Abstract
Fluctuating environments can modulate host–pathogen interactions by providing a temporary advantage to one of the interacting organisms. However, we know very little about how environmental conditions facilitate beneficial interactions between hosts and their microbial communities, resulting in individual persistence with a particular pathogen. Here, we experimentally infected Eleutherodactylus coqui frogs with the fungal pathogen Batrachochytrium dendrobatidis (Bd) under environmental conditions known to confer the survival advantage to the host during the warm-wet season, or alternatively to the pathogen during the cool-dry season. We used 16S rRNA amplicon sequencing to quantify changes in bacterial richness and phylogenetic diversity, and identified operational taxonomic units (OTUs) that became overrepresented or suppressed as a consequence of Bd infection. During the warm-wet season, frogs limited Bd infections, recruited putatively beneficial bacteria and returned to pre-infection levels of richness and phylogenetic diversity. In contrast, during the cool-dry season, Bd infections kept increasing through time, and bacterial diversity remained constant. Our findings confirm that infection outcome not only depends on abiotic factors, but also on biotic interactions between hosts and their associated bacterial communities.
Introduction
Fungal pathogens currently pose a global emerging threat to plants, animals, and ecosystems (Fisher et al., 2012). These pathogens are extremely effective at colonizing external surfaces (for example, integuments, epidermis, skin), which are already inhabited by other microorganisms. Bacteria can affect fungal colonization, survival, growth, and virulence by altering physiological and chemical processes including inhibition of colonization sites (Franks et al., 2006), regulation of nutrient availability, production of antifungal compounds (Brucker et al., 2008), or by providing precursors for fungal virulence factors (Wargo and Hogan, 2006). Furthermore, host external surfaces are continuously exposed to changes in abiotic factors, providing an additional source of selection that may act differently for bacteria and fungi. Therefore, quantifying fungal–bacterial interactions—and considering fluctuations in environmental conditions—has the potential to advance our understanding of infection outcomes that range from full disease resistance to mortality.
The relationship between the chytrid fungus Batrachochytrium dendrobatidis (Bd) and skin microbial communities in amphibians provides an excellent system to test the strength and direction of fungal–bacterial interactions. Bd is a pathogenic fungus that infects the stratum corneum and deeper cell layers of amphibian skin (Greenspan et al., 2012), disrupting osmotic balance, and often causing host mortality (Voyles et al., 2009). Epibiotic bacteria also inhabit the outer layers of the skin, and often accumulate around secretory glands (Lauer et al., 2007). Over the last few years, we have seen a rise in studies quantifying the effects of Bd infection on skin microbiomes (Jani and Briggs, 2014; Becker et al., 2015; Longo et al., 2015; Walke et al., 2015). Fungal infection disrupts the stability of bacterial communities in susceptible amphibians in both field and lab (Jani and Briggs, 2014; Longo et al., 2015). However, bacterial community structure before infection may also determine host survival; frogs with the ability to clear Bd infection carried a different bacterial community compared with individuals that died (Becker et al., 2015). Microbial communities of these Bd-resistant frogs did not exhibit any variation in structure, suggesting that stability provide resistance against Bd (Becker et al., 2015). However, unaccounted factors such as abiotic environmental fluctuations, seasonal environmental bacterial reservoirs and the standing host bacterial assemblage at time of infection, might also underlie differences in microbial communities. Therefore, to achieve a better understanding of disease as a disturbance process at the individual-level and how it translates into recovery at the population-level, we need to quantify Bd–microbiome interactions under variable and biologically realistic scenarios as would be experienced by wild amphibian populations.
In this study, we measured beneficial and antagonistic interactions between fungi and bacteria using field mesocosm experiments. Mesocosms offer the advantage of exposing hosts to natural conditions (that is, variable daily temperatures and precipitation, changes in shade, wind) that might differentially affect fungal infection and microbial communities. Considering the environmental heterogeneity that amphibians experience in their natural habitats, experimental inoculations in the lab may render an unrealistic turnover of microbial species. It is not surprising that skin microbiomes of wild-caught amphibians experience large shifts in community structure and composition when brought to the lab (Loudon et al., 2013; Becker et al., 2014), especially because in the lab they lack the environmental reservoirs that permit natural microbial recruitment (Loudon et al., 2013; Walke et al., 2014), thus promoting the establishment of skin bacterial communities very different to those originally found in nature.
We performed controlled inoculations in which hosts were exposed to Bd under natural field conditions, thereby minimizing potential artificial selection of bacteria. We challenged amphibians under conditions known to confer the advantage to the host—or alternatively to the pathogen (Longo et al., 2010), by performing the experiment over two seasons. During the warm-wet season, host immune function is at its maximum (Maniero and Carey, 1997; Raffel et al., 2006), beneficial symbionts are likely present and abundant on the amphibian skin and infection risk is low (Longo et al., 2010). Thus, we predict that these three factors will promote resistance strategies (reduced Bd load), resulting in rapid recovery after infection. As both temperature and precipitation decrease during the cool-dry season, the advantage shifts to the pathogen. Cool temperatures promote the release of zoospores from sporangia (Woodhams et al., 2008), host immune function is reduced (Maniero and Carey, 1997; Raffel et al., 2006) and beneficial symbionts are less common or lost. At these stressful times, we predict that hosts will not be able to control infections, and consequently die with high infection intensities (Longo et al., 2013). Understanding how the environment modulates these interactions is crucial to explain why some amphibians can persist with Bd infection, yet cannot fully recover after disease-mediated declines.
Materials and methods
Experimental design
We captured a total of 98 adult Eleutherodactylus coqui at El Yunque National Forest (18.299°N, 65.779°W, 649 m) during the warm-wet and cool-dry season (see Supplementary Table S1 for sample sizes and dates). We placed frogs in individual plastic bags and determined sex, body size (snout-vent-length, SVL in mm) and weight (g). Using new pairs of gloves for each individual, we swabbed frogs using a standard number of strokes (five times per foot, each side of the belly, and drink patch, for a total of 40 strokes). Swabs remained frozen at −20 °C until DNA extraction.
In the field, we housed each frog individually in a perforated 23-cm2 plastic mesh basket (HydroFarm, Petaluma, CA, USA) that was bleached before the experiment. Lids consisted of tulle fabric and fastened with antimicrobial rubber bands (Alliance Rubber Company, Hot Springs, AR, USA; size 117B). Inside the plastic basket (Supplementary Figure S1), we added dried leaves of Prestoea montana, a common palm found in the forest that is used by frogs as a calling and retreat site (Beard et al., 2003). We moistened the leaves in each unit by spraying with sterile distilled water. All leaves were collected from a single palm at the location of the experiment. Experimental containers were kept together under the tree canopy on the forest floor in a 10 m2 area.
Because we did not know the infection status of each individual frog on collection, our experimental design tests for the effect of Bd addition on natural skin bacterial communities. We designed a fully factorial mesocosm experiment with two factors (Bd and skin bacterial communities) and two levels each (manipulation and no manipulation), and assigned individuals randomly to experimental treatments. To manipulate skin bacterial communities, we applied a 1.5% hydrogen peroxide (HP) bath for 1 min, followed by rinsing with sterile distilled water. This procedure artificially reduces the abundance of skin microbes without hindering host health (Becker and Harris, 2010; Muletz et al., 2012). To manipulate Bd infection rates, we cultured Bd JEL427 in 1% tryptone agar plates and collected zoospores for use in the inoculation experiments. This strain of Bd was isolated from an infected E. coqui collected at the same locality in 2004. We experimentally infected individuals with 106 zoospores in a total of 5 ml in sterile sample containers for 24hrs (Cole Parmer, Vernon Hills, IL, USA; EW-060490-40). Frogs in the Bd− treatments were sham-infected with sterile distilled water. Thus, our four experimental treatments consisted of: Bd addition with reduced bacterial community manipulation (Bd+/HP+), Bd addition with intact bacterial community (Bd+/HP−), no Bd added but bacterial community manipulated (Bd−/HP+) and negative controls with no Bd infection or bacterial community manipulation (Bd−/HP−). We swabbed and weighed each frog every 2 days post-inoculation for 2 weeks using the same procedure described above. We recorded any instances of individuals showing the typical signs of chytridiomycosis, including lethargy, inappetence, changes in skin coloration or sloughing, abnormal postures and loss of righting reflex (Voyles et al., 2009). Skin sloughing was the most obvious sign of infection when swabbing (Supplementary Figure S2), thus we considered it as a binary predictor in our statistical analyses.
