Version Changes
Revised. Amendments from Version 1
Z-factor estimation of the assay is added. Sources for obtaining experimental materials, vectors and antibody, are indicated. Figure 6 and Supplementary Figure 2 are re-organised. Datasets for figures 1-9 and supplementary figure were converted to Excel file format.
Abstract
The DYRK1A (dual specificity tyrosine phosphorylation-regulated kinase 1A) gene encodes a proline-directed Ser/Thr kinase. Elevated expression and/or altered distribution of the kinase have been implicated in the neurological impairments associated with Down syndrome (DS) and Alzheimer’s disease (AD). Consequently, DYRK1A inhibition has been of significant interest as a potential strategy for therapeutic intervention of DS and AD. Many classes of novel inhibitors have been described in the past decade. Although non-radioactive methods for analyzing DYRK1A inhibition have been developed, methods employing radioactive tracers are still commonly used for quantitative characterization of DYRK1A inhibitors. Here, we present a non-radioactive ELISA assay based on the detection of DYRK1A-phosphorylated dynamin 1a fragment using a phosphorylation site-specific antibody. The assay was verified by the use of two well-characterized DYRK1A inhibitors, epigallocatechin gallate (EGCG) and harmine. The IC 50s for EGCG and harmine determined by the ELISA method were found to be comparable to those previously measured by radioactive tracing methods. Furthermore, we determined the mode of inhibition for EGCG and harmine by a modification of the ELISA assay. This assay confirms the mode of inhibition of EGCG (non-ATP-competitive) and harmine (ATP-competitive), as previously determined. We conclude that the ELISA platform demonstrated here is a viable alternative to the traditional radioactive tracer assays for analyzing DYRK1A inhibitors.
Keywords: non-radioactive kinase assay, EGCG, harmine, inhibitor screening
Introduction
The human DYRK1A gene 1 is mapped to a region of chromosome 21 implicated in Down syndrome (DS) 2. DS, the most common chromosomal abnormality associated with birth defects and developmental disabilities, is caused by full or partial trisomy of chromosome 21 3. Almost all DS cases inevitably lead to the development of Alzheimer’s disease (AD)-type pathology 4. Transgenic mice carrying an extra copy of DYRK1A have been shown to exhibit symptoms similar to DS, including brain abnormalities, neurodevelopmental delay, and memory impairments 5– 7. DYRK1A in the brain displays a distinct structure-specific distribution pattern 8, and it interacts with an array of factors involved in neuronal development, proliferation, and differentiation 9. The level of DYRK1A is elevated in a gene dosage-dependent manner in DS, suggesting that the protein not only plays an important role in regulating normal brain functions, but also in the etiology of DS 8, 10.
DYRK1A has been linked to neurofibrillary degeneration and β-amyloidosis of AD 11. DYRK1A was shown to phosphorylate microtubule-associated protein tau at T212 to prime tau for subsequent phosphorylation by GSK-3β at S308 12, 13. This inhibits tau’s ability to stimulate microtubule assembly and promotes self-aggregation, like abnormally hyperphosphorylated tau in AD brain 13. DYRK1A was also found to phosphorylate amyloid precursor protein (APP) at T668 14 and presenilin-1 at T354 15, which correlated with increased cleavage of APP by β and γ secretases 15, 16 respectively, and leads to the formation of neurotoxic β-amyloid peptides (Aβ). Moreover, Aβ is shown to be involved in a positive feedback loop for promoting DYRK1A expression, which may further accelerate production of Aβ 17.
The collected evidence suggests that DYRK1A is a potential drug target for the treatment of DS and AD. To this end, many classes of DYRK1A inhibitors, both natural and synthetic, have been tested 18– 20. The potency of such inhibitors has mostly been analyzed using radioactive tracer methods despite the availability of non-radioactive assays 21, 22. It may be that these methods typically require multiple steps, which is undesirable for screening. Here, we describe a simple ELISA assay for DYRK1A inhibition using dynamin 1a and its phosphorylation site antibody for detection 23, 24. The ELISA assay has been verified using two known DYRK1A inhibitors and found to be consistent with radioactive tracer methods.
Methods
Materials
Epigallocatechin gallate (EGCG; #70935) and harmine (#10010324) were obtained from Cayman Chemical. Para-nitrophenyl phosphate (PNPP) tablets and diethanolamine substrate buffer were purchased from Thermo-Fisher Scientific. EGCG and harmine were initially prepared as 50 mM stock in 100% DMSO. Working solutions of EGCG and harmine (0.01 μM – 3.2 μM) were prepared from stocks in 2% DMSO by serial dilution. Dynamin 1a pS857-specific mouse mAb 3D3 (RRID: AB_2631263, the antibody has been deposited with the Developmental Studies of Hybridoma Bank) was prepared as described 24. The antibody was partially purified from ascites using Bakerbond ABx resins (#7269-02) before use. Anti-dynamin mAb Hudy-1 (RRID: AB_309677) 25 was obtained from EMD Millipore. Alkaline phosphatase (AP) conjugated goat anti-mouse IgG secondary antibody (#115-055-146) was purchased from Jackson ImmunoResearch Laboratories, Inc.
