Summary
Elevated inflammation in the female genital tract is associated with increased HIV risk. Cervicovaginal bacteria modulate genital inflammation, however their role in HIV susceptibility has not been elucidated. In a prospective cohort of young, healthy South African women, we found that individuals with diverse genital bacterial communities dominated by anaerobes other than Gardnerella were at over 4-fold higher risk of acquiring HIV and had increased numbers of activated mucosal CD4+ T cells compared to those with Lactobacillus crispatus-dominant communities. We identified specific bacterial taxa linked with reduced (L. crispatus) or elevated (Prevotella, Sneathia, and other anaerobes) inflammation and HIV infection and found that high-risk bacteria increased numbers of activated genital CD4+ T cells in a murine model. Our results suggest that highly prevalent genital bacteria increase HIV risk by inducing mucosal HIV target cells. These findings may be leveraged to reduce HIV acquisition in women living in sub-Saharan Africa.
Keywords: female genital tract (FGT), vaginal microbiome, mucosal immunology, HIV susceptibility, sub-Saharan Africa, HIV acquisition
Graphical Abstract

Introduction
24 million people in sub-Saharan Africa are infected with HIV, with the majority of new transmission events occurring following heterosexual sex. Young African women have an up to 8-fold increased HIV prevalence compared to young men (UNAIDS, 2014), emphasizing the need for a better understanding of the factors in the female genital tract (FGT) that influence HIV acquisition.
Elevated inflammation is associated with an increased risk of HIV infection (Masson et al., 2015), and cervicovaginal bacteria have been shown to impact baseline inflammation in the FGT (Anahtar et al., 2015), suggesting a potential role for FGT bacteria in modulating acquisition risk. A small number of prospective studies indicate that women with cervicovaginal bacteria deficient in lactobacilli, a bacterial genus considered beneficial for vaginal health (Spurbeck and Arvidson, 2011), are at higher risk of acquiring HIV (Low et al., 2011; Martin et al., 1999; Myer et al., 2005). However, the depth of these studies has been limited by the use of Gram-stained vaginal smears to assess the genital bacterial composition, and by investigation of sex-worker and clinical cohorts not representative of the average healthy female population.
The recent development of high throughput sequencing technologies to characterize the human microbiome has significantly improved our understanding of the role of the microbiome in diseases such as obesity, inflammatory bowel disease, colorectal cancer (Marchesi et al., 2016) and HIV-induced AIDS (Handley et al., 2016; Monaco et al., 2016). While these studies have focused on the enteric microbiome, less is understood regarding the role of the cervicovaginal microbiome in disease susceptibility, particularly in the developing world. Additionally, the majority of microbiome studies have focused on bacteria while viruses have remained incompletely characterized, particularly in the FGT. Here we performed bacterial 16S ribosomal (r)RNA gene and viral shotgun sequencing of cervicovaginal microbiota and assessed their role in HIV acquisition in participants of the FRESH (Females Rising through Education, Support and Health) study, a prospective cohort study of healthy HIV-uninfected black South African women aged 18–23 years, monitored with high frequency testing for incident HIV infection. We demonstrated a differential association of four distinct bacterial community structures with genital inflammation, cervical HIV target cell numbers, and HIV acquisition. We additionally identified specific bacterial taxa that were closely associated with genital inflammation and HIV acquisition and showed that these same bacteria induced inflammation in in vitro and in vivo models, thus establishing their mechanistic potential for directly increasing HIV acquisition.
Results
South African women have distinct bacterial but not viral cervicovaginal communities
We followed 236 HIV-uninfected women prospectively with a median follow-up time of 336 days (IQR: 178.5 to 347 days) and a total follow-up time of 198.2 person-years. Despite intensive HIV prevention counseling, 31 women acquired HIV while 205 remained HIV-uninfected with an overall HIV incidence in the FRESH cohort of 8.43 per 100 person-years.
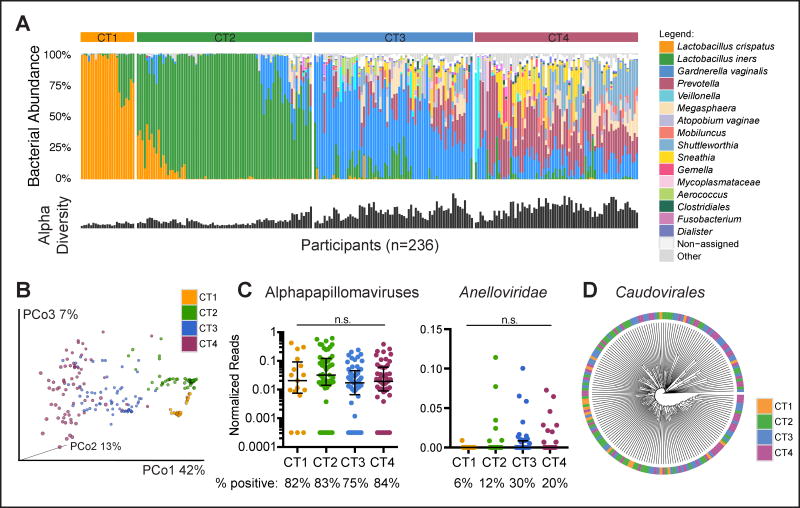
Sequencing of the V4 region of the bacterial 16S rRNA gene in these 236 individuals demonstrated the presence of distinct bacterial community types that, based on composition and diversity, clustered into 4 basic groups, which we referred to as “cervicotypes” (CTs). Similar to our previous findings in a smaller cohort (Anahtar et al., 2015), 10% of women had low diversity communities dominated by Lactobacillus crispatus (CT1, n=23), and 32% were dominated by Lactobacillus iners (CT2, n=74) (Fig. 1A). This contrasts with white women in developed countries, 90% of whom have Lactobacillus-dominant cervicovaginal communities (Ravel et al., 2011). The cervicovaginal bacterial microbiome of the remaining 58% of FRESH participants consisted of high-diversity communities with low Lactobacillus abundance, with either Gardnerella vaginalis dominance (CT3, n=68) or a bacterial genus other than Lactobacillus or Gardnerella as the dominant taxon (CT4, n=70). The most abundant taxa in CT4 were Prevotella, Gardnerella, Shuttleworthia, Sneathia and Megasphaera (Fig. 1A). Principal coordinates analysis confirmed distinct clustering of these CT groups (Fig. 1B).
