Abstract
The family Paramyxoviridae includes many viruses that significantly affect human and animal health. An essential step in the paramyxovirus life cycle is viral entry into host cells, mediated by virus-cell membrane fusion. Upon viral entry, infection results in expression of the paramyxoviral glycoproteins on the infected cell surface. This can lead to cell-cell fusion (syncytia formation), often linked to pathogenesis. Thus membrane fusion is essential for both viral entry and cell-cell fusion and an attractive target for therapeutic development. While there are important differences between viral-cell and cell-cell membrane fusion, many aspects are conserved. The paramyxoviruses generally utilize two envelope glycoproteins to orchestrate membrane fusion. Here, we discuss the roles of these glycoproteins in distinct steps of the membrane fusion process. These findings can offer insights into evolutionary relationships among Paramyxoviridae genera and offer future targets for prophylactic and therapeutic development.
Keywords: Paramyxoviridae, Paramyxovirus, attachment glycoprotein, fusion glycoprotein, viral receptors, viral entry, F-triggering, fusion cascade, membrane fusion, association model, dissociation model, fusion, attachment, fusion model, syncytia, prefusion, postfusion, prehairpin intermediate, hexamer of trimers, fusion pore formation, Nipah, Hendra, Measles, RSV, NDV, Mumps, hMPV, Hemifusion
Introduction
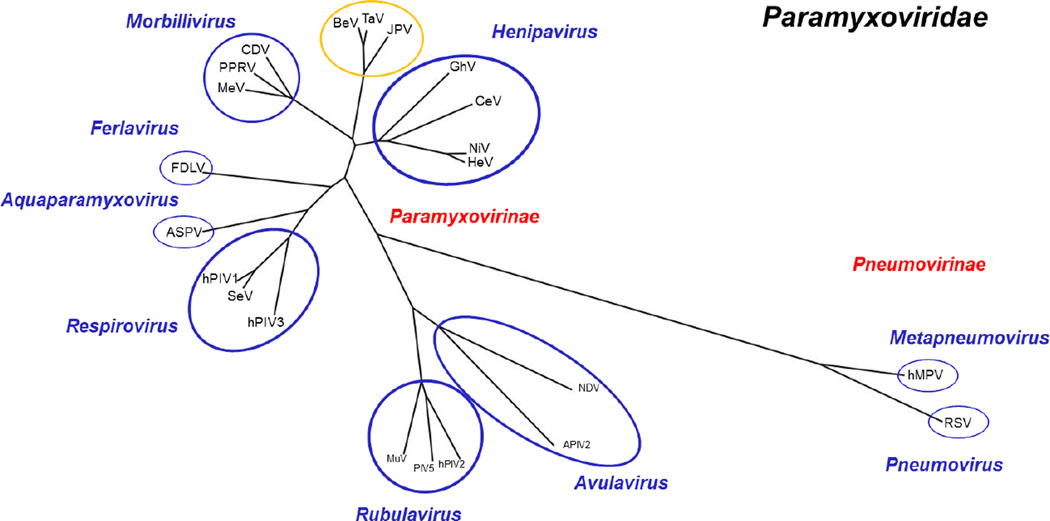
The Paramyxoviridae family of viruses contains many important pathogens that infect a wide variety of hosts including pandas, hyenas, whales, anacondas, bats, dogs, rats, avians, and humans. Members of this virus family include measles virus (MeV), respiratory syncytial virus (RSV), human metapneumovirus (hMPV), human parainfluenza viruses (e.g. hPIV3, PIV5), mumps virus (MuV), canine distemper virus (CDV), and Newcastle disease virus (NDV), as well as the deadlier zoonotic Hendra and Nipah viruses (HeV and NiV, respectively) (1–3) (Fig. 1). For example, NiV infection causes 40–90% mortality rates in humans (4–9). The Paramyxoviridae family already includes an extensive repertoire, however recent discoveries support the existence of over 60 additional paramyxoviruses in bats from South America and Africa, for example Ghana virus (GhV) (10–13).
Fig. 1. Paramyxoviridae polymerase protein phylogeny tree.
Polymerase/Large protein sequences of selected viruses were acquired from the NCBI Protein Database and Virus Pathogen Resource (ViPR). L protein sequences were aligned using the COBALT Multiple Alignment Tool-NCBI. Aligned sequences were then used to generate a phylogenic tree using COBALT NCBI TreeView1.8. The generated tree was visualized and modified using the FigTree Program. Abbreviations: APIV-2, avian parainfluenza virus 2; ASPV, Atlantic salmon paramyxovirus; BeV, Beilong virus; CDV, canine distemper virus; CeV, Cedar Virus; FDLV, Fer-de-Lance virus; GhV, Ghana virus; HeV, Hendra virus; hMPV, Human metapneumovirus; hPIV1, Human parainfluenza virus 1; hPIV2, Human parainfluenza virus 2; hPIV3, Human parainfluenza virus 3; JPV, J Paramyxovirus; PIV5, Parainfluenza virus 5; RSV, respiratory syncytial virus; MeV, Measles virus; MuV, Mumps virus; NDV, Newcastle disease virus; NiV, Nipah Virus; PPRV, Peste-des-petits ruminants virus; TaV, Tailam Virus. Paramyxoviridae family (black); sub-families (red); genus (blue); unclassified (yellow).
The Paramyxoviridae family is further subdivided into two sub-families: Paramyxovirinae and Pneumovirinae. Paramyxovirinae contains multiple genera: Avulavirus, Rubulavirus, Respirovirus, Morbillivirus, Henipavirus, Aquaparamyxovirus and Ferlavirus (3, 14–16). Pneumoviruses, on the other hand, include only two genera: Metapneumovirus and Pneumovirus. Furthermore, recently discovered viruses, such as Beilong virus (BeV), Tailam virus (TaV) and J paramyxovirus (JPV), have yet to be classified into sub-families (15, 17, 18). The evolutionary relationships between representative paramyxoviruses, as based on their relatively highly-conserved polymerase protein sequences, are outlined in Fig. 1.
Paramyxoviruses are membrane enveloped with receptor-binding (or attachment) glycoproteins HN, H, or G, and fusion glycoproteins,F, anchored in their viral membranes. The designation of the attachment glycoprotein as hemagglutinin-neuraminidase (HN), hemagglutinin (H), or glycoprotein (G) depends on protein function and differs between genera (19–26). Specifically, the HN attachment glycoprotein is characteristic of the Avulavirus, Respirovirus, and Rubulavirus genera (2, 19, 20, 26–33). HN consists of “H,” hemagglutinin activity, which mediates binding to sialic acid, and “N,” neuraminidase activity, which cleaves sialic acid. This activity assists budding virions in detaching from the infected cell and reduces re-entry of virions into other infected cells. In comparison, the H attachment glycoprotein is found in members of the Morbillivirus genus and only has hemagglutinin activity (34, 35). The G attachment glycoprotein is utilized by viruses in the genus Henipavirus and the Pneumovirinae subfamily and does not have either hemagglutinin or neuraminidase activities. Instead, the G attachment glycoproteins bind protein receptors.
For the paramyxoviruses, interactions between the viral attachment glycoprotein and the host cell receptor is generally sufficient to trigger the fusion protein, allowing virus entry. The genera Avulavirus, Rubulavirus, and Respirovirus HN proteins (e.g. those of PIV5, NDV, and hPIV1–3) utilize sialic acid as a receptor. Morbillivirus H glycoproteins (e.g. those of MeV, PPRV and CDV) can utilize CD46/MCP, CD150/SLAM, or nectin 4/PVRL4 as receptors (34, 36–43). The G glycoproteins of pneumoviruses (e.g. RSV or hMPV) can utilize nucleolin, ICAM1, heparan sulfate or other glycosaminoglycans (19, 22, 44–49). Henipavirus (e.g. NiV or HeV) G glycoproteins utilize protein receptors such as ephrinB2 and/or ephrinB3 (23, 50–57). Interestingly, there are some viruses that do not require the attachment protein to trigger fusion, such as the pneumoviruses, hMPV and RSV (20, 22, 48, 58–60). In the cases of parainfluenza virus 5 (PIV5) and Sendai virus (SeV), the fusion protein can be triggered, though less efficiently, without the presence of the attachment glycoprotein (33, 61).
