Abstract
Members of the marine genus Rhodopirellula are attached living bacteria and studies based on cultured Rhodopirellula strains suggested that three closely related species R. baltica, ‘R. europaea' and ‘R. islandica' have a limited geographic distribution in Europe. To address this hypothesis, we developed a nested PCR for a single gene copy detection of a partial acetyl CoA synthetase (acsA) from intertidal sediments collected all around Europe. Furthermore, we performed growth experiments in a range of temperature, salinity and light conditions. A combination of Basic Local Alignment Search Tool (BLAST) and Minimum Entropy Decomposition (MED) was used to analyze the sequences with the aim to explore the geographical distribution of the species and subspecies. MED has been mainly used for the analysis of the 16S rRNA gene and here we propose a protocol for the analysis of protein-coding genes taking into account the degeneracy of the codons and a possible overestimation of functional diversity. The high-resolution analysis revealed differences in the intraspecies community structure in different geographic regions. However, we found all three species present in all regions sampled and in agreement with growth experiments we demonstrated that Rhodopirellula species do not have a limited geographic distribution in Europe.
Introduction
The genus Rhodopirellula is a marine representative of a globally distributed and environmentally important bacterial phylum, Planctomycetes, in the Planctomycetes-Verrucomicrobia-Chlamydiae super-phylum (Wagner and Horn, 2006). Members of this group show some exceptional properties, such as budding reproduction and species-specific intracellular membrane-separated compartmentalization (Fuerst and Sagulenko, 2011; Speth et al., 2012) and they are proposed to have an important role in the global carbon and nitrogen cycles (Strous et al., 2002; Glöckner et al., 2003). The genus Rhodopirellula and the type strain Rhodopirellula baltica SH1T were first described by Schlesner et al. (2004). This aerobic chemoorganotrophic bacterium attaches and then reproduces by budding, which results in motile and sessile periods of life. The adult cells can be found attached on sediment particles (Winkelmann and Harder, 2009) and in association with organisms, for example, phytoplankton, sponges, and macroalgae (DeLong et al., 1993; Fuerst et al., 1997; Pimentel-Elardo et al., 2003; Bengtsson and Øvreås, 2010). Besides R. baltica, the genus comprises many strains awaiting a full taxonomic description (Winkelmann and Harder, 2009) and four validly described species isolated from different habitats around the world (Roh et al., 2013; Bondoso et al., 2014; Yoon et al., 2015). A systematic isolation resulted in a strain collection of the genus Rhodopirellula from European coastal seas and sediments, with many strains affiliating according to 16S rRNA gene sequence identity of 99.5–100% to R. baltica SH1T (Winkelmann and Harder, 2009). However, a multilocus sequence analysis and DNA–DNA hybridization experiments revealed the presence of several species (Winkelmann et al., 2010). This observation was confirmed by draft genome sequences: the average nucleotide identity of shared genes between the strains revealed three species; R. baltica (strains SH1T, SH28, WH47, SWK14 (Richter et al., 2014)), ‘R. europaea' (strains SH398 and 6C (Richter-Heitmann et al., 2014)) and ‘R. islandica' (strain K833 (Kizina et al., 2015)).
Cultivation-independent molecular approaches employing next-generation sequencing technologies enable an in-depth coverage of occurrence and diversity of microorganisms in environmental samples, well beyond the study of strains and type strains. However, an adequate gene marker is necessary to circumvent the low resolution of the 16S rRNA gene and to resolve fine-scale biogeographic structures in environmental studies (Ramette and Tiedje, 2007). So far, rpoB coding for the beta subunit of the RNA polymerase (Bondoso et al., 2013) and carB coding for the large subunit of carbamoylphosphate synthetase (Žure et al., 2015) have been used for the genus Rhodopirellula. These amplicons were designed to cover the whole genus. In this study with a phylogenetically more closely related target group of organisms, we analyzed the genomes, designed primers for a nested PCR and amplified a part of the acetyl CoA synthetase gene (acsA) to investigate the hypothesis of Winkelmann et al. (2010) that Rhodopirellula species have a limited geographic distribution in European coastal areas, with R. baltica limited to parts of the Baltic Sea and the east coast of the North Sea, ‘R. europaea' to southern North Sea to the Mediterranean Sea and ‘R. islandica' to North European Atlantic regions. Furthermore, we analyzed the microdiversity in the acsA gene sequences using the Minimum Entropy Decomposition (MED), an automated oligotyping method (Eren et al., 2013, 2014b). The ‘oligotyping' utilizes Shannon entropy (Shannon, 1948) to identify the variability in each nucleotide position and uses the high-entropy positions to partition sequence data sets into high-resolution operational taxonomic units. Position-specific variations up to a single-nucleotide resolution in the sequenced region can resolve ecologically meaningful differences between closely related organisms (McLellan et al., 2013; Eren et al., 2014a, 2015; Buttigieg and Ramette, 2015). In addition, we performed growth experiments in a wide range of temperature, salinity and light conditions to demonstrate that strains of these three species are capable to grow at environmental relevant conditions.
Materials and methods
Sampling and DNA extraction
The upper 2 cm of intertidal sandy sediments were sampled in a period from 2005 to 2013 from 147 locations around the European coast and frozen at −80 °C until further processing. Genomic DNA was extracted from 500 mg of sediment using the FastDNA Spin Kit for Soil (MP Biomedicals, Santa Ana, CA, USA) and characterized by spectroscopy (NanoDrop 100, Thermo Fisher Scientific, Waltham, Massachusetts, USA) and agarose gel electrophoresis with ethidium bromide staining.
