Abstract
The hydrothermal vent mussel Bathymodiolus azoricus lives in an intimate symbiosis with two types of chemosynthetic Gammaproteobacteria in its gills: a sulfur oxidizer and a methane oxidizer. Despite numerous investigations over the last decades, the degree of interdependence between the three symbiotic partners, their individual metabolic contributions, as well as the mechanism of carbon transfer from the symbionts to the host are poorly understood. We used a combination of proteomics and genomics to investigate the physiology and metabolism of the individual symbiotic partners. Our study revealed that key metabolic functions are most likely accomplished jointly by B. azoricus and its symbionts: (1) CO2 is pre-concentrated by the host for carbon fixation by the sulfur-oxidizing symbiont, and (2) the host replenishes essential biosynthetic TCA cycle intermediates for the sulfur-oxidizing symbiont. In return (3), the sulfur oxidizer may compensate for the host's putative deficiency in amino acid and cofactor biosynthesis. We also identified numerous ‘symbiosis-specific' host proteins by comparing symbiont-containing and symbiont-free host tissues and symbiont fractions. These proteins included a large complement of host digestive enzymes in the gill that are likely involved in symbiont digestion and carbon transfer from the symbionts to the host.
Introduction
Life at deep-sea hydrothermal vents is powered by chemosynthetic bacteria, many of which live in symbiosis with animals. Hydrothermal vents of the Mid-Atlantic Ridge (MAR) are home to the mytilid bivalve Bathymodiolus azoricus, which harbours two types of gammaproteobacterial endosymbionts (von Cosel et al., 1999; Van Dover, 2000; Duperron et al., 2006). Unlike other vent symbioses, such as the giant tube worm Riftia pachyptila and the clam Calyptogena spp., which display extreme dependence on their chemoautotrophic endosymbionts for nourishment (Childress and Fisher, 1992), B. azoricus are mixotrophs: They possess a functional gut and feeding groove for filter feeding in addition to the chemosynthetic symbionts in their gills (Page et al., 1991; Gustafson et al., 1998; von Cosel et al., 1999; Riou et al., 2010). Filter-feeding may supplement the symbiotic diet of the host with particulate and organic matter from their surroundings (Le Pennec et al., 1990) and may also enable them to survive without symbionts for limited periods of time (Colaco et al., 2011). However, aposymbiotic B. azoricus in laboratory aquaria display an overall reduction in their fitness and health (Raulfs et al., 2004; Kádár et al., 2005), suggesting that despite their nutritional flexibility, B. azoricus mussels depend on their symbionts for long-term sustenance.
B. azoricus maintains a stable symbiotic partnership with two distinct gammaproteobacterial phylotypes living within its gills (Figure 1). The co-occurrence of a thiotrophic symbiont, which oxidizes reduced sulfur compounds, and a methanotroph, which oxidizes methane (Fiala-Médioni et al., 2002), enables B. azoricus to simultaneously tap the energy from two of the most abundant reductants in the MAR vent fluids. The symbionts are housed in vacuoles within specialized gill epithelial cells called bacteriocytes. The bacteriocytes face the apical side of the gill filament, which is in contact with the vent fluids (Figure 1; Distel et al., 1995). The physical proximity of the bacteriocytes to the ambient vent fluids facilitates direct exchange of dissolved gases and substrates (Childress and Fisher, 1992) and may also allow quick responses of symbionts to changes in environmental conditions. The host presumably acquires nutrients from the symbionts by digesting them using lysosomal degradation enzymes (Streams et al., 1997; Fiala-Médioni et al., 2002; Kádár et al., 2008).
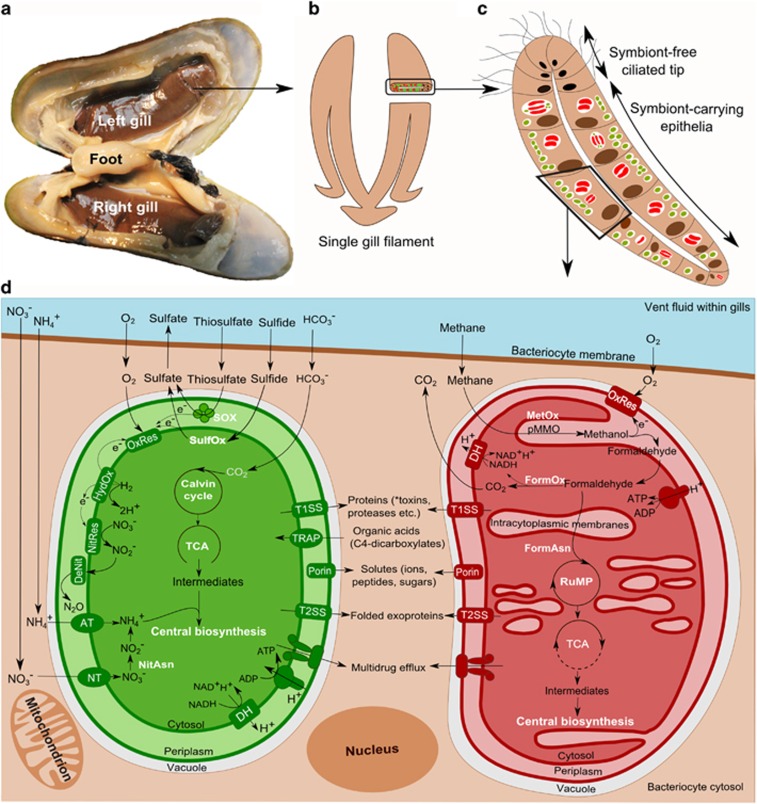
Figure 1.
Schema of a B. azoricus gill bacteriocyte. (a) Dissected B. azoricus specimen showing gills and gill filaments. (b) Schema of single gill filament. (c) Schematic cross-section of a gill filament showing bacteriocytes. (d) A single bacteriocyte showing the central pathways of the symbionts. Symbionts (thiotroph: green, methanotroph: red) are located inside vacuoles (white) surrounded by the bacteriocyte cytosol (brown) with gases and substrates exchanged between vent fluids (blue) flushing the gills. An overview of the basic metabolic processes occurring in the symbionts is shown. AT: ammonium transporter, DH: dehydrogenase, DeNit: denitrification, FormAsn: formaldehyde assimilation, FormOx: formaldehyde oxidation to formate and CO2, HydOx: hydrogen oxidation, MetOx: methane oxidation by pMMO (particulate methane-monooxygenase enzyme complex) to methanol and then to formaldehyde (see Supplementary Figure S2B for details), NitAsn: nitrogen assimilation, NitRes: nitrate respiration (see Supplementary Figure S4 for details on nitrogen metabolism), NT: nitrate transporter, OxRes: Oxidative phosphorylation with oxygen as terminal electron acceptor, SOX: thiosulfate oxidation, SulfOx: sulfide oxidation via the rDSR-APS-Sat pathway (see Supplementary Figure S2A), TCA: tricarboxylic acid cycle, TRAP: tripartite ATP-independent periplasmic transporter, T1SS: Type I secretion system, T2SS: Type II secretion system. *toxins are known to be secreted by the thiotrophs (Sayavedra et al., 2015).
The symbionts' location within host bacteriocytes allows for intricate metabolic interactions between the host and the symbionts. So far, only a handful of whole gill-based ‘omics' studies have examined this complex association, most of them focusing on the physiology of the host (Bettencourt et al., 2010; Company et al., 2011; Petersen et al., 2011; Sayavedra et al., 2015). The fact that the symbionts are uncultivable as yet and the mussels can only be maintained temporarily under controlled laboratory conditions (Kádár et al., 2005) has rendered detailed physiological investigations difficult. We therefore conducted a culture-independent proteogenomic study, which – for the first time – provided a detailed and comprehensive picture of host–symbiont interaction dynamics and metabolic interdependencies in B. azoricus. To detect host proteins potentially involved in symbiosis-specific functions, we compared protein expression patterns between symbiont-containing and symbiont-free host tissues.
Experimental procedures
Sampling of Bathymodiolus mussels and differential enrichment for proteomics
Sampling details and biological replicate numbers for all analyses performed in this study are summarized in Supplementary Table S1A. For proteome analyses, B. azoricus mussels were collected from the Menez Gwen vent field on the MAR at 37°50'41'' N, 31°31'10'' W during the RV Meteor cruise M82-3 as described by Sayavedra and colleagues (2015). The mussels were dissected, gill tissue was removed and homogenized in 1x PBS (Figure 2, see Supplementary Methods for a detailed description of the enrichment procedure). The homogenate was subjected to a combination of differential pelleting and rate-zonal centrifugation using density gradients on board the research vessel to separate the symbionts and host components from each other (Figure 2). The composition of putative host- and symbiont-enriched fractions after centrifugation was analysed using catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH, see Supplementary Information). Based on the CARD-FISH evaluation, the host-enriched supernatant (containing soluble host proteins) and the symbiont-enriched gradient pellet (containing both symbionts, but particularly enriched in the thiotroph) were chosen for further proteomic analysis (Figure 2). Additionally, complete gill and foot tissue samples from the same animals that were processed by centrifugation were also frozen at −80 °C to enable comparisons between the proteomes of whole tissues and of host- and symbiont-enriched fractions.