Figure 1. Bacterial, but not viral, cervicovaginal community structures are distinct in HIV-uninfected women.
(A) Stacked bar plot indicating the bacterial composition of the cervicovaginal microbiome of 236 HIV-uninfected women. Samples were grouped according to 4 cervicotypes (CTs), and the alpha diversity is shown below for each sample.
(B) Principal coordinates analysis (PCoA) plot of the 236 individuals, distinguished by CT.
(C) Normalized reads of alphapapillomaviruses and Anelloviridae detected in CVL fluid, grouped by CT (n=180 individuals). Groups were compared using Kruskal-Wallis test. The percentage of samples in which the indicated virus was detected is shown below.
(D) Average linkage clustering of the Bray-Curtis dissimilarity of relative abundances of Caudovirales sequences detected in CVL fluid (n=180 individuals). Lines in the plots indicate the median and the interquartile range (IQR) of the dataset.
Given the paucity of information regarding the community of viruses (the “virome”) in the FGT, we used metagenomic sequencing to characterize DNA and RNA viral populations in 180 study participants. We identified alphapapillomaviruses and Anelloviridae as the predominant eukaryotic viral taxa present in the FGT in this cohort (Fig. 1C). Numerous species of Caudovirales, the tailed bacteriophages, were also observed (Fig. 1D). In contrast to our findings for bacteria, we identified no distinct viral community structures within our cohort, nor did we detect a difference in viral composition among different bacterial communities (Fig. 1C, D). In summary, we observed four distinct cervicovaginal bacterial community types, the most prevalent ones being characterized by high-diversity and low Lactobacillus abundance, and no distinct viral community structures in the study participants.
HIV acquisition is increased in women with high-diversity, low Lactobacillus abundance FGT bacterial communities
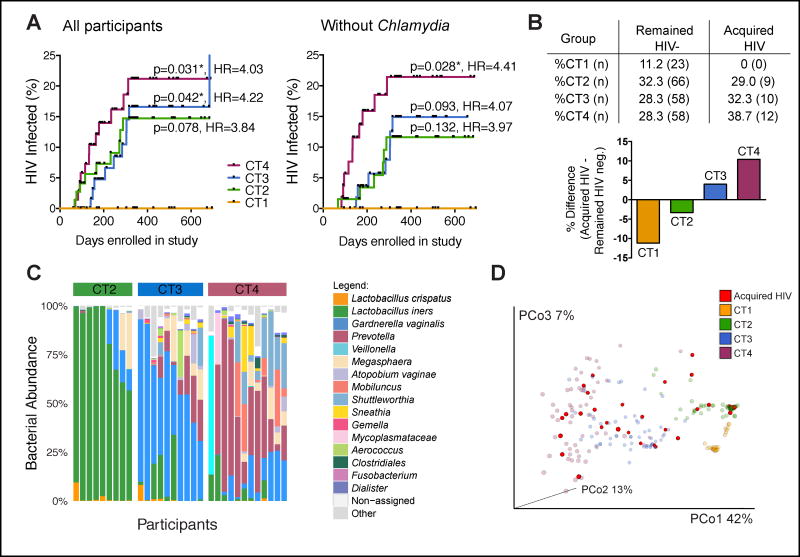
We next examined associations between cervicovaginal bacteria and HIV acquisition. We observed a greater than 4-fold increase in HIV acquisition in women with high-diversity, low Lactobacillus abundance communities compared to women with L. crispatus dominance (CT4: HR=4.03, 95% CI: 1.14 to 14.27, p=0.031, and CT3: HR=4.22, 95% CI: 1.06 to 16.88, p=0.042) (Fig. 2A). None of the women who acquired HIV had an L. crispatus-dominant community. Women with CT2 communities were also underrepresented among participants who subsequently acquired HIV relative to the uninfected group, while those with CT3 and CT4 communities were more prevalent among women who went on to become infected (Fig. 2B). The genital bacterial communities of women who subsequently acquired HIV were comparable to those of their respective CT group who remained HIV negative (Fig. 2C, Fig. S1A), with no acquisition-specific clustering within a CT (Fig. 2D), suggesting a similar HIV infection risk for women within the same CT group. In contrast to cervicovaginal bacteria, viral taxa identified in this study were not different between women who became infected and those who remained uninfected (Fig. S1B).
Figure 2. Cervicovaginal bacteria impact HIV acquisition.
