Abstract
Terrestrial ecosystem models assume that microbial communities respond instantaneously, or are immediately resilient, to environmental change. Here we tested this assumption by quantifying the resilience of a leaf litter community to changes in precipitation or nitrogen availability. By manipulating composition within a global change experiment, we decoupled the legacies of abiotic parameters versus that of the microbial community itself. After one rainy season, more variation in fungal composition could be explained by the original microbial inoculum than the litterbag environment (18% versus 5.5% of total variation). This compositional legacy persisted for 3 years, when 6% of the variability in fungal composition was still explained by the microbial origin. In contrast, bacterial composition was generally more resilient than fungal composition. Microbial functioning (measured as decomposition rate) was not immediately resilient to the global change manipulations; decomposition depended on both the contemporary environment and rainfall the year prior. Finally, using metagenomic sequencing, we showed that changes in precipitation, but not nitrogen availability, altered the potential for bacterial carbohydrate degradation, suggesting why the functional consequences of the two experiments may have differed. Predictions of how terrestrial ecosystem processes respond to environmental change may thus be improved by considering the legacies of microbial communities.
Introduction
The resilience of microbial communities—the rate at which they respond to environment change—is key to understanding their role in human, engineered and natural systems (McMahon et al., 2007; Allison and Martiny, 2008; Costello et al., 2012; Lozupone et al., 2012; Shade et al., 2012). Human activities are altering the variability and long-term trends in temperature, precipitation and nitrogen availability, factors known to alter microbial composition in natural ecosystems (Allison and Martiny, 2008; Evans and Wallenstein, 2012; Rousk et al., 2013). However, terrestrial ecosystem models assume that microbial communities respond instantaneously, or are immediately resilient, to environmental change (Schimel and Gulledge, 1998; Lawrence et al., 2009; Treseder et al., 2012; Hawkes and Keitt, 2015).
In the face of shifting conditions, it is useful to consider resilience broadly, without regard to an equilibrium state (Loreau et al., 2002). Thus, a highly resilient community responds quickly to environmental change, and its composition will be determined by contemporary conditions. In contrast, a non-resilient community will be a function of historical, as well as contemporary, environmental conditions (Hawkes and Keitt, 2015). Indeed, plant and animal species often lag for decades in their response to climate change (Parmesan, 2006; Wilczek et al., 2014). However, microorganisms are generally expected to be more resilient than larger organisms because of faster growth rates, greater dispersal capabilities, and their potential for phenotypic plasticity and rapid evolution (Shade et al., 2012).
If microbial composition is not immediately resilient, a key question is whether such delays matter for ecosystem functioning, which could lead to climate change feedbacks in natural ecosystems (Treseder et al., 2012). Prior studies testing for the effects of the historical environment on microbial functioning have yielded mixed results (for example, Updegraff et al., 1998; Waldrop and Firestone, 2006; Evans and Wallenstein, 2012; Rousk et al., 2013; Li et al., 2015). Crucially, however, these studies do not tease apart the legacies of abiotic parameters versus those of the microbial community itself. Hence, it remains unclear whether functional predictions would be improved by incorporating the response of the microbial community itself.
The Loma Ridge Global Change Experiment manipulates rainfall and nitrogen, two factors subject to anthropogenic change in southern California (Wilson et al., 2003; Fenn et al., 2003). Previously, we have shown that the leaf litter microbial community is sensitive to simulated drought and nitrogen addition in this grassland (Amend et al., 2015; Matulich et al., 2015). Bacteria and fungi decompose most plant detritus (litter) in terrestrial ecosystems (Swift et al., 1979), and this process is a key step in the carbon cycle, as it mediates the balance of carbon being respired as CO2 into the atmosphere or stored in the soil (Adair et al., 2008).
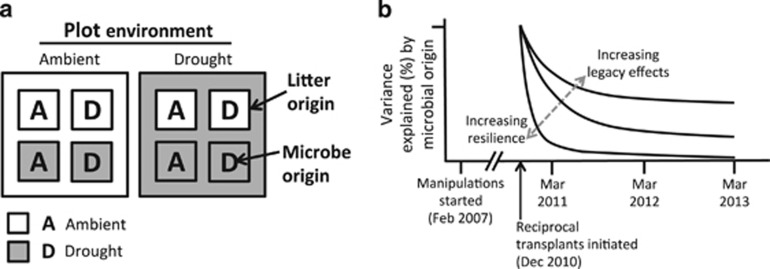
In this study, we tested the resilience of a microbial community—its composition and functioning—in its natural environment using microbial ‘cages' (Reed and Martiny, 2007; Allison et al., 2013) in the largest and longest experiment of its kind. The cages inhibit the exchange of microorganisms, allowing separate manipulation of the microbial community, litter substrate and contemporary abiotic environment (Figure 1a). In earlier studies, we reported some results from the first year of this experiment, including extracellular enzyme potential and litter mass loss (Allison et al., 2013; Alster et al., 2013). Here, we assayed the taxonomic composition of bacteria and fungi in the litterbags and followed composition and litter mass loss over 3 years. We then compared these metrics with those of control communities remaining in their original environment to quantify how long the origin of the microbial community, as opposed to its contemporary environment or litter substrate, explained variation in microbial composition or functioning (Figure 1b).
Figure 1.
(a) Experimental design of the drought reciprocal transplant within the global change experiment. A parallel design was carried out for the nitrogen reciprocal transplant. Global change manipulations were initiated in February 2007. In December 2010, microbial cage litterbags were filled with sterilized leaf litter, collected from either ambient rainfall or drought treatment plots (litter origin). Each litterbag was then inoculated with a microbial inoculum collected from ambient or drought plots (microbial origin). Finally, sealed litterbags were placed back into ambient or drought plots (plot environment). One bag from each of eight replicate blocks was destructively sampled at the end of each rainy season (March) for 3 years. (b) To assay resilience of the microbial community, we quantified how well microbial origin explained either microbial composition or functioning (litter mass loss) at each time point. A steep decline in variance explained over time indicates high resilience; a slow decline over time indicates strong legacy effects (dashed gray arrow).