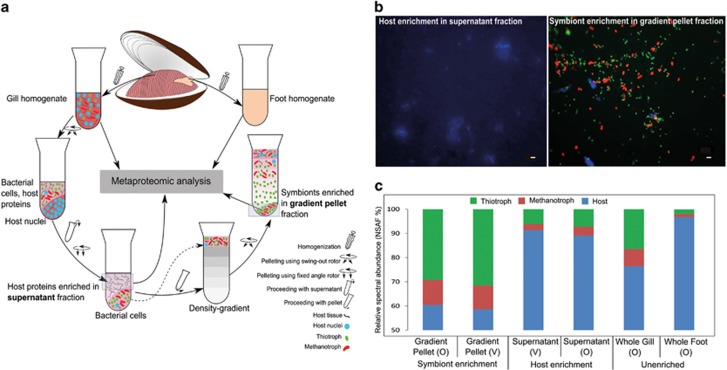
Figure 2.
Density gradient enrichment of symbiont cells and host components followed by CARD-FISH. (a) Step-wise workflow of the density gradient enrichment method for physical separation of B. azoricus host and symbiont cells. Sampling for tissue-based metaproteomic analysis of whole gill and foot tissue is also shown. Bold font: enriched fractions elaborated in panel b and c. (b) Epifluorescence micrographs of supernatant (left) and gradient pellet (right) fractions obtained from the enrichment method shown in panel a. Green CARD-FISH signal: thiotrophs; red CARD-FISH signal: methanotrophs; blue DAPI signal: host. Scale bar: 2 μm. (c) Enhanced spectral abundance of symbiont- and host-associated proteins in density gradient-based enrichments (symbiont-enriched gradient pellet and host-enriched supernatant) as compared to unenriched (gill and foot) tissues. Stacked bars show the % of spectral abundance (calculated by summing up NSAF% and averaging it across biological replicates) contributed by thiotrophs, methanotrophs and host, respectively, to the total spectral abundance of each sample type. O: proteins identified by LTQ-Orbitrap Classic MS/MS analysis (n=3), V: proteins identified by LTQ-Orbitrap Velos MS/MS analysis (n=2, see Methods).
Proteomic analysis
Specific details of the protein extraction procedure, 1D-PAGE, LC-MS/MS, protein identification, validation and quantitation can be found in the Supplementary Methods section. Briefly, soluble proteins and membrane-associated proteins were extracted from enriched gradient fractions in biological triplicates (that is, from three B. azoricus individuals) and from whole tissue samples in biological duplicates (for replicate numbers of all MS measurements see Supplementary Table S1B). Proteins were separated using 1D-PAGE and in-gel digested with trypsin. Resulting peptides were separated by liquid chromatography and analysed simultaneously by coupling the liquid chromatography online to a mass spectrometer. For identification of host and symbiont proteins, all MS/MS spectra were searched against a comprehensive protein database (see Supplementary Methods and Supplementary Table S2A for a list of all organisms whose protein sequences are included in the database). For relative semi-quantitative analysis of proteins, normalized spectral abundance factor (NSAF) values were calculated for each sample (NSAF%, Florens et al., 2006). NSAFs were subsequently also normalized for each organism individually (OrgNSAF%, Mueller et al., 2010).
Statistical analyses for the identification of putative symbiosis-relevant proteins
To identify potentially symbiosis-relevant host and symbiont proteins we did a statistical comparison of protein abundances in different sample types. For host proteins that may play a direct role in the symbiosis we tested for proteins whose NSAF values differ significantly between the symbiont-containing gill samples (whole gill and gill supernatant) versus the symbiont-free foot tissue. Host proteins that are potentially involved in direct interactions with the symbionts (that is, physically linked to the symbionts) were 'pulled down' by the symbionts during centrifugation-based symbiont enrichment. We identified these proteins by testing for host protein abundances that were significantly higher in the symbiont-enriched gradient pellet versus the whole gill samples and the gill supernatant. Statistical evaluation of differences between samples involved Log transformations of NSAF%/OrgNSAF% values followed by t-test, all of which were done using the Perseus software (version 1.4.1.3, http://www.perseus-framework.org/doku.php) as described in the Supplementary Methods.
Data accessibility
The mass spectrometry proteomics data and the protein sequence database were uploaded to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al., 2016) partner repository with the dataset identifier PXD004061 (DOI: 10.6019/PXD004061). Nucleotide and protein sequence information for the methanotrophic symbionts of B. azoricus and of Bathymodiolus sp. was deposited in GenBank under the BioProject accession numbers PRJEB13769 and PRJEB13047, respectively.
Results and discussion
Enhanced protein identification following density gradient enrichment in B. azoricus
We used a multistep centrifugation technique to physically separate host components and thiotrophic and methanotrophic symbiont cells of B. azoricus. This enrichment substantially enhanced the rate of spectral identifications for the respective individual organisms, as compared to plain, unenriched tissue. Our centrifugation procedure involved differential pelleting of host nuclei followed by rate-zonal density gradient centrifugation for symbiont enrichment (Figure 2a). We analysed the resulting fractions with CARD-FISH and DAPI staining to identify fractions where either host material or symbiont cells were enriched (Figure 2b). Relative cell counts revealed that 93% of the cells in the gradient pellet were thiotrophic and methanotrophic endosymbionts and that 97% of the solid material in the supernatant consisted of host nuclei and host cell fragments (see Supplementary Figure S1). This enrichment was clearly reflected in our proteomic results, where we found a 17.7% increase in symbiont-associated spectral identifications (NSAF) in the symbiont-enriched gradient pellet, and a 15% increase in host spectral identifications in the host-enriched supernatant, as compared to the gill tissue (Figure 2c). The enhanced spectral identification rate enabled better quantification of the identified proteins and substantially improved the proteome coverage: The number of identified proteins in enriched samples increased by 7.4% (host proteins) and 9.3% (symbiont proteins) compared to those from unenriched tissues (Supplementary Table S2C, see Supplementary Results and Discussion for absolute numbers of identified proteins). Grouping of the most abundant proteins (in terms of NSAF%) of the symbiotic partners into metabolic categories allowed us to trace the metabolic network of the B. azoricus consortium (Figure 3).
Figure 3.
The 75 most abundant proteins of the B. azoricus host and symbionts as identified in our study in LTQ-Orbitrap Velos MS/MS analyses. Each bubble represents an identified protein in the respective sample and its abundance in terms of average NSAF% (relative abundance in % of all proteins in the sample). Sample types are indicated on top, protein names are listed on the left (see Supplementary Table S3 for protein functions). Proteins were grouped based on their respective metabolic categories inferred from the KEGG and MetaCyc pathway databases. *Functionally redundant symbiont protein identifications and proteins of unknown function were excluded in this figure. **Host proteins with functions beyond the scope of this study were not included in this figure. Note: This figure illustrates only protein data from LTQ-Orbitrap Velos MS/MS analyses, but not from parallel LTQ-Orbitrap Classic measurements of the same samples (see Methods for details). The complete data set including all measurements is shown in Supplementary Table S3.