(A) Kaplan-Meier curve showing HIV infections in each CT group over time, including or excluding individuals with Chlamydia infection. Acquisition curves for CT2 (n=74/65 individuals), CT3 (n=68/57) and CT4 (n=68/54) were compared with the acquisition curve for CT1 (n=23/23). Log-rank (Mantel-Cox) test-based p values and Mantel-Haenszel hazard ratios (HR) are displayed.
(B) Distribution of CT groups in participants who remained HIV-uninfected (n=205) versus those who acquired HIV (n=31), shown as % of total, and the difference in distribution between these two groups.
(C) Stacked bar plot indicating the bacterial composition of the cervicovaginal microbiome of 31 women who subsequently acquired HIV. Samples were grouped by CT.
(D) Bacterial PCoA plot indicating study participants who acquired HIV (in red, n=31) and CT groups for those who remained HIV-uninfected (faded coloring, n=205). See also Figure S1 and Table S1.
We next assessed factors associated with different genital bacterial communities in our cohort, with particular attention to those that might increase HIV susceptibility (Byrne et al., 2016; McKinnon and Karim, 2016; Ward and Ronn, 2010) (Table S1). We observed no differences between bacterial communities and demographic factors, condom use, frequency or type of sexual acts, or number of sexual partners. The use of injectable progestin contraceptives (IPCs), a known HIV risk factor in the FRESH cohort, was also similar among women with different CTs, which is consistent with our previous finding that cervicovaginal bacterial communities did not vary with IPC use (Byrne et al., 2016). Sexually transmitted infections (STIs) were equally distributed among all groups, except for an increase in Chlamydia trachomatis infection from CT1 through CT4 (Table S1). After removing all Chlamydia-positive samples from the analysis, HIV acquisition in women with CT4 but not CT3 communities was still significantly increased compared to women with L. crispatus-dominant communities (HR: 4.41, 95% CI: 1.17 to 16.61, p=0.028) (Fig. 2A). Similar results were obtained after adjusting for Chlamydia infection. Collectively, these results demonstrate that women with high-diversity anaerobic communities have a more than 4-fold increase in HIV acquisition compared to women with an L. crispatus-dominant cervicovaginal microbiome.
Women with anaerobic FGT bacteria have increased genital HIV target cell numbers and Th17-associated cytokines
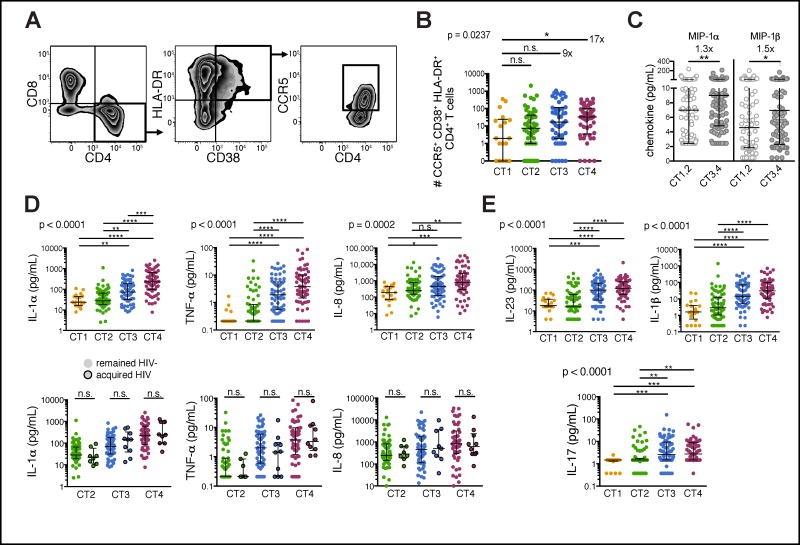
In most women, HIV infection is established following viral replication in CD4+ T cells present in the genital mucosa (Haase, 2011). Activated CD4+ T cells expressing the HIV co-receptor CCR5 (“HIV target cells”) are not only more prone to infection but also support higher degrees of viral replication than resting cells (Meditz et al., 2011; Stevenson et al., 1990), thus increasing HIV susceptibility (Haase, 2011; Koning et al., 2005). We examined the relationship between the bacterial microbiome and HIV target cell numbers in the genital tract and observed a 17-fold increase in HIV target cells in the FGT of women with CT4 versus L. crispatus-dominant communities (Fig. 3A, B; Fig. S2). Consistent with this finding, we demonstrated elevated levels of the chemokines MIP-1α and MIP-1β, which attract CCR5-expressing cells, in women with high-diversity bacterial communities (Fig. 3C). We also observed higher concentrations of cytokines and chemokines associated with increased HIV acquisition (Masson et al., 2015), such as interleukin (IL)-1α, tumor necrosis factor (TNF)-α, IL-8 (Fig. 3D; upper row), IL-12p70, interferon (IFN)-γ, and IL-10 (Fig. S3A), which confirmed our previous findings in a smaller cohort (Anahtar et al., 2015). Women who went on to become infected with HIV had similar pro-inflammatory cytokine levels as those who remained HIV-uninfected within the same CT group. This suggests comparable HIV susceptibility among women with similar genital communities (Fig. 3D; lower row).
Figure 3. High-risk genital bacterial communities are associated with increased HIV target cell numbers and pro-inflammatory cytokines.