Kinase and substrate preparation
6xHis tagged rat truncated DYRK1A containing residues 1-497 (HT-497) was used for all assays. This truncation preserves activity of DYRK1A 26, 27. A bacterial HT-497 expression vector was constructed as follows. The truncated DYRK1A gene was first obtained by PCR from the GST-DYRK1A vector 23 using a pair of primers for producing the DYRK1A fragment with a Cla I site plus a 6XHis tag (cctatcgatgcatcatcatcatcatcaccatacaggaggagagacttc) at the start codon and an in-frame termination at codon 498 plus a Xho I site (ggactcgagtcaagggctggtggacacactgtt), respectively, at 5’ and 3’ ends. PCR was performed in 50 μl mixture containing 10 ng template, 0.2 μg of each primer, 0.2 mM ATP, and PfuUltra (Agilent Technologies), as recommended by the supplier. Amplification was conducted with 20 cycles of the following steps: 94°C, 30-sec, 72°C 90-sec, and 62°C 30-sec. Custom primers were purchased from Integrated DNA Technologies. The resulting amplicon was then cloned into a modified T7 promoter-driven vector pND1 28 via the Cla I and Xho I sites, as described 29. Proline rich domain (PRD, residues 746–864) of dynamin 1a was also prepared as N-terminal tagged 6xHis fusion protein (HT-PRD) exactly as described above. The PRD fragment was first produced from the semi-synthetic dynamin 1a gene 24 by PCR using a pair of primers, (aggatcgatgcatcatcatcatcatcataacacgaccaccgtcagcacg) and (aggctcgagtcataggtcaaaaggtggtcg) for subsequent cloning into expression vector pND1, like HT-497. It should be noted that the HT-PRD construct carries an inadvertent F862L substitution. Both pHT-497 and pHT-PRD have been deposited with Addgene.
Both HT-497 and HT-PRD were expressed and purified using TALON metal affinity resin (Clontech Laboratories) under native conditions as described 29. Proteins were quantified by Bradford method 30 and stored at -80°C until use.
ELISA assay
Substrate, HT-PRD, was diluted in dilution buffer (25 mM Tris-HCl, pH 7.4 and 100 mM NaCl) to a concentration of 2 ng/μl (or higher as in Figure 1 and Figure 2) and used to coat a 96-well plate (BD Falcon #353072) with 100 μl per well (200 ng/well unless otherwise indicated) at 4°C overnight. Unbound materials were washed away with dilution buffer and wells were blocked with 150 μl blocking buffer (2% BSA, 1X PBS, and 0.25% Tween 20) at room temperature for 60 min. After blocking, wells were washed extensively with dilution buffer before subjecting to phosphorylation. DYRK1A phosphorylation was performed in wells with 100 μl reaction mix containing 25 mM HEPES, pH7.4, 100 mM NaCl, 5 mM MgCl 2, 100 μM ATP (Sigma-Aldrich Chemicals), inhibitor if needed, and 5 ng HT-497 (unless otherwise indicated). Reactions were initiated by adding HT-497 and continued for 30 min (unless otherwise indicated) at 30°C. For time course experiments, reactions were terminated by the addition of 20 mM EDTA at the indicated time points. A set of inhibition experiments typically consists of a no-inhibitor control plus a series of eight inhibitor concentrations (0.001 μM - 3.2 μM final). Each point was run in duplicate with DMSO present in all assays at 0.2% final concentration. DMSO, up to 2%, does not affect the potency of EGCG and harmine. HT-PRD phosphorylation was subsequently determined by the sandwich antibody staining protocol, first with 100 μl mAb (60 min at room temperature) then with 100 μl AP-linked anti-mouse secondary antibody (60 min at room temperature), followed by colorimetric reaction with 100 μl PNPP solution. The extent of AP reaction was monitored at λ=405 nm. For Hudy-1 staining, wells were coated, blocked, and then stained with the antibody (1:3000 dilution) for colorimetric detection as described above.
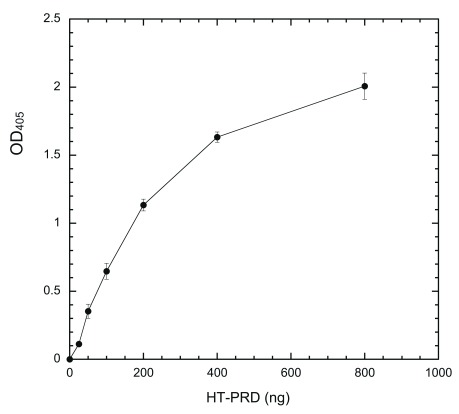
Figure 1. Coating ELISA plate with HT-PRD.
Wells were incubated with indicated amounts of HT-PRD (0, 25, 50, 100, 200, 400, and 800 ng/well) at 4°C overnight and the level of coated proteins was then detected with anti-dynamin mAb Hudy-1 by following the sandwich ELISA protocol, as described in Methods (n = 4 for each data point).
Dilution factors for both mAb 3D3 and secondary antibody were pre-determined for each batch of antibody to ensure that neither antibody was limiting in the assay. A stock to be determined was serially diluted (from 1000 to 256,000-fold) and each dilution was used together with a non-limiting concentration of the other antibody to assess the level of HT-PRD phosphorylated under standard ELISA reaction conditions without inhibitor (see Results and Discussion). OD 405 readings were normalized to the 1000-fold dilution and plotted against the dilutions of the testing antibody. Dilutions in the normalized OD 405 plateau can be used for the assay. We routinely use 1:3000 dilutions for ABx purified 3D3 stock (~1.5 mg/ml) and 1:2000 dilutions of commercial secondary antibody for the assay.