The B. azoricus symbiosis is fuelled by a versatile array of energy sources
Thiosulfate oxidation
In the thiotrophic B. azoricus symbiont, thiosulfate as an energy source seems to play a more prominent role than in other chemoautotrophic symbioses. We found the thiotroph's SoxYZ proteins for thiosulfate oxidation (Sox) to be more abundant than the dissimilatory sulfite reductase (Dsr) enzymes DsrAB for hydrogen sulfide oxidation: With 1.104 OrgNSAF% (SoxYZ) and 0.894 OrgNSAF% (DsrAB), the enzyme complexes were present in a DsrAB:SoxYZ ratio of 0.81 (soluble protein fraction of gradient pellet samples, Velos analysis, see Supplementary Table S3). In contrast, the sulfur-oxidizing symbionts of R. pachyptila and Olavius algarvensis express DsrAB at much higher levels than SoxYZ. The DsrAB:SoxYZ ratio in the Riftia symbiont was 2.5 (n=3; S. Markert, unpublished results), and 5.2 in the Olavius Gamma 1 symbiont (Kleiner et al., 2012b). In addition to the Sox enzyme complex, we identified two sulfur/thiosulfate carrier protein homologs, a TusA domain-containing protein (Rhd1, BazSymB_scaffold00007_23) and a Rhodanese-like domain-containing protein (Rhd2, BazSymB_scaffold00002_24) in the B. azoricus thiotroph (Supplementary Figure S2A). In fact, Rhd1 was one of the most abundant proteins in the thiotroph's membrane proteome (membrane OrgNSAF% from gradient pellet, Supplementary Table S3). Rhodaneses cleave thiosulfate to sulfite and sulfide (Brune, 1995, Supplementary Figure S2A), while TusA has been shown to mediate thiosulfate transfer in an acidothermophilic sulfur- and tetrathionate-oxidizing archaeon (Liu et al., 2014). Both rhodanese proteins together constituted 2.11 OrgNSAF% in the thiotrophic B. azoricus symbiont (for comparison: the Riftia symbiont's rhodanese proteins make up only 0.15 OrgNSAF %, n=3; S. Markert, unpublished). The B. azoricus thiotroph clearly oxidizes sulfide, as indicated by the detection of most components of the rDSR-APS-Sat pathway (see Supplementary Results and Discussion). However, the relatively high expression of thiosulfate-metabolizing and -transfer enzymes (compared to the sulfur-oxidizing symbionts of other host animals) indicates that thiosulfate oxidation may be particularly important for the B. azoricus symbiosis in its specific habitat. This is in agreement with previous studies, in which thiosulfate was observed to stimulate carbon fixation much more than sulfide in the thiotrophic symbionts of related Bathymodiolus species (Belkin et al., 1986; Fisher et al., 1987).
Thiosulfate is more stable than sulfide and is less toxic to aerobic respiration (Harada et al., 2009). In other symbiotic species such as vestimentiferan tubeworms and vesicomyid clams, sulfide is transported in a less harmful form, bound to the host's hemoglobin (Arp et al., 1984, 1987; Doeller et al., 1988). However, no dedicated host proteins for sulfide transport are known in Bathymodiolus mussels (Powell and Somero, 1986). High concentrations of thiosulfate (0.178 mM in gills versus 0.079 mM in other tissues) in the closely related B. thermophilus led Fisher et al (1988) to propose that the host may detoxify sulfide from the environment to the less toxic thiosulfate. Host-mediated thiosulfate production as a means of sulfide detoxification was recently also suggested for B. brevior (Beinart et al., 2015). If B. azoricus uses a similar mechanism of sulfide detoxification, then its symbionts would be exposed to thiosulfate concentrations that are much higher than those in the environment. This may stimulate higher expression of thiosulfate-metabolizing enzymes in the B. azoricus thiotroph as compared to other symbionts, whose hosts do not produce thiosulfate as a means of sulfide detoxification.
Hydrogen oxidation
In addition to its capacity for thiotrophy, the B. azoricus thiotroph is also capable of using hydrogen (H2) as an electron acceptor (Petersen et al., 2011). Genes for hydrogen oxidation were clearly expressed, but at lower abundance compared with enzymes involved in sulfide or thiosulfate oxidation. H2 oxidation may therefore not be as important as the oxidation of reduced sulfur compounds for the thiotroph under the conditions prevailing at the sampling site in this study (see Supplementary Figure S2A and Supplementary Results and Discussion).
Methane oxidation
The energy-generating methane oxidation process is the most prominent metabolic pathway in the methanotrophic B. azoricus symbiont. Its key enzymes, the particulate methane monooxygenase PmoCAB and the methanol dehydrogenase XoxF that catalyze the oxidation of methane to formaldehyde, constituted 28.6% of the membrane OrgNSAF in the gradient pellet samples and 2.4% of the gill OrgNSAF, respectively (Supplementary Figure S2B, Supplementary Results and Discussion). Considering the high catalytic efficiency of both enzymes (Chan et al., 2013; Keltjens et al., 2014), these high expression levels likely correspond to very high methane oxidation rates in the B. azoricus methanotroph. As indicated by our results, the symbiont can accomplish the subsequent energy-producing oxidation of formaldehyde to formate and CO2 via two parallel metabolic routes, that is, the tetrahydrofolate (H4F) pathway and the dephospho-tetrahydromethanopterin (H4MPT) pathway (see Supplementary Figure S3 and Supplementary Results and Discussion for details). We identified enzymes of both pathways in the methanotroph's proteome. The presence of multiple routes for formaldehyde metabolism may offer enhanced metabolic flexibility to the symbiont under various environmental conditions. Also, since accumulation of formaldehyde is toxic to the cells, the two formaldehyde oxidation pathways may operate as overflow valves for controlling excess formaldehyde levels, while simultaneously generating electrons and reducing equivalents during the process (Crowther et al., 2008).
Pathways for carbon assimilation are highly expressed in both symbionts while crucial TCA cycle enzymes are missing in the thiotroph
Assimilation of C1 compounds
In the proteomes of both the thiotrophic and the methanotrophic B. azoricus symbiont, carbon fixation pathways were highly abundant, reflecting their importance in the symbiotic association. The thiotroph uses the Calvin-Benson-Bassham cycle for autotrophic fixation of CO2 (see Supplementary Results and Discussion and Figure 4a). All Calvin-Benson-Bassham pathway enzymes were highly abundant in the thiotroph. The large subunit of ribulose bisphosphate carboxylase/oxygenase (RuBisCO form I, CbbL, BazSymA_Acontig00018_18), the key enzyme of the Calvin-Benson-Bassham cycle, was consistently the most abundant enzyme of the thiotroph's carbon metabolism in all samples analysed in our study (Figures 3 and 4a, Supplementary Table S3). The methanotroph expressed a complete ribulose monophosphate (RuMP) pathway for the assimilation of carbon from formaldehyde, as well as parts of a second carbon assimilation pathway, the serine cycle (see Figure 4b, Supplementary Figure S3 and Supplementary Results and Discussion). The high abundance of the two RuMP pathway key enzymes hexulose-6-phosphate formaldehyde lyase (Hps, BAZMOX_41472_2) and 3-hexulose-6-phosphate isomerase (Hpi1, BAGiLS_016202) indicates that the methanotroph primarily uses this pathway for formaldehyde assimilation. The serine cycle seems to play a minor role in carbon assimilation as compared to the dominant RuMP pathway under the conditions in this study (see Supplementary Results and Discussion).
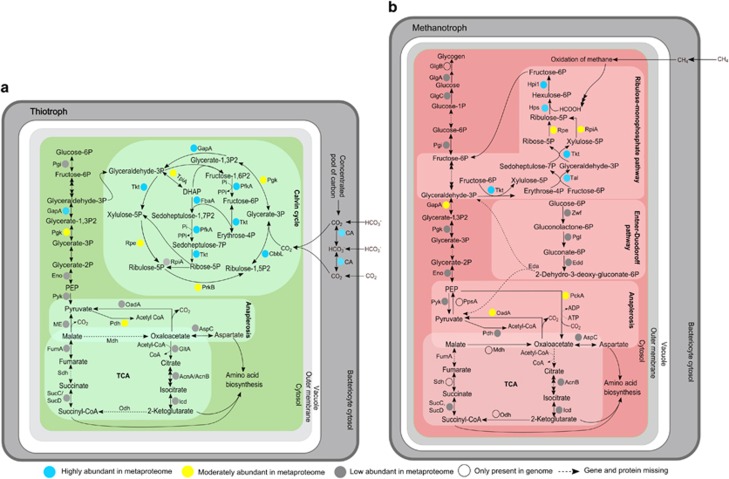
Figure 4.
Central carbon metabolism in B. azoricus symbionts. (a) In the thiotrophic symbiont. (b) In the methanotrophic symbiont. Eda: KHG/KDPG aldolase, GlgB: 1,4-alpha-glucan branching enzyme, Mdh: Malate dehydrogenase, ME: malic enzyme, Odh: 2-oxoglutarate dehydrogenase, PpsA: PEP synthase, Sdh: succinate dehydrogenase. For functions of enzymes that were detected as proteins in this study refer to Supplementary Table S3. Protein abundances correspond to average OrgNSAF% in the respective organism from whole gill tissue samples (in Velos MS/MS measurements, see Supplementary Table S3). See Supplementary Table S2B for genome assemblies used for gene prediction. Multiple arrows indicate multiple enzymatic steps. Dotted arrows indicate enzymes that are missing in the respective symbiont's genome. Sizes are not drawn to scale.