Primer design and amplification of acsA gene
Amplicon sequences (Winkelmann et al., 2010) and genomes of nine Rhodopirellula strains and six other planctomycetes (Blastopirellula marina, Gemmata obscuriglobus, Planctomyces maris, Planctomyces limnophilus, Planctomyces staleyi and Candidatus Kuenenia stuttgartiensis) were used to design degenerated nested PCR primers. Partial gene amplicons of acsA, carB, fumC, glyA, guaA and icd were in silico specific for R. baltica, ‘R. europaea' and ‘R. islandica' and had several mismatches to other Rhodopirellula strains. Optimisation of PCR conditions gave the best results for nested primers of the acsA gene (NCBI GenID:1792396 in SeqID NC_005027.1). The external acsA primers targeted a 586 bp long amplicon from the position 1079 to 1663: forward primer 5′-GACDTGYTTGATGTACGAAG-3′ and reverse primer 5′-TCSACYTCGATCGTGCTCA-3′. The internal acsA primers targeted a 487 bp long amplicon from the position 1139 to 1625: forward primer 5′-GTYGAGCGKCACAARGTCA-3′ and reverse primer 5′-GATCACATCRTCGATGCGT-3′. Fifteen barcoded internal primers were used for Illumina MiSeq sequencing (Supplementary Table S1). In the first amplification, the PCR mixture contained 10–15 ng of genomic DNA, 3.3 μm of each acsA external primer, and a 2 × PCR master mix (Promega, Madison, Wisconsin, USA) in 30 μl volume. The template DNA was denatured for 4 min at 94 °C, followed by 30 cycles of 1 min at 94 °C, 1 min at 58 °C, 3 min at 72 °C, and a final extension of 10 min at 72 °C. Amplicons were purified with a PCR purification kit (Qiagen, Hilden, Germany), quantified by spectroscopy and visualized on 1% agarose gel stained with ethidium bromide. Depending on the amplicon quantity, 1 μl of non-diluted or diluted (1:10, 1:100) purified PCR amplicon served as template for amplification with internal acsA primers (3.3 μm) and a 2 × PCR master mix (Promega) in 30 μl volume. The PCR protocol was 4 min at 94 °C, followed by 20 or 30 cycles of 1 min at 94 °C, 1 min at 58 °C, 3 min at 72 °C and a final extension of 10 min at 72 °C (Supplementary Table S2). After amplicon purification and quantification, 43 out of 95 amplicons obtained after the second PCR were selected for sequencing on a MiSeq platform (Illumina, San Diego, California, USA) at the Max Planck-Genome-Centre, Cologne, Germany (Table 1).
Table 1. Overview on sample sites in this study.
| Sample ID | Sample location | Latitude, longitude (°) | Sampling date |
|---|---|---|---|
| 2 | Spain | 39.51722, 2.47806 | 05.03.2005 |
| 6 | Poland | 54.45028, 18.57250 | 03.2005 |
| 9 | Scotland | 56.36917, −2.82833 | 03.2005 |
| 11 | Netherlands | 51.49000, 4.05500 | 03.2005 |
| 19 | Greece | 38.95583, 20.76083 | 04.2005 |
| 23 | Iceland | 64.03556, −22.69861 | 18.05.2005 |
| 25 | Germany | 54.27500, 13.66833 | 24.06.2005 |
| 26 | Ireland | 53.25972, −6.11028 | 29.06.2005 |
| 28 | Germany | 54.17300, 7.88933 | 06.07.2005 |
| 32 | Sweden | 58.25000, 11.45167 | 23.09.2005 |
| 35 | Italy | 42.58944, 10.09667 | 08.08.2005 |
| 38 | France | 48.64972, −1.50889 | 11.09.2005 |
| 49 | Portugal | 36.999586, −7.98027 | 20.06.2013 |
| 54 | Belgium | 51.166643, 2.761859 | 27.06.2013 |
| 57 | Netherlands | 51.833974, 3.924392 | 28.06.2013 |
| 58 | Netherlands | 51.87514, 4.040154 | 28.06.2013 |
| 60 | Netherlands | 52.621679, 4.618224 | 28.06.2013 |
| 61 | Netherlands | 52.842044, 4.691182 | 26.06.2013 |
| 65 | France | 48.719873, −3 995452 | 28.06.2013 |
| 70 | Norway | 58.94198, 5.67152 | 07.2012 |
| 76 | Norway | 64.76812, 11.62288 | 07.2012 |
| 80 | Norway | 66.48755, 13.22710 | 07.2012 |
| 86 | Norway | 69.68903, 18.86810 | 07.2012 |
| 92 | England | 50.417368, −5.075652 | 07.08.2013 |
| 94 | England | 50.043092, −5.642209 | 08.07.2013 |
| 96 | England | 50.35077, −4.26656 | 03.09.2013 |
| 101 | Scotland | 55.848596, −6.090192 | 02.09.2013 |
| 104 | Scotland | 58.256017, −6.327359 | 09.09.2013 |
| 109 | Scotland | 56.93383, −7.494062 | 24.07.2013 |
| 112 | France | 48.641524, −2.308327 | 29.06.2013 |
| 116 | France | 48.102158, −4.352176 | 23.08.2013 |
| 117 | France | 48.09098, −4.299953 | 24.08.2013 |
| 120 | Italy | 40.744985, 13.945478 | 13.07.2013 |
| 123 | Spain | 38.16703, −0.48596 | 16.06.2013 |
| 126 | Greece | 37.583647, 23.394953 | 09.09.2013 |
| 129 | Ireland | 52.678646, −9.65115 | 02.08.2013 |
| 131 | Croatia | 43.502523, 16.447045 | 10.04.2012 |
| 142 | Germany | 55.03432, 8.42428 | 04.2013 |
| 143 | Germany | 53.73668, 7.69893 | 04.2013 |
| 144 | Netherlands | 51.45950, 4.08026 | 10.2012 |
| 145 | France | 50.99695, 1.98212 | 10.2012 |
| 146 | France | 49.33955, −0.46952 | 10.2012 |
| 147 | France | 48.61387, −1.74368 | 10.2012 |
Preprocessing of Illumina raw reads
MiSeq sequencing yielded 4 713 544 paired-end reads. Linker and primer sequences were removed using Cutadapt 1.8 (https://github.com/marcelm/cutadapt; (Martin, 2011)) and the reads were demultiplexed using Sabre 1.000 (https://github.com/najoshi/sabre). Pair-end reads were merged with PEAR 0.9.5 (Zhang et al., 2014) and quality trimmed to a phred quality of 20 using the BBDuk program from the BBMAP 34.00 suite (http://sourceforge.net/projects/bbmap/). The open-reading frames were identified using the getorf program from the EMBOSS suite 6.6.0 (Rice et al., 2000) and sequences with stop codons were removed. Furthermore, the acsA sequences were translated and aligned using MAFFT version 7 (Katoh and Standley, 2013). The amino-acid alignment was back-translated to nucleotides and visually inspected and refined using Seaview version 4 (Gouy et al., 2009). The preprocessing steps resulted in 1 493 643 high-quality sequences (483 bp) from 43 sample sites (Supplementary Table S2). Raw sequence reads were deposited at the NCBI (http://www.ncbi.nlm.nih.gov), Bioproject ID: PRJNA293505.