Most paramyxoviruses are thought to enter host cells via direct fusion between the host cell plasma membrane and the viral membrane. Unlike other viruses, such as Ebola or Influenza viruses, paramyxoviruses are generally believed to not require endocytosis and low-pH to initiate viral and host cell membrane fusion (62–64). Once a cell is infected, the attachment and fusion glycoproteins are expressed at the cell surface (65–70). Thus, cells infected with paramyxoviruses can fuse with naïve receptor-containing cells, forming multi-nucleated cells (syncytia) (5, 7, 59, 71–75). This is considered a mode of viral spread from infected to naïve cells.
During cell infection, other viral proteins are synthesized and the genome is replicated. Paramyxoviruses contain non-segmented, single-stranded RNA genomes with negative-sense polarity (-ssRNA). Genome sizes vary among different paramyxoviruses, ranging between near 15kbp to near 19kbp. Since paramyxoviruses are -ssRNA viruses, virions must package their own RNA-dependent-RNA-polymerase (large protein), encoded by the L gene, to allow for transcription and replication. The genome also encodes for the nucleoprotein (N) that coats the RNA genome and protects it from degradation, forming ribonucleoprotein (RNP) complexes, and assists viral genome replication and virion packaging (76–80). Additionally, RNP complexes also contain the viral phosphoprotein (P) (76). The phosphoprotein complexes with the large protein to form a functioning RNA-dependent-RNA-polymerase. The P gene transcript can undergo further processing to form the V and W proteins, while C is translated via frame shift. These proteins play important roles in blocking innate immune responses, enhancing virulence, and managing viral RNA synthesis in many paramyxoviruses (81–88). Finally, the matrix protein (M) is generally required for complete viral assembly and budding (69, 79, 89–95). The presence of the N, P, L, M proteins and the surface glycoproteins is conserved across the Paramyxoviridae family.
The glycoproteins
The attachment glycoproteins of paramyxoviruses form tetramers, specifically dimers of dimers (21, 24–26, 34, 35, 53). Each monomer is capable of binding a receptor molecule. Once bound by the appropriate receptor, conformational changes in the attachment glycoprotein are generally thought to expose the stalk region, triggering the fusion glycoprotein to undergo the membrane fusion cascade (21, 96–104) (Fig. 2A).
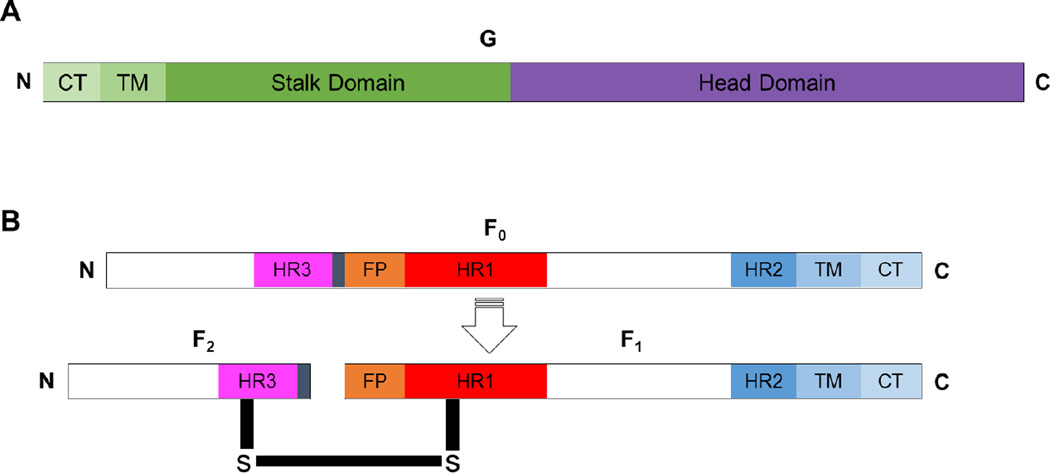
Fig. 2. Paramyxovirus attachment and fusion glycoproteins.
A) Diagram depicting the attachment glycoprotein. B) Diagram depicting the precursor of the paramyxovirus fusion glycoprotein (F0, top) and the cleaved, biologically active and disulfide linked paramyxovirus protein (F1-F2, bottom). The fusion peptide (FP), heptad repeats HR1, HR2, and HR3, transmembrane (TM); and cytoplasmic tail (CT) domains are shown, and the N- and C-termini are indicated.
The fusion glycoprotein of paramyxoviruses is classified as a trimeric Class I fusion protein (105–111). Class I fusion proteins exhibit high alpha-helical secondary structural content, and typically require processing to expose the intramolecular hydrophobic fusion peptide (FP). Before the fusion protein is biologically active, it is made as a precursor (F0) that is transported to the cell surface and then endocytosed. In the endosomal compartment, the precursor glycoprotein (F0) is cleaved by either cathepsins (e.g. for HeV or NiV) or furin-like proteases (e.g. for RSV, NDV) (112–115). The processed precursor is composed of F1 and F2 subunits that are disulfide linked and transported back to the plasma membrane as a fusion-active unit (Fig. 2B).
The fusion core of an F trimer consists of the heptad repeat one and two (HR1 and HR2) domains (105, 116, 117). The HR1 domain is located proximal to the N-terminus of the F1 subunit and is designated as HR1, HRA, or HRN. Conversely, the HR2 domain is proximal to the C-terminus and designated as HR2, HRB, or HRC (105, 116, 117). Importantly, an additional helical region is located upstream from the fusion peptide, as a part of F2, and designated as HR3 or HRC (111, 118). As can be seen from these designations, two completely distinct regions can be called HRC. Therefore, to avoid confusion, we will use the nomenclature HR1, HR2, and HR3 to refer to these 3 helical regions (Fig. 2B).
The HR1 and HR2 alpha-helical domains have a high propensity to bind each other and form antiparallel helical bundles (105, 106, 116, 117). The HR1 domain, adjacent to the FP is located towards the head of the fusion protein, is relatively hydrophobic, and is shielded in the metastable, pre-fusion conformation. On the other hand, the amphipathic HR2 region is exposed to aqueous solutions and exists in a “stalk-like” conformation. The binding of HR1 and HR2 provides the energy for viral and cellular membrane merging. The HR3 domain has been shown to not interact with the HR1 or HR2 domains in the case of RSV, suggesting other structural roles (106, 111, 118, 119). For PIV5, an analogous domain in the F2 subunit affects the fusion capabilities of F (120). The role of the HR3 domain remains a knowledge gap for most paramyxoviruses.
Structural information for the H, HN, and G Glycoproteins
Crystal structures for the extracellular, globular domains of the H, HN, and G glycoproteins have been solved for a substantial portion of the paramyxoviruses, including at least one virus from each genus within the Paramyxovirinae sub-family (26, 28, 53, 121, 122). While there are some differences between the receptor attachment proteins of different paramyxoviruses, considerable structural similarities stand out. First, paramyxovirus receptor attachment proteins are transmembrane type II proteins comprised of an N-terminal cytoplasmic tail followed by a transmembrane domain, a stalk, and a globular head. Additionally, attachment glycoproteins tetramerize into dimers of dimers (67). Until somewhat recently, the structures of paramyxoviral attachment protein stalks were unknown; however, recent studies with Newcastle disease virus have shown that the helical stalks of each monomer cluster to form a four-helix bundle (4HB) (25). Beyond the stalk, the C-terminal globular head of each monomer contains a β-propeller with six blades, each with four β-sheets running anti-parallel to each other (1). Close structural analysis of the monomers in complex reveals that the interactions in the stalk and across the β-1 and β-6 blades are critical to the formation of the tetramer (53, 67). The geometries of HN, H, or G glycoproteins are highly similar. Relatively lower similarity between the structures of G and H, as well as the existence of an enlarged, nonfunctional sialic acid binding motif within the MeV H protein, supports a model where G and H proteins diverged from HN towards binding proteinaceous receptors (67, 123).