Data analysis
Data transformation, calculation, plotting, curve fitting, and IC 50 calculation were performed in KaleidaGraph ( http://www.synergy.com/wordpress_650164087; Mac version 4.1). Data was corrected for background (readings from wells with only PNPP) before subsequent manipulations. To determine IC 50, the residual DYRK1A activity was first calculated as the ratio to the no-inhibitor control in that set. The resulting residual activity was then plotted against its corresponding inhibitor concentrations in semi-log graph and the plot was fit to the sigmoidal equation, y = a+(b-a)/(1+(x/c) d), for IC 50 calculation. Data throughout the article are presented as mean ± standard error of mean.
ATP competition assay
The standard ELISA protocol was modified to run under conditions allowing a constant inhibitor to compete against varying ATP concentrations in inhibiting DYRK1A. Briefly, a set of competition experiments had four DYRK1A assays in the presence of different ATP concentrations (100, 200, 400, or 800 μM) with a single fixed concentration of the inhibitor to be tested. An identical set, except without inhibitor, was performed in parallel (no-inhibitor controls). The inhibitor concentration used was roughly twice the IC 50 of the inhibitor. All other procedures of the assay are unchanged. Residual kinase activity with the inhibitor at each ATP concentration was first calculated as a percentage of the corresponding no-inhibitor control. The residual kinase activity was subsequently converted to inhibition potency as the difference from 1. The value for each ATP concentration was then normalized to the inhibition potency at 100 μM ATP and plotted.
Results and discussion
Development of the non-radioactive DYRK1A ELISA assay
We chose to follow the ELISA-based protocol 31, 32 in developing our assay, by immobilizing the substrate followed by kinase phosphorylation in the wells, as this format offers the advantage of a simple, proven design versus other non-radioactive approaches 33. Like many non-radioactive approaches 33, our assay relies on a phospho-specific antibody to differentiate between phosphorylated from un-phosphorylated substrates. The antibody used in the assay, mAb 3D3, was raised against DYRK1A-phosphorylated Dynatide 3; a peptide derived from the DYRK1A phosphorylation site of dynamin 1a at S857 24. 3D3 has been shown to recognize only pS857-dynamin 1a in rat brain extracts upon extensive phosphorylation 24.
Dynatide 3, which is used routinely as a substrate to measure DYRK1A activity by the radioisotope/filter binding method 27, 34, 35, was first tested as a substrate. Unfortunately, it failed to produce any signal upon phosphorylating coated Dynatide 3 with DYRK1A, presumably due to a lack of peptide coating. To circumvent this problem, we used a 6X histidine-tagged PRD of dynamin 1a (HT-PRD) as a DYRK1A substrate. This fragment coats wells in a concentration-dependent manner and the amount of immobilized proteins, as revealed by mAb Hudy-1 staining, are proportional to input proteins up to 200 ng/well of HT-PRD (15 pmole/well) ( Figure 1).
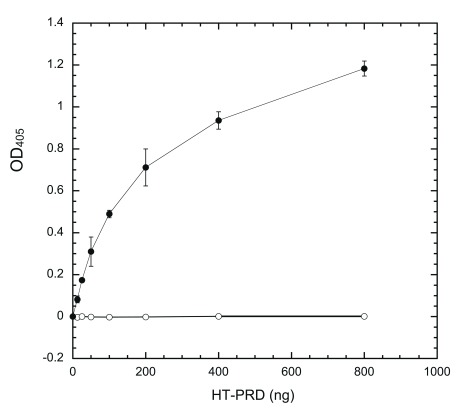
We then examined whether immobilized HT-PRD is accessible to DYRK1A. Wells coated with varying amounts of HT-PRD were subjected to exhaustive phosphorylation in situ with excess DYRK1A (HT-497) (see Methods) for 60 min and probed with excess (non-limiting) mAb 3D3 and secondary antibodies (see below in Figure 5). Phosphorylated immobilized HT-PRD was recognized by 3D3. The 3D3 signal was elevated in response to increasing input of HT-PRD ( Figure 2, filled circles) initially, then leveled off, closely resembling the response of substrate coating ( Figure 1). As controls, uncoated wells phosphorylated by HT-497 ( Figure 1) and coated HT-PRD, processed without HT-497, produced no detectable signals ( Figure 2, empty circles). These results indicate that immobilized HT-PRD is phosphorylatable by DYRK1A and that the output of the assay requires DYRK1A phosphorylation.