Taxonomic classification of partial acsA gene sequences
The high-quality reads were taxonomically assigned using the Basic Local Alignment Search Tool (BLAST) version 2.2.30+ (Camacho et al., 2009). Sequences were compared with a database containing acsA sequences of Rhodopirellula strains (Supplementary Table S3) using nucleotide–nucleotide BLAST (blastn). Sequences that had >94% identity to strains of R. baltica, ‘R. europaea' or ‘R. islandica' were assigned to these species. The threshold was inferred from the intraspecies pairwise identities of the acsA amplicon sequence (483 bp) of four R. baltica strains (94.8–97.3%) and the interspecies identities (<92%). Sequences with identities lower than 94% to strains of the three Rhodopirellula species were further analyzed using the non-redundant sequence database of NCBI (June 2015 version).
MED analysis
The subspecies diversity (microdiversity) in the acsA gene sequences affiliated to R. baltica, ‘R. europaea' or ‘R. islandica' was studied using the MED pipeline version 2.2. MED is an automated oligotyping method that partitions the whole data sets into homogenous operational taxonomic units (MED nodes). To account for singularities of the protein-coding genes such as the degeneracy of the genetic code, we developed a protocol and set of diagnostics prior to the application of oligotyping. First, we calculated the entropy of each position as described in the oligotyping pipeline. Then we analyzed the distribution of the entropy values for each codon position to observe a possible entropy bias towards the third codon position owing to the much faster evolution rate of the third position in comparison with the first and the second codon position. Afterwards, we calculated the frequencies of transitions and transversions to unravel the nature of the nucleotide variations observed in the third codon position and plotted the frequencies against the K80 genetic distance (Kimura's 2-parameters distance (Kimura, 1980)) in the so-called saturation plot. Owing to the large size of the next-generation sequencing data sets, we estimated the saturation plot by performing 100 random subsamples of 10 000 sequences each. Those diagnostics have been implemented in the R package oligo4fun (http://genomewalker.github.io/oligo4fun/). Oligo4fun relies on the R packages SPIDER (Brown et al., 2012) and APE (Paradis et al., 2004). The data set of sequences affiliating to R. baltica, ‘R. europaea' and ‘R. islandica' were analyzed with this protocol. We studied the effect of the higher entropy in the third codon position on the final number of nodes by MED analyses using two data sets: one using all base positions of the R. baltica-affiliated sequences and the same data set where we removed the third codon position. The representative nucleotide sequences of the final nodes obtained in both MED analyses were translated to amino acids (the third codon position was restored). The resulting proteins were clustered at the 100% identity level using CD-HIT v4.5.6 (Li and Godzik, 2006) to reveal the effect of silent substitutions on the MED partitioning at the functional level. A comparison of the cluster representatives with CD-HIT-2d allowed us to identify shared fractions between both MED analyses.
Based on results of these preliminary MED analyses, the third position in the codon was removed from all sequences and MED analyses were performed for each species-affiliated sequence data set separately. Different absolute sequence numbers required different M values, which serve as noise filter: a node will be eliminated if the abundance of the sequence is below the M value. The M values were calculated dividing the number of sequences by 10 000 as recommended by Eren et al., (2014b). For R. baltica, ‘R. europaea', and ‘R. islandica', M values of 75, 17 and 22 were used, respectively. The number of discriminant locations used for entropy decomposition discriminant (d) was set to 1 for all species. Maximum variation allowed in each node (V) was computed in the MED pipeline and was set to three nucleotides. A summary of MED analyses is given in Supplementary Table S4.
Diversity within Rhodopirellula species
The sequence counts within the data sets of R. baltica, ‘R. europaea' and ‘R. islandica' were normalized using a cumulative sum scaling, which divides the counts by the cumulative sum of counts up to a percentile determined by a data-driven approach (Paulson et al., 2013; Barberán et al., 2015). We estimated the inverse Simpson diversity index for each Rhodopirellula species using the interpolation and extrapolation with Hill numbers methodology as described by Chao et al. (2014) and implemented in the iNEXT package for the R environment (https://github.com/JohnsonHsieh/iNEXT). For the sample size-based rarefaction approach and extrapolation we discarded all samples with abundance smaller than the median and we defined a base sample size two times larger than the mean abundance of the remaining samples. The 95% confidence intervals were obtained using the bootstrap method developed by Chao et al. (2014) with 200 replicates. In addition, species richness and Shannon diversity indices were estimated for each sample. Sample size-based rarefaction curves for R. baltica, ‘R. europaea' and ‘R. islandica' are shown in Supplementary Figure S1. Estimated species richness and Shannon diversity indices are provided in Supplementary Table S5.
Species community analysis
For the analysis, we discarded samples where the number of sequences were less than the median value of all samples and all the MED nodes with prevalence lower than 10%. After the filtering, 22 samples remained for R. baltica, 20 for ‘R. europaea' and 22 for ‘R. islandica'. Using the cumulative sum scaling normalized counts (see diversity section), we log2 transformed the normalized counts (Paulson et al., 2013). We calculated the species community similarity using the Bray-Curtis dissimilarity distance implemented in the vegan R package (http://vegan.r-forge.r-project.org/). We used the Sørensen similarity coefficients (1—Bray-Curtis dissimilarity) to construct a graph using the samples as nodes and the similarities as edges, then we superimposed the graph on a map. To simplify the graph, we implemented a trimming algorithm that tries to maintain the connectivity of the graph with the least number of edges, to highlight the most similar associations without the need of applying any arbitrary threshold.
Growth experiments
R. baltica SH1T, ‘R. europaea' 6C and ‘R. islandica' K833 were grown in three replicates at different temperature, salinity and light conditions in 250 ml flasks containing 50 ml of liquid M13a medium (DSMZ medium 600a; Schlesner, 1994), with modifications described by Winkelmann and Harder (2009) and shaken at 50 rpm. Standard conditions for growth were dark, 28 °C and salinity 35 practical salinity unit (100% artificial sea water). Experimental conditions included four temperatures (14, 21, 28, 37 °C), four salinities (25, 50, 100, 125, 150% artificial sea water) and three light conditions (24 h dark, 24 h light and 12 h light/12 h dark). Cultures were sampled daily for optical density measurements at 600 nm (OD600) to generate growth curves and determine growth rates.