We further investigated whether the resilience of microbial functioning in the litterbags was linked to changes in a community's potential for carbohydrate degradation. Microorganisms carry different suites of glycoside hydrolase (GH) genes that encode enzymes that hydrolyze the glycosidic bond between the carbohydrate moieties of polymers such as cellulose (Berlemont and Martiny, 2015). We used metagenomic sequencing to compare GH composition among the transplanted communities.
Materials and methods
Field site
The Loma Ridge Global Change Experiment was established in February 2007 in the foothills of Santa Ana Mountains within the Irvine Ranch National Landmark in Orange County, California (117.704°W, 33.742°N; Potts et al., 2012). Annual mean temperature is 17 °C, and mean precipitation is 325 mm, falling primarily during a ‘wet' season from November to April (Kimball et al., 2014). The grassland is dominated by the annual grass genera Avena, Bromus and Lolium; the annual forb genera Erodium and Lupinus; and the native perennial grass Nassella pulchra.
We used a subset of the global change manipulations: two levels of precipitation (ambient and ~40% reduction) and two levels of nitrogen (ambient or 60 kg N ha−1 year−1 added as slow-release calcium nitrate) applied to subplots within precipitation treatments (Allison et al., 2013). Drought was imposed by covering the plots only when rainfall was forecast. The covers (polyethylene sheeting over steel arch frames) were removed again soon after rainfall, reducing rainfall by half with negligible effects on temperature or sunlight in the plots (Potts et al., 2012). Both treatments altered litter chemistry (Allison et al., 2013).
Reciprocal transplant
After the global change treatments were in place for 3.5 years, we set up reciprocal transplants within the larger manipulations. The first year of the transplant experiments are described in Allison et al. (2013), but for this study, we continued the experiments for a total of 3 years. To manipulate litter origin, we collected senesced plant material from the three types of plots (ambient, drought and added nitrogen). To keep the litter treatment constant across years, the litter from this collection was stored dry at room temperature and used each year of the experiment. Homogenized dry litter (2 g) was placed into nylon membrane bags with 0.45 μm pores (GE Healthcare Life Sciences, Pittsburgh, PA, USA). The bags allowed water, nutrients and possibly small bacteria to pass through. We sterilized all bags and their contents with at least 22 kGy gamma irradiation. To manipulate microbe origin, we inoculated the sterile litterbags with 50 mg non-sterile, dried and ground (Wiley mill, 1 mm mesh) litter. To manipulate the plot environment, we distributed these litterbags (N=360) into the treatment plots on 15 December 2010. We retrieved the litterbags in batches of 120 (15 treatment combinations × 8 blocks) on 3 March, 14 June and 14 November 2011. For the second year, new sterile litterbags were made, inoculated with 50 mg from each of the litterbags collected in November 2011, and placed in the field on 7 December 2011. Litterbags were collected on 5 March, 4 June and 19 November 2012. For the third year, new sterile litterbags were inoculated with the November 2012 samples and placed in the field on 3 December 2012. A final collection was taken on 4 March 2013.
Each litterbag was destructively sampled for litter mass, bacterial and fungal abundance, and DNA extraction. Before placing the litterbags in the field, we weighed the fresh litter and dried a subsample to constant mass at 65 °C to obtain dry mass. All mass losses are reported as percentage initial dry mass.
Microbial abundance and biomass
Bacterial cell densities were measured by flow cytometry (Allison et al., 2013). A 0.1 g subsample of fresh litter was ground and fixed within 8 h of collection. Cells were extracted from litter by adding tetrasodium pyrophosphate to a final concentration of 0.01 m and gently sonicated for 30 min at 4 °C. The extract was filtered, stained with SYBR Green, and incubated for 15 min at room temperature. Particles in stained extracts and unstained controls were counted by flow cytometry (BD Accuri C6; BD Biosciences, San Jose, CA, USA), and cell densities were calculated as the number of stained counts minus unstained counts per g dry litter. We converted bacterial cell counts to biomass (ug C g−1 dry litter) assuming spherical cells with radius 0.6 μm and C density of 2.2 × 10−13 g mm−3 (Bratbak, 1985).
Fungal hyphal lengths were determined by modifying a procedure described previously (Allison et al., 2013). Briefly, another 0.1 g subsample of fresh litter was ground, dried, and dispersed in 10 ml sodium hexametaphosphate solution (0.395% mass/volume) with vigorous stirring. This solution was filtered and stained with acid fuchsin and filtered onto two filters. Both filters were dried and mounted on a microscope slide. Hyphal lengths (m g−1 dry litter) were determined on phase contrast microscope under × 100 magnification using the grid-intercept method (Giovannetti and Mosse, 1980) and 50 grids per filter. Hyphal lengths were converted to biomass (μg C g−1 dry litter) assuming a fresh density of 1.1 g cm−3, 33% dry mass, 40% C in dry mass and hyphal diameter of 5.2 μm (Paul and Clark, 1996). Hyphal diameter was estimated as described in Alster et al. (2013) and did not vary by treatment or date.
Genetic analyses
From each litterbag, 0.05 g of ground litter was flash frozen using liquid nitrogen and stored at −80 °C for DNA extraction. Samples were extracted using a previously described procedure (Matulich et al., 2015), and cleaned using an AllPrep Kit (Qiagen, Valencia, CA, USA). To characterize bacterial composition, the V4 region of the 16S ribosomal RNA (rRNA) gene and the D1 and D2 region of the 28S rRNA gene was PCR amplified as in (Matulich et al., 2015). PCR products were cleaned using the Agencourt AMPure XP PCR Purification System (Beckman Coulter Inc., Indianapolis, IN, USA), quantified using a Quant-iT (Life Technologies, Grand Island, NY, USA) Assay Kit and a Synergy 4 microplate reader (BioTek, Winooski, VT, USA), pooled into equimolar concentrations and pyrosequenced at the Duke University ISGP Sequencing Facility on a 454 Life Sciences FLX sequencer using Titanium chemistry (454 Life Sciences, Branford, CT, USA).