Frogs remained in enclosures for 15 days and were fed crickets every 2 days after data collection and swabbing. Our experimental design captures the bacterial community in its entirety, and does not differentiate between transient and resident bacteria contributed by environmental sources. We performed the experiment two times: once during the warm-wet season (November 2014) and once during the cool-dry season (January 2015). During November 2014, the average temperature was 23.4 °C (min. 17.2 °C, max. 29.4 °C) and total precipitation was 349 mm. In contrast, the average temperature in January 2015 was 22 °C (min. 18.3 °C, max. 26.7 °C) and total precipitation was 163 mm (Palma Sola Station, NOAA Online Weather Data). The Institutional Animal Care and Use Committee of Cornell University (2013-0074), Puerto Rico's Department of Natural Resources (2014-IC-74), and the US Forest Service approved experimental animal use. Frogs were euthanized at the end of the experiment, and archived at the Cornell University Museum of Vertebrates.
Quantification of Bd infection intensity
We extracted DNA from swabs using the MO BIO Power Soil DNA isolation kit (MO BIO, Carlsbad, CA, USA). All DNA extraction plates contained negative controls (blanks). To determine each individual's infection intensity, we used standard methods of quantitative PCR (qPCR) for Bd diagnosis (Boyle et al., 2004) and ran duplicate qPCR reactions for each sample using a Viia7 qPCR machine (Life Technologies, Carlsbad, CA, USA). We used Bd isolate JEL427 to generate the standard curve of zoospores for the qPCR runs. Individual infection intensity was measured as the mean number of zoospore genomic equivalents estimated by qPCR from the two well reactions.
16S rRNA amplicon sequencing
We followed the Earth Microbiome Project protocol to quantify diversity of bacterial communities from skin swabs (Caporaso et al., 2012). Briefly, we PCR-amplified the V4 region of the 16S rRNA gene using universal primers 515F and 806R. We ran reactions in triplicate plus a negative water control. Amplicons were visualized in 2% agarose gel, pooled, and quantified using QuantiFluor (Promega, Madison, WI, USA). Equimolar quantities from each sample (100 ng) were combined to generate the amplicon library, and then cleaned using Agencourt AMPure XP beads (Beckman Coulter, Brea, CA, USA). We prepared three libraries for Illumina MiSeq (2 × 250 bp) sequencing at the Biotechnology Research Center (BRC) Genomics Facility at Cornell University.
Sequence processing and OTU assignment
We used Quantitative Insights into Microbial Ecology v 1.9.0 (QIIME) to split libraries and assign sequences into operational taxonomic units (OTUs) (Caporaso et al., 2010b). We assigned OTUs using the subsampled open reference protocol in QIIME, which clusters sequences against the Greengenes database (May 2013) using uclust (DeSantis et al., 2006; Edgar, 2010). Briefly, 0.1% of the reads that did not match the Greengenes database are subsampled and clustered de novo (Caporaso et al., 2010a; Rideout et al., 2014). We excluded OTUs with fewer than 100 reads (Bokulich et al., 2013; Loudon et al., 2013), removed samples with less than 10 000 reads in coverage and normalized the OTU table using cumulative sum scaling in QIIME (McMurdie and Holmes, 2014). The number of reads in the remaining samples ranged from 10 030 to 273 016 (average: 49 510). Raw reads have been deposited in the NCBI Sequence Read Archive (SRR4010786).
Statistical analyses of Bd infection intensity and alpha diversity of bacteria
We log-transformed Bd infection intensity [log10(x+1)] for all analyses performed in R version 3.2.3 (R Core Team, 2014). To determine differences in infection at baseline (day 0), we compared initial Bd prevalence and intensity between seasons by using a χ2 test of independence and a t-test, respectively. We used three separate linear mixed models to estimate changes in Bd infection intensity, OTU richness and phylogenetic diversity over time for each treatment, and treated individual ID as a random effect. We used R package lme4 (Bates et al., 2014) to design the mixed models with the general equation: lmer(y~treatment+day+season+skin slough+weight+initial load (y+1)+treatment*day+treatment*season+day*season+treatment*slough+day*slough+season*slough+(1|individual_ID). For these models, we were interested in understanding the interactions of treatment, day and season effects, which could provide insights into the progression of the infection under different environmental scenarios. We used individual-level variables such as weight and evidence of skin sloughing (binary coded) to understand changes in host responses through time. We took into account previous exposure of each individual by incorporating their initial load (infection intensity, richness, or phylogenetic diversity) at day 0 as a fixed effect. We calculated least squares means, confidence intervals and Tukey's comparisons for the fixed effects and interactions using package lsmeans in R (Lenth and Hervé, 2016).
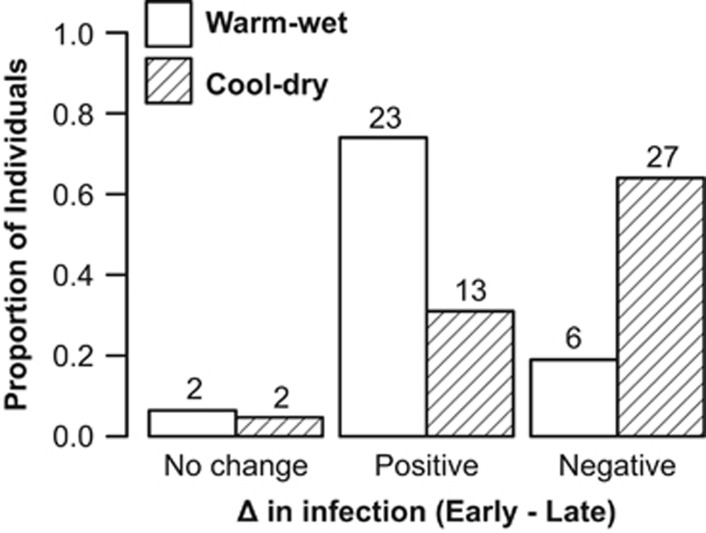
To determine if host ability to respond to infection changed between seasons, we compared the relative change in infection intensity between two stages (early vs. late). We considered their average infection intensity during days 0 to 3 as the early stage, and days 6 to 15 as the late stage. Individuals showing a positive response (early–late>0), possibly developed the infection, and were able to clear or reduce it. In contrast, individuals showing a negative response (early–late<0) were unable to control infections by the end of the experiment. We used a χ2 test to determine whether seasons were associated to the ability to clear/reduce infection, remain infected or uninfected.
Differential abundance of bacteria
For the analyses of differential bacterial abundance, we filtered our OTU table to include only samples for which individual frogs persisted throughout the entire experiment (73 frogs × 6 sampling days (1 initial+5 post-inoculation)=438 samples). By doing this filtering step, we avoided including effects related to host mortality. We used R packages phyloseq and DESeq2 (McMurdie and Holmes, 2013; Love et al., 2014) to import the OTU table to the R environment and identify differences in abundance of bacterial taxa between treatment groups with respect to the negative controls (Bd−/HP−). We calculated the geometric means of bacterial abundance counts before estimating size factors. Our design in DESeq2 tested the effects of day, treatment, season, Bd infection intensity, interactions between treatment*day, season*day and treatment*season on the abundance of each bacterial OTU. Specifically, our objective was to answer three questions about the most common host-associated bacteria: (1) which bacterial taxa (if any) were significantly over abundant (or suppressed) when comparing between wet and dry seasons? (2) which OTUs increased or decreased in abundance after the addition of Bd, clearing of bacteria (HP+) or both and (3) do these responses depend on the season? For these analyses, we used an alpha of 0.01 to decrease Type I error. We filtered taxa using this cutoff in the adjusted P-values, and narrowed our focus to the top 30 most abundant taxa based on mean number of reads.