Evidence for specific attachment-fusion protein interaction sites
For both HN and G glycoproteins, receptor interactions have been shown to occur within binding pockets in the head. Two examples of this are shown in Fig. 3 (26, 52). This is contrasted with findings that the receptors for H bind to the side of the head, as reviewed in (123, 124). While it has not always been the case, current understanding is that upon receptor binding, the attachment protein undergoes conformational changes that, in turn, trigger the fusion protein to undergo its own series of conformational changes, ultimately leading to membrane fusion executed by F (53). Examples of active site changes upon receptor binding have been seen for the NiV G, NDV HN, and PIV3 HN proteins (25, 53, 122). In the case of NiV G, structural and biochemical assays support two specific and sequential conformational changes in the head domain that act to reveal the stalk for F-triggering (97). Similarly, MeV H heads are suggested to shift during F-triggering (125). At least one study has identified F-HN sites of interaction for NDV (126). While the interactions between the fusion and attachment glycoproteins are not well understood, studies have shown the importance of the attachment protein stalk domain in such interactions (21, 97). For some paramyxoviruses, the globular head may also be involved in interactions with F, specifically during F-triggering (127, 128). Models of structural interactions between F and H/HN/G exist, yet the absence of any co-crystallization of these proteins remains a significant objective for the field.
Fig. 3. Receptor binding with attachment glycoproteins occurs at the head domain.
The attachment protein binds the cellular receptor within a binding pocket(s) in the globular head. As illustrated, the glycoprotein monomer heads have separate β-blades distinguished by different colors for structures of the PIV5 HN (A) or NiV G (B). In both of these ribbon models sialic acid (A; orange cluster) or ephrinB2 (B; colored grey) binds the attachment protein towards the center of the β-propeller. This figure was adapted from (123). The PDB code for the NiV G-ephrin B2 complex is 2VSM (52).
Conserved structural patterns in paramyxovirus fusion proteins
There are several knowledge gaps concerning the structures of the paramyxovirus fusion protein. Obstacles include its transition through several extremely different conformations, protease-dependent processing that occurs during fusion protein maturation, and the transient nature of the pre-hairpin intermediate conformation (PHI). In contrast to the attachment glycoproteins, fusion glycoproteins are type I transmembrane proteins with their extracellular regions at the N-terminus and their transmembrane (TM) and cytoplasmic tail (CT) domains at the C-terminus (Fig. 2).
Structures have been solved for fusion proteins from most of the genera in the Paramyxoviridae family. Specifically, pre-fusion structures are available for HeV, NDV, NiV, and PIV5 (107, 109–111, 119, 129–133). Work for PIV5 has demonstrated that the F pre-fusion conformational structure does not significantly change after cleavage (107). While no paramyxovirus fusion protein has been effectively crystallized at the PHI, its existence is supported by many biochemical studies and by recent findings using transmission electron microscopy (107). The post-fusion six-helix bundle (6HB) conformation has been resolved for RSV, NDV, and hPIV3 (119, 131, 132). Examples for the pre- and post-fusion conformations of F are shown in Fig. 4 (119, 131). The least understood conformation of F is the transient PHI. Determination of this structure through traditional crystallographic means is unlikely to occur. Thus the field must find alternative methods to study this conformation.
Fig. 4. The pre- and post-fusion conformations of paramyxovirus F glycoproteins.
Substantial structural rearrangements occur in the progression from pre-fusion to post-fusion F glycoprotein conformations. Several domains are distinguished by color including the large HR1-containing region (magenta) and the HR2 domain (blue). In the pre-fusion structure from PIV5 (A), a trimeric stabilization domain was added to support crystallization (grey). The six-helix bundle conformation is apparent in the hPIV3 structure (B). This figure was adapted from (111). The PDB codes for A and B are 2B9B and 1ZTM, respectively (111, 119).
The paramyxovirus membrane fusion process
Several steps are recognized as important during paramyxoviral viral-cell or cell-cell membrane fusion. Early steps include: binding of the attachment protein HN/H/G to its cognate cell-surface receptor, the resulting activation of the HN/H/G protein to undergo its own set of conformational rearrangements, and the subsequent triggering of the F protein to undergo extensive conformational rearrangements that result in membrane merging. Later steps in the membrane fusion process include fusion pore formation and expansion. Noteworthy, although these are very well recognized membrane fusion steps, additional steps in this complex process may remain to be discovered (108, 134, 135).
Early steps in the paramyxovirus membrane fusion process
Triggering of F by the attachment protein HN, H, or G
Despite the lack of structural data for paramyxoviral attachment-fusion glycoprotein interactions, all functional data available points to such interactions being essential for members of the Paramyxovirinae subfamily, and for some members of the Pneumovirinae subfamily. Our knowledge of both the spatial and temporal nature of these interactions is relatively poor. However, we know that these interactions can occur at different times for different paramyxoviruses. This has given rise to the Association vs. Dissociation models of attachment-fusion protein interactions (31).
Association vs. dissociation models
At least two models can explain the differences in the timing of the attachment-fusion glycoprotein interactions. The HN glycoproteins of paramyxoviruses that bind sialic acid, such as PIV5, hPIV3, NDV, etc., do not appear to bind their homologous fusion glycoproteins until after HN has bound its respective receptor, appearing to follow the association or provocateur model. In contrast, the H or G glycoproteins that attach to membrane protein receptors, such as MeV or CDV H, which can bind SLAM or CD46, or NiV or HeV, which can bind ephrinB2 and/or ephrinB3, appear to interact with the F protein prior to receptor binding. Then, upon receptor binding the glycoproteins relatively dissociate, following the dissociation or clamp model. These models have been previously reviewed at length (27, 31, 32). Multiple publications have shown strong evidence to support either the association model for the sialic acid binding paramyxoviruses, such as NDV (103), or to support the dissociation model for protein-binding viruses, such as MeV, NiV, or HeV (127, 136–140).
Five models of F activation
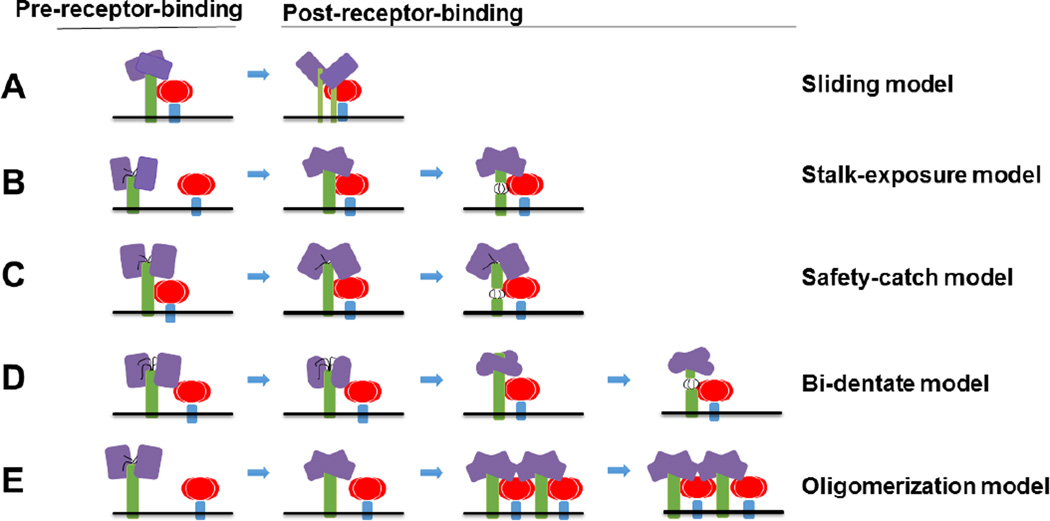
We know a substantial amount regarding how Class I fusion proteins execute membrane fusion, from studying not only paramyxoviral F proteins, but also other RNA viral glycoproteins such as influenza virus HA and HIV gp41. However, we know relatively little regarding how the paramyxoviral F protein receives the signal from G, and how G couples receptor binding with F-triggering. There are at least five proposed modes of F-triggering by G. Namely, the sliding, stalk-exposure, safety-catch, bi-dentate, and oligomerization models (Fig. 5).
Fig. 5. Five models of F activation (A–E).
Attachment glycoprotein tetramers are colored in purple (heads) and green (stalks, transmembrane, and cytoplasmic tail domains). F is highlighted in red (heads) and blue (stalks, transmembrane, and cytoplasmic tail domains). In the bi-dentate model, the conformational changes of the head are shown as slightly distinct shapes. F-triggering regions in the stalk are shown as black lines (B–D).
The sliding model proposes that upon receptor binding, the heads of the attachment glycoprotein slide laterally from a planar to a more staggered configuration (Fig. 5A). In turn the stalk of the protein would move from a compact closed 4HB to a partially dissociated conformation, leading to disengagement from and “freedom” of F. This model infers that the heads, upon receptor binding, generate specific signals that travel down to the stalks for F activation. Since head/stalk hybrid attachment proteins have been reported to preserve F-triggering activity for example for MeV, and mutants that assume the activated attachment protein conformation still trigger membrane fusion, this model may at best not fit all paramyxoviruses (37, 141).