Incomplete TCA cycle in the thiotroph
The thiotrophic symbiont of B. azoricus appears to be unable to replenish the crucial carbon metabolism intermediates oxaloacetate and succinate. As expected in an obligate autotroph, the thiotroph lacks the tricarboxylic acid (TCA) cycle enzyme 2-oxoglutarate dehydrogenase (Odh; Wood et al., 2004; Kleiner et al., 2012a). The absence of Odh effectively prevents wasteful re-oxidation of autotrophically fixed organic carbon, while still allowing for the production of carbon precursors for amino acid biosynthesis and other anabolic pathways. Intermediates of this ‘horse shoe' TCA cycle are usually replenished by anaplerotic pathways such as the glyoxylate bypass. Surprisingly, our proteogenomic analysis revealed that not only the gene encoding Odh is missing in the thiotroph, but also the genes for the TCA cycle enzymes malate dehydrogenase (Mdh) and succinate dehydrogenase (Sdh, Figure 4a). Moreover, the thiotroph apparently lacks all known genes encoding anaplerotic enzymes for the replenishment of the essential TCA cycle intermediates oxaloacetate and succinate (see Supplementary Results and Discussion), although it expresses enzymes involved in oxaloacetate and succinate consumption (Figure 4a). Oxaloacetate is an important precursor for biosynthesis of essential amino acids of the aspartate family, while succinate, or rather its derivative succinyl-CoA, is required for the biosynthesis of porphyrins and amino acids. Replenishment of oxaloacetate and succinate is therefore crucial for cellular metabolism and alternate mechanisms for refilling both intermediates must exist (see below).
Complete TCA cycle in the methanotroph
Unlike the thiotrophic symbiont of B. azoricus, the methanotroph has a complete set of genes encoding TCA cycle enzymes, including odh (BAZMOX_25028_0). Odh, Mdh and Sdh were, however, not detected in the methanotroph's proteome (Figure 4b). In the facultative methylotroph Methylobacterium extorquens AM1, odh expression is repressed during growth on C1-compounds (Chistoserdova et al., 2003). It is therefore possible that the methanotrophic symbiont of B. azoricus expresses a complete, energy-producing TCA cycle during growth on multicarbon compounds when the preferred carbon source methane is not available, and an incomplete version of the TCA cycle that produces anabolic intermediates when methane is plentiful (Zhao and Hanson, 1984; Chistoserdova et al., 2003; Wood et al., 2004; Dedysh et al., 2005). In the absence of methane, cellular storage polymers such as glycogen may serve as readily available multicarbon compounds for energy generation. This idea is supported by the detection of glycogen synthesis enzymes in the methanotroph's proteome (Figure 4b, Supplementary Table S3, Supplementary Results and Discussion).
Some metabolic tasks are accomplished jointly by the symbiotic partners
CO2 concentration by carbonic anhydrase in gills
In the B. azoricus gill metaproteome, host-derived carbonic anhydrase (CA) is one of the most abundant proteins expressed (Figures 3 and 4a, Supplementary Table S3, BAGiLS_000922). The enzyme was ~10 times more abundant in gills and ~19 times more abundant in the supernatant, respectively, as compared to symbiont-free foot tissue (Table 1), suggesting a symbiosis-specific role for the host CA in B. azoricus. In marine invertebrates such as anemones, corals, tubeworms and clams harbouring carbon-fixing bacteria, the ubiquitous enzyme CA facilitates the reversible conversion of bicarbonate, the dominant form of seawater carbon, to CO2 for transport to symbiont-bearing tissues (Kochevar and Childress, 1996). The B. azoricus CA therefore likely represents a CO2-concentrating mechanism, which provides elevated levels of inorganic carbon for fixation by the thiotrophic symbiont (Figure 4a). High activity and protein expression of host CA was also described in gills of Bathymodiolus mussels hosting only methanotrophic symbionts, where CA may be involved in the elimination of CO2 produced as an end product of methane oxidation (Hongo et al., 2013). In addition to the host CA, we also found the CAs of the thiotroph and of the methanotroph expressed (BAT01672, BAZMOX_02303_3), albeit in very low abundances. It is unclear whether either of the CAs from host, methanotroph or thiotroph participate in the elimination of methanotroph-derived CO2. We speculate that in B. azoricus this methanotroph-derived CO2, instead of being eliminated, may be recycled by the thiotrophic symbiont for CO2 fixation (Nelson and Fisher, 1995).
Table 1. Host proteins with putative symbiosis-specific roles.
| Protein accession number | Protein function |
Statistical significance
|
Expression ratios
|
||||||
|---|---|---|---|---|---|---|---|---|---|
|
Group A
|
Group B
|
Group A
|
Group B
|
||||||
| Gill vs. Foot | Sup vs. Foot | GP vs. Sup | GP vs. Gill | Gill:Foot | Sup:Foot | GP:Sup | GP:Gill | ||
| Proteins with putative digestive functions | |||||||||
| BAGiLS_012512 | Glycoside hydrolase, family 18, catalytic domain (similar to chitinase) | X | Gill only | Sup only | |||||
| BAGiLS_002552 | Glycoside hydrolase, family 18, catalytic domain (similar to chitinase) | X | Gill only | Sup only | |||||
| BAGiLS_000898 | Peptidase M28 (Aminopeptidase Y) | X | 26.82 | 9.38 | |||||
| BAGiLS_003294 | Peptidase, cysteine peptidase active site (Cathepsin protein) | X | X | 13.34 | 23.35 | ||||
| BAGiLS_021119 | Putative beta-glycosidase | X | Gill only | Sup only | |||||
| BAGiLS_000621 | Saposin B | X | 88.4 | 114.34 | |||||
| BAGiLS_005809 | Palmitoyl protein thioesterase | X | 3.53 | 3.21 | |||||
| BAGiLS_000071 | Peptidase M16, C-terminal | X | 15.2 | 2.39 | |||||
| BAGiLS_000588 | Peptidase family M28 (aminopeptidase) | X | 1.3 | 11.92 | |||||
| BAGiLS_000416 | Peptidase M16, C-terminal | X | 8.14 | 2.56 | |||||
| BAGiLS_001815 | Peptidase M16, C-terminal | X | 15.65 | 5.89 | |||||
| BAGiLS_009112 | Peptidase M16, core | X | 9.75 | 1.22 | |||||
| Carbon transport and metabolism | |||||||||
| BAGiLS_001414 | CA, carbonic anhydrase, alpha-class, catalytic domain | X | X | 10.65 | 19.21 | ||||
| BAGiLS_000922 | CA, carbonic anhydrase, alpha-class, catalytic domain | X | 10.1 | 15.4 | |||||
| BAGiLS_005913 | Odh, dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex | X | 24.94 | 3.16 | |||||
| BAGiLS_010531 | Sdh, fumarate reductase/succinate dehydrogenase flavoprotein, C-terminal | X | 14.1 | 1.81 | |||||
| Immune-related functions | |||||||||
| BAGiLS_002519 | Sea anemone cytolysin | X | X | Gill only | Sup only | 1.35 | 5.57 | ||
| BAGiLS_009564 | Coagulation factor 5/8-type domain protein | X | X | Gill only | Sup only | ||||
| BAGiLS_003790 | C-type lectin | Gill only | Sup only | ||||||
| BAGiLS_008830 | sushi domain-containing protein (involved in cell adhesion) | X | Gill only | Sup only | |||||
| BAGiLS_005314 | MD-2-related lipid-recognition domain protein (putative pathogen recognition protein) | X | X | 25.36 | 12.56 | ||||
| BAGiLS_003036 | Putative adhesin/invasin | X | 41.44 | 7.21 | |||||
| BAGiLS_019106 | Complement C1q protein | X | 6.97 | 13.67 | |||||
Samples shown in this table were chosen for comparison on the following basis: Group A: to identify putative host proteins that may play a direct role in the symbiosis, gill tissue and supernatant samples (Sup, enriched in host cytosolic proteins from gill tissue) were compared against the symbiont-free foot tissue. Group B: to detect putative host proteins involved in direct interactions with the symbionts (that is, physically linked to the symbionts or the symbiont-surrounding vacuolar membrane), gradient pellet samples (GP, enriched in symbionts) were compared against gill tissue and supernatant samples. Expression ratios were calculated from OrgNSAF% values (averages across all replicates) of the two respective sample types, for example, Gill : Foot, GP : Sup, etc. Proteins detected in only one of the two samples in comparison are indicated by ‘Gill only', ‘Sup only' and ‘GP only'. Significant differences in protein expression levels between two sample types are indicated by ‘X' in the Statistical significance columns. A t-test with permutation-based false discovery rate (FDR) calculation (FDR=5%) was used to test for statistical significance (see methods for details). This table only includes host proteins putatively involved in symbiont digestion, immune-related functions and carbon supply for the thiotrophic symbiont. For additional proteins with putative symbiosis-relevant functions refer to Supplementary Tables S4A and S4B. For protein accession numbers refer to the DeepSeaVent database (http://transcriptomics.biocant.pt/deepSeaVent/).