Changes in beta diversity of bacteria
We measured changes in beta diversity by comparing skin bacterial communities using unweighted and weighted Unifrac distances, as well as Bray–Curtis distances. We used function adonis in R package vegan (Oksanen et al., 2015) to perform permutational multivariate analyses of variance (PERMANOVA). These analyses tested for differences in bacterial community structure across seasons, treatments and Bd infection status at three time points (initial: Day 0, after inoculation: Day 3, and end of experiment: Day 15). We report changes in beta diversity over time using a detrended correspondence analysis plot of unweighted-Unifrac distances.
Results
Interactions between time, season and, skin sloughing determined Bd infection intensity
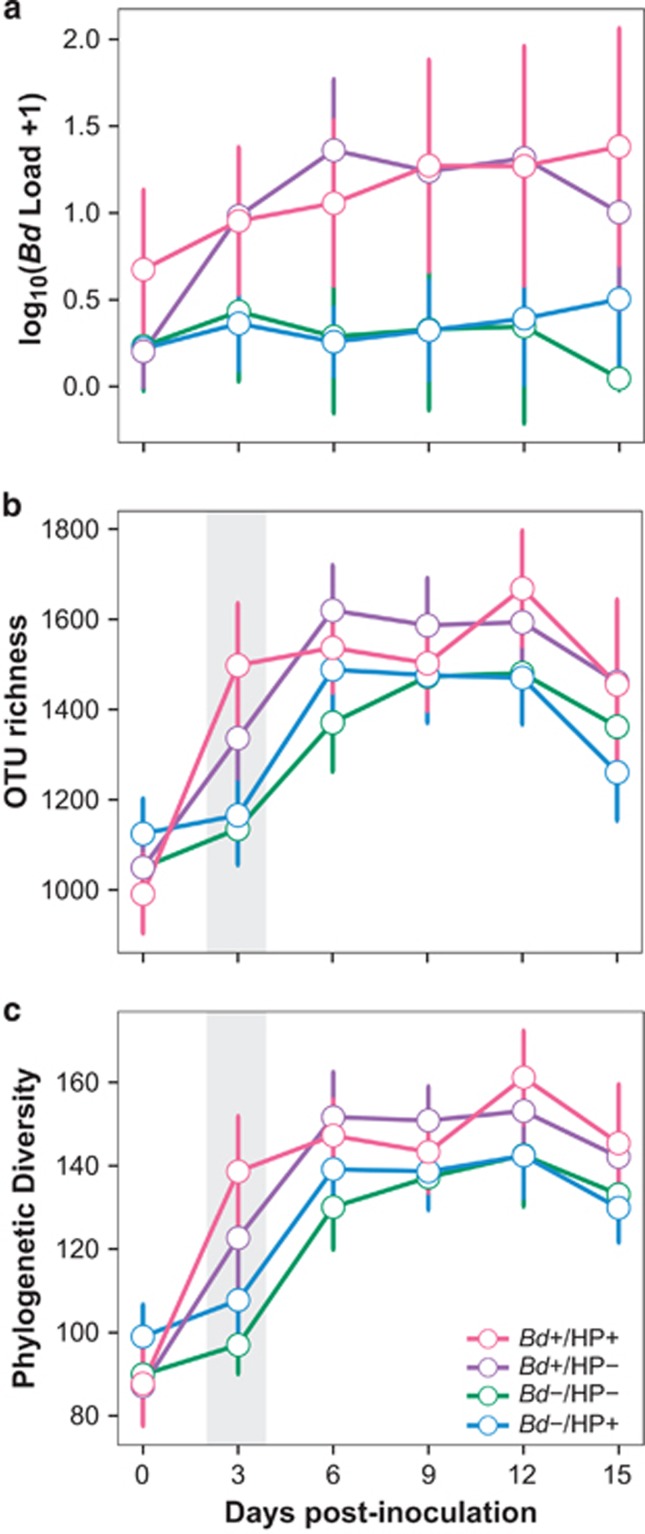
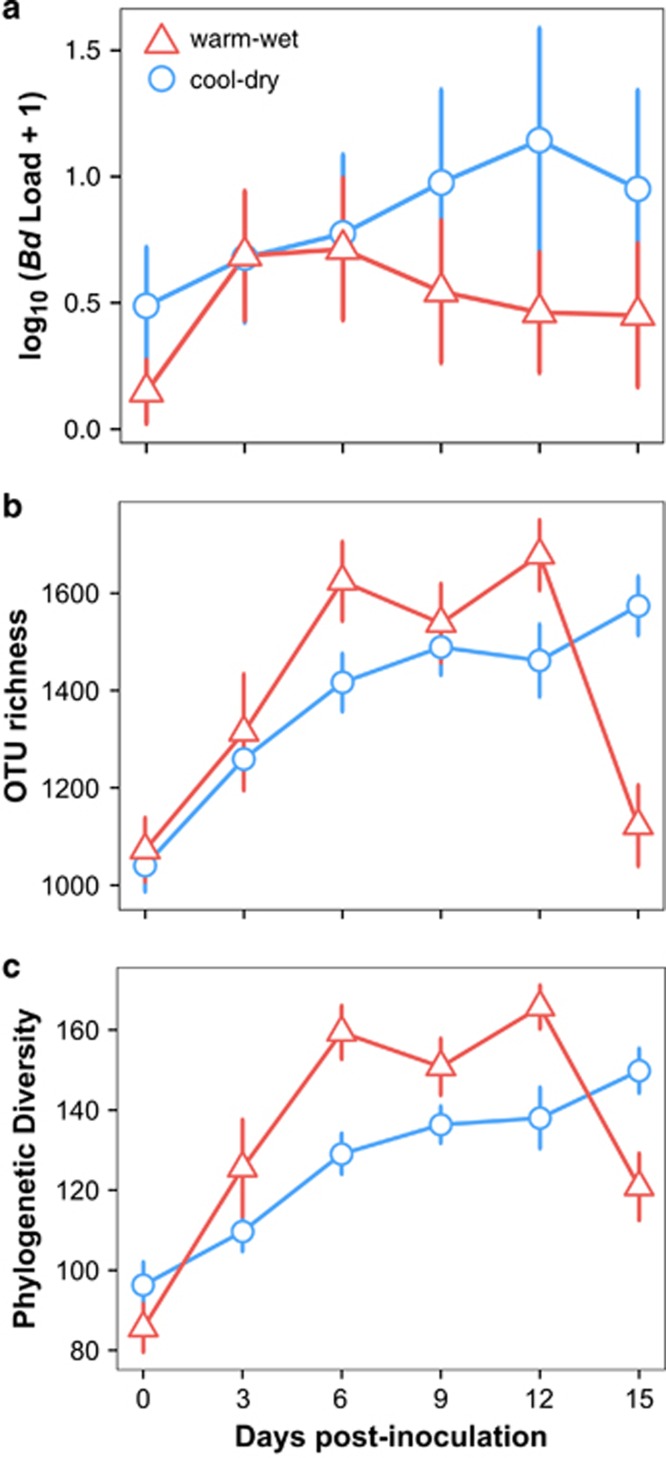
At capture, Bd prevalence among individuals was similar across seasons (wet: 50% vs dry: 46%, χ2=0.157, df=1, P=0.692, Supplementary Table S1), but Bd infection intensity was significantly higher during the cool-dry season (t=2.67, df=93, P<0.01, Supplementary Table S1), confirming previous findings of seasonal patterns of infection (Longo et al., 2010). Our mixed model of Bd infection intensity revealed significant effects of all interaction terms except treatment*season, which indicated that treatment effects did not vary between the two experimental trials (Supplementary Table S2; Supplementary Figure S3). As expected after inoculation, individuals in Bd+ treatments had higher Bd infection intensities than individuals in Bd− treatments (Figure 1a; Supplementary Table S3). However, inoculation with Bd and application of the HP bath (Bd+/HP+ treatment) did not augment Bd infection intensity as predicted if bacterial communities buffered pathogen growth compared with Bd+/HP− treatments (day 0 to day 3: β=0.20, df=113.33, t=0.822, P=0.84, Figure 1a). Furthermore, HP did not increase or decrease average infection intensity in control treatments (Bd−/HP+ vs Bd−/HP−, P>0.05, Supplementary Table S3). The interaction term day*season revealed that average Bd infection intensity showed fluctuations through time that depended on seasons (Figure 2a). More specifically, frogs maintained relatively constant infection intensities through time only during the warm-wet season, as shown by the lack of significance among daily pairwise comparisons (Figure 2a; Supplementary Table S3). In contrast, during the cool-dry season Bd infection intensity increased significantly over time and peaked at day 12 (Figure 2a; Supplementary Table S3).