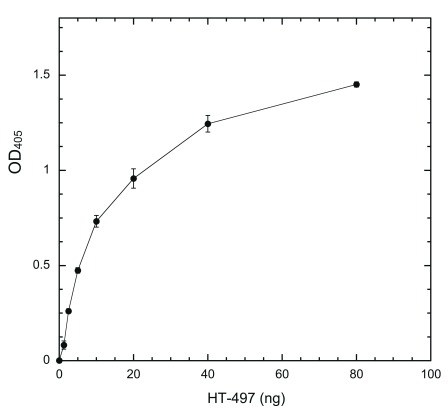
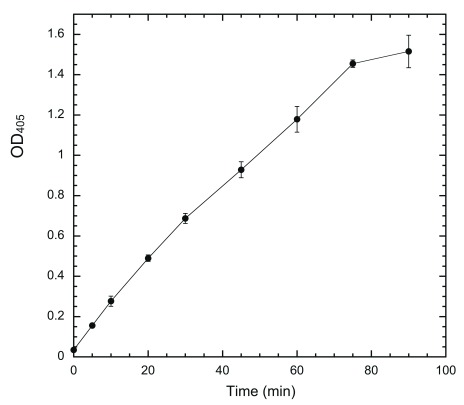
If a system is to be useful in determining inhibitor potency quantitatively, the output of the system must be solely dependent on DYRK1A activity in a linear fashion. We used a fixed amount of coated HT-PRD (200 ng/well) to identify the proper conditions. The system response to changes of HT-497 was first examined ( Figure 3). Our ELISA system produces sufficient signal to be readily distinguished from the noise of no-kinase control, with ~1 ng HT-497 (~17 fmole) phosphorylation at 30°C for 30 min. The output (the equivalent of reaction rate) is elevated accordingly as enzyme concentration increased, but the ratio of elevation to enzyme concentration, in proportion to enzyme, is progressively reduced ( Figure 3). This is a typical enzyme concentration-dependent reaction profile when the substrate becomes the limiting factor 36. Time-course experiments were subsequently conducted with 5 ng HT-497, as the highest enzyme concentration producing a near-linear enzyme-dependent response. The output was found to be linear with reaction times up to about 75 min ( Figure 4). Therefore, we use the following standard conditions [200 ng of substrate, 5 ng HT-497 (0.82 nM), 100 μM ATP, and 30 min kinase reaction at 30°C] for all subsequent experiments. The Z’-factor for the assay performed under standard conditions was estimated and found to be greater than 0.7 ( Supplementary Table).
Figure 2. Phosphorylation of coated HT-PRD by DYRK1A.
Wells were coated with indicated amounts of HT-PRD (0, 25, 50, 100, 200, 400, and 800 ng/well) and then subjected to extensive DYRK1A phosphorylation in situ by incubation with 80 ng of HT-497 at 30°C for 60 min. The level of S857 phosphorylation was then detected with 3D3 following the sandwich ELISA protocol, as described in Methods (n = 4 for each data point). Filled circles (●), with kinase; empty circles (○), without kinase.
Figure 3. DYRK1A concentration-dependent phosphorylation of coated HT-PRD.
Wells were coated with 200 ng/well HT-PRD and then subjected to DYRK1A phosphorylation with varying amounts of HT-497 (1.25, 2.5, 5, 10, 20, 40, and 80 ng) at 30°C for 30 min. The level of S857 phosphorylation was then detected with 3D3 as described in Methods (n = 6 for each data point).
Figure 4. Time-course phosphorylation of coated HT-PRD by DYRK1A.
Wells were coated with 200 ng/well of HT-PRD and then subjected to DYRK1A phosphorylation with 5 ng HT-497 at 30°C. The reactions were terminated at the indicated time points (0, 5, 10, 20, 30, 45, 60, 75, and 90 min) by the addition of 20 mM EDTA. The level of S857 phosphorylation was then detected with 3D3 as described (n = 3 for each data point).
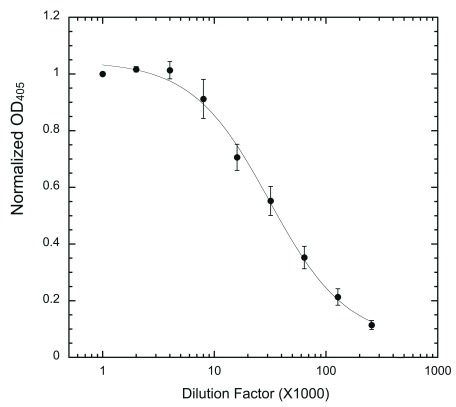
Figure 5. 3D3 dilution factor determination.
Wells were coated with 200 ng/well HT-PRD and then subjected to phosphorylation with 5 ng HT-497 under the standard reaction conditions. 3D3 to be tested was serially diluted (from 1000 to 256,000x) and used to probe the phosphorylated wells, followed by secondary antibody as described. Normalized OD 405 was calculated (see Methods) and used for plotting (n = 9 for each data point).
To support accurate measurement of IC 50, the amounts of antibody, both 3D3 and secondary antibody, must not be limiting. Otherwise, immunostaining will most likely under-report the actual phosphorylation level at lower concentrations of inhibitor, which could skew the IC 50 calculation. Therefore, each batch of antibody was titered to determine the maximal dilution can be used. As shown for titering of 3D3, when the antibody is limiting (provided that a non-limiting concentration of secondary antibody is used), the readout will increase upon addition of 3D3 until a plateau indicating saturation ( Figure 5). Only dilutions that produce readout in the plateau (non-limiting) region should be used for the assay ( Figure 5). Dilution factors for the secondary antibody were similarly determined ( Supplementary Figure).
