The large N-terminal region of the Brr2 RNA helicase guides productive spliceosome activation. Genes Dev 2015; 29:2576-87; PMID:26637280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Absmeier E, Rosenberger L, Apelt L, Becke C, Santos KF, Stelzl U, Wahl MC. A noncanonical PWI domain in the N-terminal helicase-associated region of the spliceosomal Brr2 protein. Acta Crystallogr D 2015; 71:762-71; PMID:25849387; http://dx.doi.org/ 10.1107/S1399004715001005 [DOI] [PubMed] [Google Scholar]
- [22].Mozaffari-Jovin S, Wandersleben T, Santos KF, Will CL, Lührmann R, Wahl MC. Novel regulatory principles of the spliceosomal Brr2 RNA helicase and links to retinal disease in humans. RNA Biol 2014; 11:298-312; PMID:24643059; http://dx.doi.org/ 10.4161/rna.28353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mozaffari-Jovin S, Wandersleben T, Santos KF, Will CL, Lührmann R, Wahl MC. Inhibition of RNA helicase Brr2 by the C-terminal tail of the spliceosomal protein Prp8. Science 2013; 341:80-4; PMID:23704370; http://dx.doi.org/ 10.1126/science.1237515 [DOI] [PubMed] [Google Scholar]
- [24].Nguyen TH, Li J, Galej WP, Oshikane H, Newman AJ, Nagai K. Structural basis of Brr2-Prp8 interactions and implications for U5 snRNP biogenesis and the spliceosome active site. Structure 2013; 21:910-19; PMID:23727230; http://dx.doi.org/ 10.1016/j.str.2013.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Maeder C, Kutach AK, Guthrie C. ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nat Struct Mol Biol 2009; 16:42-8; PMID:19098916; http://dx.doi.org/ 10.1038/nsmb.1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mozaffari-Jovin S, Santos KF, Hsiao HH, Will CL, Urlaub H, Wahl MC, Lührmann R. The Prp8 RNase H-like domain inhibits Brr2-mediated U4/U6 snRNA unwinding by blocking Brr2 loading onto the U4 snRNA. Genes Dev 2012; 26:2422-34; PMID:23124066; http://dx.doi.org/ 10.1101/gad.200949.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nguyen TH, Galej WP, Bai XC, Oubridge C, Newman AJ, Scheres SH, Nagai K. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 A resolution. Nature 2016; 530:298-302; PMID:26829225; http://dx.doi.org/ 10.1038/nature16940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Walbott H, Mouffok S, Capeyrou R, Lebaron S, Humbert O, van Tilbeurgh H, Henry Y, Leulliot N. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J 2010; 29:2194-204; PMID:20512115; http://dx.doi.org/ 10.1038/emboj.2010.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tanaka N, Aronova A, Schwer B. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev 2007; 21:2312-25; PMID:17875666; http://dx.doi.org/ 10.1101/gad.1580507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Christian H, Hofele RV, Urlaub H, Ficner R. Insights into the activation of the helicase Prp43 by biochemical studies and structural mass spectrometry. Nucleic Acids Res 2014; 42:1162-79; PMID:24165877; http://dx.doi.org/ 10.1093/nar/gkt985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Silverman EJ, Maeda A, Wei J, Smith P, Beggs JD, Lin RJ.-- Interaction between a G-patch protein and a spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol Cell Biol 2004; 24:10101-10; PMID:15542821; http://dx.doi.org/ 10.1128/MCB.24.23.10101-10110.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Warkocki Z, Schneider C, Mozaffari-Jovin S, Schmitzova J, -Hobartner C, Fabrizio P, Lührmann R. The G-patch protein Spp2 couples the spliceosome-stimulated ATPase activity of the DEAH-box protein Prp2 to catalytic activation of the spliceosome. Genes Dev 2015; 29:94-107; PMID:25561498; http://dx.doi.org/ 10.1101/gad.253070.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].van Nues RW, Beggs JD. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics 2001; 157:1451-67; PMID:11290703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wan RX, Yan CY, Bai R, Wang L, Huang M, Wong CC, Shi Y. The 3.8 Å structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis. Science 2016; 351:466-75; PMID:26743623; http://dx.doi.org/ 10.1126/science.aad6466 [DOI] [PubMed] [Google Scholar]