The stalk-exposure/induce fit model proposes that receptor binding may induce conformational changes of the attachment protein from a four-heads-down conformation to a (at least) some-heads-up conformation. Such heads-up conformation would free a stalk domain in the attachment protein to interact with F (Fig. 5B). Both the latest structural and mechanistic data for NDV and PIV5 HN proteins (25, 100, 142), as well as the fact that the headless attachment proteins of PIV5, MeV, and NiV can trigger F, are consistent with the stalk-exposure model (97, 101, 102). It is important to note, however, that for several viruses, including MeV, CDV, hPIV3, and NiV, attachment protein-F interactions exist prior to receptor binding, so variations of this model are likely.
The safety-catch model proposes that the attachment protein heads, for example in the four-heads-down conformation would fold into an auto-repressed conformation, as in “locking” the ability of their stalks to trigger F (Fig. 5C). The “safety lock” has been proposed to be provided by head-stalk connecting domains or spacers, which can also be found solely in the stalk domains (98, 102, 143). This safety lock would be removed by movement of the heads upon receptor binding. This model suggests that F can interact with the attachment protein prior to receptor binding without premature triggering because of the auto-repressed conformation of the attachment protein. This model may be seen as a variation of the stalk-exposure/induce fit model just described above.
The bi-dentate model has been originally suggested for NiV. This model proposes that F interacts with both the head and stalk of the attachment protein, and that receptor binding switches the major interactions with the fusion protein from the head to the stalk of the attachment protein (Fig. 5D). Evidence to support this model include the strong interactions of G and F prior to receptor binding (127, 136, 137), the interactions of G and F when N-glycans were introduced to the stalk (144), and the three receptor-induced conformational changes observed in G, two in the head and one in the stalk, prior to F-triggering (97).
The Oligomerization model proposes that receptor binding may lead to increased attachment protein oligomerization, which in turn triggers F (Fig. 5E). This model has been suggested based on the fact that the heads-up conformation may not be sufficient for F-triggering for hPIV3 (145). However, there is no direct evidence for the higher-order formation of oligomers upon receptor binding. Noteworthy, NiV F has been recently reported to form hexamers of trimers, and such oligomeric formations appear to have a role in membrane fusion modulation (110). The role of G oligomerization in the activation of such F oligomeric complexes remains to be determined.
It is important to note that these models are not necessarily mutually exclusive. There is a level of redundancy among the models, and two or more of them may apply to a single paramyxovirus. More structural and functional data, particularly for attachment fusion protein interactions, will likely help further describe and corroborate these models. Cryo-TEM and single particle 3D reconstruction from electron micrographs using direct electron detection (DED) technology might be helpful for this endeavor.
Late steps in the paramyxovirus membrane fusion process
Fusion of membrane bilayers is not spontaneous or trivial. Only by expending the energy stored in the metastable pre-fusion F trimer (Fig. 6A), through irreversible conformational changes, can the paramyxoviruses gain access to its host cell machinery (146). Additionally, this process may even require several F proteins undergoing synchronized conformational changes to overcome the high-energy barrier of membrane fusion, with the henipaviruses potentially using six F trimers to execute membrane fusion (33, 110). F mediated membrane fusion typically occurs at a neutral pH, shown for PIV5 (147); although low pH does not inhibit entry in PIV5, RSV, or NDV (147–149). SER virus, as well as certain hMPV strains, are known exceptions and do have low pH requirements (150, 151). Most Paramyxoviridae virions likely directly fuse with their target cell membrane, but there is evidence that endocytic processes may occur for NiV, NDV, and RSV (150, 152–155). MuV has been shown to be sensitive to calcium concentrations and a calcium-channel agonist has shown inhibition, while SeV is unaffected by either (156, 157). Other requirements may also exist.
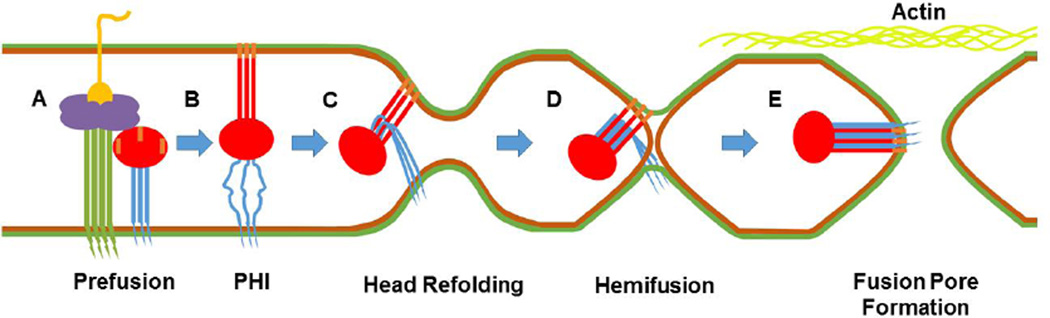
Fig. 6. Model of the late steps in paramyxovirus membrane fusion.
Inner and outer membrane leaflets of viral and cellular membranes are shown as green and brown, respectively. The attachment protein is shown with a green stalk and purple head, while yellow depicts its bound receptor. For F, the cytoplasmic tail (CT), transmembrane (TM) domain, and HR2 are shown in blue. The head region through HR1 is shown in red and the fusion peptide in orange. A) The attachment glycoprotein binds its receptor and triggers prefusion F. B) To reach the prehairpin intermediate (PHI), HR2 melts and forms an open stalk. HR1 extends and projects the fusion peptide into the target membrane. (C) The head continues to refold, bringing HR1 and HR2 into close proximity, and pulls the membranes in. (D) HR1 and HR2 “zipper” together and the outer membranes fuse to form the hemifusion intermediate. (E) Inner membranes fuse as zippering continues through the transmembrane domain, forming the 6-helix bundle. F’s CT is now exposed to intracellular proteins and the fusion pore expands. The actin cytoskeleton (shown in light green) is likely involved in fusion pore expansion.
A recent crystal structure of pre-fusion NiV F trimers displays a hexamer-of-trimer architecture. Using this structure to establish a hypothetical, hexameric model for F, keeping in mind that evidence for this is NiV-specific, individual trimers could be interacting with a part of HR1, the FP, and a part of F2. Two monomers in each F trimer appear to be aligned for interactions with adjacent trimers, while the third monomer faces away and remains exposed. This exposed monomer could be the site where G initiates triggering (110). With FPs and their surrounding regions interacting with neighboring F trimers, and with dramatic conformational changes occurring as F transitions into the PHI, the transition, itself, of F into a PHI could facilitate triggering of neighboring trimers. Destabilization of a neighbor’s pre-fusion conformation by “pulling” up the FP could result in a triggering event. The signal would then continue trimer to trimer within the hexamer. This scheme of cooperative triggering provides an intermolecular basis for how a single triggered F may kinetically relay a signal from the attachment protein to other F proteins and may address the need for multiple F proteins to be involved in membrane fusion by spatiotemporally synchronizing multiple (six?) fusion cascades.
Although it has not been formally shown to exist for all members of the Paramyxoviridae family, the PHI is known to form after F triggering (Fig. 6B). To reach this conformation, HR2 melts, losing its coiled-coil structure, forming an open stalk. HR1 refolds into an elongated helix and harpoons outwards towards the target membrane forming a new, trimeric coiled-coil stalk. The FP, attached to HR1, is projected towards and inserted into the target membrane. Hydrophobicity anchors the FP within the target membrane as a helix and the FP may further oligomerize, as PIV5 FPs have shown the capacity to form hexameric bundles (158). While the PHI’s existence has been most demonstrated for PIV5, there are significant similarities in membrane fusion across the family (130).