Biosynthesis of amino acids and vitamins/cofactors in B. azoricus
Only seven genes associated with amino acid biosynthesis were found in the B. azoricus host EST library, of which four were identified as proteins (Supplementary Figure S5). In contrast, the genome of the thiotrophic symbiont contains essentially complete gene sets for the biosynthesis of all 20 proteinogenic amino acids and of 11 vitamins and cofactors. Ninety-five per cent of the thiotroph's amino acid synthesis-related genes (that is, 63 of 66) and 43% (26 out of 60) of the genes associated to cofactor biosynthesis were identified as proteins (Supplementary Figure S5). In the methanotroph, 43 amino acid synthesis genes and 30 cofactor synthesis genes were detected in the genome, of which 30 (70%) and 11 (37%), respectively, were detected at the protein level (Supplementary Figure S5). As amino acids are indispensable for protein biosynthesis in the host, the sparse presence of amino acid synthesis-related genes in B. azoricus (even when considering the incompleteness of the available transcriptome information) may indicate that the bivalve depends on its symbionts for supply with amino acids and cofactors. This putative deficiency in amino acid biosynthetic enzymes seems to be uncommon in non-symbiotic marine mussels as several species are capable of de novo synthesizing most amino acids from TCA pathway intermediates (Ellis et al., 1985). Bathymodiolus mussels can acquire amino acids by direct uptake from seawater or from breakdown of organic matter ingested through filter feeding (Riou et al., 2010). But whether this can satisfy all of their nutritional requirements is unclear (Martins et al., 2008), particularly given the unsteady availability of organic nutrients in the dynamic vent habitats. Our results suggest that the B. azoricus symbionts are capable of providing their host with all required amino acids and prosthetic groups, a scenario that was previously also suggested for the thiotrophic vesicomyid symbioses of Calyptogena magnifica and C. okutanii (Newton et al., 2008). Amino acids that are synthesized by the B. azoricus symbionts may be made available to the host through intracellular symbiont digestion, as indicated by the detection of abundant host-derived digestive enzymes in the gill tissue (see below).
Replenishment of oxaloacetate and succinate
As mentioned above, the thiotroph seems not to be able to restore its oxaloacetate and succinate pools autonomously, as several TCA cycle enzymes are missing in its genome, and potential anaplerotic routes that could replace the missing enzymes are not encoded (Figures 4a and 5a, see Supplementary Results and Discussion for details). The required intermediates may instead be provided by the host, as suggested by the results of our proteome analysis: The host enzyme phosphoenolpyruvate carboxykinase (PckA, BAGiLS_012326) was abundantly expressed in the B. azoricus gill metaproteome and its expression was 3.7 fold higher in the gill as compared to the symbiont-free foot tissue (calculated from host gill and foot OrgNSAF% Supplementary Table S3). PckA mediates the reversible carboxylation of phosphoenolpyruvate to oxaloacetate (Prichard and Schofield, 1968; Moon et al., 1977; Lee, 2002) and may thus provide oxaloacetate for import into the thiotroph in B. azoricus (Figure 5a and b). This would effectively mean that a part of the carbon fixation is done on the host side. Such an intimate metabolic integration of host and symbiont on the level of central carbon metabolism would provide the host with a direct way to control symbiont metabolism.
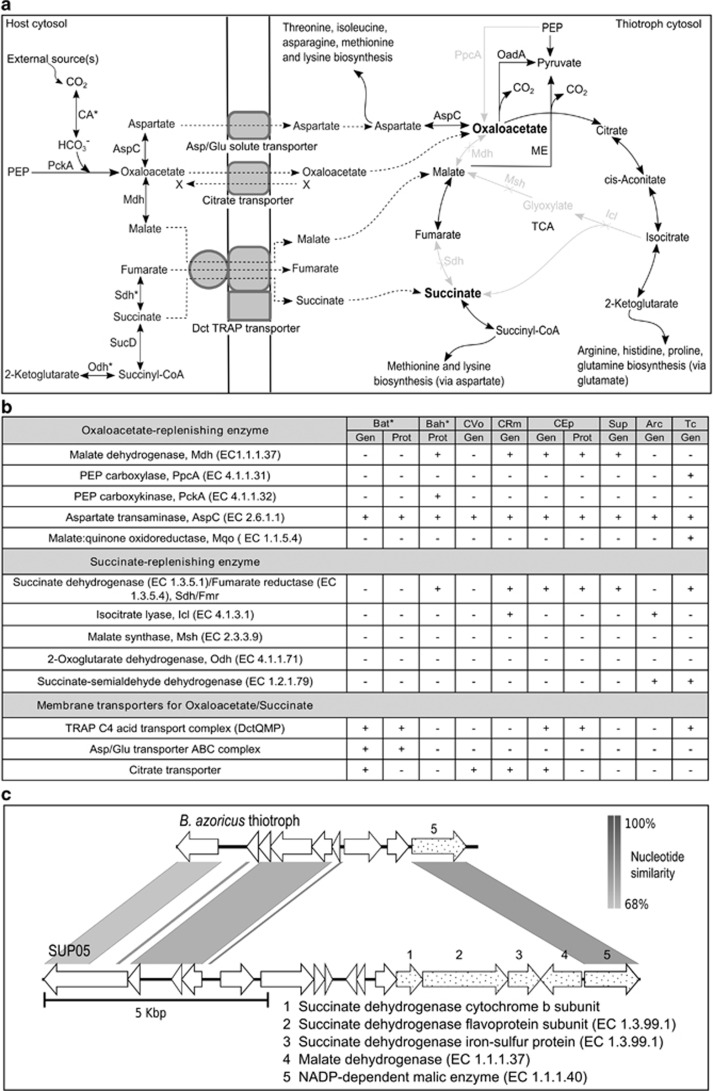
Figure 5.
Speculated routes of oxaloacetate and succinate replenishment in the thiotrophic B. azoricus symbiont. (a) Incomplete TCA cycle in thiotrophs and possible mechanism of oxaloacetate and succinate replenishment by host enzymes. Enzymes catalyzing reactions shown in grey are not encoded in the B. azoricus thiotroph genome. All host enzymes indicated here except AspC are highly abundant in the gill proteome of B. azoricus (*these host proteins were detected with significantly elevated expression levels in the symbiont-containing samples, as compared to the symbiont-free foot tissue, see also Table 1). For abundance of the thiotroph's TCA cycle enzymes refer to Figure 4a. A citrate transporter, which might potentially import oxaloacetate (in exchange for an unknown substrate such as pyruvate or acetate indicated in the figure by an X), was not detected in the proteome but is encoded in the thiotroph's genome. The depleted TCA intermediates oxaloacetate and succinate/succinyl-CoA are marked in bold. AspC: aspartate transaminase, Icl: isocitrate lyase, Mdh: malate dehydrogenase, Msh: malate synthase, ME: malic enzyme, OadA: oxaloacetate decarboxylase, Odh: 2-oxoglutarate dehydrogenase, PEP: phosphoenolpyruvate, PckA: PEP carboxykinase, Sdh: succinate dehydrogenase. Note that amino acid biosynthesis from oxaloacetate also involves succinyl-CoA, but that this interconnection of both pathways is not shown in the interest of clarity. (b) Overview of enzymes that can potentially transport or replenish oxaloacetate and succinate in different sulfur-oxidizing Gammaproteobacteria. Presence or absence of the corresponding enzyme gene (Gen) or protein (Prot) is indicated by + and −, respectively. *Data from this study. Bat: B. azoricus thiotroph, Bah: B. azoricus host (Bettencourt et al., 2010), CVo: Ca. Vesicomyosocius okutanii (Calyptogena okutanii symbiont, Kuwahara et al., 2007), CRm: Ca. Ruthia magnifica (C. magnifica symbiont, Newton et al., 2007), CEp: Ca. Endoriftia persephone (Riftia pachyptila symbiont, Markert et al., 2011), Sup: SUP05 (free-living chemoautotroph, Walsh et al., 2009), Arc: ARCTIC96BD-19 (Swan et al., 2011), Tc: Thiomicrospira crunogena Xcl-2 (Scott et al., 2006). (c) Some TCA cycle enzymes (dotted arrows 1–4) are missing in the B. azoricus thiotroph, in contrast to SUP05. See Supplementary Table S2B for genome assemblies included in the gene prediction for this figure.