The amplicon sequences were processed using the QIIME (version 1.6.0) toolkit (Caporaso et al., 2010) with the following parameters: quality score >50, sequence length >300 and <700 for fungi and >200 and <550 for bacteria, maximum homopolymer of 6, 6 maximum ambiguous bases and 0 mismatched bases in the primer. Sequences were denoised using Denoiser (Reeder and Knight, 2010), and operational taxonomic units (OTUs) were picked at the 97% identity level using UCLUST (Edgar, 2010) in QIIME. The taxonomic identity of the OTUs was assigned to bacteria by reference to SILVA 108 (Quast et al., 2013) and the Greengenes database (DeSantis et al., 2006) using the naive RDP classifier within QIIME (Wang et al., 2007). Taxonomic identity of the fungal OTUs were assigned by reference to the 28S LSU RDP database (Cole et al., 2013) using BLAST. Nucleotide sequences are available from NCBI SRA (deposition SRP067657).
To assay GH composition, we created metagenomic libraries from the March 2011 samples. We first pooled equal concentrations of DNA extracts from four litterbags undergoing the same treatment yielding two replicates for each of the combinations of factors (microbial origin, plot environment and litter origin) for the two experiments (drought and added nitrogen). This pooling was necessary to yield enough DNA for some of the samples. We fragmented the DNA of the pooled samples to 300 bp using a hydroshear (Covaris, Woburn, MA, USA) and prepared 30 metagenomic libraries using a Truseq library kit (Illumina, San Diego, CA, USA). (The same ambient-ambient-ambient (microbe-litter-plot) samples were used for the two experiments.) The libraries were sequenced with on the Illumina HiSeq 2000 platform (100- bp paired ends; treated as single reads for downstream analysis). Sequences were uploaded onto the MG-RAST server (Glass and Meyer, 2011) (accessions 4514321-31, 4535613-25 and 4537557-62). In total, 44.4 Gbp (passed QC) were obtained. Taxonomic annotation, using the M5NR database, at the genus level, was considered for sequences with e-value⩽10−5.
We used a custom bioinformatics pipeline to detect and assign GHs to particular families and in the case of the GH8 family, further discriminated between functions (Berlemont et al., 2014; Berlemont and Martiny, 2015). GH families were assigned to substrate target categories according to the substrate specificities of characterized enzymes, as stated in the CAZy database (Lombard et al., 2014). In addition, biosynthetic cellulases (BcsZ) from GH8 were separated from hydrolytic enzymes (Berlemont and Martiny, 2013).
Statistics
Multivariate analysis was conducted using PRIMER 6.0 and PERMANOVA+ (Clarke and Gorley, 2006; Anderson et al., 2008). Before these tests, samples with poor quality data were excluded, after which 332 bacterial and 343 fungal samples remained. For both the bacteria and fungi, we then generated a rarefied distance matrix among the remaining samples. We created 100 OTU tables from the original data, randomly drawing the lowest common number of sequences from each of the remaining samples (1265 sequences for bacteria; 2527 for fungi). To weight the rarer taxa more heavily, we transformed each table by taking the square root of each cell value and rounding to the nearest integer. We then calculated a Bray–Curtis distance matrix for each of the 100 OTU tables. Finally, for each pairwise comparison between samples (that is, each cell in the distance table), we chose the median Bray–Curtis value among the 100 distance matrices.
To test for the effects of the experimental factors on bacterial and fungal composition, we performed three-way permutational multivariate analyses of variance (PERMANOVA) using the default settings, including all three factors as fixed effects. Except in one case, there were no significant interactions between the factors on microbial composition. Therefore, to improve the estimates of the variation explained by the significant factors, we re-ran the tests with only the main factors. In the one case where the plot-by-microbe interaction was significant (for the fungi in the drought experiment in 2011; P=0.038), we included this variation in the calculations. To identify the taxa contributing to significant treatment differences, we used similarity percentage (SIMPER) analysis. Non-metric multidimensional scaling (MDS) ordination was used to visualize patterns in community composition.
We estimated the effect size—the relative importance of the three manipulated factors—on microbial composition using the estimates of the components of variation from the PERMANOVA. We divided each factor's estimate by the sum of the estimates for all significant factors plus the residual variation (Anderson et al., 2008), then multiplied by 100 to report this value as a percentage. This approach is analogous to the unbiased univariate analysis of variance (ANOVA) estimators of variance components (Searle et al., 1992). We estimated the effect size of the factors on mass loss by using a standard three-way ANOVA (Gotelli and Ellison, 2004). To parallel the composition analyses, an ANOVA was performed for each year (averaged over the sampling dates). Although variance partitioning approaches have limitations, they are especially useful for comparing effects within an experiment, as we do here (Olejnik and Algina, 2003).
To test the effects of global change on GH gene composition, we performed a three-way PERMANOVA on the relative abundance of bacterial GH genes in the metagenomic libraries, lumping the genes into 11 substrate target categories. Similar results were obtained when the genes were categorized into finer categories, defined by the GH family or by GH family and genus-level taxonomy. Given the minimal replication of the metagenomic libraries, we limited our analysis to tests of the three main terms (that is, we did not test for interactions between the terms).
Finally, we analyzed percentage mass loss and microbial abundances using a factorial mixed-model ANOVA with repeated measures, transforming the data when necessary to improve normality and homogeneity of variances. The model included microbe origin, litter origin, plot environment, and date as fixed effects (along with their interactions) and two random effects: block and subject nested within block. We defined subject as the batch of litterbags with the same block, plot, litter origin, and microbe origin.