After reaching the PHI, it has been stated that the globular head of F undergoes conformational changes aligning HR1 and HR2 antiparallel to one another. We speculate that conformational changes in the globular head of F pull the N-terminal end of the open stalk, amphipathic HR2 to the C-terminal end of the newly formed trimeric HR1 stalk (Fig. 6C). These two domains have a high avidity for one another, so the initial meeting of HR1 and HR2 may lead to a “zippering” event along the grooves in the HR1 stalk. As HR2 zippers down the HR1 stalk, it regains structure, being pulled back into a helix, as seen in post-fusion crystal structures. Zippering progresses and the membranes bound proximally to each HR are then brought closer together. Once HR1 and HR2 have fully bound, the membranes have been pressed together and the outer leaflets fuse. The instability of a strong positive curvature brought on by pulling the membranes together and with continued force from HR1:HR2 interactions, may cause lipids in the outer leaflet to reorient into a more stable, partially fused structure: hemifusion (Fig. 6D). With a theoretical length of 25–48 amino acids, for PIV5, it is hypothesized that the TM domain of F may help stabilize hemifusion by spanning its membrane and the connecting region between the two fused outer leaflets, although this has yet to be confirmed (159). To our knowledge there is no direct evidence to support the slow HR1-HR2 zippering process; however, kinetic evidence that the transition from PHI to 6HB takes on the order of a few minutes, not seconds or milliseconds (at least for NiV F), makes this model plausible.
Fusion pore formation is thought to be mediated by the ectodomain of F. It has been shown that truncations of the PIV5 CT can progress through hemifusion and fuse the inner leaflets, forming a small pore between the virus and its host (160, 161). In contrast, truncations of SER virus CT rescued full syncytium formation, despite the wild-type F pH requirement (151, 162). Zippering likely continues into the membrane itself by interactions between the FP and TM domains (158). The FPs form the bundle’s core and TM domains lodge along grooves formed by the three clustered helices, as shown for RSV, NDV, and hPIV3 post-fusion crystal structures (119, 131–133). This final conformation of F is the 6HB (Fig 6E): a low energy, irreversible state indicative of a fully expended high-energy protein (163).
While the fusion pore has formed, viral RNA cannot pass through, as the estimated 1–2nm pore is not large enough (164). Subsequent expansion of the fusion pore is thought to be mediated by the F CT (161). Also, for PIV5, actin has been shown to inhibit fusion pore expansion (165). This may indicate that a major obstacle in pore expansion could be the cytoskeleton. While fusion pore expansion has not been well studied, intracellular proteins, such as Rho GTPases for RSV and HeV, have been shown to affect syncytium formation (166, 167). Interactions with intracellular proteins by the F CT, or by unknown functional domains of these viral proteins, somehow lead to cytoskeletal rearrangements or other processes necessary to enlarge the fusion pore (168, 169).
Conclusions
In summary, membrane fusion is accomplished by two paramyxovirus glycoproteins: the attachment and fusion glycoproteins. While there are differences in how this crucial aspect of the paramyxoviral life cycle is accomplished, there are many commonalities. All steps of the membrane fusion process, from receptor binding to fusion pore expansion, may offer future targets for therapeutic development. The more we understand each of these steps, the better we will be prepared to target them.
Footnotes
Compliance with Ethics Guidelines
Conflict of Interest
Hector Aquilar-Carrenos, Bryce Henderson, Juana Zamora declare no conflicts of interest
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
REFERENCES
Papers of particular interest, published recently, have been highlighted as:
•Of importance
••Of major importance
- 1.Bossart KN, Fusco DL, Broder CC. Paramyxovirus Entry. Viral Entry into Host Cells. 2013;790:95–127. doi: 10.1007/978-1-4614-7651-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang A, Dutch RE. Paramyxovirus fusion and entry: multiple paths to a common end. Viruses. 2012;4:613–636. doi: 10.3390/v4040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enders G. Paramyxoviruses. In: Baron S, editor. Medical Microbiology. 4th. Galveston (TX): 1996. [Google Scholar]
- 4.Ching PK, de los Reyes VC, Sucaldito MN, Tayag E, Columna-Vingno AB, Malbas FF, Jr, Bolo GC, Jr, Sejvar JJ, Eagles D, Playford G, Dueger E, Kaku Y, Morikawa S, Kuroda M, Marsh GA, McCullough S, Foxwell AR. Outbreak of henipavirus infection, Philippines, 2014. Emerg Infect Dis. 2015;21:328–331. doi: 10.3201/eid2102.141433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goh KJ, Tan CT, Chew NK, Tan PS, Kamarulzaman A, Sarji SA, Wong KT, Abdullah BJ, Chua KB, Lam SK. Clinical features of Nipah virus encephalitis among pig farmers in Malaysia. N Engl J Med. 2000;342:1229–1235. doi: 10.1056/NEJM200004273421701. [DOI] [PubMed] [Google Scholar]
- 6.Harit AK, Ichhpujani SG, Gill KS, Lal S, Ganguly NK, Agarwal SP. Nipah/Hendra virus outbreak in Siliguri, West Bengal, India in 2001. Indian J Med Res. 2006;123:553–560. [PubMed] [Google Scholar]
- 7.Hossain MJ, Gurley ES, Montgomery JM, Bell M, Carroll DS, Hsu VP, Formenty P, Croisier A, Bertherat E, Faiz MA, Azad AK, Islam R, Molla MA, Ksiazek TG, Rota PA, Comer JA, Rollin PE, Luby SP, Breiman RF. Clinical presentation of nipah virus infection in Bangladesh. Clin Infect Dis. 2008;46:977–984. doi: 10.1086/529147. [DOI] [PubMed] [Google Scholar]
- 8.Luby SP, Hossain MJ, Gurley ES, Ahmed BN, Banu S, Khan SU, Homaira N, Rota PA, Rollin PE, Comer JA, Kenah E, Ksiazek TG, Rahman M. Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001–2007. Emerg Infect Dis. 2009;15:1229–1235. doi: 10.3201/eid1508.081237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parashar UD, Sunn LM, Ong F, Mounts AW, Arif MT, Ksiazek TG, et al. Case-control study of risk factors for human infection with the new zoonotic paramyxoviruses, Nipah virus, during a 1998–1999 outbreak of severe encephalitis in Malaysia. J Infect Dis. 2000;181:1755–1759. doi: 10.1086/315457. [DOI] [PubMed] [Google Scholar]
- 10.Drexler JF, Corman VM, Gloza-Rausch F, Seebens A, Annan A, Ipsen A, Kruppa T, Muller MA, Kalko EK, Adu-Sarkodie Y, Oppong S, Drosten C. Henipavirus RNA in African bats. PLoS One. 2009;4:e6367. doi: 10.1371/journal.pone.0006367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peel AJ, Baker KS, Crameri G, Barr JA, Hayman DT, Wright E, Broder CC, Fernandez-Loras A, Fooks AR, Wang LF, Cunningham AA, Wood JL. Henipavirus neutralising antibodies in an isolated island population of African fruit bats. PLoS One. 2012;7:e30346. doi: 10.1371/journal.pone.0030346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pernet O, Schneider BS, Beaty SM, LeBreton M, Yun TE, Park A, Zachariah TT, Bowden TA, Hitchens P, Ramirez CM, Daszak P, Mazet J, Freiberg AN, Wolfe ND, Lee B. Evidence for henipavirus spillover into human populations in Africa. Nature Communications. 2014:5. doi: 10.1038/ncomms6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Drexler JF, Corman VM, Muller MA, Maganga GD, Vallo P, Binger T, Gloza-Rausch F, Cottontail VM, Rasche A, Yordanov S, Seebens A, Knornschild M, Oppong S, Adu Sarkodie Y, Pongombo C, Lukashev AN, Schmidt-Chanasit J, Stocker A, Carneiro AJ, Erbar S, Maisner A, Fronhoffs F, Buettner R, Kalko EK, Kruppa T, Franke CR, Kallies R, Yandoko ER, Herrler G, Reusken C, Hassanin A, Kruger DH, Matthee S, Ulrich RG, Leroy EM, Drosten C. Bats host major mammalian paramyxoviruses. Nat Commun. 2012;3:796. doi: 10.1038/ncomms1796. This study surveyed approximately 5,000 bat samples from several bat species across continents and has led to the discoveries of novel Paramyxoviruses in South America and Africa.