Results
Taxonomic classification of acsA gene sequences
BLAST analysis revealed that 75% of 1 493 643 amplicon sequences in the whole data set affiliated with the species R. baltica, ‘R. europaea' and ‘R. islandica', thus they had >94% identity to a reference sequence (Supplementary Table S2). The rest of the sequences were assigned to Planctomycetaceae affiliating to other Rhodopirellula (18% of all sequences, no affiliation to known strains on the species level) or to the genera Blastopirellula (5%) and Pirellula (2%) (data not shown).
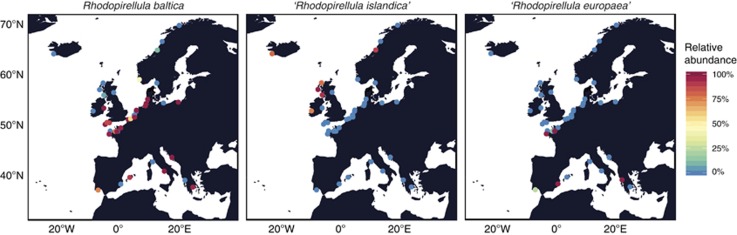
In the 43 sample sites, sequences of R. baltica and ‘R. islandica' were found in all samples and sequences of ‘R. europaea' were found in 40 samples, with relative sequence abundances ranging from 0.01% to 99% in each sample (Figure 1). R. baltica was detected with high relative abundance (43–99%) in 23 samples covering all regions sampled. In samples from Scotland (101 (sample ID, see Table 1)) and Norway (76) this species accounted for 7 and 10%, respectively, and in the remaining 19 samples the relative abundances were <1%. High abundances of ‘R. islandica' were found in six samples, from Iceland (23), Ireland (129), Scotland (101, 104 and 109) and Norway (76), with relative abundances of 74–90%. In other samples, ‘R. islandica' accounted for 0.02–2% of total sequences. ‘R. europaea' dominated in four samples, two from the Mediterranean Sea (19 and 123) and two from France (117 and 38) with relative abundances of over 97%. ‘R. europaea' was also found in samples from the Mediterranean Sea (49), France (112) and North Sea (11), with relative abundances of 24, 14 and 5%, respectively. In the rest of the data set, ‘R. europaea'-related sequences accounted for 0.02–2%.
Figure 1.
Distribution and relative abundance of acsA gene sequences of R. baltica, 'R. islandica' and 'R. europaea' in each sampling site identified by BLAST.
MED analysis
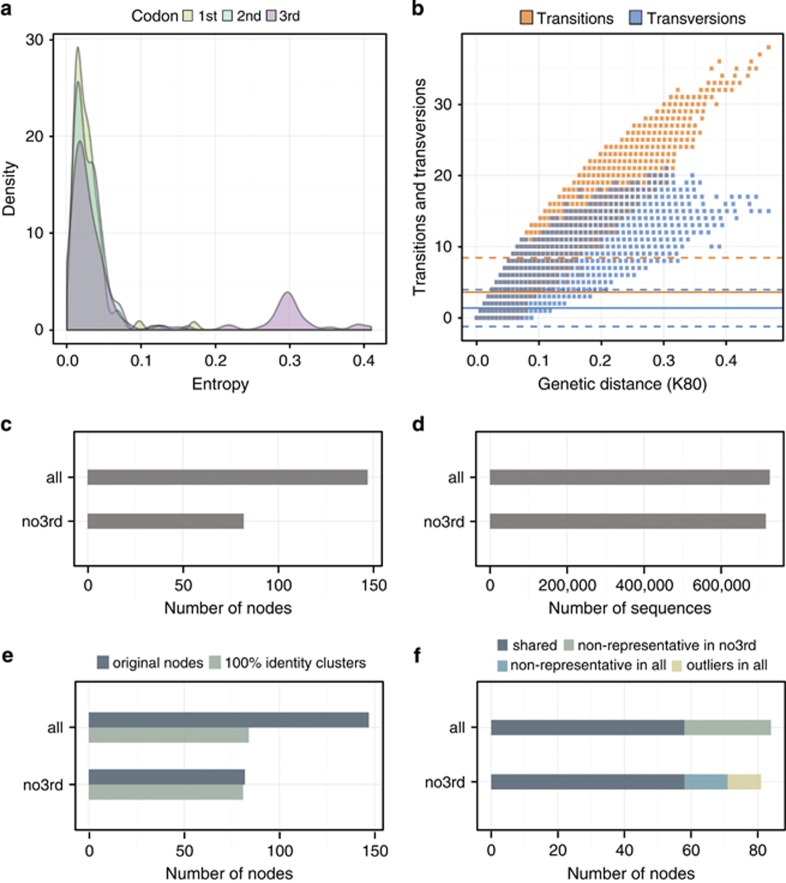
The distribution of the entropy values for each codon position showed a clear bias toward the third position in the codon for the whole data set (Supplementary Figure S2) and the R. baltica data set (Figure 2a). The saturation substitution analyses revealed that transitions are more frequent than transversions in the third position of the codon for the whole data set (Supplementary Figure S3) and the R. baltica data set (Figure 2b). MED analysis of R. baltica sequences with the third position (‘all') in the codon gave ~1.8 times more nodes than MED analysis of the data set with the third position removed (‘no3rd'), although both approaches harbored a similar number of sequences (Figures 2c and d). Translation of the node representative sequences yielded clusters at 100% protein sequence identity. This revealed the effect of higher frequencies of transitions in the third codon position resulting in silent substitutions. As shown in Figure 2e, MED analysis of the ‘all' data set showed that 40% of the node representatives had the same amino-acid sequence (84 non-identical MED nodes). In contrast, only 1% of the translated node representatives were identical (81 non-identical MED nodes) when the MED analysis was performed with only the first two bases of each codon (‘no3rd'). The composition of the 100% identity clusters showed that both approaches shared 58 of the final node representative sequences (Figure 2f). All of the 26 non-shared representative nodes from the ‘all' data set were included in one of the ‘no3rd' representative nodes. The fraction not shared in the ‘no3rd' data set corresponds to non-representative sequences (13 MED nodes, 16%) and outliers due the M parameter (10 MED nodes, 12%) in the ‘all' data set. Those results showed how the removal of the third codon position increased the resolution of the MED analyses obtaining a higher number of functionally non-redundant MED nodes, hence, the removal of the third codon position to account for the bias of the third codon position and a possible overestimation of the diversity was well justified.
Figure 2.