Oxaloacetate could furthermore be converted to malate or aspartate before transfer from host to thiotroph (Figure 5a), as indicated by high concentrations of host Mdh (BAGiLS_010003, 0.55% gill OrgNSAF) in the gill metaproteome. The host's aspartate transaminase AspC (CAD42721.2) was also expressed, although at lower abundance (0.012% gill OrgNSAF; Figures 5a and b, Supplementary Table S3). Succinate/succinyl-CoA might be produced or interconverted to fumarate before transfer to the thiotroph by the two host enzymes fumarate reductase/succinate dehydrogenase, Sdh (BAGiLS_010531, Figure 5a), and dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase, Odh (BAGiLS_005913). Both proteins showed significantly elevated expression levels in the symbiont-enriched gradient pellet as compared to the host-enriched supernatant in our study (Table 1).
Uptake of malate or succinate by the thiotrophic symbiont requires the presence of the TRAP-type C4 dicarboxylate transporter complex DctQMP, whereas aspartate can be imported through the glutamate/aspartate ABC transporter complex, GltIJKL, respectively (Figure 5a). Complete gene sets for both these transporter complexes are encoded in the thiotrophic symbiont's genome. The solute receptor component DctP (BazSymB_scaffold00004_53), a component of the TRAP-type C4 dicarboxylate transporter complex, and the periplasmic binding component GltI (BazSymA_Acontig00019_5) of the glutamate/aspartate ABC transporter complex were also identified in the thiotroph's proteome (Figure 5b and Supplementary Table S3). Transporters for oxaloacetate are rare in bacteria, but a citrate transporter (BazSymA_Acontig02368_3), which is encoded in the thiotroph's genome, shows promiscuity towards oxaloacetate as its substrate and may therefore be used for oxaloacetate uptake in the thiotroph (Pudlik and Lolkema, 2011). Oxaloacetate may furthermore also be imported through the above-mentioned TRAP-type C4 dicarboxylate transporter, which is poorly characterized with respect to its specificity.
Is the thiotroph turning into an obligate symbiont?
The mdh and sdh genes, which are missing from the thiotroph's genome, are present in a single gene cluster in the symbiont's free-living relative SUP05 (Walsh et al., 2009; Figure 5c). This might indicate a selective loss of these genes during the thiotroph's transition from a free-living to a symbiotic lifestyle, possibly driven by the presence of functional substitutes in the host (as demonstrated in our study). The thiotrophic symbiont may hence even be obligately dependent on the host for its metabolic needs. The fact that no active free-living stage of the thiotrophic B. azoricus symbiont has ever been detected in the vent environment might support this speculation. However, as the mdh gene (but not the sdh gene) is also absent from the recently published genome of the free-living SUP05 member ‘Candidatus Thioglobus autotrophica' (Shah and Morris, 2015), the possibility of an obligate symbiosis in B. azoricus remains hypothetical. It might be speculated that a putative free-living stage of the thiotroph – if there is one – may rely on the uptake of oxaloacetate or other small organic molecules from the environment, circumventing the need for the missing TCA cycle enzyme functions.
Adaptations to a symbiotic lifestyle
To identify candidate genes and proteins that are potentially involved in symbiosis-specific functions we pursued two complementary approaches: (i) We compared symbiont genomes and proteomes with those of their free-living relatives. This approach revealed that chaperones and DNA-binding proteins are extraordinarily abundant in both symbionts (see below). It furthermore showed the unexpected presence of genes and proteins involved in CRISPR-Cas and restriction-modification systems in the thiotrophic B. azoricus symbiont, which may hint at a yet to be determined role of these proteins in host-symbiont interactions (see Supplementary Results and Discussion for details). (ii) We compared protein expression in symbiont-enriched B. azoricus samples versus symbiont-free samples, and in symbiont-containing tissue versus symbiont-free tissue to identify host proteins involved in interactions with the symbionts (see Methods for details and Supplementary Table S4 for a comprehensive list of putative symbiosis-relevant proteins identified in our analysis). With this approach we identified a broad repertoire of digestive enzymes, which are likely involved in symbiont digestion (see below). We also detected 23 immune-related host proteins, of which seven had significantly higher expression levels in symbiont-containing samples, and whose exact function in the symbiosis is unclear (see Supplementary Results and Discussion).
High expression of digestive enzymes in gill tissue
In our proteome analysis, we identified 58 host proteins with putative proteolytic and carbohydrate-degrading functions (Supplementary Table S3), of which 12 were significantly more abundant in symbiont-containing samples (whole gills and gradient pellet) compared to symbiont-free (foot tissue) and host-enriched samples (host-enriched supernatant, Table 1). These abundant host enzymes are likely involved in the digestion of the symbionts. In chemoautotrophic invertebrate symbioses, two modes of nutrient transfer from symbiont to host have been proposed: (i) direct digestion of symbionts by the host, and/or (ii) translocation of nutrients from symbiont tissue to the host cells, termed ‘milking' (Streams et al., 1997). Although previous observations indicated that symbiont digestion might be the major mode of nutrient transfer in B. azoricus (Fisher and Childress, 1992; Streams et al., 1997; Fiala-Médioni et al., 2002), no direct evidence existed so far.
Seven of the host proteins with significantly higher abundances in symbiont-containing samples were putative proteases and peptidases. Among them were lysosomal proteases such as saposin B (BAGiLS_000621) and cathepsin (BAGiLS_003294), which showed >80 fold and ~13 fold higher abundance, respectively, in gills compared to foot samples (Table 1). Lysosomes have been implicated in symbiont digestion in gills of B. azoricus (Fiala-Médioni et al., 2002) and other bivalves (Boetius and Felbeck, 1995) and our results further corroborate the idea that lysosomal host proteases may facilitate the degradation of symbiont proteins during symbiont digestion. We furthermore identified two glycosidases (glycoside hydrolases), which were significantly enriched in the gills (Table 1): BAGiLS_012512 was most abundant in the gill membrane fraction, indicating a possible involvement in the hydrolysis of bacterial cell-surface polysaccharides (Davies and Henrissat, 1995; see Supplementary Table S3). During symbiont digestion, some of the particularly abundant host glycosidases in the gill tissue may also target the methanotrophic symbiont's glycogen reserves (see above).
Abundant chaperones and bacterial nucleoid proteins in B. azoricus symbionts
Our proteome data revealed remarkably high expression levels of molecular chaperones in both symbionts and of histone-like DNA-binding proteins in the thiotrophic symbiont: The chaperones GroEL, GroES, DnaJ and DnaK together constituted 3.02% and 7.16% (gill OrgNSAF) of the total protein abundance in the thiotroph and in the methanotroph, respectively (Velos analysis, see Supplementary Table S3 and Supplementary Results and Discussion). Bacterial nucleoid-associated proteins, such as the histone-like bacterial DNA-binding protein (Hns) and the DNA-binding protein HU beta (HupB), constituted 16.87% (gill OrgNSAF) in the thiotroph, the latter being the most abundant protein of the entire gill metaproteome (Figure 3, Supplementary Table S3). In the thiotroph, GroEL and HupB were even more abundant than the most abundant sulfur oxidation-related protein DsrH and the carbon-fixing RuBisCO. High expression levels of chaperones and DNA-binding proteins have also been observed in other symbionts (Baumann et al., 1996) and in pathogenic bacteria (Neckers and Tatu, 2008), indicating that these proteins might have a symbiosis-specific function in the B. azoricus symbionts. As the thiotroph might be evolving into an obligate symbiont (see above), enhanced mutation rates and corresponding increased protein misfolding might be a possible explanation for the observed high chaperone concentrations (see Supplementary Results and Discussion for details).
Conclusion
Our proteogenomic anaylsis of the deep-sea mussel B. azoricus and its two uncultured symbionts provided detailed insights into the molecular mechanisms and strategies that underpin this symbiosis, which is otherwise difficult to access by cultivation-based approaches. The ability of the B. azoricus symbionts to use an extensive range of energy sources including sulfide, thiosulfate, methane and hydrogen to fuel chemosynthesis may enable the host to exploit a wide range of environmental conditions. Complementing metabolic pathways between the host and its symbionts point to metabolic interdependence between the individual partners, which may nevertheless enhance the metabolic efficiency of the consortium as a whole. Particularly, the inability of the thiotrophic symbiont to replenish its oxaloacetate and succinate pools and the high abundance of chaperones in their proteome may indicate that this bacterium could be on the verge of becoming an obligate symbiont. The potential integration of host and symbiont metabolism at the level of TCA cycle intermediates furthermore raises some intriguing questions on how this metabolic interconnection evolved and if it provides a mechanism for the host to exert control on symbiont metabolism. Our study forms a comprehensive basis for future investigations to examine how obligate associations could evolve from facultative symbiotic partnerships.