- 14.Falk K, Batts WN, Kvellestad A, Kurath G, Wiik-Nielsen J, Winton JR. Molecular characterisation of Atlantic salmon paramyxovirus (ASPV): a novel paramyxovirus associated with proliferative gill inflammation. Virus Res. 2008;133:218–227. doi: 10.1016/j.virusres.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Kurath G, Batts WN, Ahne W, Winton JR. Complete genome sequence of Fer-de-Lance virus reveals a novel gene in reptilian paramyxoviruses. J Virol. 2004;78:2045–2056. doi: 10.1128/JVI.78.4.2045-2056.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basler CF, Garcia-Sastre A, Palese P. A novel paramyxovirus? Emerg Infect Dis. 2005;11:108–112. doi: 10.3201/eid1101.040653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magoffin DE, Mackenzie JS, Wang LF. Genetic analysis of J-virus and Beilong virus using minireplicons. Virology. 2007;364:103–111. doi: 10.1016/j.virol.2007.01.045. [DOI] [PubMed] [Google Scholar]
- 18.Woo PC, Lau SK, Wong BH, Wong AY, Poon RW, Yuen KY. Complete genome sequence of a novel paramyxovirus, Tailam virus, discovered in Sikkim rats. J Virol. 2011;85:13473–13474. doi: 10.1128/JVI.06356-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adamson P, Thammawat S, Muchondo G, Sadlon T, Gordon D. Diversity in glycosaminoglycan binding amongst hMPV G protein lineages. Viruses. 2012;4:3785–3803. doi: 10.3390/v4123785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biacchesi S, Pham QN, Skiadopoulos MH, Murphy BR, Collins PL, Buchholz UJ. Infection of nonhuman primates with recombinant human metapneumovirus lacking the SH, G, or M2–2 protein categorizes each as a nonessential accessory protein and identifies vaccine candidates. J Virol. 2005;79:12608–12613. doi: 10.1128/JVI.79.19.12608-12613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bose S, Welch BD, Kors CA, Yuan P, Jardetzky TS, Lamb RA. Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. Journal of virology. 2011;85:12855–12866. doi: 10.1128/JVI.06350-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang A, Masante C, Buchholz UJ, Dutch RE. Human metapneumovirus (HMPV) binding and infection are mediated by interactions between the HMPV fusion protein and heparan sulfate. Journal of virology. 2012;86:3230–3243. doi: 10.1128/JVI.06706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee B, Pernet O, Ahmed AA, Zeltina A, Beaty SM, Bowden TA. Molecular recognition of human ephrinB2 cell surface receptor by an emergent African henipavirus. Proc Natl Acad Sci U S A. 2015;112:E2156–E2165. doi: 10.1073/pnas.1501690112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan P, Leser GP, Demeler B, Lamb RA, Jardetzky TS. Domain architecture and oligomerization properties of the paramyxovirus PIV 5 hemagglutinin-neuraminidase (HN) protein. Virology. 2008;378:282–291. doi: 10.1016/j.virol.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yuan P, Swanson KA, Leser GP, Paterson RG, Lamb RA, Jardetzky TS. Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14920–14925. doi: 10.1073/pnas.1111691108. This study offered the first tructural understanding of an attachment glycoprotein stalk. Since the stalk is often associated with F-triggering, structural data is valuable to gain insight into this process.
- 26.Yuan P, Thompson TB, Wurzburg BA, Paterson RG, Lamb RA, Jardetzky TS. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13:803–815. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 27.Aguilar HC, Iorio RM. Henipavirus Membrane Fusion and Viral Entry. Current topics in microbiology and immunology. 2012;359:79–94. doi: 10.1007/82_2012_200. [DOI] [PubMed] [Google Scholar]
- 28.Crennell S, Takimoto T, Portner A, Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 29.Iorio RM, Field GM, Sauvron JM, Mirza AM, Deng R, Mahon PJ, Langedijk JP. Structural and functional relationship between the receptor recognition and neuraminidase activities of the Newcastle disease virus hemagglutinin-neuraminidase protein: receptor recognition is dependent on neuraminidase activity. J Virol. 2001;75:1918–1927. doi: 10.1128/JVI.75.4.1918-1927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iorio RM, Glickman RL, Sheehan JP. Inhibition of fusion by neutralizing monoclonal antibodies to the haemagglutinin-neuraminidase glycoprotein of Newcastle disease virus. The Journal of general virology. 1992;73(Pt 5):1167–1176. doi: 10.1099/0022-1317-73-5-1167. [DOI] [PubMed] [Google Scholar]
- 31.Iorio RM, Mahon PJ. Paramyxoviruses: different receptors - different mechanisms of fusion. Trends Microbiol. 2008;16:135–137. doi: 10.1016/j.tim.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iorio RM, Melanson VR, Mahon PJ. Glycoprotein interactions in paramyxovirus fusion. Future Virol. 2009;4:335–351. doi: 10.2217/fvl.09.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dutch RE, Joshi SB, Lamb RA. Membrane Fusion Promoted by Increasing Surface Densities of the Paramyxovirus F and HN Proteins: Comparison of Fusion Reactions Mediated by Simian Virus 5 F, Human Parainfluenza Virus Type 3 F, and Influenza HA. Journal of Virology. 1998;72:7745–7753. doi: 10.1128/jvi.72.10.7745-7753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santiago C, Celma ML, Stehle T, Casasnovas JM. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat Struct Mol Biol. 2010;17:124–129. doi: 10.1038/nsmb.1726. [DOI] [PubMed] [Google Scholar]
- 35.Brindley MA, Plemper RK. Blue native PAGE and biomolecular complementation reveal a tetrameric or higher-order oligomer organization of the physiological measles virus attachment protein H. J Virol. 2010;84:12174–12184. doi: 10.1128/JVI.01222-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hashiguchi T, Ose T, Kubota M, Maita N, Kamishikiryo J, Maenaka K, Yanagi Y. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nature structural & molecular biology. 2011;18:135–141. doi: 10.1038/nsmb.1969. [DOI] [PubMed] [Google Scholar]
- 37.Khosravi M, Bringolf F, Rothlisberger S, Bieringer M, Schneider-Schaulies J, Zurbriggen A, Origgi F, Plattet P. Canine Distemper Virus Fusion Activation: Critical Role of Residue E123 of CD150/SLAM. Journal of Virology. 2016;90:1622–1637. doi: 10.1128/JVI.02405-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melia MM, Earle JP, Abdullah H, Reaney K, Tangy F, Cosby SL. Use of SLAM and PVRL4 and identification of pro-HB-EGF as cell entry receptors for wild type phocine distemper virus. PLoS One. 2014;9:e106281. doi: 10.1371/journal.pone.0106281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muhlebach MD, Mateo M, Sinn PL, Prufer S, Uhlig KM, Leonard VH, Navaratnarajah CK, Frenzke M, Wong XX, Sawatsky B, Ramachandran S, McCray PB, Cichutek K, von Messling V, Lopez M, Cattaneo R. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature. 2011 doi: 10.1038/nature10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noyce RS, Bondre DG, Ha MN, Lin LT, Sisson G, Tsao MS, Richardson CD. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS pathogens. 2011;7:e1002240. doi: 10.1371/journal.ppat.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noyce RS, Richardson CD. Nectin 4 is the epithelial cell receptor for measles virus. Trends Microbiol. 2012;20:429–439. doi: 10.1016/j.tim.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Sellin CI, Davoust N, Guillaume V, Baas D, Belin MF, Buckland R, Wild TF, Horvat B. High pathogenicity of wild-type measles virus infection in CD150 (SLAM) transgenic mice. J Virol. 2006;80:6420–6429. doi: 10.1128/JVI.00209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welstead GG, Hsu EC, Iorio C, Bolotiin S, Richardson CD. Mechanism of CD150 (SLAM) Down Regulation from the Host Cell Surface by Measles Virus Hemagglutinin Protein. Journal of Virology. 2004;78:9666–9674. doi: 10.1128/JVI.78.18.9666-9674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tayyari F, Marchant D, Moraes TJ, Duan W, Mastrangelo P, Hegele RG. Identification of nucleolin as a cellular receptor for human respiratory syncytial virus. Nat Med. 2011;17:1132–1135. doi: 10.1038/nm.2444. [DOI] [PubMed] [Google Scholar]