MED partitioning analysis of R. baltica sequence dataset: distribution of the entropies values in each codon position (a); substitution saturation plots for the third codon position from one of the sub-sampling iterations with 10000 sequences (b); number of MED nodes for sequences with all codon positions (all) and sequences where the third codon position was removed (no3rd) (c); number of sequences contained in the final MED nodes (d); clustering of the final MED nodes on the aminoacid level at 100% identity for MED analysis when using all coding positions (all) compared to when the third codon position was removed (no3rd) (e); overlap between the nodes with and without the third codon position after the clustering at 100% identity on aminoacid level (f).
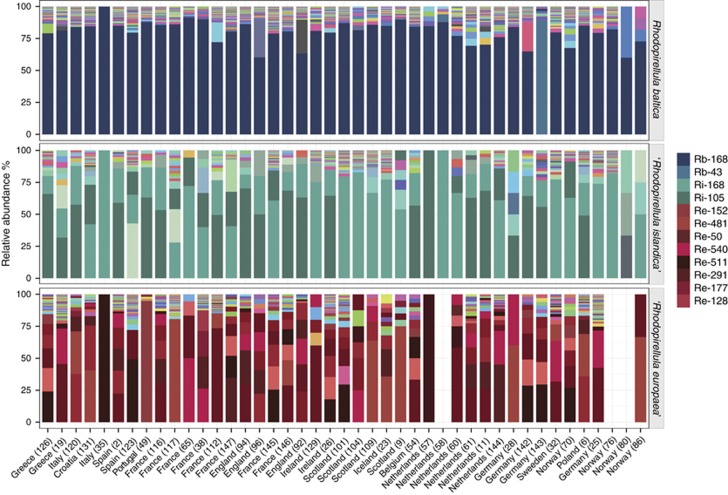
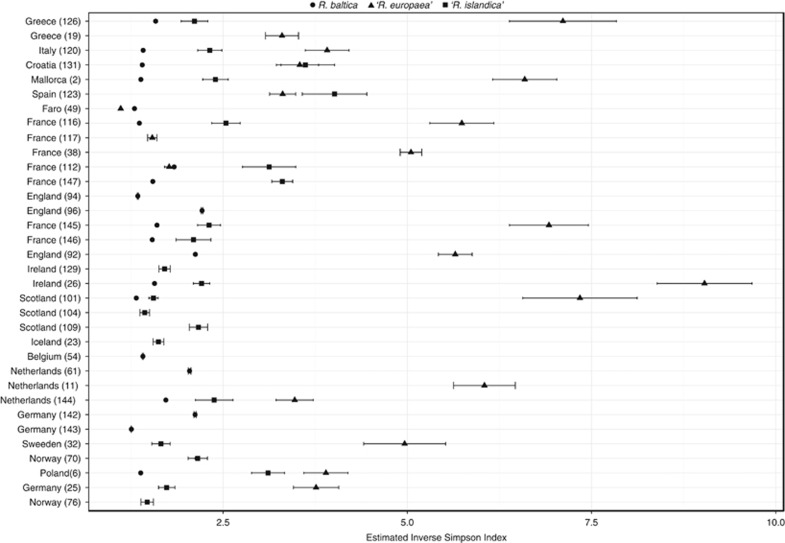
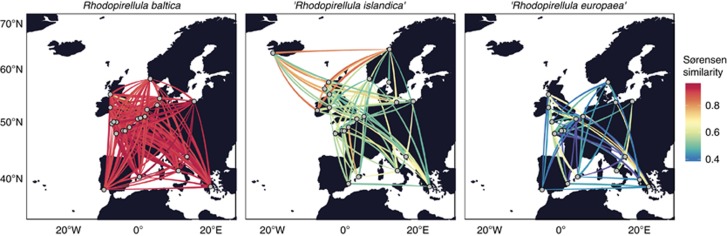
MED analysis of the individual species R. baltica, ‘R. europaea' and ‘R. islandica' resulted in 82, 221 and 82 final nodes, respectively (Figure 3). The total node abundance in each sample site for each species is given in Supplementary Table S6. For the R. baltica sequence data set, one node (Rb-168) was dominant in all samples with relative abundance of 60–100%, except in sample 143 from the North Sea, where node Rb-43 was dominant with a relative abundance of 89%. Node Rb-43 was also found in 13 other samples of the regions sampled. Other nodes were present with lower relative abundances; 14 nodes were found in >30 samples and 46 nodes in >20 samples. The rest were nodes present in <20 samples. R. baltica had the lowest acsA gene diversity in comparison with the other two species and had a relatively low diversity regardless of the sequence abundance of the species in the sample. ‘R. islandica' and ‘R. europaea' had a lower genetic diversity in the samples where those species were abundant (Figure 4). The low values of diversity on R. baltica are in concordance with the community structure along the European coasts; Sørensen similarity coefficient suggests that all European regions exhibit a similar community structure for R. baltica (Figure 5).
Figure 3.
Relative abundance of final MED nodes in all sampling sites sorted by different species. Nodes with highest abundances and described in the text are colorcoded.
Figure 4.
Diversity of the three Rhodopirellula species per sampling site, as measured by the estimation of the inverse Simpson index of the final MED nodes.
Figure 5.
Graphical representation of the community structure similarity estimated by the Sørensen similarity coefficient for the three Rhodopirellula species. Low Sørensen similarity edges have been trimmed using the algorithm described in Materials and Methods.
According to BLAST analysis, ‘R. islandica' was abundant in samples from Iceland (23), Ireland (129), Scotland (101, 104, 109) and Norway (76). One node, Ri-168, was dominant in those samples with relative abundances of 66–82%. In total, this node was found in 41 samples with relative abundance of 10–82%. The second most abundant node (Ri-105) was found in 38 samples with relative abundance of 1–70%. From 82 final nodes, 37 were present in at least 10 different samples (10–41). Although ‘R. islandica' showed no habitat limitation, the species community analysis revealed a clear geographical clustering, separating the communities from the Northern Atlantic from the rest of Europe (Figure 5).
‘R. europaea' is the species with the largest diversity values (Figure 4), and different nodes were dominant in different samples with high abundances of the species (19, 123, 38, 117, 49, 112), in contrast to ‘R. islandica'. Eight nodes found abundant in those samples were also abundant in 12–36 samples, being well distributed on the European coasts. Furthermore, the low values of similarities of ‘R. europaea' communities suggest that this species with its subspecies is highly adapted to its geographical location, presenting a limited dispersion (Figure 5).