Measuring IC 50 and the mode of inhibition for DYRK1A inhibitors
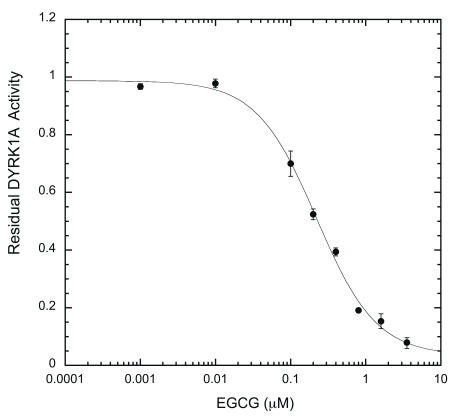
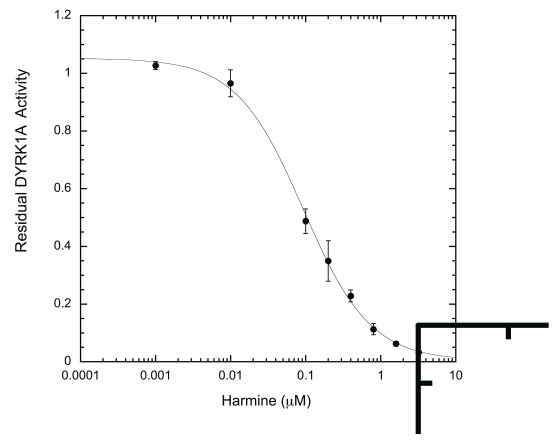
We subsequently tested the system by examining two well-characterized inhibitors, EGCG and harmine 27, 35. A typical inhibition profile conducted by the ELISA method for EGCG ( Figure 6A) and harmine ( Figure 6B) follows a sigmoidal function. IC 50s for EGCG and harmine determined by the ELISA method were 0.215 ± 0.024 μM and 0.107 ± 0.018 μM, respectively. These values are comparable to those obtained earlier by us and others with different substrates and protocols, including the radioisotope/filter binding assay, generally regarded as the gold standard for kinase inhibition assays ( Table 1) 22, 37– 39. The results obtained from this ELISA assay appear to be as reproducible as any given enzymatic assay. These results confirm that our ELISA platform is a valid system for quantitative characterization of DYRK1A inhibitors.
Figure 6A. Epigallocatechin gallate (EGCG) inhibition profile.
EGCG inhibition assays were performed in the presence of serially diluted EGCG (0.001 – 3.2 μM) under the standard reaction conditions. DYRK1A activity at any given EGCG concentration was calculated as a ratio to the activity of the no-inhibitor control and plotted on the Y-axis, versus EGCG concentration. IC 50 was calculated from the plot after curve fitting, as described in Methods (n = 6 for each data point).
Figure 6B. Harmine inhibition profile.
Harmine inhibition was conducted and analyzed exactly as described in Figure 6A for EGCG (n = 6 for each data point).
Table 1. List of IC 50 for epigallocatechin gallate (EGCG) and harmine obtained by different approaches.
| Inhibitor IC 50 (μM) | DYRK1A * | Substrate | [ATP] (μM) | Detection |
|---|---|---|---|---|
| EGCG | ||||
| 0.33 | 1 | Woodtide 40 | 100 | Radioisotope 37 |
| 0.31 (K I) | 1 | FAM-Woodtide | 100 | HPLC/Fluorescence 22 |
| 0.43 | 2 | Dynatide 3 24 | 50 | Radioisotope 27 |
| 0.215 # | 3 | HT-PRD | 100 | ELISA (this study) |
| Harmine | ||||
| 0.08 | 1 | Woodtide | 100 | Radioisotope 38 |
| 0.033 | 1 | DYRKtide 41 | 100 | Radioisotope 39 |
| 0.20 (K I) | 1 | FAM-Woodtide | 50 | HPLC/Fluorescence 22 |
| 0.075 | 2 | Dynatide 3 | 50 | Radioisotope 35 |
| 0.107 # | 3 | HT-PRD | 100 | ELISA (this study) |
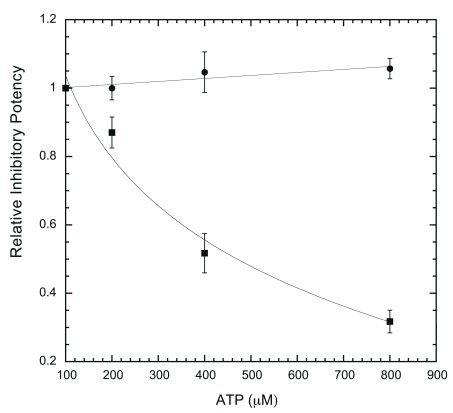
We then modified the ELISA protocol to run the assays under a single concentration of inhibitor with varying ATP concentrations, to determine whether ATP is competitive against the inhibitor in question. This allows the efficacy of inhibition to be evaluated with changing ATP. ATP is expected to influence the potency of competitive inhibitors, but not that of non-competitive inhibitors. As shown in Figure 7, harmine loses potency against DYRK1A when ATP is increased from 100 to 800 μM, indicating an ATP-competitive mode, while EGCG potency remains essentially unchanged (non-ATP-competitive). The inhibitory modes for harmine and EGCG revealed by the ELISA assay are the same as previously reported by the radioisotope/filter binding method 27, 35. This further validates the ELISA assay.
Figure 7. ATP competition assay.