- 45.Behera AK, Matsuse H, Kumar M, Kong X, Lockey RF, Mohapatra SS. Blocking intercellular adhesion molecule-1 on human epithelial cells decreases respiratory syncytial virus infection. Biochem Biophys Res Commun. 2001;280:188–195. doi: 10.1006/bbrc.2000.4093. [DOI] [PubMed] [Google Scholar]
- 46.Schildgen V, van den Hoogen B, Fouchier R, Tripp RA, Alvarez R, Manoha C, Williams J, Schildgen O. Human Metapneumovirus: lessons learned over the first decade. Clinical microbiology reviews. 2011;24:734–754. doi: 10.1128/CMR.00015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schowalter RM, Chang A, Robach JG, Buchholz UJ, Dutch RE. Low-pH Triggering of Human Metapneumovirus Fusion: Essential Residues and Importance in Entry. Journal of Virology. 2009;83:1511–1522. doi: 10.1128/JVI.01381-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thammawat S, Sadlon TA, Hallsworth PG, Gordon DL. Role of cellular glycosaminoglycans and charged regions of viral G protein in human metapneumovirus infection. J Virol. 2008;82:11767–11774. doi: 10.1128/JVI.01208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Bleek GM, Osterhaus AD, de Swart RL. RSV 2010: Recent advances in research on respiratory syncytial virus and other pneumoviruses. Vaccine. 2011;29:7285–7291. doi: 10.1016/j.vaccine.2011.07.114. [DOI] [PubMed] [Google Scholar]
- 50.Aguilar HC, Fang AQ, Aspericueta V, Negrete OA, Akyol Z, Lee B. EphrinB2 receptor-induced conformational changes in the Nipah virus attachment glycoprotein [Google Scholar]
- 51.Bonaparte MI, Dimitrov AS, Bossart KN, Crameri G, Mungall BA, Bishop KA, Choudhry V, Dimitrov DS, Wang L, Eaton BT, Broder CC. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci U S A. 2005;102:10652–10657. doi: 10.1073/pnas.0504887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bowden TA, Aricescu AR, Gilbert RJ, Grimes JM, Jones EY, Stuart DI. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat Struct Mol Biol. 2008;15:567–572. doi: 10.1038/nsmb.1435. [DOI] [PubMed] [Google Scholar]
- 53.Bowden TA, Crispin M, Harvey DJ, Aricescu AR, Grimes JM, Jones EY, Stuart DI. Crystal structure and carbohydrate analysis of Nipah virus attachment glycoprotein: a template for antiviral and vaccine design. J Virol. 2008;82:11628–11636. doi: 10.1128/JVI.01344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marsh GA, de Jong C, Barr JA, Tachedjian M, Smith C, Middleton D, Yu M, Todd S, Foord AJ, Haring V, Payne J, Robinson R, Broz I, Crameri G, Field HE, Wang LF. Cedar virus: a novel Henipavirus isolated from Australian bats. PLoS Pathog. 2012;8:e1002836. doi: 10.1371/journal.ppat.1002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Negrete OA, Chu D, Aguilar HC, Lee B. Single Amino Acid Changes in the Nipah and Hendra Virus Attachment Glycoproteins Distinguish EphrinB2 from EphrinB3 Usage. J Virol. 2007;81:10804–10814. doi: 10.1128/JVI.00999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Negrete OA, Levroney EL, Aguilar HC, Bertolotti-Ciarlet A, Nazarian R, Tajyar S, Lee B. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature. 2005;436:401–405. doi: 10.1038/nature03838. [DOI] [PubMed] [Google Scholar]
- 57.Negrete OA, Wolf MC, Aguilar HC, Enterlein S, Wang W, Muhlberger E, Su SV, Bertolotti-Ciarlet A, Flick R, Lee B. Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog. 2006;2:e7. doi: 10.1371/journal.ppat.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karron RA, Buonagurio DA, Georgiu AF, Whitehead SS, Adamus JE, Clements-Mann ML, Harris DO, Randolph VB, Udem SA, Murphy BR, Sidhu MS. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci U S A. 1997;94:13961–13966. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kahn JS, Schnell MJ, Buonocore L, Rose JK. Recombinant vesicular stomatitis virus expressing respiratory syncytial virus (RSV) glycoproteins: RSV fusion protein can mediate infection and cell fusion. Virology. 1999;254:81–91. doi: 10.1006/viro.1998.9535. [DOI] [PubMed] [Google Scholar]
- 60.Karger A, Schmidt U, Buchholz UJ. Recombinant bovine respiratory syncytial virus with deletions of the G or SH genes: G and F proteins bind heparin. Journal of General Virology. 2001;82:631–640. doi: 10.1099/0022-1317-82-3-631. [DOI] [PubMed] [Google Scholar]
- 61.Leyrer S, Bitzer M, Lauer U, Kramer J, Neubert WJ, Sedlmeier R. Sendai virus-like particles devoid of haemagglutinin-neuraminidase protein infect cells via the human asialoglycoprotein receptor. J Gen Virol. 1998;79(Pt 4):683–687. doi: 10.1099/0022-1317-79-4-683. [DOI] [PubMed] [Google Scholar]
- 62.Aleksandrowicz P, Marzi A, Biedenkopf N, Beimforde N, Becker S, Hoenen T, Feldmann H, Schnittler HJ. Ebola virus enters host cells by macropinocytosis and clathrin-mediated endocytosis. J Infect Dis 204 Suppl. 2011;3:S957–S967. doi: 10.1093/infdis/jir326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sakurai Y, Kolokoltsov AA, Chen C, Tidwell MW, Bauta WE, Klugbauer N, Grimm C, Wahl-Schott C, Biel M, Davey RA. Two-pore channels control Ebola virus host cell entry and are drug targets for disease treatment. Science. 2015;347:995–998. doi: 10.1126/science.1258758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lakadamyali M, Rust MJ, Zhuang X. Endocytosis of influenza viruses. Microbes Infect. 2004;6:929–936. doi: 10.1016/j.micinf.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berghall H, Wallen C, Hyypia T, Vainionpaa R. Role of cytoskeleton components in measles virus replication. Arch Virol. 2004;149:891–901. doi: 10.1007/s00705-003-0264-9. [DOI] [PubMed] [Google Scholar]
- 66.Bohn W, Ciampor F, Rutter R, Mannweiler K. Localization of nucleocapsid associated polypeptides in measles virus-infected cells by immunogold labelling after resin embedding. Arch Virol. 1990;114:53–64. doi: 10.1007/BF01311011. [DOI] [PubMed] [Google Scholar]
- 67.Bowden TA, Crispin M, Harvey DJ, Jones EY, Stuart DI. Dimeric architecture of the Hendra virus attachment glycoprotein: evidence for a conserved mode of assembly. J Virol. 2010;84:6208–6217. doi: 10.1128/JVI.00317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Campbell SM, Crowe SM, Mak J. Lipid rafts and HIV-1: from viral entry to assembly of progeny virions. J Clin Virol. 2001;22:217–227. doi: 10.1016/s1386-6532(01)00193-7. [DOI] [PubMed] [Google Scholar]
- 69.Ciancanelli MJ, Basler CF. Mutation of YMYL in the Nipah virus matrix protein abrogates budding and alters subcellular localization. J Virol. 2006;80:12070–12078. doi: 10.1128/JVI.01743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Coronel EC, Murti KG, Takimoto T, Portner A. Human parainfluenza virus type 1 matrix and nucleoprotein genes transiently expressed in mammalian cells induce the release of virus-like particles containing nucleocapsid-like structures. J Virol. 1999;73:7035–7038. doi: 10.1128/jvi.73.8.7035-7038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Horvath CM, Paterson RG, Shaughnessy MA, Wood R, Lamb RA. Biological activity of paramyxovirus fusion proteins: factors influencing formation of syncytia. J Virol. 1992;66:4564–4569. doi: 10.1128/jvi.66.7.4564-4569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kruger N, Hoffmann M, Weis M, Drexler JF, Muller MA, Winter C, Corman VM, Gutzkow T, Drosten C, Maisner A, Herrler G. Surface glycoproteins of an African henipavirus induce syncytium formation in a cell line derived from an African fruit bat, Hypsignathus monstrosus. J Virol. 2013;87:13889–13891. doi: 10.1128/JVI.02458-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Okamoto K, Ohgimoto S, Nishio M, Tsurudome M, Kawano M, Komada H, Ito M, Sakakura Y, Ito Y. Paramyxovirus-induced syncytium cell formation is suppressed by a dominant negative fusion regulatory protein-1 (FRP-1)/CD98 mutated construct: an important role of FRP-1 in virus-induced cell fusion. Journal of General Virology. 1997;78:775–783. doi: 10.1099/0022-1317-78-4-775. [DOI] [PubMed] [Google Scholar]