Figure 1.
Significant interactions between treatment and time in: (a) Bd infection intensity, (b) OTU richness and (c) phylogenetic diversity of skin bacterial communities. Circles represent the mean of each treatment by day, lines across the mean represent the 95% confidence intervals. Significance for the interaction terms is available in Supplementary Tables S2–S5. Gray shading indicates significant differences between Bd+ and Bd− treatments.
Figure 2.
Significant interactions between season and time in: (a) Bd infection intensity, (b) OTU richness and (c) phylogenetic diversity of skin bacterial communities. Each circle represents the mean (across all treatments) for the cool-dry season. Triangles denote the mean for the warm-wet season. Lines represent the 95% confidence intervals. Significance for the interaction terms is available in Supplementary Tables S2–S5.
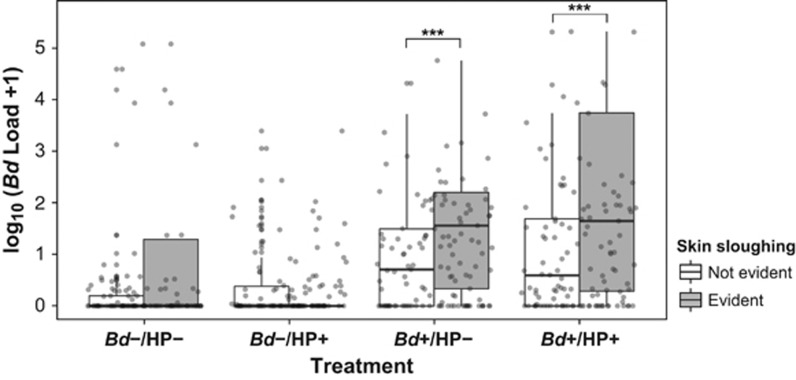
Changes in Bd infection intensity were also correlated with the magnitude of skin sloughing, which varied within treatments, seasons, and with time (all interactions P<0.04, Supplementary Figure S4; Supplementary Table S2). Consistent with the signs of chytridiomycosis, the proportion of individuals with skin sloughing was significantly higher in Bd+ treatments and during the cool-dry season (Supplementary Figure S4). However, skin sloughing did not reduce pathogen growth, because individuals with this clinical sign had significantly higher infection intensities in Bd+ treatments (P<0.001, Figure 3; Supplementary Table S2) and in particular on day 12 (β=−1.12, df=313.67, t=−3.428, P<0.001).
Figure 3.
Boxplot of log10(Bd load+1) classified by treatment and skin sloughing. Points represent the raw data. Asterisks denote significance of Tukey's pairwise comparisons from the mixed model term Treatment × Slough (Supplementary Table S2). An example of skin sloughing is shown in Supplementary Figure S2.
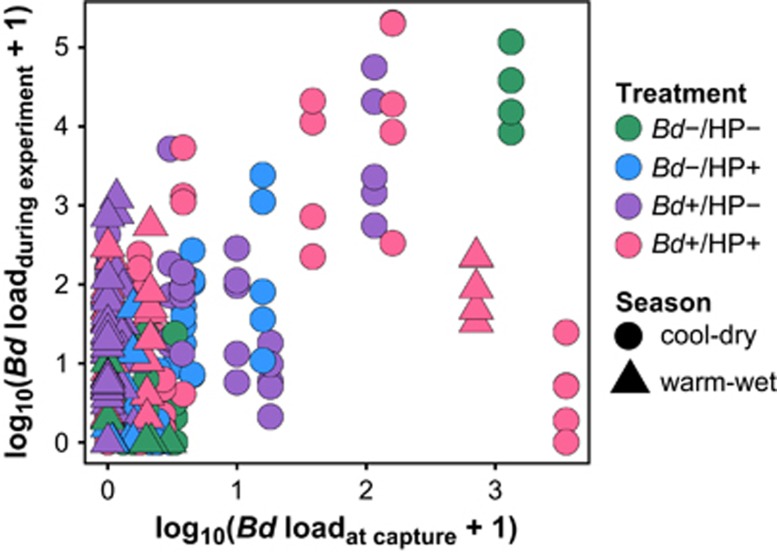
Our data showed that previous exposure to Bd was positively associated with the average Bd infection intensities during the experiment (Figure 4; Supplementary Table S2). Therefore, infection status at the beginning of the experiment predicted host responses to infection. During the warm-wet season, a significantly higher proportion of frogs cleared or reduced the infection at earlier stages of the experiment (Figure 5), indicating that hosts were favored during this season. In contrast, this advantage shifted to the pathogen during the cool-dry season, and 64% of frogs showed negative responses to infection (Figure 5).
Figure 4.
Relationship between initial Bd infection intensity (at capture) and Bd infection intensity during the experiment (GLM (initial infection intensity): β=0.76±0.12, F=77.2, t=6.316, P<0.001, Supplementary Table S2).
Figure 5.
Proportion of individual frogs categorized by their ability to recover after experimental inoculation with Bd. At the end of the experiment, an individual frog could either remain uninfected, clear or reduce the infection (positive responses) or remain infected (negative responses). Number of frogs is reported on top of the bars.
Only six individuals died during the course of the experiment (1 in warm-wet season and 5 in cool-dry season, Supplementary Table S4). All of them were infected during the experiment, including several individuals in the Bd−treatments (Supplementary Figure S3). Some natural infections also resulted in host mortality. For example, one individual from the Bd−/HP− treatment in cool-dry season died 13 days after collection (initial load: 1323 zoospores, load at death: 117 846 zoospores; Supplementary Figure S3).
Interactions between time, treatment, season, and skin sloughing also modulated alpha diversity of skin microbes
With respect to alpha diversity of skin bacterial communities, the mixed models showed significant effects of interactions between treatment*day, day*season and day*skin sloughing. Despite overall increase in OTU richness and phylogenetic diversity through time (Figures 1b and c; Supplementary Tables S5 and S6), we found delayed responses dependent on the addition of Bd. Individuals in Bd+ treatments experienced an increase in bacterial diversity three days post-infection, whereas individuals in Bd− treatments only exhibited a comparable increase after six days (Figures 1b and c). The application of both Bd and HP resulted in a higher rate of change in richness and phylogenetic diversity from day 0 to day 3 (Figures 1b and c), which stabilized after day 6. On day 3, OTU richness of individuals in the Bd+/HP− treatment tended to be lower than Bd+/HP+ treatments (Figure 1b), but the comparison fell short of significance (β=−205.15, df=373.90, t=−2.381, P=0.08). The interaction between season and day revealed a steady increase in OTU richness and phylogenetic diversity during the cool-dry season (Figures 2b and c; Supplementary Tables S5 and S6). Remarkably, skin bacterial communities returned to their initial OTU richness before inoculation only during the warm-wet season (Figure 2b, day 3 to 15: β=78.7, df=365.76, t=0.717, P=0.95). Phylogenetic diversity also showed a similar pattern from day 3 to day 15 (Figure 2c, β=−10.7, df=362.76, t=−1.11, P=0.80).
Although we did not detect a significant interaction between season and sloughing that could help explain this reduction in diversity, we found a significant effect of the interaction between day and skin sloughing on bacterial diversity (F=3.32, P=0.01, Supplementary Table S5; Supplementary Figure S5), indicating that the few individuals shedding skin significantly reduced OTU richness from day 9 to day 12. This change was mostly driven by individuals from the cool-dry season, because during the warm-wet season only one individual showed skin sloughing during day 9 (Supplementary Figure S5). In addition, phylogenetic diversity also dropped significantly from day 9 to day 12 for individuals that were experiencing skin sloughing (diff=−48.8, df=368.52, t=−2.898, P=0.03), independent of season.