Growth experiments
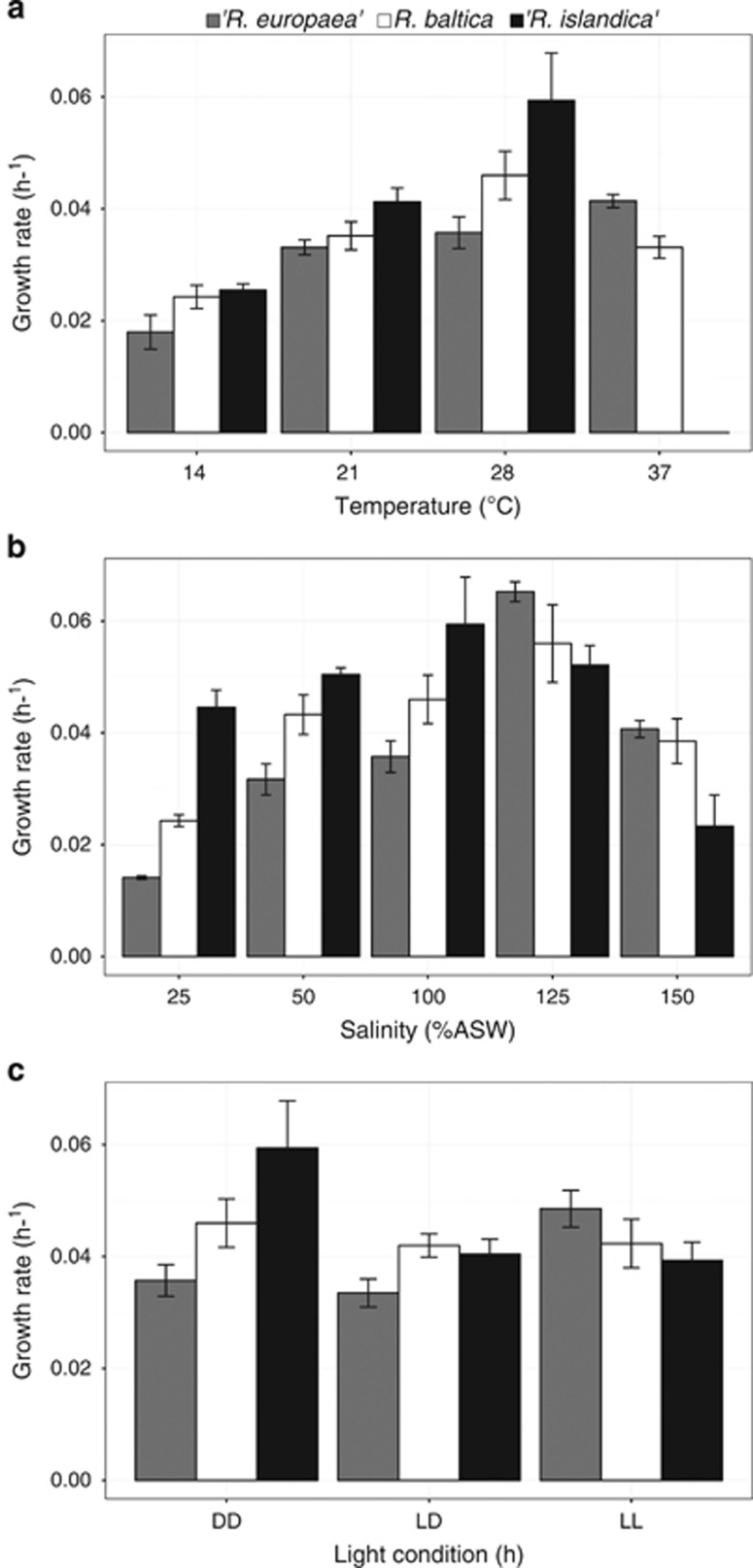
Under the standard growth conditions the growth rates of species R. baltica, ‘R. europaea' and ‘R. islandica' were 0.045, 0.035 and 0.059 h−1, with doubling times of 15, 19 and 12 h, respectively. All strains grew in all the conditions tested with different rates, except for ‘R. islandica', which did not grow at 37 °C (Figure 6).
Figure 6.
Effect of temperature (a), salinity (b) and light (c) on growth rates of R. baltica, 'R. islandica', and 'R. europaea'. Error bars indicate the standard deviation of the mean of growth rates for three cultures.
Discussion
The occurrence of three attached living, closely related species of the genus Rhodopirellula, R. baltica, ‘R. europaea' and ‘R. islandica', in European coastal regions was investigated by BLAST and MED analysis of a partial acsA amplicon, enabling an in-depth coverage of the diversity of these three species in intertidal sediments. Cultivation-independent studies based on protein-coding genes need a number of genomes as reference basis, which enables the development of primers for a small group of taxa with a discriminative power against the rest of the organisms (Bondoso et al., 2013; Žure et al., 2015). In this case, acsA gene resulted as a good candidate to delimitate the amplification to three species with a 16S rRNA gene identity of >99.5%. Furthermore, 75% of the obtained sequences affiliated to the targeted species and 18% were distantly related to other Rhodopirellula strains, suggesting a highly selective amplification, as cells of Rhodopirellula represented only 2% of planctomycetal cells in a coastal sediment (Žure et al., 2015).
Oligotyping was originally developed to detect subtle nucleotide differences in the 16S rRNA gene, which is a non-protein-coding gene and functional selection is characterized by strongly conserved regions flanking hypervariable regions that have diverged over evolutionary time. Different patterns of functional selection are observed in protein-coding genes, where changes at the third position in the codon are often non-selective due to the degenerated nature of the genetic code, whereas changes in the first or second position are likely to produce a change in amino acidic sequence. Using a high-resolution, entropy-based method like MED on coding sequences can result in an overestimation of the effective genetic diversity in the environment as a result of the neutral character of changes in the third codon position. Indeed, the largest entropy values were observed in the third codon position. We also observed that transitions outnumbered transversions on the third codon positions, resulting in silent substitutions on the acsA gene. MED analysis of sequences including the third position in the codon gave almost twice as many nodes, but on the amino-acid sequence level, 40% of node representative sequences were identical. Furthermore, with the removal of the third codon position we obtained a higher number of non-redundant MED nodes. For functional studies involving oligotyping where the ecologically meaningful unit is the protein sequence, we recommend to perform a pre-study including either the first two or all three codon positions to reveal the ecological importance of the third codon position in the gene under study. In our case study, we removed the third codon position of partial acsA amplicons to avoid the overestimation of the functional diversity.