- [35].Agafonov DE, Kastner B, Dybkov O, Hofele RV, Liu WT, Urlaub H, Lührmann R, Stark H. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science 2016; 351:1416-20; PMID:26912367; http://dx.doi.org/ 10.1126/science.aad2085 [DOI] [PubMed] [Google Scholar]
- [36].Galej WP, Wilkinson ME, Fica SM, Oubridge C, Newman AJ, Nagai K. Cryo-EM structure of the spliceosome immediately after branching. Nature 2016; 537:197-201; PMID:27459055; http://dx.doi.org/ 10.1038/nature19316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yan C, Wan R, Bai R, Huang G, Shi Y. Structure of a yeast activated spliceosome at 3.5 A resolution. Science 2016; 353:904-11; PMID:27445306; http://dx.doi.org/ 10.1126/science.aag0291 [DOI] [PubMed] [Google Scholar]
- [38].Rauhut R, Fabrizio P, Dybkov O, Hartmuth K, Pena V, Chari A, Kumar V, Lee CT, Urlaub H, Kastner B, et al.. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science 2016; 353:1399-1405; PMID:27562955 [DOI] [PubMed] [Google Scholar]
- [39].Wan R, Yan C, Bai R, Huang G, Shi Y. Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution. Science 2016; 353:895-904; PMID:27445308; http://dx.doi.org/ 10.1126/science.aag2235 [DOI] [PubMed] [Google Scholar]
- [40].Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y. Structure of a yeast spliceosome at 3.6-Å resolution. Science 2015; 349:1182-91; PMID:26292707; http://dx.doi.org/ 10.1126/science.aac7629 [DOI] [PubMed] [Google Scholar]
- [41].Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Express Purif 2005; 41:207-34; http://dx.doi.org/ 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- [42].Mueller U, Forster R, Hellmig M, Huschmann FU, Kastner A, Malecki P, Pühringer S, Röwer M, Sparta K, Steffien M, Ühlein M, Wilk P, Weiss MS. The macromolecular crystallography beamlines at BESSY II of the Helmholtz-Zentrum Berlin: Current status and perspectives. Eur Phys J Plus 2015; 130:141; http://dx.doi.org/ 10.1140/epjp/i2015-15141-2 [DOI] [Google Scholar]
- [43].Kabsch W. XDS. Acta Crystallogr D 2010; 66:125-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D 2007; 63:32-41; PMID:17164524; http://dx.doi.org/ 10.1107/S0907444906045975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D 2004; 60:2126-32; PMID:15572765; http://dx.doi.org/ 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- [46].Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Nicholls RA, Winn MD, Long F, Vagin AA. Refmac5 for the refinement of macromolecular crystal structures. Acta Crystallogr D 2011; 67:355-67; PMID:21460454; http://dx.doi.org/ 10.1107/S0907444911001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Schroder GF, Levitt M, Brunger AT. Deformable elastic network refinement for low-resolution macromolecular crystallography. Acta Crystallogr D 2014; 70:2241-55; PMID:25195739; http://dx.doi.org/ 10.1107/S1399004714016496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D 2012; 68:352-67; PMID:22505256; http://dx.doi.org/ 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bunkoczi G, Read RJ. Improvement of molecular-replacement models with Sculptor. Acta Crystallogr D 2011; 67:303-12; PMID:21460448; http://dx.doi.org/ 10.1107/S0907444910051218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ryder SP, Recht MI, Williamson JR. Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods Mol Biol 2008; 488:99-115; PMID:18982286; http://dx.doi.org/ 10.1007/978-1-60327-475-3_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