(A) Representative flow cytometry plots to identify HIV target cells isolated from cervical cytobrushes. Live T cells (CD45+CD3+) were gated on cells expressing CD4, HLA-DR, CD38 and CCR5.
(B) Flow cytometry analysis of HIV target cell numbers in cytobrushes from 169 individuals, grouped by CT.
(C,D,E) Chemokine and cytokine concentrations in CVL fluid from 219 individuals, grouped by CT. The bottom figure panel in (D) further distinguishes between samples of women who became infected with HIV and those who remained HIV-uninfected. Lines indicate the median and the interquartile range (IQR) of each dataset. Groups were compared using Kruskal-Wallis test with Dunn’s post-hoc analyses. See also Figures S2 and S3.
We further examined cytokines associated with specific T helper (Th) subsets as recent work suggests that Th17 cells are the first targets of SIV and HIV infection in the FGT (Rodriguez-Garcia et al., 2014; Stieh et al., 2016). While we detected no or little difference in Th2-associated cytokines IL-4, IL-5 and IL-13 between CT groups (Fig. S3B), we measured a notable increase in IL-17 and IL-17-inducing cytokines IL-23 and IL-1β in the cervicovaginal lavage (CVL) fluid of women with CT3 and CT4 structures (Fig. 3E; Fig. S3E). This finding suggested that increased numbers of Th17 cells, the predominant producers of IL-17 in the genital mucosa (Gosmann et al., 2014; Stieh et al., 2016), were induced in the presence of CT3 and CT4 bacteria. Taken together, these results reveal an association of high-diversity genital bacterial communities with increased production of local pro-inflammatory cytokines and recruitment and activation of mucosal CD4+ T cells.
Specific bacterial taxa are differentially associated with genital inflammation and HIV acquisition
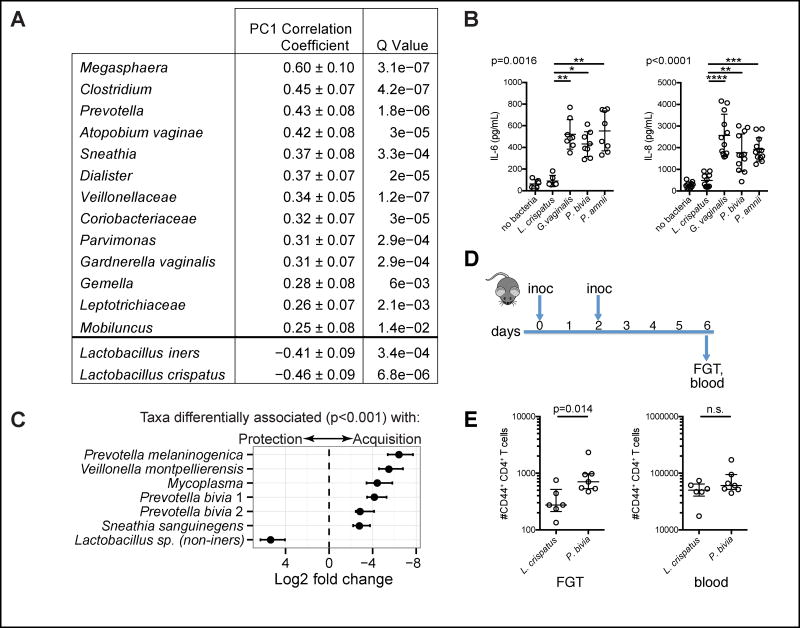
We next investigated whether specific bacterial taxa were associated with genital inflammation and HIV infection. We found that L. crispatus and, to a lesser extent, L. iners were associated with reduced inflammation as indicated by an inverse correlation with the first cytokine principal component (PC1), which explained 51% of the variation in cytokines (Fig. 4A, Fig. S3C, D). In contrast, anaerobic taxa were associated with increased inflammation, the five taxa most strongly correlated with cytokine PC1 being Megasphaera, Clostridium, Prevotella, Atopobium vaginae and Sneathia (Fig. 4A). Consistent with this finding, cervical epithelial cells produced higher concentrations of IL-6 and IL-8 when co-cultured with Gardnerella vaginalis, Prevotella bivia and Prevotella amnii compared to L. crispatus (Fig. 4B). To more closely examine bacterial taxa associated with HIV acquisition, we identified resolved sequence variants (RSV) from 16S rRNA gene amplicons. Non-iners lactobacilli were significantly more abundant in women who remained HIV-uninfected (Fig. 4C). In contrast, Prevotella melaninogenica, Veillonella montpellierensis, Mycoplasma, Prevotella bivia and Sneathia sanguinegens, most of which were associated with increased genital inflammation, were all positively associated with HIV acquisition (Fig. 4C). In summary, we identified specific bacterial taxa associated with reduced (e.g. L. crispatus) or increased (e.g. Prevotella and Sneathia) inflammation and HIV acquisition.
Figure 4. Specific bacterial taxa are associated with genital inflammation and HIV infection.
(A) Bacterial taxa in 219 individuals positively or negatively correlated with cytokine principal component 1 (PC1), an indicator of the presence of genital inflammation. Q-values reflect corrected p-values after multiple testing.
(B) Cytokine and chemokine concentrations measured in the supernatant of cervical epithelial cells co-cultured with the indicated bacteria for 24 h (pooled results from 2–3 independent experiments; Kruskal-Wallis test).