Results and Discussion
The taxonomic composition and diversity in the litterbags indicated that the cages allowed a natural grassland litter community to persist while preventing microbial immigration. The dominant taxa (defined by 97% sequence similarity of 16S and 28S rRNA genes) were also abundant outside the litterbags at this site (Matulich et al., 2015). Fourteen of the 20 most abundant bacterial taxa (phyla Actinobacteria, Bacteriodetes and Proteobacteria) were shared in both the litterbags and in naturally occurring leaf litter, and their relative frequencies were highly correlated (r=0.72; P=0.004; Supplementary Figure S1). Similar patterns were observed for fungi, with the dominant taxa belonging to the Ascomycota and Basidiomycota phyla (r=0.77, P=0.002; Supplementary Figure S1). At the same time, bacterial and fungal richness was highest in the inoculum and declined over time in the litterbags (Supplementary Figure S2). Such a decline is expected, as the initial inoculum would comprise litter specialists as well as transients from nearby habitats (for example, soil and live plants) that would be outcompeted on leaf litter.
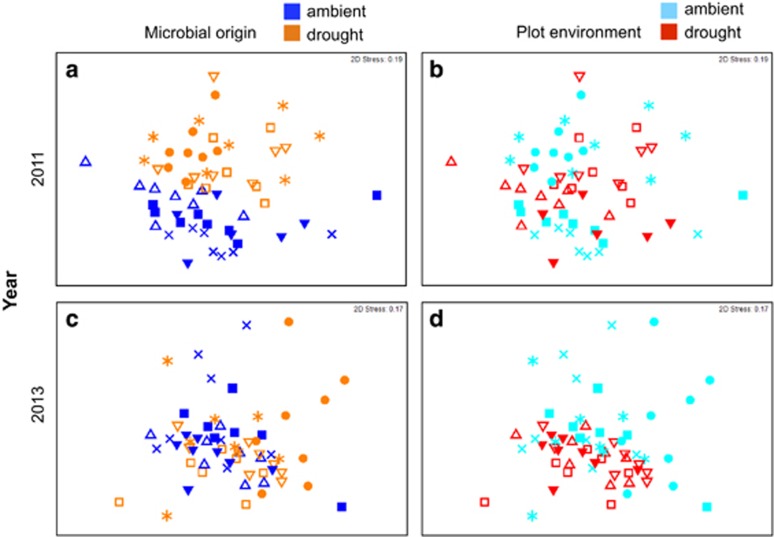
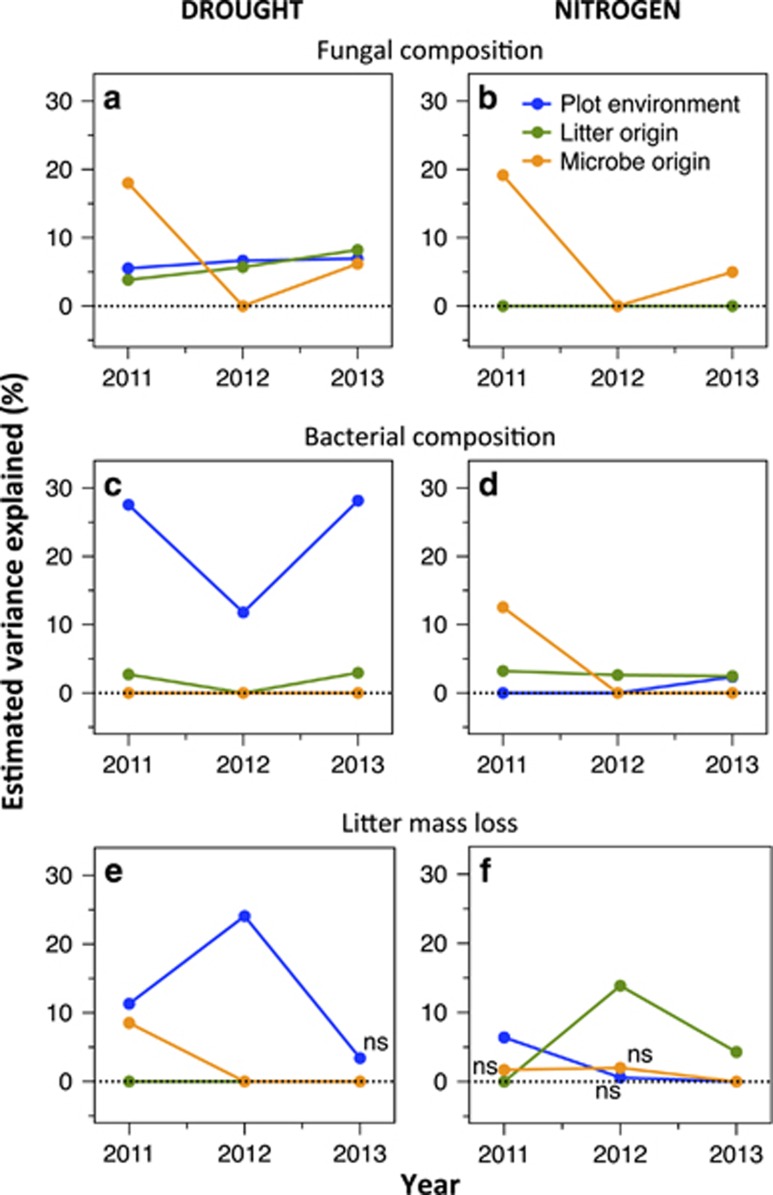
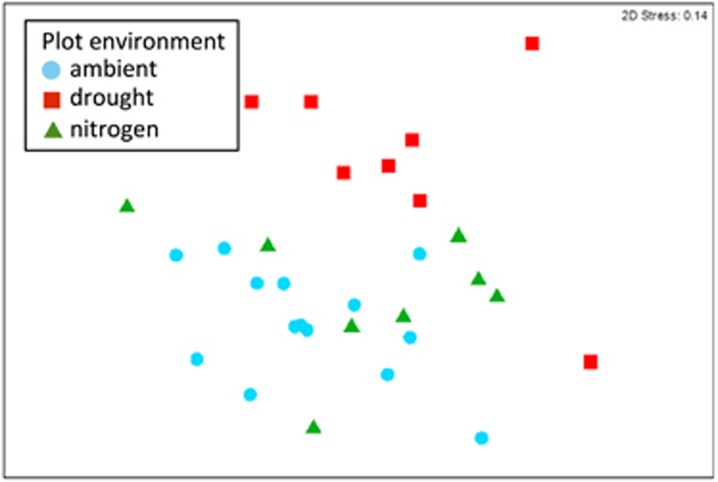
To test for a legacy of microbial composition, we compared the effect size (estimated variance explained in a PERMANOVA model; Anderson, 2001) of microbial origin (the initial inoculum in the litterbag) over time (Figure 1b). This comparison reveals how long a legacy of initial composition was apparent after being transferred to a new litter or plot environment. To illustrate this analysis, we first plot the results of the fungi in the drought experiment using non-metric MDS (Figure 2). In 2011, just 3 months after the initial inoculation of the litterbags, all three factors significantly altered fungal composition (P<0.001; Table 1). However, most (18%) of the variation in fungal composition could be explained by whether the inoculum originated from an ambient or drought plot (Figure 2a) rather than the origin of the litter (4%) or the plot environment (5.5%) of the litterbag (Figure 2b; Table 1). By 2013, the microbial origin effect was much less apparent (6%), whereas the effects of litter origin and plot environment were slightly clearer (Figures 2c and d). Plotting the effect sizes over time (orange line in Figure 3a) reveals that fungal composition was in part determined by the original inoculum even after 3 years.
Figure 2.
Non-metric MDS plots of the fungal communities in the drought experiment litterbags in 2011 (a, b) and 2013 (c, d). Within a year, the samples are color coded on the left-hand column by the origin of the initial microbial inoculum (dark blue and orange) or on the right-hand column by the plot environment in which the litterbags were placed (light blue and red). The different symbols denote the particular combination of the three experimental factors. In order of plot environment, litter origin, and microbial origin: ▪ is AAA, ● is AAD, × is ADA, * is ADD, ▵ is DAA, ▿is DAD, □ is DDD, and ▾ is DDA, where A is ambient and D is drought. Stress values indicate goodness of fit in two dimensions.
Table 1. The effect of plot environment, litter origin and microbe origin on microbial composition.
| Factor | 2011 | 2012 | 2013 | |
|---|---|---|---|---|
| Drought | ||||
| Bacteria | Plot environment | 0.001 | 0.001 | 0.001 |
| Litter origin | 0.011 | 0.170 | 0.008 | |
| Microbe origin | 0.158 | 0.158 | 0.078 | |
| Fungi | Plot environment | 0.002 | 0.001 | 0.001 |