Acknowledgments
Thanks to Captain and Crew of RV Meteor and to C Borowski and D Fink for sampling during the cruise. We appreciate the excellent technical assistance of S Wetzel, J Matulla and S Grund, and are grateful to F Bonn for protein database searches. We thank Antony CP for discussion regarding the methanotroph metabolism. This study was supported by the EU-funded Marie Curie Initial Training Network ‘Symbiomics' (project no. 264774). Manuel Kleiner was supported by an NSERC Banting Postdoctoral Fellowship, Lizbeth Sayavedra was supported by a DAAD scholarship, and Jillian M Petersen was supported by the Max Planck Society, and the Vienna Science and Technology Fund (WWTF) through project VRG14-021.
Author contributions
RP performed the FISH analyses, sample preparation for all MS analyses, analysed the proteomes and related genomic data, and wrote the manuscript. MK developed the symbiont enrichment procedure, designed and performed the statistical analysis to identify symbiosis-specific proteins and contributed to the manuscript writing and revision. RP and MK established the protein databases. LS and JP conducted the genome analyses and revised the manuscript. MM, AO and DB performed the MS analyses and AO also conducted database searches. ND supervised and coordinated sampling and genome analysis. TS and StM designed, supervised and coordinated the proteomic analyses, and StM contributed substantially to the manuscript writing and revision.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Arp AJ, Childress JJ, Fisher CR. (1984). Metabolic and blood-gas transport characteristics of the hydrothermal vent bivalve Calyptogena magnifica. Physiol Zool 57: 648–662. [Google Scholar]
- Arp AJ, Childress JJ, Vetter RD. (1987). The sulphide-binding protein in the blood of the vestimentiferan tube-worm, Riftia pachyptila, is the extracellular haemoglobin. J Exp Biol 128: 139–158. [Google Scholar]
- Baumann P, Baumann L, Clark MA. (1996). Levels of Buchnera aphidicola chaperonin GroEL during growth of the aphid Schizaphis graminum. Curr Microbiol 32: 279–285. [Google Scholar]
- Beinart R, Gartman A, Sanders J, Luther G, Girguis P. (2015). The uptake and excretion of partially oxidized sulfur expands the repertoire of energy resources metabolized by hydrothermal vent symbioses. Proc Roy Soc B 282: 20142811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkin S, Nelson DC, Jannasch HW. (1986). Symbiotic assimilation of CO2 in 2 hydrothermal vent animals, the mussel Bathymodiolus thermophilus and the tube worm Riftia pachyptila. Biol Bull 170: 110–121. [Google Scholar]
- Bettencourt R, Pinheiro M, Egas C, Gomes P, Afonso M, Shank T et al. (2010). High-throughput sequencing and analysis of the gill tissue transcriptome from the deep-sea hydrothermal vent mussel Bathymodiolus azoricus. BMC Genomics 11: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetius A, Felbeck H. (1995). Digestive enzymes in marine invertebrates from hydrothermal vents and other reducing environments. Mar Biol 122: 105–113. [Google Scholar]
- Brune DC. (1995). Sulfur compounds as photosynthetic electron donors. In: Blankenship RE, Madigan MT, Bauer CE (eds), Anoxygenic Photosynthetic Bacteria. Kluver: Dordrecht, The Netherlands, pp 847–870. [Google Scholar]
- Chan SI, Lu YJ, Nagababu P, Maji S, Hung MC, Lee MM et al. (2013). Efficient oxidation of methane to methanol by dioxygen mediated by tricopper clusters. Angew Chem Int Ed 52: 3731–3735. [DOI] [PubMed] [Google Scholar]
- Childress JJ, Fisher CR. (1992). The biology of hydrothermal vent animals – physiology, biochemistry, and autotrophic symbioses. Oceanogr Mar Biol 30: 337–441. [Google Scholar]
- Chistoserdova L, Chen SW, Lapidus A, Lidstrom ME. (2003). Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J Bacteriol 185: 2980–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaco A, Bettencourt R, Costa V, Lino S, Lopes H, Martins I et al. (2011). LabHorta: a controlled aquarium system for monitoring physiological characteristics of the hydrothermal vent mussel Bathymodiolus azoricus. ICES J Mar Sci 68: 349–356. [Google Scholar]
- Company R, Antúnez O, Bebianno MJ, Cajaraville MP, Torreblanca A. (2011). 2-D difference gel electrophoresis approach to assess protein expression profiles in Bathymodiolus azoricus from Mid-Atlantic Ridge hydrothermal vents. J Proteomics 74: 2909–2919. [DOI] [PubMed] [Google Scholar]
- Crowther GJ, Kosaly G, Lidstrom ME. (2008). Formate as the main branch point for methylotrophic metabolism in Methylobacterium extorquens AM1. J Bacteriol 190: 5057–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Henrissat B. (1995). Structures and mechanisms of glycosyl hydrolases. Structure 3: 853–859. [DOI] [PubMed] [Google Scholar]
- Dedysh SN, Knief C, Dunfield PF. (2005). Methylocella species are facultatively methanotrophic. J Bacteriol 187: 4665–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel DL, Lee HK, Cavanaugh CM. (1995). Intracellular coexistence of methano- and thioautotrophic bacteria in a hydrothermal vent mussel. Proc Natl Acad Sci USA 92: 9598–9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doeller JE, Kraus DW, Colacino JM, Wittenberg JB. (1988). Gill hemoglobin may deliver sulfide to bacterial symbionts of Solemya velum (Bivalvia, Mollusca). Biol Bull 175: 388–396. [Google Scholar]
- Duperron S, Bergin C, Zielinski F, Blazejak A, Pernthaler A, McKiness ZP et al. (2006). A dual symbiosis shared by two mussel species, Bathymodiolus azoricus and Bathymodiolus puteoserpentis (Bivalvia: Mytilidae), from hydrothermal vents along the northern Mid-Atlantic Ridge. Environ Microbiol 8: 1441–1447. [DOI] [PubMed] [Google Scholar]
- Ellis LL, Burcham JM, Paynter KT, Bishop SH. (1985). Amino acid metabolism in euryhaline bivalves: regulation of glycine accumulation in ribbed mussel gills. J Exp Zool 233: 347–358. [DOI] [PubMed] [Google Scholar]
- Fiala-Médioni A, McKiness ZP, Dando P, Boulegue J, Mariotti A, Alayse-Danet AM et al. (2002). Ultrastructural, biochemical, and immunological characterization of two populations of the mytilid mussel Bathymodiolus azoricus from the Mid-Atlantic Ridge: evidence for a dual symbiosis. Mar Biol 141: 1035–1043. [Google Scholar]
- Fisher CR, Childress JJ, Oremland RS, Bidigare RR. (1987). The importance of methane and thiosulfate in the metabolism of the bacterial symbionts of two deep-sea mussels. Mar Biol 96: 59–71. [Google Scholar]
- Fisher CR, Childress JJ, Arp AJ, Brooks JM, Distel D, Favuzzi JA et al. (1988). Microhabitat variation in the hydrothermal vent mussel, Bathymodiolus thermophilus, at the Rose Garden vent on the Galapagos Rift. Deep-Sea Res 35: 1769–1791. [Google Scholar]
- Fisher CR, Childress JJ. (1992). Organic-carbon transfer from methanotrophic symbionts to the host hydrocarbon-seep mussel. Symbiosis 12: 221–235. [Google Scholar]
- Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, Workman JL et al. (2006). Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 40: 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson RG, Turner RD, Lutz RA, Vrijenhoek RC. (1998). A new genus and five new species of mussels (Bivalvia, Mytilidae) from deep-sea sulfide/hydrocarbon seeps in the Gulf of Mexico. Malacologia 40: 63–112. [Google Scholar]
- Harada M, Yoshida T, Kuwahara H, Shimamura S, Takaki Y, Kato C et al. (2009). Expression of genes for sulfur oxidation in the intracellular chemoautotrophic symbiont of the deep-sea bivalve Calyptogena okutanii. Extremophiles 13: 895–903. [DOI] [PubMed] [Google Scholar]
- Hongo Y, Nakamura Y, Shimamura S, Takaki Y, Uematsu K, Toyofuku T et al. (2013). Exclusive localization of carbonic anhydrase in bacteriocytes of the deep-sea clam Calyptogena okutanii with thioautotrophic symbiotic bacteria. J Exp Biol 216: 4403–4414. [DOI] [PubMed] [Google Scholar]