Seasons and treatments differentially affected particular members of skin bacterial communities
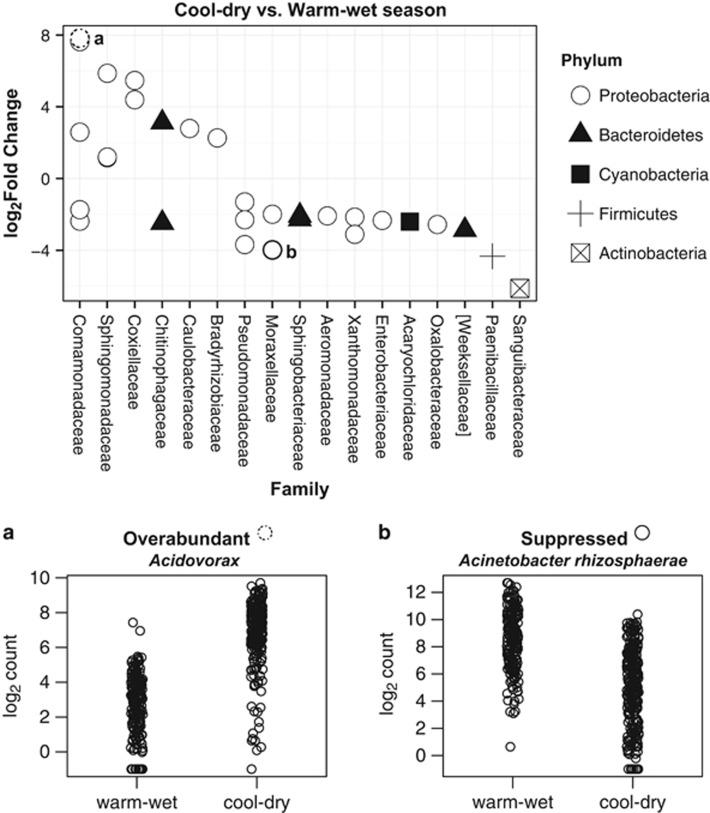
The analyses of fold change abundance of bacteria from the cool-dry season to the warm-wet season revealed strong shifts in at least 420 bacterial taxa after FDR correction (Supplementary Tables S7 and S8). In general, 241 taxa exhibited positive fold changes during the cool-dry season (above the ‘0' log2 fold change threshold), the season in which frogs carried higher Bd infection intensities (Supplementary Table S7). By ordering all OTUs with significant fold changes (241+179 OTUs) with respect to their average counts (Figure 6; Supplementary Table S8), we found that 63% of the top 30 OTUs were suppressed during the cool-dry season. For example, Acinetobacter rhizosphaerae, a coccobacilli with putative anti-Bd function (Woodhams et al., 2015), was overrepresented during the wet-warm season and decreased during the cool-dry season (Figure 6). In contrast, we detected OTUs such as Acidovorax that were abundant during the cool-dry season, but less common during the wet-warm season (Figure 6).
Figure 6.
Differential abundance of bacteria between the cool-dry season and the warm-wet season. Taxa above the zero line are significantly over abundant during the cool-dry season (a). In contrast, taxa below the zero line are significantly suppressed during the cool-dry season (b). We show two examples to illustrate the changes. Identities of each OTU can be found in Supplementary Table S7.
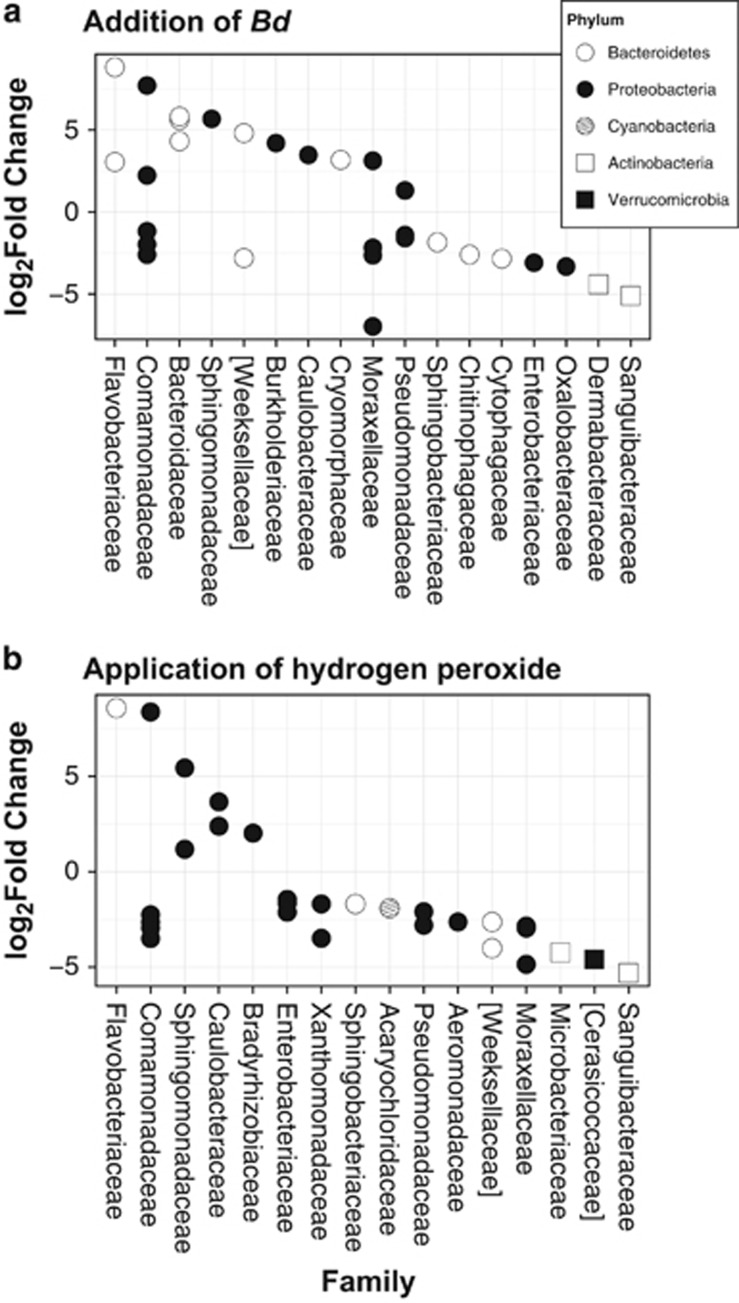
The addition of Bd changed the abundance of 82-1638 bacterial taxa, depending on whether or not season was considered (Supplementary Tables S7–S9). For the most abundant OTUs, Bd typically resulted in the increase of Proteobacteria and Bacteroidetes (Figure 7a). Myroides odoratimimus, an opportunistic pathogen member of the family Flavobacteriaceae (Maraki et al., 2012), was the most abundant bacterium enhanced by the introduction of Bd (Figure 7a; Supplementary Table S9). Interestingly, A. rhizosphaerae (family Moraxellaceae, log2 fold change −6.97) was the least common bacterial taxon overall (Figure 7a; Supplementary Table S9).
Figure 7.
Differential abundance of bacteria between (a) Bd+/HP− treatment and Bd−/HP− treatment, and (b) Bd−/HP+ treatment and Bd−/HP− treatment. Taxa above the zero line are significantly over abundant, whereas taxa below the zero line are significantly suppressed. Identities of each OTU can be found in Supplementary Table S8.
The application of HP caused an overall reduction in bacterial abundance as expected, with 999 taxa significantly suppressed compared with 754 that became over abundant (Supplementary Table S7). HP reduced 77% of the top 30 most abundant taxa (Supplementary Table S9), in particular the members of the phylum Proteobacteria (Figure 7b). Similar to the application of Bd, M. odoratimimus was the most abundant taxa followed by Acidovorax (family Comamonadaceae, Figure 7b), indicating the possible role of M. odoratimimus as an opportunistic pathogen. Finally, the addition of both Bd and HP doubled the log2 fold change of M. odoratimimus when compared with Bd or HP alone (Supplementary Table S9).