For each inhibitor (epigallocatechin gallate (EGCG) and harmine), assays were conducted with a single concentration of inhibitor in four different ATP concentrations (100, 200, 400, 800 μM) and quantified as described in Methods. The inhibitory potency at three other ATP concentrations was calculated as relative to that at 100 μM ATP and used to plot against ATP concentrations. Inhibitor concentrations used in the assays were 0.4 μM for EGCG (●) and 0.2 μM for harmine (▪) (n = 6 for each data point).
Copyright: © 2017 Liu Y et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
As noted, non-radioactive DYRK1A assays have been described 21, 22. These assays employ similar solution DYRK1A reactions at first stage and then different approaches for measuring the phosphorylated products. One used a phospho-specific antibody to capture products for subsequent immuno/colorimetric detection 21, while another used a fluorescein-tagged substrate and analyzed products by high performance liquid chromatography/fluorescence detection 22. The above methods have been optimized for sensitivity to measure cellular DYRK1A activity. We do not know whether our ELISA method, at the current stage, affords such level of sensitivity. Nevertheless, as we have demonstrated, our ELISA assay provides sufficient sensitivity for analyzing inhibitor activity with recombinant DYRK1A. Furthermore, because of the standard ELISA protocol, our assay is straightforward to perform with the entire process carried out in a single well. The tools and equipment for adapting this plate-based assay for high throughput automation are widely available, and if necessary, the assay can be refined to further improve the sensitivity. We believe that our assay offers a simple, rapid, and reliable non-radioactive method suitable for replacing the radioactive trace assays in quantifying and screening DYRK1A inhibitors.
Data availability
The data referenced by this article are under copyright with the following copyright statement: Copyright: © 2017 Liu Y et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication). http://creativecommons.org/publicdomain/zero/1.0/
Dataset 1.
Raw data for Figures 1– 7 and Supplementary Figure are supplied in Excel format.
10.5256/f1000research.10582.d155317 42 Zipped file, containing the following:
Data for Figure 1. Coating ELISA plate with HT-PRD. Data (OD 405) for 0 – 800 ng of coated HT-PRD per well were shown. Background measurements for all Figures (1– 7 and Supplementary Figure) were obtained using wells with PNPP only (no coating, no phosphorylation, and no antibodies), which were performed in parallel with each experimental replicate/triplicate for background correction. The data shown in the files for all Figures have been corrected using averaged background from each set.
Data for Figure 2. Phosphorylation of coated HT-PRD by DYRK1A. Data (OD 405) for 0 – 800 ng coated HT-PRD per well were shown.
Data for Figure 3. DYRK1A concentration-dependent phosphorylation of coated HT-PRD. Data (OD 405) for phosphorylation with 0 – 80 ng HT-497 were shown.
Data for Figure 4. Time-course phosphorylation of coated HT-PRD by DYRK1A. Time-course phosphorylation data (OD 405) with 0–90 min incubation time were shown.
Data for Figure 5. 3D3 dilution factor determination. Data (OD 405) for 3D3 dilution 1:1,000 – 256,000 were shown.
Data for Figure 6A. Epigallocatechin gallate ( EGCG) inhibition profile. Data (OD 405) for EGCG 0 – 3.2 μM were shown.
Data for Figure 6B. Harmine inhibition profile. Data (OD 405) for harmine 0 – 3.2 μM were shown.
Data for Figure 7. ATP competition assay. Data (OD 405) for ATP 100 – 800 μM were shown.
Data for Supplementary Figure. Secondary antibody dilution factor determination. Data (OD 405) for secondary antibody dilution 1:1,000 – 256,000 were shown.
Acknowledgements
We thank Dr. Kevin Hwang for critical reading of the manuscript.
Funding Statement
This work is supported by the New York State Office for People with Developmental Disabilities, the parent agency of New York State Institute for Basic Research in Developmental Disabilities. No extramural fund was used to support this research.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; referees: 2 approved]
Supplementary materials
Supplementary Figure. Secondary antibody dilution factor determination. Phosphorylated HT-PRD in ELISA wells were prepared and probed with 3D3 (1:3000 dilution) followed by serially diluted secondary antibody (from 1000 to 256,000x) similarly as described in Figure 5. Like 3D3 titering, normalized OD 405 was calculated for plotting (n = 6 for each data point).
.
Supplementary Table. Estimation of Z’-factor for the ELISA assay performed under standard conditions..