(C) Bacterial taxa significantly differentially abundant in women who became infected with HIV (n=31) and those who remained uninfected (n=205), including two strains of P. bivia. Error bars indicate the standard error estimate for the log2 fold change in abundance. (D) Germ-free mice were intravaginally inoculated with L. crispatus or P. bivia on day 0 and day 2 and sacrificed on day 6.
(E) Numbers of activated (CD44+) CD4+ T cells detected in the female genital tract (FGT) and in 100 μl blood of the gnotobiotic mice on day 6 of the experiment (pooled results from 3 independent experiments; Mann-Whitney test). Lines in the plots indicate the median and the interquartile range (IQR) of the dataset. See also Figure S4.
Vaginal inoculation of germ-free mice with anaerobic bacteria increases numbers of activated CD4+ T cells in the FGT
To more definitively assess a mechanistic role for these bacteria in inducing increased numbers of FGT HIV target cells, we inoculated germ-free mice intravaginally with L. crispatus or P. bivia (Fig. 4D, E). We detected higher numbers of activated (CD44+) CD4+ T cells in the genital mucosa, but not in the blood, of mice inoculated with P. bivia compared to mice that received L. crispatus (Fig. 4E). Mice treated with P. bivia further had increased numbers of mucosal CCR5+ CD4+ T cells (Fig. S4A) and showed a trend towards elevated genital TNF-α concentrations (Fig. S4B) compared to mice inoculated with L. crispatus. Together, these results indicate that specific bacterial taxa may increase HIV infection risk by inducing elevated numbers of HIV target cells in the FGT.
Discussion
Here we presented a comprehensive characterization of the genital bacterial and viral microbiome in healthy young women in sub-Saharan Africa, and identified a significant association of distinct cervicovaginal bacterial communities and specific bacterial taxa with HIV acquisition. The sequencing-based identification of cervicovaginal bacteria and viruses provides a substantial advancement over previous prospective studies that used Gram-stained vaginal smears (Low et al., 2011; Martin et al., 1999; Myer et al., 2005).
Our results indicate that high-diversity, low Lactobacillus abundance bacterial communities in the FGT negatively impact vaginal health by increasing the risk of HIV acquisition. In addition, prior studies have reported associations between Lactobacillus-deficient genital microbiota and other poor reproductive outcomes such as preterm delivery, late miscarriage (Hay et al., 1994; Lamont et al., 2011) and cervicitis (Gorgos et al., 2015). The increased rate of HIV acquisition in women with high-diversity, low Lactobacillus abundance bacterial communities observed in our study could be explained by the sensing of specific bacterial antigens or components such as lipopolysaccharide (LPS); we previously demonstrated that CT4 communities were enriched in LPS relative to CT1, and that LPS signaling pathways were significantly upregulated in cervical antigen-presenting cells of women with high-diversity communities (Anahtar et al., 2015). This likely results in activation and recruitment of HIV target cells to the female genital mucosa. In support of this hypothesis, we show a 17-fold increase in HIV target cell numbers in the FGT mucosa of women with CT4 vs. CT1 communities. This finding is further corroborated by the presence of increased numbers of activated CD4+ T cells in the genital mucosa of germ-free mice intravaginally inoculated with P. bivia compared to mice inoculated with L. crispatus, providing direct evidence of the impact of these bacteria on the recruitment of activated CD4+ T cells to the FGT. Increased concentrations of Th17-inducing cytokines IL-23 and IL-1β, and of IL-17 in the presence of CT3 and CT4 bacteria further indicated the presence of elevated numbers of Th17 cells, which are particularly susceptible to HIV infection and have been described as the first cells to be infected by lentivirus in the FGT (Christensen-Quick et al., 2016; Stieh et al., 2016). While the development of the Th17 subset in human epithelial tissues is not fully understood, in murine models, Th17 cells are specifically induced by bacteria in the gut (Ivanov et al., 2009). Thus, the bacterial composition of the FGT microbiome may influence HIV susceptibility not only by increasing local HIV target cell numbers, but also by influencing T helper cell differentiation within the genital mucosa. Whether FGT bacteria contribute to shaping mucosal T helper cell subsets will need to be addressed in future studies.
Reduced HIV acquisition in women with L. crispatus-dominant communities may be based on their ability to impair the growth of inflammatory bacteria and pathogens through the production of lactic acid, hydrogen peroxide and bactericidal compounds (Spurbeck and Arvidson, 2011), thus diminishing inflammation and HIV target cell numbers. Consistent with this hypothesis, L. crispatus did not co-exist with the more diverse anaerobic communities associated with HIV acquisition. In contrast, L. iners was frequently found in CT3 and CT4 communities, and 29% of the women who became infected with HIV had L. iners-dominant communities while none had L. crispatus dominance. This observation may further be explained by reduced HIV virion diffusion rates in the mucus from women with L. crispatus dominated vaginal microbiota compared to women with L. iners dominance (Nunn et al., 2015). These findings identify L. crispatus as a Lactobacillus species associated with decreased HIV infection and highlight the importance of distinguishing between different lactobacilli in the context of HIV susceptibility. Our findings further indicate that individuals within a particular CT group are at similar risk of acquiring HIV. This suggests that more than half the population of young black South African women is at increased HIV infection risk based on the composition of their cervicovaginal microbiome.