| Litter origin | 0.009 | 0.001 | 0.001 | |
| Microbe origin | 0.002 | 0.072 | 0.005 | |
| Nitrogen | ||||
| Bacteria | Plot environment | 0.569 | 0.570 | 0.038 |
| Litter origin | 0.001 | 0.055 | 0.033 | |
| Microbe origin | 0.001 | 0.462 | 0.171 | |
| Fungi | Plot environment | 0.229 | 0.292 | 0.314 |
| Litter origin | 0.117 | 0.161 | 0.323 | |
| Microbe origin | 0.001 | 0.180 | 0.013 | |
The results of PERMANOVA tests for bacterial or fungal composition for each year in each experiment (drought or added nitrogen). Values are P-values and bolded when <0.05.
Figure 3.
Percent estimated variance explained by plot environment (blue), litter origin (green) and microbe origin (orange) on microbial composition and litter mass loss each year. As in Figure 1b, a steep decline in the variance explained by microbial origin over time indicates high resilience. Top panels (a and b) are fungal composition, middle (c and d) are bacterial composition and bottom (e and f) are mass loss. Panels on the left-hand side are for the drought experiment and on the right-hand side, for the nitrogen experiment. For fungal and bacterial composition, estimated variance is plotted at zero if the factor was not significant (P>0.05, except one case where P=0.055) in the PERMANOVA (Table 1). For litter mass loss, all positive estimates are plotted and marked by ‘ns' if not significant.
In the nitrogen experiment, fungal composition was also still affected by the legacy of the microbial community after 3 years (Figure 3b; Table 1). In contrast, bacterial composition was more resilient than fungal composition. In the drought experiment, microbial origin did not affect bacterial composition even after the first rainy season (Figure 3c). In the nitrogen experiment, microbial origin explained a significant amount (12.5%) of bacterial composition during the first year, but was an insignificant factor by the second year (Figure 3d; Table 1).
Notably, the legacies of the microbial origin were driven by changes in a relatively small number of taxa. Just 10 fungal taxa explained >45% of the compositional variation between the ambient and treatment communities (Supplementary Table S1). Similarly, 10 bacterial taxa explained >20% of the compositional variation in the N experiment (bacterial composition was resilient by the first sampling pointin drought experiment; Supplementary Table S2). The slower response of fungi versus bacteria could be due to slower growth and turnover rates of fungal spores and hyphae than bacterial cells. Indeed, although bacterial density fluctuated by orders of magnitude between sampling points, fungal density was less dynamic (Supplementary Figure S3).
A potential methodological reason for the differential responses of the bacterial and fungal communities could be that very small bacteria are immigrating into the litterbags; however, we do not think this possibility explains this result for two reasons. First, even if a few bacteria (and perhaps even fungal hyphae) immigrate through the litterbag mesh, they would make up a small fraction of the total community. Given that all but the rarest taxa are likely already present in the inoculum to all litterbags, this contamination would have a negligible effect on the abundance of any particular taxon and therefore community composition. Further, if unwanted immigration is much higher than expected, size selection should result in different litterbag communities than those observed outside the bags. In fact, the communities inside and outside of the litterbags were highly correlated (Supplementary Figure S1).