Skin bacterial community structure differed by seasons and treatments
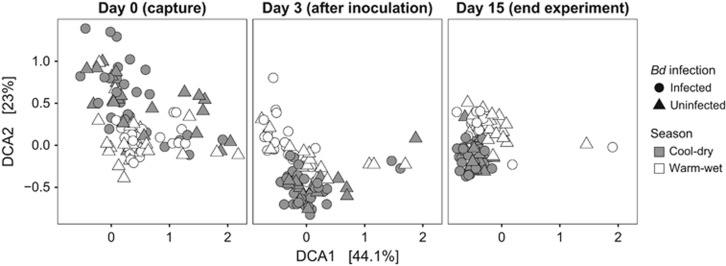
At capture (Day 0), frogs collected during the warm-wet season carried a different bacterial community structure than those collected during the cool-dry season (Figure 8, PERMANOVA Unweighted Unifrac: F1,72=4.03, R2=0.054, P<0.001; Weighted Unifrac: F1,72=8.81, R2=0.113, P<0.001; and Bray–Curtis: F1,72=5.89, R2=0.077, P<0.001). As expected, treatment, Bd infection status, and the interaction between seasons and treatments were not significant during Day 0. However, 3 days after inoculation, we detected significant changes in community structure by seasons (Unweighted Unifrac: F1,85=9.66, R2=0.096, P<0.001), treatments (F1,85=2.33, R2=0.069, P<0.001), their interaction (Unweighted Unifrac: F1,85=1.91, R2=0.056, P<0.001), but these were unrelated to Bd infection status (Unweighted Unifrac: F1,85=1.18, R2=0.012, P=0.178). The same permutational tests using weighted Unifrac and Bray–Curtis distances matrices showed similar results, thus are not presented here. By the end of the experiment, we recovered very similar patterns to Day 3, in which seasons (Unweighted Unifrac: F1,77=10.25, R2=0.114, P<0.001), treatments (F1,77=1.64, R2=0.055, P<0.001), their interaction (Unweighted Unifrac: F1,77=1.50, R2=0.050, P<0.001) still influenced bacterial community structure, but not individual Bd infection status (Unweighted Unifrac: F1,77=1.08, R2=0.012, P=0.269).
Figure 8.
Detrended correspondence analysis plot of unweighted-Unifrac distances of skin bacterial communities at three different time points (capture, 3 days after inoculation, and at the end of the experiment). To facilitate visualization of points, we show each day in a separate panel.
Discussion
By accounting for the variability in environmental conditions influencing host responses, not only our findings provide evidence that fungal diseases disrupt host health and skin bacterial communities, but also that amphibians show contrasting recovery patterns. We quantified, for the first time, the progression of fungal–bacterial interactions and how these correlated with disease outcome under natural environmental conditions, providing a novel perspective on the function of skin microbiome in amphibians. Challenging frogs with the same dose of pathogen provoked strikingly different host responses depending on environmental conditions, confirming our previous hypothesis of lability in host defenses (Longo et al., 2010, 2013). During the warm-wet season, frogs were able to control infections significantly better than during the cool-dry season (Figures 2a and 5; Supplementary Figure S4) possibly due to increased temperatures and high humidity, resulting in elevated immune function (Maniero and Carey, 1997; Raffel et al., 2006) and different symbiotic community (Figure 8). All frogs in both seasonal experiments showed an increase in bacterial diversity independent of Bd treatment. However, in the warm-wet season, this increase in bacterial richness and phylogenetic diversity was followed by a sudden drop at the end of experiment (Figures 2b and c). This finding indicates that the increase in diversity resulted from recruitment of new taxa to the skin microbiome, either through colonization of beneficial or opportunistic bacteria, or both. Only in the warm-wet season, did E. coqui frogs limit Bd, return to initial bacterial diversity found when captured, and benefit from the availability of putatively beneficial microbes such as A. rhizosphaerae (Figure 6). In contrast, as we predicted, frogs could not recover during the cool-dry season because beneficial microbes were suppressed, and the skin was colonized by putative opportunistic microbes such as Acidovorax and M. odoratimimus, which became more abundant as a secondary result of infection. Our results from the simultaneous application of both Bd and HP showed additive effects on M. odoratimimus, indicating that this OTU required special conditions to increase its abundance. Whether or not this OTU is an opportunistic pathogens of frogs still needs to be determined. Combined, our results confirm that natural fluctuations in environmental conditions shift the survival advantage from the pathogen to the host, and back again, and that this shift includes interactions with host bacterial communities.
Coping with Bd under optimal environmental conditions
As ectotherms, amphibians rely on environmental temperatures to achieve essential physiological processes including digestion, immunity, metabolism and reproduction. Our findings showed that just a slight increase in temperatures experienced during the warm-wet season provides frogs with a significant advantage that helps minimize the impact of infections (Figure 5). Our results corroborate many others that show that higher temperatures decrease the severity of infection (for example, Woodhams et al., 2008; Murphy et al., 2011; Becker et al., 2012; Rowley and Alford, 2013). In many of these cases, behavioral fever (that is, the process by which ectotherms seek areas of warmer temperatures to increase body temperatures) was the mechanism providing resistance to hosts. Behavioral fever did not occur during our experiment, because our mesocosm setting did not provide frogs the opportunity to achieve higher body temperatures and clear infection (Woodhams et al., 2003; Rowley and Alford, 2013). Instead, in our experiment the combination of higher overall environmental temperatures and downstream processes, such as recruitment of bacterial diversity and enhanced immune capacity, may have modulated the observed infection outcomes.
Host immunity and pathogen infectivity likely interacted with temperature to lower Bd infection intensities during the warm-wet season. On one hand, natural variation in temperature impacts the expression of amphibian immunity and influences susceptibility to disease (Maniero and Carey, 1997; Raffel et al., 2006). On the other hand, warmer temperatures decrease the activity period and lifespan of Bd zoospores (Woodhams et al., 2008; Voyles et al., 2012), possibly affecting their dispersal and transmission. In terms of the possible immune mechanisms increasing resistance to infection during the warm-wet season, coqui frogs lack antimicrobial peptides against Bd (Rollins-Smith et al., 2015), but they may rely on adaptive immunity, as has been shown for other frogs (Savage and Zamudio, 2011; Ellison et al., 2014; McMahon et al., 2014; Savage et al., 2016). The fact that we found a positive relationship between previous exposure and Bd infection intensity during the experiment indicates that acquired immunity may not confer full resistance to coqui frogs (Figure 4), because individuals still get infected when challenged with the pathogen at high doses. Nonetheless, previous exposure may still offer partial protection, and that protection is enhanced during the warm-wet season by both abiotic variables and beneficial symbionts, allowing for seasonal population recovery.
In our experiment, a very small percentage (four frogs, Figure 5) of frogs remained uninfected throughout the experiment, regardless of treatment (Supplementary Table S4). Once inoculated, frogs had a high probability of clearing or reducing Bd infection intensity only during the warm-wet season (Figure 5). This finding suggests that pre-inoculation status does not reflect a naive state and could be considered an honest indicator of pathogen resistance across seasons, because frogs were significantly less infected and carried lower infection intensities than during the cool-dry season. In contrast, the likelihood of showing positive responses after infection decreased by half (from 74% in the warm-wet season to 31% in the cool-dry season, Figure 5), revealing that previously infected individuals lost any potential immune protection when environmental conditions became cooler and drier. Our data suggest that host immune function may be at maximum performance during the warm-wet season because frogs were able to reduce Bd, OTU richness and phylogenetic diversity in a period of 2 weeks. This result corroborates earlier findings that some amphibian immune factors might become ineffective at cooler temperatures (Maniero and Carey, 1997; Ribas et al., 2009). Additional research on the identity of immune factors, and exactly how they are environmentally mediated, is clearly necessary.
Advantages of diverse skin microbiota during the warm-wet season
During the warm-wet season, harboring a diverse pool of putatively beneficial taxa may offer frogs an additional advantage to decrease infection intensity. Frogs at this time point had a different community structure than frogs collected during the cool-dry season (Figure 8). Our data demonstrated that many OTUs from families Xanthomonadaceae, Oxalobacteraceae, Pseudomonadaceae, among others, were significantly more abundant during the warm-wet season (Figure 6). However, during the cool-dry season some of these bacterial taxa remained present, albeit at lower levels, as part of the normal microbiota (Figure 6, Supplementary Table S6). One such example is A. rhizosphaerae, an OTU that was suppressed during the cool-dry season in our experiment (Figure 8). Non-susceptible species have bacterial communities enriched with Acinetobacter (Rebollar et al., 2016). Over the last few years, researchers have tested 27 isolates of A. rhizosphaerae against Bd, yet only 60% of them were able to inhibit the pathogen (Woodhams et al., 2015). By testing candidate probiotics against several Bd isolates, Antwis et al. (2015) demonstrated that only a small proportion of bacteria have a broad-spectrum capacity. Thus, our results extend these recent findings by indicating that some candidate probiotics such as A. rhizosphaerae might also be inefficient at lower temperatures (Daskin et al., 2014).