References
- 1. Becker W, Sippl W: Activation, regulation, and inhibition of DYRK1A. FEBS J. 2011;278(2):246–256. 10.1111/j.1742-4658.2010.07956.x [DOI] [PubMed] [Google Scholar]
- 2. Song WJ, Sternberg LR, Kasten-Sportès C, et al. : Isolation of human and murine homologues of the Drosophila minibrain gene: human homologue maps to 21q22.2 in the Down syndrome "critical region". Genomics. 1996;38(3):331–339. 10.1006/geno.1996.0636 [DOI] [PubMed] [Google Scholar]
- 3. Rahmani Z, Blouin JL, Creau-Goldberg N, et al. : Critical role of the D21S55 region on chromosome 21 in the pathogenesis of Down syndrome. Proc Natl Acad Sci U S A. 1989;86(15):5958–5962. 10.1073/pnas.86.15.5958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wegiel J, Wisniewski HM, Dziewiatkowski J, et al. : Differential susceptibility to neurofibrillary pathology among patients with Down syndrome. Dementia. 1996;7(3):135–141. 10.1159/000106868 [DOI] [PubMed] [Google Scholar]
- 5. Smith DJ, Stevens ME, Sudanagunta SP, et al. : Functional screening of 2 Mb of human chromosome 21q22.2 in transgenic mice implicates minibrain in learning defects associated with Down syndrome. Nat Genet. 1997;16(1):28–36. 10.1038/ng0597-28 [DOI] [PubMed] [Google Scholar]
- 6. Branchi I, Bichler Z, Minghetti L, et al. : Transgenic mouse in vivo library of human Down syndrome critical region 1: association between DYRK1A overexpression, brain development abnormalities, and cell cycle protein alteration. J Neuropathol Exp Neurol. 2004;63(5):429–440. 10.1093/jnen/63.5.429 [DOI] [PubMed] [Google Scholar]
- 7. Ahn KJ, Jeong HK, Choi HS, et al. : DYRK1A BAC transgenic mice show altered synaptic plasticity with learning and memory defects. Neurobiol Dis. 2006;22(3):463–472. 10.1016/j.nbd.2005.12.006 [DOI] [PubMed] [Google Scholar]
- 8. Wegiel J, Kuchna I, Nowicki K, et al. : Cell type- and brain structure-specific patterns of distribution of minibrain kinase in human brain. Brain Res. 2004;1010(1–2):69–80. 10.1016/j.brainres.2004.03.008 [DOI] [PubMed] [Google Scholar]
- 9. Tejedor FJ, Hämmerle B: MNB/DYRK1A as a multiple regulator of neuronal development. FEBS J. 2011;278(2):223–235. 10.1111/j.1742-4658.2010.07954.x [DOI] [PubMed] [Google Scholar]
- 10. Dowjat WK, Adayev T, Kuchna I, et al. : Trisomy-driven overexpression of DYRK1A kinase in the brain of subjects with Down syndrome. Neurosci Lett. 2007;413(1):77–81. 10.1016/j.neulet.2006.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wegiel J, Gong CX, Hwang YW: The role of DYRK1A in neurodegenerative diseases. FEBS J. 2011;278(2):236–245. 10.1111/j.1742-4658.2010.07955.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Woods YL, Cohen P, Becker W, et al. : The kinase DYRK phosphorylates protein-synthesis initiation factor eIF2Bepsilon at Ser539 and the microtubule-associated protein tau at Thr212: potential role for DYRK as a glycogen synthase kinase 3-priming kinase. Biochem J. 2001;355(Pt 3):609–615. 10.1042/bj3550609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu F, Liang Z, Wegiel J, et al. : Overexpression of Dyrk1A contributes to neurofibrillary degeneration in Down syndrome. FASEB J. 2008;22(9):3224–3233. 10.1096/fj.07-104539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ryoo SR, Cho HJ, Lee HW, et al. : Dual-specificity tyrosine(Y)-phosphorylation regulated kinase 1A-mediated phosphorylation of amyloid precursor protein: evidence for a functional link between Down syndrome and Alzheimer's disease. J Neurochem. 2008;104(5):1333–44. 10.1111/j.1471-4159.2007.05075.x [DOI] [PubMed] [Google Scholar]
- 15. Ryu YS, Park SY, Jung MS, et al. : Dyrk1A-mediated phosphorylation of Presenilin 1: a functional link between Down syndrome and Alzheimer's disease. J Neurochem. 2010;115(3):574–584. 10.1111/j.1471-4159.2010.06769.x [DOI] [PubMed] [Google Scholar]
- 16. Lee MS, Kao SC, Lemere CA, et al. : APP processing is regulated by cytoplasmic phosphorylation. J Cell Biol. 2003;163(1):83–95. 10.1083/jcb.200301115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kimura R, Kamino K, Yamamoto M, et al. : The DYRK1A gene, encoded in chromosome 21 Down syndrome critical region, bridges between beta-amyloid production and tau phosphorylation in Alzheimer disease. Hum Mol Genet. 2007;16(1):15–23. 10.1093/hmg/ddl437 [DOI] [PubMed] [Google Scholar]
- 18. Becker W, Soppa U, Tejedor FJ: DYRK1A: a potential drug target for multiple Down syndrome neuropathologies. CNS Neurol Disord Drug Targets. 2014;13(1):26–33. 10.2174/18715273113126660186 [DOI] [PubMed] [Google Scholar]
- 19. Duchon A, Herault Y: DYRK1A, a Dosage-Sensitive Gene Involved in Neurodevelopmental Disorders, Is a Target for Drug Development in Down Syndrome. Front Behav Neurosci. 2016;10:104. 10.3389/fnbeh.2016.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stotani S, Giordanetto F, Medda F: DYRK1A inhibition as potential treatment for Alzheimer's disease. Future Med Chem. 2016;8(6):681–696. 10.4155/fmc-2016-0013 [DOI] [PubMed] [Google Scholar]