In contrast to the dissipating effect of the microbial origin over time, plot environment and litter origin had a relatively consistent effect on microbial composition over the 3 years (blue and green lines in Figures 3a–d). This result was expected as the plot and litter treatments were maintained over the course of the experiment (new litter from a single collection was added yearly). Precipitation treatment was a particularly strong determinant of bacterial composition (explaining between 12% and 28% of composition variation across the 3 years). The reduced effect in 2012 (Figure 3c) is likely due the timing of the precipitation manipulations that year; both ambient and drought plots received the same rainfall during the 6 weeks before sampling (Supplementary Figure S4).
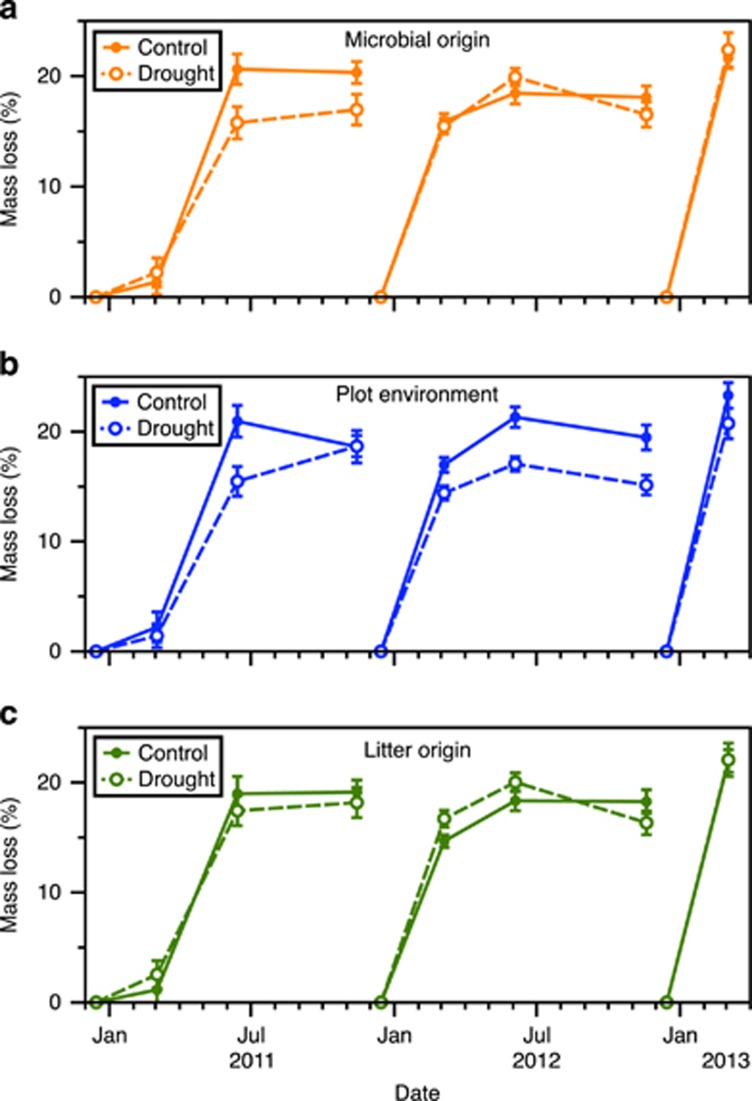
Microbial functioning was also not immediately resilient to the global change manipulations. In the drought experiment, litterbags inoculated with microbial communities from the drought treatment experienced 16.5% slower decomposition by the end of the first rainy season (Figure 4a; microbe origin first year: P=0.009 (Allison et al., 2013); microbial origin all 3 years: P=0.056; microbe × year: P=0.032; Supplementary Table S3). The strength of this microbial origin effect was similar to, or even stronger than, that of the plot environment (Figure 4b). However, by the second rainy season, microbial origin no longer influenced litter mass loss in the drought experiment (Figures 3e and 4). In the nitrogen experiment, microbial origin only weakly affected mass loss (Supplementary Figure S5); the high nitrogen community increased mass loss by an average of 6% over the 3 years, but this effect was not significant in the post hoc individual year tests (Figure 3f). Thus, the compositional differences between the ambient and added nitrogen communities appeared nearly functionally redundant, at least in terms of the decomposition rate.
Figure 4.
Effect of (a) microbial origin, (b) plot environment and (c) litter origin on percentage mass loss in the drought experiment. N=32 for each treatment at each date. Error bars are ±1 s.e.m. See Supplementary Table S3 and Figure 3e for significance and effect sizes, respectively.
What explains the lag in the decomposition response? As inoculum communities were collected within tens of meters of one another, we assume that the same taxa were present in all litterbags even though immigration was restricted. Also, the differences between the drought and ambient communities were caused by moderate changes in the abundance of common taxa, rather than major shifts in the identity of common taxa (Supplementary Figure S1; Matulich et al., 2015). We therefore hypothesize that the lag in microbial composition and its functioning was due to slow growth and turnover of individual taxa. However, we cannot rule out the possibility that the convergence in the community response was also partly due to de novo point mutations and horizontal gene transfer among cells during the experiment.
We also examined the possibility that decomposition rates were altered by changes in microbial abundance rather than microbial composition. Bacterial abundance was measured by flow cytometry and fungal abundance by microscopy. We then converted these metrics to biomass to compare the groups. Drought directly reduced bacterial mass by 30% (P=0.0001; Supplementary Table S3; Supplementary Figure S3), which was accompanied by lower mass loss. However, bacterial mass was also 58% lower when growing on litter from the drought environment (Supplementary Figure S3), and this reduction was not accompanied by a change in mass loss (Figure 3e). Across the entire experiment, bacterial abundance (the vast majority of microbial biomass) had only a marginal effect on litter mass loss by the next sampling date (P=0.054) and no significant interaction between bacterial mass and sampling date (P=0.73; overall analysis of covariance model: F3, 472=60.07, adjusted R2=0.272). Together, these results suggest that microbial composition, not abundance, was primarily responsible for the community influence on decomposition rate (Figure 4a).