In vitro and in vivo antifungal activity of skin bacteria should differ as a consequence of environmental conditions and host immune factors. So far, most cases of in vitro anti-Bd activity has been quantified using bacterial metabolites and cell factors found in cell-free supernatants at a single temperature (Bell et al., 2013). However, the quantity and identity of these metabolites may depend on temperature and bacterial density (Daskin et al., 2014; Yasumiba et al., 2016). Therefore, understanding environmental limits to Bd inhibition will be important if we are to consider any bacteria as potential probiotics for conservation management.
Skin sloughing as a tolerance strategy under suboptimal environmental conditions
Our results demonstrated that frogs could not limit Bd infections or colonization by new bacterial taxa during the cool-dry season (Figure 2). Nonetheless, only 5 of the 50 individuals in the experiment died during this season (Supplementary Table S4) and most frogs persisted despite higher infections (Supplementary Figure S3). Recent studies have demonstrated that increased skin sloughing does not have a measurable effect on Bd infection intensity in Australian green tree frogs (Ohmer et al., 2014), yet reduces the amount of culturable microbes (Meyer et al., 2012; Cramp et al., 2014), and is more common at higher temperatures (Cramp et al., 2014). Taken together, these results suggest a potential adaptive strategy for reducing opportunistic bacteria and other pathogens.
Our findings significantly differ from those studies in that we detected higher skin sloughing in the cool-dry season (Supplementary Figure S4). In addition, sloughing in coqui frogs did not measurably reduce overall Bd infection intensity and bacterial diversity (Figure 4; Supplementary Figure S5). The skin sloughing that we document here is not a natural skin shedding process, but part of the pathology associated with chytridiomycosis (Voyles et al., 2009). Whether sloughing was a cause or a consequence of Bd infection dynamics still needs to be determined. Because skin sloughing is a costly potential mechanism of defense, and we saw no reduction in pathogen load, we hypothesize that this mechanism contributes to tolerance in a different way. If sloughing in coqui frogs serves to maintain infections at bay for a fixed amount of time (cool-dry season), then this strategy could protect the host during a time that immune responses are slow, ineffective, or potentially damaging (Ellison et al., 2015). Sloughing should be further explored using microscopy or molecular techniques to determine the viability of bacteria and Bd. In this way, we will be able to understand the functional consequences and the timing of this response to infection.
Conclusions
After a pathogen invades naive hosts, interacting species need to find a balance to persist. Limiting the severity of the damage caused by a pathogen, rather than eliminating them, is an advantageous strategy (Roy and Kirchner, 2000) not only for the host but also for the pathogen. In this paper, we described how a frog species, that persists in populations where Bd is enzootic, survives despite relatively high infection prevalence and intensity. Our results indicate that when the environment offers a survival advantage to the host, both increased temperatures and host-associated bacteria contribute to resistance. In contrast, during the cool-dry season, the environment favors the pathogen, thus frogs shift their defense strategy to tolerance. We propose that an important avenue for future research will be to obtain comparative and quantitative data on defense strategies that allow other species to persist with this infectious disease. Quantifying defense mechanisms and identifying the ones that provide the highest fitness benefit can significantly improve our management strategies for the reintroduction of susceptible and threatened species.
Acknowledgments
We thank AE Ellison, MM Gray and C McDonald for harvesting and shipping Bd zoospores, and help in the lab. AL López-Torres, D Rodriguez, BB Johnson, CA Rodríguez-Gómez, M Longo, D Cancel and M Rodríguez-Medina provided invaluable help in the field by capturing frogs or by assisting during the mesocosm experiment. We also thank PA Burrowes and RL Joglar for access to the lab at the University of Puerto Rico. LM Johnson (Cornell University Statistical Consulting Unit) helped develop the regression models for this study. The Lips Lab at University of Maryland, I Hewson at Cornell University and three anonymous referees provided valuable comments and edits on earlier versions of this manuscript. Grants from the National Science Foundation Evolutionary Processes (DEB-1120249 to KRZ) and Doctoral Dissertation Improvement (DEB-1310036 to KRZ and AVL) programs provided support for this study. AVL was supported by the following fellowships and grants: Ford Foundation Predoctoral Fellowship, Cornell University (CU) SUNY/Sage Diversity Fellowships, CU Department of Ecology and Evolutionary Biology Grants, CU Graduate School Travel Grant, Andrew W. Mellon Graduate Student Grants and Atkinson's Center for Sustainable Biodiversity Fund.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Antwis RE, Preziosi RF, Harrison XA, Garner TW. (2015). Amphibian symbiotic bacteria do not show universal ability to inhibit growth of the global pandemic lineage of Batrachochytrium dendrobatidis. Appl Environ Microbiol 81: 3706–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S. (2015). Fitting linear mixed-effects models using lme4. J Stat Softw 67: 1–48. [Google Scholar]
- Beard KH, McCullough S, Eschtruth AK. (2003). Quantitative assessment of habitat preferences for the Puerto Rican terrestrial frog, Eleutherodactylus coqui. J Herpetol 37: 10–17. [Google Scholar]
- Becker CG, Rodriguez D, Longo AV, Talaba AL, Zamudio KR. (2012). Disease risk in temperate amphibian populations is higher at closed-canopy sites. PLoS One 7: e48205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker MH, Harris RN. (2010). Cutaneous bacteria of the redback salamander prevent morbidity associated with a lethal disease. PLoS One 5: e10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker MH, Richards-Zawacki CL, Gratwicke B, Belden LK. (2014). The effect of captivity on the cutaneous bacterial community of the critically endangered Panamanian golden frog (Atelopus zeteki. Biol Conserv 176: 199–206. [Google Scholar]
- Becker MH, Walke JB, Cikanek S, Savage AE, Mattheus N, Santiago CN et al. (2015). Composition of symbiotic bacteria predicts survival in Panamanian golden frogs infected with a lethal fungus. Proc R Soc B 282: 20142881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SC, Alford RA, Garland S, Padilla G, Thomas AD. (2013). Screening bacterial metabolites for inhibitory effects against Batrachochytrium dendrobatidis using a spectrophotometric assay. Dis Aquat Org 103: 77–85. [DOI] [PubMed] [Google Scholar]
- Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10: 57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle DG, Boyle DB, Olsen V, Morgan JA, Hyatt AD. (2004). Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis in amphibian samples using real-time Taqman PCR assay. Dis Aquat Org 60: 141–148. [DOI] [PubMed] [Google Scholar]
- Brucker RM, Harris RN, Schwantes CR, Gallaher TN, Flaherty DC, Lam BA et al. (2008). Amphibian chemical defense: antifungal metabolites of the microsymbiont Janthinobacterium lividum on the salamander Plethodon cinereus. J Chem Ecol 34: 1422–1429. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. (2010a). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010b). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6: 1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramp RL, McPhee RK, Meyer EA, Ohmer ME, Franklin CE. (2014). First line of defence: the role of sloughing in the regulation of cutaneous microbes in frogs. Conserv Physiol 2: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskin JH, Bell SC, Schwarzkopf L, Alford RA. (2014). Cool temperatures reduce antifungal activity of symbiotic bacteria of threatened amphibians—implications for disease management and patterns of decline. PLoS One 9: e100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Ellison AR, Savage AE, DiRenzo GV, Langhammer P, Lips KR, Zamudio KR. (2014). Fighting a losing battle: vigorous immune response countered by pathogen suppression of host defenses in the chytridiomycosis-susceptible frog Atelopus zeteki. G3 4: 1275–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison AR, Tunstall T, DiRenzo GV, Hughey MC, Rebollar EA, Belden LK et al. (2015). More than skin deep: functional genomic basis for resistance to amphibian chytridiomycosis. Genome Biol Evol 7: 286–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL et al. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484: 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks A, Egan S, Holmström C, James S, Lappin-Scott H, Kjelleberg S. (2006). Inhibition of fungal colonization by Pseudoalteromonas tunicata provides a competitive advantage during surface colonization. Appl Environ Microbiol 72: 6079–6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan SE, Longcore JE, Calhoun AJ. (2012). Host invasion by Batrachochytrium dendrobatidis: fungal and epidermal ultrastructure in model anurans. Dis Aquat Org 100: 201–210. [DOI] [PubMed] [Google Scholar]
- Jani AJ, Briggs CJ. (2014). The pathogen Batrachochytrium dendrobatidis disturbs the frog skin microbiome during a natural epidemic and experimental infection. Proc Natl Acad Sci USA 111: E5049–E5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer A, Simon MA, Banning JL, Andre E, Duncan K, Harris RN. (2007). Common cutaneous bacteria from the eastern red-backed salamander can inhibit pathogenic fungi. Copeia, 630–640.