Oligotyping can also be used for analysing amino-acid sequences (Sintes et al., 2016). However protein alignments do not have the resolution to resolve relationships on the species level or below, and using oligotyping on amino-acid sequences of acsA gene of Rhodopirellula species demonstrated an underestimation of the diversity in the environment. For example, oligotyping analysis of the ‘R. islandica' data set on amino-acid sequences resulted in ten times fewer oligotypes with one node dominating in all regions (data not shown).
Based on the BLAST results, the three species showed a wide geographical distribution with high abundances of R. baltica found in samples covering all European regions, whereas high abundances of ‘R. europaea' and ‘R. islandica' were found only in the samples from the regions where Winkelmann et al. (2010) had isolated the strains and which had been suggested as a preferable habitat. However, as we found these species in almost all samples it is likely that upon favorable change in the environment they could become abundant in other regions. Intertidal marine systems are highly dynamic systems and microorganisms living there are exposed to high fluctuations in environmental conditions so it is likely that observed abundances of different Rhodopirellula species in different samples reflect the conditions present in the environment at the time of the sampling. Changes in the environment may result in the shift of the rare and abundant species in a particular environmental condition (Caporaso et al., 2012). As the present results deal only with the diversity and distribution of Rhodopirellula species from samples taken at a single time point, future studies should address temporal variability and in situ measurements of environmental parameters. The experimental determination of growth rates of three Rhodopirellula species demonstrated the broad range of temperatures, salinities and light conditions in which these species are able to grow, which supports our finding that three Rhodopirellula species were present in all regions sampled, from the Mediterranean Sea, which is characterized by high salinity (~38 practical salinity unit) and surface water temperature (~16–26 °C), to the North Europe Atlantic regions, which are characterized by lower salinity (~34 practical salinity unit) and temperature (~6–16 °C) (http://www.nodc.noaa.gov/).
Although all species were found in all European regions sampled, MED analysis revealed a difference in community composition of the individual species. The MED nodes may be interpreted as subspecies. ‘R. baltica' had the lowest diversity in all regions, and our analysis showed that all sampling sites present a similar community structure of nodes, suggesting this species does not have any habitat limitation and is highly adapted to changing environmental conditions. A transcriptomic study of R. baltica by Wecker et al. (2009) demonstrated that the species is highly responsive to its environment, adjusting the expression of >3000 genes in response to changes of temperature or salinity. On the contrary, ‘R. europaea' had the highest diversity in all samples and the low values of the Sørensen similarity coefficient suggest that the different ecotypes (MED nodes) are specific to each geographical location. In the case of ‘R. islandica' we observed a clear clustering of the samples located in North Europe where water temperatures are lower, suggesting that this species has ecotypes adapted to cold environments. These differences could be explained by differences in genomes of these three species (Glöckner et al., 2003; Richter-Heitmann et al., 2014; Richter et al., 2014; Kizina et al., 2015). The size of the genomes is >7 Mb with over 6000 predicted open-reading frames, which reflects the complex lifestyle of Rhodopirellula species and ability to colonize highly dynamic intertidal coastal sediments. The gene-content comparison revealed that species share ~55% genes, whereas ~45% are species and strain-specific. The high number of shared genes may explain that we found co-occurrence of the species in different regions as the species shared common features allowing them to colonize the same habitats, whereas the species-specific genes may explain the difference in community structure and adaptation.
Taken together, we demonstrated that a MED analysis of protein-coding genes can be a powerful tool to study the within species community structure and this high-resolution analysis revealed differences within Rhodopirellula species in different geographic regions. However, we found all three species present in all regions sampled and in agreement with growth experiments, we demonstrated that R. baltica, ‘R. europaea' and ‘R. islandica' do not have a limited geographic distribution in European coastal sediments as previously suggested.
Acknowledgments
We would like to thank all sample contributors from EMBC (http://www.embcplus.org), MARES (http://www.mares-eu.org), Plymouth University and Max Planck-Institute, Bremen. We thank Maryia Khomich and Christina Probian for technical assistance. This work was supported by the Max Planck Society and MARES (Erasmus Mundus Joint Doctorate programme coordinated by Ghent University, FPA 2011-0016). AFG was supported by the Micro B3 project, which is funded from the European Union's Seventh Framework Programme (FP7; Joint Call OCEAN.2011-2: Marine microbial diversity – new insights into marine ecosystems functioning and its biotechnological potential) under grant agreement no. 287589.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Bengtsson MM, Øvreås L. (2010). Planctomycetes dominate biofilms on surfaces of the kelp Laminaria hyperborea. BMC Microbiol 10: 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberán A, Ladau J, Leff JW, Pollard KS, Menninger HL, Dunn RR, Fierer N. (2015). Continental-scale distributions of dust-associated bacteria and fungi. Proc Natl Acad Sci USA 112: 5756–5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondoso J, Albuquerque L, Lobo-da-Cunha A, Da Costa MS, Harder J, Lage OM. (2014). Rhodopirellula lusitana sp. nov. and Rhodopirellula rubra sp. nov., isolated from the surface of macroalgae. Syst Appl Microbiol 37: 157–164. [DOI] [PubMed] [Google Scholar]
- Bondoso J, Harder J, Lage OM. (2013). rpoB gene as a novel molecular marker to infer phylogeny in Planctomycetales. Antonie Van Leeuwenhoek 104: 477–488. [DOI] [PubMed] [Google Scholar]
- Brown SDJ, Collins RA, Boyer S, Lefort M-C, Malumbres-Olarte J, Vink CJ et al. (2012). Spider: An R package for the analysis of species identity and evolution, with particular reference to DNA barcoding. Mol Ecol Resour 12: 562–565. [DOI] [PubMed] [Google Scholar]