To investigate why microbial functioning may be more resilient to changes in nitrogen than to changes in rainfall, we characterized how the global change treatments altered the potential for carbohydrate degradation. Metagenomic sequencing revealed that the litter communities carried an abundant and diverse collection of GH genes. Most (>98%) of the >710 000 identified GH sequences were bacterial, and GH sequences accounted for 0.31% of all annotated bacterial sequences. GH genes can be further classified into different GH families that target different substrates (Lombard et al., 2014; Berlemont and Martiny, 2015). The sequences fell into 67 different GH families that could be divided into 5805 unique types based on both GH family and genus-level taxonomy. Many of the GH genes target oligosaccharides, but others revealed the potential for structural polysaccharide deconstruction, including cellulose, xylan and chitin (Supplementary Figure S6).
The composition of bacterial GH genes in the litterbags was significantly altered by the plot environment (PERMANOVA: P<0.001, variance explained: 27.8%) and specifically, by changes in precipitation (Figure 5). Drought selected for an increased abundance of GH genes that targeted starch, xylan, mixed polysaccharides, and ‘other animal' (for example, non-chitin) polysaccharides, and a decreased abundance of those targeting oligosaccharides and ‘other plant' (for example, non-cellulose or xylan) polysaccharides (Supplementary Table S4). In contrast, added nitrogen did not alter GH composition (Figure 5), in agreement with previous work at this site (Berlemont et al., 2014). This result suggests a mechanistic reason for why microbial functioning was more resilient to changes in nitrogen than to changes in rainfall. Drought altered bacterial taxonomic composition in such a way that carbohydrate degradation traits were also altered, leading to a lower decomposition rate. In contrast, taxonomic shifts in response to nitrogen addition did not alter carbohydrate degradation traits, and the functional consequences were barely detectable. This difference highlights the correlation (or lack thereof) between effect and response traits, where an organism's effect traits affect ecosystem functioning and its response traits influence how a taxon responds to environmental change (Lavorel and Garnier, 2002; Martiny et al., 2015). In this litter community, traits that affect litter decomposition appear to be correlated with the response to changing rainfall, but not to changing nitrogen availability.
Figure 5.
Non-metric MDS plots of bacterial GH composition classified by substrate targeted in the litterbags after the first rainy season. Plot environment (ambient (circles), drought (squares) and added nitrogen (triangles)) significantly altered GH composition (P<0.001), but litter origin and microbial origin did not (P>0.05). Similar results were observed when the analysis was performed based on unique GH types or GH families.
Two additional considerations will be important to understand the consequences of microbial legacies for predictions of ecosystem processes. First, we only investigated the effect on decomposition, but the same legacies may simultaneously alter additional, unmeasured processes. Second, given that ecosystems are constantly being disturbed, it will be important to investigate how even a 1-year lag time may compound over time. Nevertheless, terrestrial microbial communities should not broadly be expected to be instantaneously resilient.
Acknowledgments
We thank J Brown, A Chase, B Khalili, M Nelson and R Puxty for comments on earlier versions of the manuscript and K Matulich for technical and statistical assistance. This work was supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research (BER), under Award Number DE-PS02-09ER09-25 and the NSF Major Research Instrumentation program.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Adair EC, Parton WJ, Del Grosso SJ, Silver WL, Harmon ME, Hall SA et al. (2008). Simple three-pool model accurately describes patterns of long-term litter decomposition in diverse climates. Global Change Biol 14: 2636–2660. [Google Scholar]
- Allison SD, Martiny JBH. (2008). Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci USA 105: 11512–11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison SD, Lu Y, Weihe C, Goulden ML, Martiny AC, Treseder KK et al. (2013). Microbial abundance and composition influence litter decomposition response to environmental change. Ecology 94: 714–725. [DOI] [PubMed] [Google Scholar]
- Alster CJ, German DP, Lu Y, Allison SD. (2013). Microbial enzymatic responses to drought and to nitrogen addition in a southern California grassland. Soil Biol Biochem 64: 68–79. [Google Scholar]
- Amend AS, Martiny AC, Allison SD, Berlemont R, Goulden ML, Lu Y et al. (2015). Microbial response to simulated global change is phylogenetically conserved and linked with functional potential. ISME J 10: 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32–46. [Google Scholar]
- Anderson MJ, GR, Clarke KR. (2008) PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods. PRIMER-E Ltd: Plymouth, UK. [Google Scholar]
- Berlemont R, Martiny AC. (2013). Phylogenetic distribution of potential cellulases in bacteria. Appl Environ Microbiol 79: 1545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlemont R, Allison SD, Weihe C, Lu Y, Brodie EL, Martiny JBH et al. (2014). Cellulolytic potential under environmental changes in microbial communities from grassland litter. Front Microbiol 5: 639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlemont R, Martiny AC. (2015). Genomic potential for polysaccharide deconstruction in bacteria. Appl Environ Microbiol 81: 1513–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratbak G. (1985). Bacterial biovolume and biomass estimations. Appl Environ Microbiol 49: 1488–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke K, Gorley R. (2006) Primer v6: User Manual/Tutorial. PRIMER-E: Plymouth, UK. [Google Scholar]
- Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y et al. (2013). Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42(Database issue): D633–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, Relman DA. (2012). The application of ecological theory toward an understanding of the human microbiome. Science 336: 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and Workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Evans SE, Wallenstein MD. (2012). Soil microbial community response to drying and rewetting stress: does historical precipitation regime matter? Biogeochemistry 109: 101–116. [Google Scholar]
- Fenn ME, Poth MA, Bytnerowicz A, Sickman JO, Takemoto BK. (2003). Effects of ozone, nitrogen deposition, and other stressors on montane ecosystems in the Sierra Nevada. Dev Environ Sci 2: 111–155. [Google Scholar]
- Giovannetti M, Mosse B. (1980). An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytologist 84: 489–500. [Google Scholar]
- Glass EM, Meyer F. (2011). The metagenomics RAST server: a public resource for the automatic phylogenetic and functional analysis of metagenomes. Handbook of Molecular Microbial Ecology I: Metagenomics and Complementary Approaches. Wiley: Hoboken, NJ, USA, pp, 325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotelli NJ, Ellison AM. (2004) A Primer of Ecological Statistics. Sinauer: Sunderland, MA, USA. [Google Scholar]
- Hawkes CV, Keitt TH. (2015). Resilience vs historical contingency in microbial responses to environmental change. Ecol Lett 18: 612–625. [DOI] [PubMed] [Google Scholar]
- Kimball S, Goulden ML, Suding KN, Parker S. (2014). Altered water and nitrogen input shifts succession in a southern California coastal sage community. Ecol Appl 24: 1390–1404. [DOI] [PubMed] [Google Scholar]
- Lavorel S, Garnier E. (2002). Predicting changes in community composition and ecosystem functioning from plant traits: revisiting the Holy Grail. Funct Ecol 16: 545–556. [Google Scholar]
- Lawrence CR, Neff JC, Schimel JP. (2009). Does adding microbial mechanisms of decomposition improve soil organic matter models? A comparison of four models using data from a pulsed rewetting experiment. Soil Biol Biochem 41: 1923–1934. [Google Scholar]
- Li Q, Yang Y, Bao XL, Liu F, Liang WJ, Zhu JG et al. (2015). Legacy effects of elevated ozone on soil biota and plant growth. Soil Biol Biochem 91: 50–57. [Google Scholar]
- Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42: D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreau M, Downing A, Emmerson M, Gonzalez A, Hughes J, Inchausti P et al. (2002). A new look at the relationship between diversity and stability In: Loreau M, Naeem S, Inchausti P (eds). Biodiversity and Ecosystem Functioning: Synthesis and Perspectives. Oxford University Press: New York, NY, USA, 79–91. [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny JBH, Jones SE, Lennon JT, Martiny AC. (2015). Microbiomes in light of traits: a phylogenetic perspective. Science 350: aac9323. [DOI] [PubMed] [Google Scholar]
- Matulich K, Weihe C, Allison SD, Amend A, Berlemont R, Goulden ML et al. (2015). Temporal variation overshadows the response of leaf litter microbial communities to simulated global change. ISME J 9: 2477–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon KD, Martin HG, Hugenholtz P. (2007). Integrating ecology into biotechnology. Curr Opin Biotech 18: 287–292. [DOI] [PubMed] [Google Scholar]
- Olejnik S, Algina J. (2003). Generalized eta and omega squared statistics: measures of effect size for some common research designs. Psychol Methods 8: 434–447. [DOI] [PubMed] [Google Scholar]
- Parmesan C. (2006). Ecological and evolutionary responses to recent climate change. Annu Rev Ecol Evol S 37: 637–669. [Google Scholar]
- Paul EA, Clark FE. (1996) Soil Microbiology and Biochemistry 2nd edn. Academic Press: San Diego, CA, USA. [Google Scholar]
- Potts DL, Suding KN, Winston GC, Rocha AV, Goulden ML. (2012). Ecological effects of experimental drought and prescribed fire in a southern California coastal grassland. J Arid Environ 81: 59–66. [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed HE, Martiny JBH. (2007). Testing the functional significance of microbial composition in natural communities. FEMS Microbiol Ecol 62: 161–170. [DOI] [PubMed] [Google Scholar]
- Reeder J, Knight R. (2010). Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods 7: 668–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousk J, Smith AR, Jones DL. (2013). Investigating the long-term legacy of drought and warming on the soil microbial community across five European shrubland ecosystems. Global Change Biol 19: 3872–3884. [DOI] [PubMed] [Google Scholar]
- Schimel JP, Gulledge J. (1998). Microbial community structure and global trace gases. Global Change Biol 4: 745–758. [Google Scholar]
- Searle SR, Casella G, McCulloch CE. (1992) Variance Components. Wiley: New York, NY, USA. [Google Scholar]
- Shade A, Peter H, Allison SD, Baho DL, Berga M, Bürgmann H et al. (2012). Fundamentals of microbial community resistance and resilience. Front Microbiol 3: 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift MJ, Heal OW, Anderson JM. (1979) Decomposition in Terrestrial Ecosystems. Blackwell Scientific Publications: Oxford, UK. [Google Scholar]
- Treseder KK, Balser TC, Bradford MA, Brodie EL, Dubinsky EA, Eviner VT et al. (2012). Integrating microbial ecology into ecosystem models: challenges and priorities. Biogeochemistry 109: 7–18. [Google Scholar]
- Updegraff K, Bridgham SD, Pastor J, Weishampel P. (1998). Hysteresis in the temperature response of carbon dioxide and methane production in peat soils. Biogeochemistry 43: 253–272. [Google Scholar]
- Waldrop MP, Firestone MK. (2006). Response of microbial community composition and function to soil climate change. Microbial Ecol 52: 716–724. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczek AM, Cooper MD, Korves TM, Schmitt J. (2014). Lagging adaptation to warming climate in Arabidopsis thaliana. Proc Natl Acad Sci USA 111: 7906–7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T, Williams L, Smith J, Mendelsohn R. (2003) Global Climate Change and California: Potential Implications for Ecosystems, Health, and the Economy—Consultant Report. California Energy Commission: USA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.