- Lenth RV, Hervé M. (2016). Least-squares means: The R package lsmeans. J Stat Softw 69: 1–33. [Google Scholar]
- Longo AV, Burrowes PA, Joglar RL. (2010). Seasonality of Batrachochytrium dendrobatidis infection in direct-developing frogs suggests a mechanism for persistence. Dis Aquat Org 92: 253–260. [DOI] [PubMed] [Google Scholar]
- Longo AV, Ossiboff RJ, Zamudio KR, Burrowes PA. (2013). Lability in host defenses: terrestrial frogs die from chytridiomycosis under enzootic conditions. J Wildl Dis 49: 197–199. [DOI] [PubMed] [Google Scholar]
- Longo AV, Savage AE, Hewson I, Zamudio KR. (2015). Seasonal and ontogenetic variation of skin microbial communities and relationships to natural disease dynamics in declining amphibians. R Soc Open Sci 2: 140377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudon AH, Woodhams DC, Parfrey LW, Archer H, Knight R, McKenzie V et al. (2013). Microbial community dynamics and effect of environmental microbial reservoirs on red-backed salamanders (Plethodon cinereus. ISME J 8: 830–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber T, Anders HJ. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniero GD, Carey C. (1997). Changes in selected aspects of immune function in the leopard frog, Rana pipiens, associated with exposure to cold. J Comp Physiol B 167: 256–263. [DOI] [PubMed] [Google Scholar]
- Maraki S, Sarchianaki E, Barbagadakis S. (2012). Myroides odoratimimus soft tissue infection in an immunocompetent child following a pig bite: case report and literature review. Braz J Infect Dis 16: 390–392. [DOI] [PubMed] [Google Scholar]
- McMahon TA, Sears BF, Venesky MD, Bessler SM, Brown JM, Deutsch K et al. (2014). Amphibians acquire resistance to live and dead fungus overcoming fungal immunosuppression. Nature 511: 224–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes EC. (2013). phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8: e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. (2014). Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10: e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EA, Cramp RL, Bernal MH, Franklin CE. (2012). Changes in cutaneous microbial abundance with sloughing: possible implications for infection and disease in amphibians. Dis Aquat Organ 101: 235–242. [DOI] [PubMed] [Google Scholar]
- Muletz CR, Myers JM, Domangue RJ, Herrick JB, Harris RN. (2012). Soil bioaugmentation with amphibian cutaneous bacteria protects amphibian hosts from infection by Batrachochytrium dendrobatidis. Biol Conserv 152: 119–126. [Google Scholar]
- Murphy PJ, St-Hilaire S, Corn PS. (2011). Temperature, hydric environment, and prior pathogen exposure alter the experimental severity of chytridiomycosis in boreal toads. Dis Aquat Organ 95: 31–42. [DOI] [PubMed] [Google Scholar]
- Ohmer MEB, Cramp RL, White CR, Franklin CE. (2014). Skin sloughing rate increases with chytrid fungus infection load in a susceptible amphibian. Funct Ecol 29: 674–682. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB et al. (2015) vegan: Community Ecology Package, R package version 22-1. Available at: https://CRAN.R-project.org/package=vegan.
- R Core Team. (2014) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- Raffel TR, Rohr JR, Kiesecker JM, Hudson PJ. (2006). Negative effects of changing temperature on amphibian immunity under field conditions. Funct Ecol 20: 819–828. [Google Scholar]
- Rebollar EA, Hughey MC, Medina D, Harris RN, Ibanez R, Belden LK. (2016). Skin bacterial diversity of Panamanian frogs is associated with host susceptibility and presence of Batrachochytrium dendrobatidis. ISME J 10: 1682–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas L, Li MS, Doddington BJ, Robert J, Seidel JA, Kroll JS et al. (2009). Expression profiling the temperature-dependent amphibian response to infection by Batrachochytrium dendrobatidis. PLoS One 4: e8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout JR, He Y, Navas-Molina JA, Walters WA, Ursell LK, Gibbons SM et al. (2014). Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2: e545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins-Smith LA, Reinert LK, Burrowes PA. (2015). Coqui frogs persist with the deadly chytrid fungus despite a lack of defensive antimicrobial peptides. Dis Aquat Org 113: 81–83. [DOI] [PubMed] [Google Scholar]
- Rowley JJL, Alford RA. (2013). Hot bodies protect amphibians against chytrid infection in nature. Sci Rep 3: 1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy BA, Kirchner JW. (2000). Evolutionary dynamics of pathogen resistance and tolerance. Evolution 54: 51–63. [DOI] [PubMed] [Google Scholar]
- Savage AE, Zamudio KR. (2011). MHC genotypes associate with resistance to a frog-killing fungus. Proc Natl Acad Sci USA 108: 16705–16710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage AE, Terrell KA, Gratwicke B, Mattheus NM, Augustine L, Fleischer RC. (2016). Reduced immune function predicts disease susceptibility in frogs infected with a deadly fungal pathogen. Conserv Physiol 4: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voyles J, Young S, Berger L, Campbell C, Voyles WF, Dinudom A et al. (2009). Pathogenesis of chytridiomycosis, a cause of catastrophic amphibian declines. Science 326: 582–585. [DOI] [PubMed] [Google Scholar]
- Voyles J, Johnson LR, Briggs CJ, Cashins SD, Alford RA, Berger L et al. (2012). Temperature alters reproductive life history patterns in Batrachochytrium dendrobatidis, a lethal pathogen associated with the global loss of amphibians. Ecol Evol 2: 2241–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walke JB, Becker MH, Loftus SC, House LL, Cormier G, Jensen RV et al. (2014). Amphibian skin may select for rare environmental microbes. ISME J 8: 2207–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walke JB, Becker MH, Loftus SC, House LL, Teotonio TL, Minbiole KPC et al. (2015). Community structure and function of amphibian skin microbes: an experiment with bullfrogs exposed to a chytrid fungus. PLoS One 10: e0139848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo MJ, Hogan DA. (2006). Fungal–bacterial interactions: a mixed bag of mingling microbes. Curr Opin Microbiol 9: 359–364. [DOI] [PubMed] [Google Scholar]
- Woodhams DC, Alford RA, Marantelli G. (2003). Emerging disease of amphibians cured by elevated body temperature. Dis Aquat Org 55: 65–67. [DOI] [PubMed] [Google Scholar]
- Woodhams DC, Alford RA, Briggs CJ, Johnson M, Rollins-Smith LA. (2008). Life-history trade-offs influence disease in changing climates: strategies of an amphibian pathogen. Ecology 89: 1627–1639. [DOI] [PubMed] [Google Scholar]
- Woodhams DC, Alford RA, Antwis RE, Archer H, Becker MH, Belden LK et al. (2015). Antifungal isolates database of amphibian skin-associated bacteria and function against emerging fungal pathogens. Ecology 96: 595–595. [Google Scholar]
- Yasumiba K, Bell S, Alford R. (2016). Cell density effects of frog skin bacteria on their capacity to inhibit growth of the chytrid fungus, Batrachochytrium dendrobatidis. Microb Ecol 71: 124–130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.