- Buttigieg PL, Ramette A. (2015). Biogeographic patterns of bacterial microdiversity in Arctic deep-sea sediments (HAUSGARTEN, Fram Strait). Front Microbiol 5: 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10: 421–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Paszkiewicz K, Field D, Knight R, Gilbert JA. (2012). The Western English Channel contains a persistent microbial seed bank. ISME J 6: 1089–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A, Gotelli NJ, Hsieh TC, Sander EL, Ma KH, Colwell RK et al. (2014). Rarefaction and extrapolation with Hill numbers: a framework for sampling and estimation in species diversity studies. Ecol Monogr 84: 45–67. [Google Scholar]
- DeLong EF, Franks DG, Alldredge AL. (1993). Phylogenetic diversity of aggregate-attached vs free-living marine bacterial assemblages. Limnol Oceanogr 38: 924–934. [Google Scholar]
- Eren AM, Borisy GG, Huse SM, Mark Welch JL. (2014. a). Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci USA 111: E2875–E2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG et al. (2013). Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol 4: 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Morrison HG, Lescault PJ, Reveillaud J, Vineis JH, Sogin ML. (2014. b). Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J 9: 968–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Sogin ML, Morrison HG, Vineis JH, Fisher JC, Newton RJ et al. (2015). A single genus in the gut microbiome reflects host preference and specificity. ISME J 9: 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst JA, Gwilliam HG, Lindsay M, Lichanska A, Belcher C, Vickers JE et al. (1997). Isolation and molecular identification of planctomycete bacteria from postlarvae of the giant tiger prawn Penaeus monodon. Appl Environ Microbiol 63: 254–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst JA, Sagulenko E. (2011). Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol 9: 403–413. [DOI] [PubMed] [Google Scholar]
- Glöckner FO, Kube M, Bauer M, Teeling H, Lombardot T, Ludwig W et al. (2003). Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc Natl Acad Sci 100: 8298–8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. (2009). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120. [DOI] [PubMed] [Google Scholar]
- Kizina J, Žure M, Rhiel E, Munn CB, Richter M, Harder J. (2015). Permanent draft genome of ‘Rhodopirellula islandica' strain K833. Mar Genomics 24: 249–251. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. [DOI] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10+. [Google Scholar]
- McLellan SL, Newton RJ, Vandewalle JL, Shanks OC, Huse SM, Eren AM et al. (2013). Sewage reflects the distribution of human faecal Lachnospiraceae. Environ Microbiol 15: 2213–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Paulson JN, Stine OC, Bravo HC, Pop M. (2013). Differential abundance analysis for microbial marker-gene surveys. Nat Methods 10: 1200–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel-Elardo S, Wehrl M, Friedrich AB, Jensen PR, Hentschel U. (2003). Isolation of planctomycetes from Aplysina sponges. Aquat Microb Ecol 33: 239–245. [Google Scholar]
- Ramette A, Tiedje JM. (2007). Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb Ecol 53: 197–207. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. (2000). EMBOSS: the european molecular biology open software suite. Trends Genet 16: 276–277. [DOI] [PubMed] [Google Scholar]
- Richter-Heitmann T, Richter M, Klindworth A, Wegner C-E-E, Frank CS, Glöckner FO et al. (2014). Permanent draft genomes of the two Rhodopirellula europaea strains 6C and SH398. Mar Genomics 13: 15–16. [DOI] [PubMed] [Google Scholar]
- Richter M, Richter-Heitmann T, Klindworth A, Wegner C-E-E, Frank CS, Harder J et al. (2014). Permanent draft genomes of the three Rhodopirellula baltica strains SH28, SWK14 and WH47. Mar Genomics 13: 13–14. [DOI] [PubMed] [Google Scholar]
- Roh SW, Lee H-W-W, Yim KJ, Shin N-R-R, Lee J, Whon TW et al. (2013). Rhodopirellula rosea sp. nov., a novel bacterium isolated from an ark clam Scapharca broughtonii. J Microbiol 51: 301–304. [DOI] [PubMed] [Google Scholar]
- Schlesner H. (1994). The development of media suitable for the microorganisms morphologically resembling Planctomyces spp., Pirellula spp., and other Planctomycetales from various aquatic habitats using dilute media. Syst Appl Microbiol 17: 135–145. [Google Scholar]
- Schlesner H, Rensmann C, Tindall BJ, Gade D, Rabus R, Pfeiffer S et al. (2004). Taxonomic heterogeneity within the Planctomycetales as derived by DNA–DNA hybridization, description of Rhodopirellula baltica gen. nov., sp. nov., transfer of Pirellula marina to the genus Blastopirellula gen. nov. as Blastopirellula marina comb. nov. and emended description of the genus Pirellula. Int J Syst Evol Microbiol 54: 1567–1580. [DOI] [PubMed] [Google Scholar]
- Shannon CE. (1948). A mathematical theory of communication. Bell Syst Tech J 27: 379–423. [Google Scholar]
- Sintes E, De Corte D, Haberleitner E, Herndl GJ. (2016). Geographic distribution of archaeal ammonia oxidizing ecotypes in the Atlantic Ocean. Front Microbiol 7: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speth DR, van Teeseling MC, Jetten MS. (2012). Genomic analysis indicates the presence of an asymmetric bilayer outer membrane in Planctomycetes and Verrucomicrobia. Front Microbiol 3: 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strous M, Kuenen JG, Fuerst JA, Wagner M, Jetten MSM. (2002). The anammox case—a new experimental manifesto for microbiological eco-physiology. Antonie Van Leeuwenhoek 81: 693–702. [DOI] [PubMed] [Google Scholar]
- Wagner M, Horn M. (2006). The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol 17: 241–249. [DOI] [PubMed] [Google Scholar]
- Wecker P, Klockow C, Ellrott A, Quast C, Langhammer P, Harder J, Glöckner FO. (2009). Transcriptional response of the model planctomycete Rhodopirellula baltica SH1T to changing environmental conditions. BMC Genomics 10: 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelmann N, Harder J. (2009). An improved isolation method for attached-living Planctomycetes of the genus Rhodopirellula. J Microbiol Methods 77: 276–284. [DOI] [PubMed] [Google Scholar]
- Winkelmann N, Jaekel U, Meyer C, Serrano W, Rachel R, Rosselló-Mora R et al. (2010). Determination of the diversity of Rhodopirellula isolates from European seas by multilocus sequence analysis. Appl Environ Microbiol 76: 776–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J, Matsuo Y, Kasai H, Lee M-K. (2015). Phylogenetic and taxonomic analyses of Rhodopirellula caenicola sp. nov., a new marine Planctomycetes species isolated from Iron Sand. J Phylogenet Evol Biol 3: 143. [Google Scholar]
- Zhang J, Kobert K, Flouri T, Stamatakis A. (2014). PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30: 614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Žure M, Munn CB, Harder J. (2015). Diversity of Rhodopirellula and related planctomycetes in a North Sea coastal sediment employing carB as molecular marker. FEMS Microbiol Lett 362: fnv127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.