- 21. Lilienthal E, Kolanowski K, Becker W: Development of a sensitive non-radioactive protein kinase assay and its application for detecting DYRK activity in Xenopus laevis oocytes. BMC Biochem. 2010;11:20. 10.1186/1471-2091-11-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bui LC, Tabouy L, Busi F, et al. : A high-performance liquid chromatography assay for Dyrk1a, a Down syndrome-associated kinase. Anal Biochem. 2014;449:172–178. 10.1016/j.ab.2013.12.024 [DOI] [PubMed] [Google Scholar]
- 23. Chen-Hwang MC, Chen HR, Elzinga M, et al. : Dynamin is a minibrain kinase/dual specificity Yak1-related kinase 1A substrate. J Biol Chem. 2002;277(20):17597–17604. 10.1074/jbc.M111101200 [DOI] [PubMed] [Google Scholar]
- 24. Huang Y, Chen-Hwang MC, Dolios G, et al. : Mnb/Dyrk1A phosphorylation regulates the interaction of dynamin 1 with SH3 domain-containing proteins. Biochemistry. 2004;43(31):10173–10185. 10.1021/bi036060+ [DOI] [PubMed] [Google Scholar]
- 25. Warnock DE, Terlecky LJ, Schmid SL: Dynamin GTPase is stimulated by crosslinking through the C-terminal proline-rich domain. EMBO J. 1995;14(7):1322–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Himpel S, Panzer P, Eirmbter K, et al. : Identification of the autophosphorylation sites and characterization of their effects in the protein kinase DYRK1A. Biochem J. 2001;359(Pt 3):497–505. 10.1042/bj3590497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adayev T, Chen-Hwang MC, Murakami N, et al. : Kinetic properties of a MNB/DYRK1A mutant suitable for the elucidation of biochemical pathways. Biochemistry. 2006;45(39):12011–12019. 10.1021/bi060632j [DOI] [PubMed] [Google Scholar]
- 28. Cunningham PR, Weitzmann CJ, Nurse K, et al. : Site-specific mutation of the conserved m 6 2Am 6 2A residues of E. coli 16S ribosomal RNA. Effects on ribosome function and activity of the KsgA methyltransferase. Biochim Biophy Acta. 1990;1050(1–3):18–26. [DOI] [PubMed] [Google Scholar]
- 29. Hwang YW, Zhong JM, Poullet P, et al. : Inhibition of SDC25 C-domain-induced guanine-nucleotide exchange by guanine ring binding domain mutants of v-H-ras. J Biol Chem. 1993;268(33):24692–24698. [PubMed] [Google Scholar]
- 30. Bradford MM: A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1–2):248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 31. Yano T, Taura C, Shibata M, et al. : A monoclonal antibody to the phosphorylated form of glial fibrillary acidic protein: application to a non-radioactive method for measuring protein kinase activities. Biochem Biophys Res Commun. 1991;175(3):1144–1151. 10.1016/0006-291X(91)91685-6 [DOI] [PubMed] [Google Scholar]
- 32. Farley K, Mett H, McGlynn E, et al. : Development of solid-phase enzyme-linked immunosorbent assays for the determination of epidermal growth factor receptor and pp60 c-src tyrosine protein kinase activity. Anal Biochem. 1992;203(1):151–157. 10.1016/0003-2697(92)90056-D [DOI] [PubMed] [Google Scholar]
- 33. Wang Y, Ma H: Protein kinase profiling assays: a technology review. Drug Discov Today Technol. 2015;18:1–8. 10.1016/j.ddtec.2015.10.007 [DOI] [PubMed] [Google Scholar]
- 34. Adayev T, Chen-Hwang MC, Murakami N, et al. : Dual-specificity tyrosine phosphorylation-regulated kinase 1A does not require tyrosine phosphorylation for activity in vitro. Biochemistry. 2007;46(25):7614–7624. 10.1021/bi700251n [DOI] [PubMed] [Google Scholar]
- 35. Adayev T, Wegiel J, Hwang YW: Harmine is an ATP-competitive inhibitor for dual-specificity tyrosine phosphorylation-regulated kinase 1A (Dyrk1A). Arch Biochem Biophys. 2011;507(2):212–218. 10.1016/j.abb.2010.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Segel IH: Enzyme Kinetics, Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. John Wiley & Sons, Inc.1975. Reference Source [Google Scholar]
- 37. Bain J, McLauchlan H, Elliott M, et al. : The specificities of protein kinase inhibitors: an update. Biochem J. 2003;371(Pt 1):199–204. 10.1042/BJ20021535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bain J, Plater L, Elliott M, et al. : The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408(3):297–315. 10.1042/BJ20070797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Göckler N, Jofre G, Papadopoulos C, et al. : Harmine specifically inhibits protein kinase DYRK1A and interferes with neurite formation. FEBS J. 2009;276(21):6324–6337. 10.1111/j.1742-4658.2009.07346.x [DOI] [PubMed] [Google Scholar]
- 40. Woods YL, Rena G, Morrice N, et al. : The kinase DYRK1A phosphorylates the transcription factor FKHR at Ser329 in vitro, a novel in vivo phosphorylation site. Biochem J. 2001;355(Pt 3):597–607. 10.1042/bj3550597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Himpel S, Tegge W, Frank R, et al. : Specificity determinants of substrate recognition by the protein kinase DYRK1A. J Biol Chem. 2000;275(4):2431–2438. 10.1074/jbc.275.4.2431 [DOI] [PubMed] [Google Scholar]
- 42. Liu Y, Adayev T, Hwang YW: Dataset 1 in: An ELISA DYRK1A non-radioactive assay suitable for the characterization of inhibitors. F1000Research. 2017. Data Source [DOI] [PMC free article] [PubMed] [Google Scholar]