The absence of distinct viral signatures in women with different bacterial communities suggests a primary role for bacteria in modulating HIV acquisition risk. However, it is possible that a greater depth of sequencing, or expansion of viral databases will reveal associations that were not observed here. The stability in bacteriophage community structure across CT groups further suggests that the observed bacterial communities were not being modified by bacteriophage predation, a mechanism that could explain the formation of distinct bacterial communities in different individuals.
Clinical studies assessing the efficacy of regimens such as antibiotics and probiotic Lactobacillus vaginal suppositories in modifying the genital microbiome of women with bacterial vaginosis showed significant recurrence rates (Bradshaw et al., 2012; Woodman, 2016). Our findings underscore the importance of developing more effective regimens that achieve a sustained alteration of the genital microbiome since this could potentially reduce HIV acquisition. In this context, strategies targeting specific bacterial taxa associated with reduced or increased HIV acquisition may be useful, although the host mechanisms that maintain specific microbial communities in the genital tract need to be better understood. Our results advance our understanding of the cervicovaginal microbiome as an HIV risk factor and demonstrate the importance of considering the microbiome in the development of new treatments and preventive strategies to reduce HIV acquisition in young women living in sub-Saharan Africa.
Experimental Procedures
Study cohort
Study participants were enrolled in the Females Rising through Education, Support, and Health (FRESH) study, a prospective observational study of 18–23 year old HIV uninfected women conducted in Umlazi, South Africa. At the time of data analysis, pre-infection mucosal samples from 31 women who became HIV-infected, and samples from 205 women who remained uninfected were available. The study protocol was approved by the Biomedical Research Ethics Committee of the University of KwaZulu-Natal and the Massachusetts General Hospital Institutional Review Board (2012P001812/MGH). Informed consent was obtained following explanation of the nature and possible consequences of the study.
Clinical procedures
Twice per week, participants had a finger prick blood draw for HIV RNA viral load testing and every 3 months had a peripheral blood draw and a pelvic exam and completed an HIV risk questionnaire administered by a counselor. The pelvic exam involved the collection of ectocervical and midvaginal swabs (Catch-All, Epicentre), a cervicovaginal lavage (CVL), and an endocervical cytobrush.
Bacterial 16S rRNA V4 gene sequencing
Nucleic acid extracted from cervical swabs was amplified and sequenced as previously described (Anahtar et al., 2016), and analysis was performed using MacQIIME.
Definition of cervicotypes
Samples with the relative majority of sequences assigned to the genus Lactobacillus (but not Lactobacillus iners) were defined as CT1. Lactobacillus iners was the predominant species in CT2, Gardnerella vaginalis in CT3, and CT4 had a dominant bacterial taxon other than Lactobacillus, L. iners or G. vaginalis.
Virome sequencing
Sequencing of viral DNA and RNA extracted from CVL fluid was performed as detailed in the Supplemental Experimental Procedures.
Sexually transmitted infection (STI) testing
STI testing was performed by Global Labs, South Africa.
Measurement of cytokines
Cytokine concentrations in CVL fluid were determined using a human cytokine/chemokine multiplexed bead assay (Millipore) and human and mouse ELISA (eBioscience, R&D Systems).
Flow cytometry
Human and mouse cells were stained with a viability dye (Invitrogen) and labeled with monoclonal antibodies as specified in the Supplemental Experimental Procedures. Samples were acquired on a FACS Aria III or LSR II (BD). Data were analyzed using FlowJo Version 9.8.5 (FlowJo Enterprise).
Detection of Differentially Abundant Bacterial Taxa
Resolved sequence variants (RSV) from 16S amplicons were identified using the dada2 procedure (Callahan et al., 2016). Differentially abundant taxa were determined using the DESeq2 package (Love et al., 2014).
Co-culture of epithelial cells and bacteria
Human endocervical epithelial cells (End1/E6E7, ATCC) were co-cultured anaerobically with Lactobacillus crispatus (provided by Dr. David N. Fredricks), Prevotella bivia (ATCC 29303), Prevotella amnii (CCUG 53648T) or Gardnerella vaginalis (ATCC 14018) for 24 h.
Mouse experiments
All animal procedures were approved by the Animal Resources at Children’s Hospital (ARCH) review committee (protocol # 16-08-3219R). CVL fluid was collected from 6–9 week old female germ-free Swiss-Webster mice housed according to an out-of-the-isolator protocol (Faith et al., 2014), followed by application of 2x108 CFU/mL to the vaginal cavity. On the day of harvest, blood, CVL fluid and, after CO2 asphyxiation, the female reproductive tract (vagina, cervix) were collected, processed and used for downstream analyses.
Statistics
For comparison of continuous data between two and more groups, Mann-Whitney test and Kruskal-Wallis test with Dunn’s post hoc analyses, respectively, were used. The time to HIV acquisition was summarized using Kaplan-Meier curves, and significance was assessed by Log-rank test. Fisher’s exact test was performed for comparison of categorical data between two or more groups. Principal component analysis (PCA) was used to obtain summary measures for the multi-variable cytokine set and performed using FactoMineR package in R. Multivariate analysis of cytokine and bacterial data was performed using MaAsLin (https://huttenhower.sph.harvard.edu/maaslin) (Morgan et al., 2012) and the Benjamini-Hochberg method to adjust for multiple testing. P values are two-sided and indicated as follows: *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; n.s.: p≥ 0.05. The analyses were performed in Prism 6 (GraphPad) and R Studio.