- Kádár E, Bettencourt R, Costa V, Santos RS, Lobo-Da-Cunha A, Dando P. (2005). Experimentally induced endosymbiont loss and re-acquirement in the hydrothermal vent bivalve Bathymodiolus azoricus. J Exp Mar Biol Ecol 318: 99–110. [Google Scholar]
- Kádár E, Davis SA, Lobo-da-Cunha A. (2008). Cytoenzymatic investigation of intracellular digestion in the symbiont-bearing hydrothermal bivalve Bathymodiolus azoricus. Mar Biol 153: 995–1004. [Google Scholar]
- Keltjens JT, Pol A, Reimann J, Op den Camp HJM. (2014). PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl Microbiol Biot 98: 6163–6183. [DOI] [PubMed] [Google Scholar]
- Kleiner M, Petersen JM, Dubilier N. (2012a). Convergent and divergent evolution of metabolism in sulfur-oxidizing symbionts and the role of horizontal gene transfer. Curr Opin Microbiol 15: 621–631. [DOI] [PubMed] [Google Scholar]
- Kleiner M, Wentrup C, Lott C, Teeling H, Wetzel S, Young J et al. (2012b). Metaproteomics of a gutless marine worm and its symbiotic microbial community reveal unusual pathways for carbon and energy use. Proc Natl Acad Sci USA 109: E1173–E1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochevar RE, Childress JJ. (1996). Carbonic anhydrase in deep-sea chemoautotrophic symbioses. Mar Biol 125: 375–383. [Google Scholar]
- Kuwahara H, Yoshida T, Takaki Y, Shimamura S, Nishi S, Harada M et al. (2007). Reduced genome of the thioautotrophic intracellular symbiont in a deep-sea clam, Calyptogena okutanii. Curr Biol 17: 881–886. [DOI] [PubMed] [Google Scholar]
- Le Pennec M, Donval A, Herry A. (1990). Nutritional strategies of the hydrothermal ecosystem bivalves. Prog Oceanogr 24: 71–80. [Google Scholar]
- Lee DL. (2002) The Biology of Nematodes. Taylor & Francis: London. [Google Scholar]
- Liu LJ, Stockdreher Y, Koch T, Sun ST, Fan Z, Josten M et al. (2014). Thiosulfate transfer mediated by DsrE/TusA homologs from acidothermophilic sulfur-oxidizing archaeon Metallosphaera cuprina. J Biol Chem 289: 26949–26959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markert S, Gardebrecht A, Felbeck H, Sievert SM, Klose J, Becher D et al. (2011). Status quo in physiological proteomics of the uncultured Riftia pachyptila endosymbiont. Proteomics 11: 3106–3117. [DOI] [PubMed] [Google Scholar]
- Martins I, Colaço A, Dando PR, Martins I, Desbruyères D, Sarradin P-M et al. (2008). Size-dependent variations on the nutritional pathway of Bathymodiolus azoricus demonstrated by a C-flux model. Ecol Model 217: 59–71. [Google Scholar]
- Moon TW, Mustafa T, Hulbert WC, Podesta RB, Mettrick DF. (1977). The phosphoenol-pyruvate branchpoint in adult Hymenolepis diminuta (Cestoda): a study of pyruvate kinase and phosphoenol-pyruvate carboxykinase. J Exp Zool A Comp Exp Biol 200: 325–336. [DOI] [PubMed] [Google Scholar]
- Mueller RS, Denef VJ, Kalnejais LH, Suttle KB, Thomas BC, Wilmes P et al. (2010). Ecological distribution and population physiology defined by proteomics in a natural microbial community. Mol Syst Biol 6: 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckers L, Tatu U. (2008). Molecular chaperones in pathogen virulence: emerging new targets for therapy. Cell Host Microbe 4: 519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Fisher CR. (1995). Chemoautotrophic and methanotrophic endosymbiotic bacteria at deep-sea vents and seeps. In: Karl DM (ed). The Microbiology of Deep-sea Hydrothermal Vents. CRC Press: Boca Raton, FL, pp 125–167. [Google Scholar]
- Newton ILG, Girguis PR, Cavanaugh CM. (2008). Comparative genomics of vesicomyid clam (Bivalvia: Mollusca) chemosynthetic symbionts. BMC Genomics 9: 585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton ILG, Woyke T, Auchtung TA, Dilly GF, Dutton RJ, Fisher MC et al. (2007). The Calyptogena magnifica chemoautotrophic symbiont genome. Science 315: 998–1000. [DOI] [PubMed] [Google Scholar]
- Page HM, Fiala-Medioni A, Fisher CR, Childress JJ. (1991). Experimental evidence for filter-feeding by the hydrothermal vent mussel, Bathymodiolus thermophilus. Deep-Sea Res 38: 1455–1461. [Google Scholar]
- Petersen JM, Zielinski FU, Pape T, Seifert R, Moraru C, Amann R et al. (2011). Hydrogen is an energy source for hydrothermal vent symbioses. Nature 476: 176–180. [DOI] [PubMed] [Google Scholar]
- Powell MA, Somero GN. (1986). Adaptations to sulfide by hydrothermal vent animals – sites and mechanisms of detoxification and metabolism. Biol Bull 171: 274–290. [Google Scholar]
- Prichard R, Schofield P. (1968). The metabolism of phosphoenolpyruvate and pyruvate in the adult liver fluke Fasciola hepatica. Biochim Biophys Acta 170: 63–76. [DOI] [PubMed] [Google Scholar]
- Pudlik AM, Lolkema JS. (2011). Mechanism of citrate metabolism by an oxaloacetate decarboxylase-deficient mutant of Lactococcus lactis IL1403. J Bacteriol 193: 4049–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulfs EC, Macko SA, Van Dover CL. (2004). Tissue and symbiont condition of mussels (Bathymodiolus thermophilus exposed to varying levels of hydrothermal activity. J Mar Biol Assoc UK 84: 229–234. [Google Scholar]
- Riou V, Colaco A, Bouillon S, Khripounoff A, Dando P, Mangion P et al. (2010). Mixotrophy in the deep sea: a dual endosymbiotic hydrothermal mytilid assimilates dissolved and particulate organic matter. Mar Ecol Prog Ser 405: 187–201. [Google Scholar]
- Sayavedra L, Kleiner M, Ponnudurai RP, Wetzel S, Pelletier E, Barbe V et al. (2015). Abundant toxin-related genes in the genomes of beneficial symbionts from deep-sea hydrothermal vent mussels. eLife 4: e07966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KM, Sievert SM, Abril FN, Ball LA, Barrett CJ, Blake RA et al. (2006). The genome of deep-sea vent chemolithoautotroph Thiomicrospira crunogena XCL-2. PLoS Biol 4: e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah V, Morris RM. (2015). Genome sequence of ‘Candidatus Thioglobus autotrophica' strain EF1, a chemoautotroph from the SUP05 clade of marine Gammaproteobacteria. Genome Announc 3: e01156–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streams ME, Fisher CR, Fiala-Médioni A. (1997). Methanotrophic symbiont location and fate of carbon incorporated from methane in a hydrocarbon seep mussel. Mar Biol 129: 465–476. [Google Scholar]
- Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D et al. (2011). Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333: 1296–1300. [DOI] [PubMed] [Google Scholar]
- Van Dover CL. (2000) The Ecology of Deep-sea Hydrothermal Vents. Princeton University Press: New Jersey. [Google Scholar]
- Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I et al. (2016). 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44: D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Cosel R, Comtet T, Krylova E. (1999). Bathymodiolus (Bivalvia: Mytilidae) from the hydrothermal vents on the Azores Triple Junction and the Logatchev hydrothermal field, Mid-Atlantic Ridge. Veliger 42: 218–248. [Google Scholar]
- Walsh DA, Zaikova E, Howes CG, Song YC, Wright JJ, Tringe SG et al. (2009). Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science 326: 578–582. [DOI] [PubMed] [Google Scholar]
- Wood AP, Aurikko JP, Kelly DP. (2004). A challenge for 21st century molecular biology and biochemistry: what are the causes of obligate autotrophy and methanotrophy? FEMS Microbiol Rev 28: 335–352. [DOI] [PubMed] [Google Scholar]
- Zhao S-J, Hanson RS. (1984). Variants of the obligate methanotroph isolate 761M capable of growth on glucose in the absence of methane. Appl Environ Microbiol 48: 807–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data and the protein sequence database were uploaded to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al., 2016) partner repository with the dataset identifier PXD004061 (DOI: 10.6019/PXD004061). Nucleotide and protein sequence information for the methanotrophic symbionts of B. azoricus and of Bathymodiolus sp. was deposited in GenBank under the BioProject accession numbers PRJEB13769 and PRJEB13047, respectively.