Supplementary Material
Acknowledgments
We would like to thank the FRESH participants; T. Cele for performing the pelvic exams; T. Sikhakhane, S. Ngcobo, and S. Zungu for clinical support; H. Shen for flow cytometry assistance; M. Karpel for performing the IL-8 ELISA. This work was supported by the Collaboration for AIDS Vaccine Discovery of the Bill and Melinda Gates Foundation, the International AIDS Vaccine Initiative (IAVI) (UKZNRSA1001), the NIAID (1R01AI111918 and R01AI067073) and the Harvard Center for AIDS Research. D.S.K. and D.R.W. received additional support from the Burroughs Wellcome Fund, M.N.A. from the National Institute of General Medical Sciences (award number T32GM007753), D.R.W. from the NIH (AI1113217) and C.M. from a Doris Duke Clinical Scientist Development Award. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIGMS or the NIH.
Footnotes
Author Contributions
C.G., M.N.A. and D.S.K. conceived the study and designed experiments. C.G., M.F., B.A.B. and M.N.A. performed the nucleic acid extraction from swabs and 16S PCR. C.G. performed 16S sequencing and analysis; L.D. extracted nucleic acid from CVL and S.A.H. and C.D. performed viral sequencing and analysis. N.P., B.A.B. and C.G. performed flow cytometry, and C.G. analyzed the data; C.G. performed cytokine measurements and data analyses; C.M. performed epithelial co-culture assays; C.G., Y.C., D.R.W. and M.F. performed or were otherwise involved in experiments using germ-free mice; G.A. and S.A.H. performed analyses using MaAsLin and Dada2/Deseq2, respectively. A.M., M.D. and B.A.B. collected behavioral data; N.I. processed and managed clinical samples; M.S.G. was involved with statistical analyses; C.H. and H.W.V. were involved with experimental design and data analysis; T.N., K.L.D., B.D.W., and D.S.K. designed and managed the clinical study; C.G. and D.S.K. wrote the manuscript, and M.N.A. and M.F. assisted in manuscript preparation. All authors discussed the results and commented on the manuscript.
Accession numbers
16S rRNA and viral sequences analyzed in this study are associated with project number PRJEB14858 in the European Nucleotide Archive.
Conflict of interest
The authors declare no competing interests.
Supplemental Information includes four figures, one table and Supplemental Experimental Procedures and can be found with this article online at
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anahtar MN, Bowman BA, Kwon DS. Efficient Nucleic Acid Extraction and 16S rRNA Gene Sequencing for Bacterial Community Characterization. Journal of visualized experiments : JoVE. 2016 doi: 10.3791/53939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anahtar MN, Byrne EH, Doherty KE, Bowman BA, Yamamoto HS, Soumillon M, Padavattan N, Ismail N, Moodley A, Sabatini ME, et al. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity. 2015;42:965–976. doi: 10.1016/j.immuni.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw CS, Pirotta M, De Guingand D, Hocking JS, Morton AN, Garland SM, Fehler G, Morrow A, Walker S, Vodstrcil LA, Fairley CK. Efficacy of oral metronidazole with vaginal clindamycin or vaginal probiotic for bacterial vaginosis: randomised placebo-controlled double-blind trial. PLoS One. 2012;7:e34540. doi: 10.1371/journal.pone.0034540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne EH, Anahtar MN, Cohen KE, Moodley A, Padavattan N, Ismail N, Bowman BA, Olson GS, Mabhula A, Leslie A, et al. Association between injectable progestin-only contraceptives and HIV acquisition and HIV target cell frequency in the female genital tract in South African women: a prospective cohort study. The Lancet Infectious Diseases. 2016;16:441–448. doi: 10.1016/S1473-3099(15)00429-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nature methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen-Quick A, Lafferty M, Sun L, Marchionni L, DeVico A, Garzino-Demo A. Human Th17 cells lack HIV-inhibitory RNases and are highly permissive to productive HIV infection. J Virol. 2016 doi: 10.1128/JVI.02869-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med. 2014:6. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgos LM, Sycuro LK, Srinivasan S, Fiedler TL, Morgan MT, Balkus JE, McClelland SR, Fredricks DN, Marrazzo JM. Relationship of Specific Bacteria in the Cervical and Vaginal Microbiotas With Cervicitis. Sex Transm Dis. 2015;42:475–481. doi: 10.1097/OLQ.0000000000000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosmann C, Mattarollo SR, Bridge JA, Frazer IH, Blumenthal A. IL-17 suppresses immune effector functions in human papillomavirus-associated epithelial hyperplasia. J Immunol. 2014;193:2248–2257. doi: 10.4049/jimmunol.1400216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AT. Early events in sexual transmission of HIV and SIV and opportunities for interventions. Annual review of medicine. 2011;62:127–139. doi: 10.1146/annurev-med-080709-124959. [DOI] [PubMed] [Google Scholar]
- Handley SA, Desai C, Zhao G, Droit L, Monaco CL, Schroeder AC, Nkolola JP, Norman ME, Miller AD, Wang D, et al. SIV Infection-Mediated Changes in Gastrointestinal Bacterial Microbiome and Virome Are Associated with Immunodeficiency and Prevented by Vaccination. Cell Host Microbe. 2016;19:323–335. doi: 10.1016/j.chom.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay PE, Lamont RF, Taylor-Robinson D, Morgan DJ, Ison C, Pearson J. Abnormal bacterial colonisation of the genital tract and subsequent preterm delivery and late miscarriage. BMJ (Clinical research ed) 1994;308:295–298. doi: 10.1136/bmj.308.6924.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning FA, Otto SA, Hazenberg MD, Dekker L, Prins M, Miedema F, Schuitemaker H. Low-level CD4+ T cell activation is associated with low susceptibility to HIV-1 infection. J Immunol. 2005;175:6117–6122. doi: 10.4049/jimmunol.175.9.6117. [DOI] [PubMed] [Google Scholar]
- Lamont RF, Nhan-Chang CL, Sobel JD, Workowski K, Conde-Agudelo A, Romero R. Treatment of abnormal vaginal flora in early pregnancy with clindamycin for the prevention of spontaneous preterm birth: a systematic review and metaanalysis. Am J Obstet Gynecol. 2011;205:177–190. doi: 10.1016/j.ajog.2011.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low N, Chersich MF, Schmidlin K, Egger M, Francis SC, van de Wijgert JH, Hayes RJ, Baeten JM, Brown J, Delany-Moretlwe S, et al. Intravaginal practices, bacterial vaginosis, and HIV infection in women: individual participant data meta-analysis. PLoS Med. 2011;8:e1000416. doi: 10.1371/journal.pmed.1000416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65:330–339. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin HL, Richardson BA, Nyange PM, Lavreys L, Hillier SL, Chohan B, Mandaliya K, Ndinya-Achola JO, Bwayo J, Kreiss J. Vaginal lactobacilli, microbial flora, and risk of human immunodeficiency virus type 1 and sexually transmitted disease acquisition. J Infect Dis. 1999;180:1863–1868. doi: 10.1086/315127. [DOI] [PubMed] [Google Scholar]
- Masson L, Passmore JA, Liebenberg LJ, Werner L, Baxter C, Arnold KB, Williamson C, Little F, Mansoor LE, Naranbhai V, et al. Genital inflammation and the risk of HIV acquisition in women. Clin Infect Dis. 2015;61:260–269. doi: 10.1093/cid/civ298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon LR, Karim QA. Factors Driving the HIV Epidemic in Southern Africa. Current HIV/AIDS reports. 2016;13:158–169. doi: 10.1007/s11904-016-0314-z. [DOI] [PubMed] [Google Scholar]
- Meditz AL, Haas MK, Folkvord JM, Melander K, Young R, McCarter M, Mawhinney S, Campbell TB, Lie Y, Coakley E, et al. HLA-DR+ CD38+ CD4+ T lymphocytes have elevated CCR5 expression and produce the majority of R5-tropic HIV-1 RNA in vivo. J Virol. 2011;85:10189–10200. doi: 10.1128/JVI.02529-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco CL, Gootenberg DB, Zhao G, Handley SA, Ghebremichael MS, Lim ES, Lankowski A, Baldridge MT, Wilen CB, Flagg M, et al. Altered Virome and Bacterial Microbiome in Human Immunodeficiency Virus-Associated Acquired Immunodeficiency Syndrome. Cell Host Microbe. 2016;19:311–322. doi: 10.1016/j.chom.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer L, Kuhn L, Stein ZA, Wright TC, Denny L. Intravaginal practices, bacterial vaginosis, and women's susceptibility to HIV infection: epidemiological evidence and biological mechanisms. The Lancet Infectious Diseases. 2005;5:786–794. doi: 10.1016/S1473-3099(05)70298-X. [DOI] [PubMed] [Google Scholar]
- Nunn KL, Wang YY, Harit D, Humphrys MS, Ma B, Cone R, Ravel J, Lai SK. Enhanced Trapping of HIV-1 by Human Cervicovaginal Mucus Is Associated with Lactobacillus crispatus-Dominant Microbiota. MBio. 2015;6:e01084–01015. doi: 10.1128/mBio.01084-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Garcia M, Barr FD, Crist SG, Fahey JV, Wira CR. Phenotype and susceptibility to HIV infection of CD4+ Th17 cells in the human female reproductive tract. Mucosal Immunol. 2014;7:1375–1385. doi: 10.1038/mi.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurbeck RR, Arvidson CG. Lactobacilli at the front line of defense against vaginally acquired infections. Future microbiology. 2011;6:567–582. doi: 10.2217/fmb.11.36. [DOI] [PubMed] [Google Scholar]
- Stevenson M, Stanwick TL, Dempsey MP, Lamonica CA. HIV-1 replication is controlled at the level of T cell activation and proviral integration. The EMBO journal. 1990;9:1551–1560. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieh DJ, Matias E, Xu H, Fought AJ, Blanchard JL, Marx PA, Veazey RS, Hope TJ. Th17 Cells Are Preferentially Infected Very Early after Vaginal Transmission of SIV in Macaques. Cell Host Microbe. 2016;19:529–540. doi: 10.1016/j.chom.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNAIDS, U.J.P.o.H.A. The gap report 2014: Adolescent girls and young women. 2014. [Google Scholar]
- Ward H, Ronn M. Contribution of sexually transmitted infections to the sexual transmission of HIV. Current opinion in HIV and AIDS. 2010;5:305–310. doi: 10.1097/COH.0b013e32833a8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodman Z. Can one size fit all? Approach to bacterial vaginosis in sub-Saharan Africa. Annals of clinical microbiology and antimicrobials. 2016;15:16. doi: 10.1186/s12941-016-0132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.