- 74.Wong KT, Shieh WJ, Kumar S, Norain K, Abdullah W, Guarner J, Goldsmith CS, Chua KB, Lam SK, Tan CT, Goh KJ, Chong HT, Jusoh R, Rollin PE, Ksiazek TG, Zaki SR. Nipah virus infection: pathology and pathogenesis of an emerging paramyxoviral zoonosis. Am J Pathol. 2002;161:2153–2167. doi: 10.1016/S0002-9440(10)64493-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Merz DC, Scheid A, Choppin PW. Importance of antibodies to the fusion glycoprotein of paramyxoviruses in the prevention of spread of infection. J Exp Med. 1980;151:275–288. doi: 10.1084/jem.151.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Habchi J, Longhi S. Structural Disorder within Paramyxoviral Nucleoproteins and Phosphoproteins in Their Free and Bound Forms: From Predictions to Experimental Assessment. Int J Mol Sci. 2015;16:15688–15726. doi: 10.3390/ijms160715688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harcourt BH, Tamin A, Ksiazek TG, Rollin PE, Anderson LJ, Bellini WJ, Rota PA. Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology. 2000;271:334–349. doi: 10.1006/viro.2000.0340. [DOI] [PubMed] [Google Scholar]
- 78.Loney C, Mottet-Osman G, Roux L, Bhella D. Paramyxovirus ultrastructure and genome packaging: cryo-electron tomography of sendai virus. J Virol. 2009;83:8191–8197. doi: 10.1128/JVI.00693-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pantua HD, McGinnes LW, Peeples ME, Morrison TG. Requirements for the assembly and release of Newcastle disease virus-like particles. J Virol. 2006;80:11062–11073. doi: 10.1128/JVI.00726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ray G, Schmitt PT, Schmitt AP. C-Terminal DxD-Containing Sequences within Paramyxovirus Nucleocapsid Proteins Determine Matrix Protein Compatibility and Can Direct Foreign Proteins into Budding Particles. J Virol. 2016;90:3650–3660. doi: 10.1128/JVI.02673-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prescott J, de Wit E, Feldmann H, Munster VJ. The immune response to Nipah virus infection. Arch Virol. 2012;157:1635–1641. doi: 10.1007/s00705-012-1352-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Childs K, Stock N, Ross C, Andrejeva J, Hilton L, Skinner M, Randall R, Goodbourn S. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology. 2007;359:190–200. doi: 10.1016/j.virol.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 83.Horikami SM, Hector RE, Smallwood S, Moyer SA. The Sendai Virus C Protein Binds the L Polymerase Protein to Inhibit Viral RNA Synthesis Virology. 1997;235:261–270. doi: 10.1006/viro.1997.8702. [DOI] [PubMed] [Google Scholar]
- 84.Kato A, Ohnishi Y, Kohase M, Saito S, Tashiro M, Nagai Y. Y2, the smallest of the Sendai virus C proteins, is fully capable of both counteracting the antiviral action of interferons and inhibiting viral RNA synthesis. J Virol. 2001;75:3802–3810. doi: 10.1128/JVI.75.8.3802-3810.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu LL, Puri M, Horvath CM, Sen GC. Select paramyxoviral V proteins inhibit IRF3 activation by acting as alternative substrates for inhibitor of kappaB kinase epsilon (IKKe)/TBK1. J Biol Chem. 2008;283:14269–14276. doi: 10.1074/jbc.M710089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Parisien JP, Bamming D, Komuro A, Ramachandran A, Rodriguez JJ, Barber G, Wojahn RD, Horvath CM. A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2. J Virol. 2009;83:7252–7260. doi: 10.1128/JVI.00153-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Patterson JB, Thomas D, Lewicki H, Billeter MA, Oldstone MB. V and C proteins of measles virus function as virulence factors in vivo. Virology. 2000;267:80–89. doi: 10.1006/viro.1999.0118. [DOI] [PubMed] [Google Scholar]
- 88.Audsley MD, Moseley GW. Paramyxovirus evasion of innate immunity: Diverse strategies for common targets. World J Virol. 2013;2:57–70. doi: 10.5501/wjv.v2.i2.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Harrison MS, Sakaguchi T, Schmitt AP. Paramyxovirus assembly and budding: building particles that transmit infections. Int J Biochem Cell Biol. 2010;42:1416–1429. doi: 10.1016/j.biocel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Patch JR, Han Z, McCarthy SE, Yan L, Wang LF, Harty RN, Broder CC. The YPLGVG sequence of the Nipah virus matrix protein is required for budding. Virol J. 2008;5:137. doi: 10.1186/1743-422X-5-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pohl C, Duprex WP, Krohne G, Rima BK, Schneider-Schaulies S. Measles virus M and F proteins associate with detergent-resistant membrane fractions and promote formation of virus-like particles. J Gen Virol. 2007;88:1243–1250. doi: 10.1099/vir.0.82578-0. [DOI] [PubMed] [Google Scholar]
- 92.Schmitt AP, Leser GP, Morita E, Sundquist WI, Lamb RA. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J Virol. 2005;79:2988–2997. doi: 10.1128/JVI.79.5.2988-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schmitt AP, Leser GP, Waning DL, Lamb RA. Requirements for budding of paramyxovirus simian virus 5 virus-like particles. J Virol. 2002;76:3952–3964. doi: 10.1128/JVI.76.8.3952-3964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Takimoto T, Murti KG, Bousse T, Scroggs RA, Portner A. Role of matrix and fusion proteins in budding of Sendai virus. J Virol. 2001;75:11384–11391. doi: 10.1128/JVI.75.23.11384-11391.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takimoto T, Portner A. Molecular mechanism of paramyxovirus budding. Virus Res. 2004;106:133–145. doi: 10.1016/j.virusres.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 96.Ader N, Brindley MA, Avila M, Origgi FC, Langedijk JP, Orvell C, Vandevelde M, Zurbriggen A, Plemper RK, Plattet P. Structural rearrangements of the central region of the morbillivirus attachment protein stalk domain trigger F protein refolding for membrane fusion. The Journal of biological chemistry. 2012;287:16324–16334. doi: 10.1074/jbc.M112.342493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu Q, Stone JA, Bradel-Tretheway B, Dabundo J, Benavides Montano JA, Santos-Montanez J, Biering SB, Nicola AV, Iorio RM, Lu X, Aguilar HC. Unraveling a three-step spatiotemporal mechanism of triggering of receptor-induced Nipah virus fusion and cell entry. PLoS Pathog. 2013;9:e1003770. doi: 10.1371/journal.ppat.1003770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ader-Ebert N, Khosravi M, Herren M, Avila M, Alves L, Bringolf F, Ouml;rvell C, Langedijk JP, Zurbriggen A, Plemper RK, Plattet P. Sequential Conformational Changes in the Morbillivirus Attachment Protein Initiate the Membrane Fusion Process. Plos Pathogens. 2015:11. doi: 10.1371/journal.ppat.1004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bishop KA, Hickey AC, Khetawat D, Patch JR, Bossart KN, Zhu Z, Wang LF, Dimitrov DS, Broder CC. Residues in the stalk domain of the Hendra virus G glycoprotein modulate conformational changes associated with receptor binding. J Virol. 2008;82:11398–11409. doi: 10.1128/JVI.02654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bose S, Song AS, Jardetzky TS, Lamb RA. Fusion Activation through Attachment Protein Stalk Domains Indicates a Conserved Core Mechanism of Paramyxovirus Entry into Cells. Journal of Virology. 2014;88:3925–3941. doi: 10.1128/JVI.03741-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bose S, Zokarkar A, Welch BD, Leser GP, Jardetzky TS, Lamb RA. Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2625–E2634. doi: 10.1073/pnas.1213813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brindley MA, Suter R, Schestak I, Kiss G, Wright ER, Plemper RK. A Stabilized Headless Measles Virus Attachment Protein Stalk Efficiently Triggers Membrane Fusion. Journal of Virology In Press. 2013 doi: 10.1128/JVI.01945-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Melanson VR, Iorio RM. Amino acid substitutions in the F-specific domain in the stalk of the newcastle disease virus HN protein modulate fusion and interfere with its interaction with the F protein. J Virol. 2004;78:13053–13061. doi: 10.1128/JVI.78.23.13053-13061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Navaratnarajah CK, Negi S, Braun W, Cattaneo R. Membrane fusion triggering: three modules with different structure and function in the upper half of the measles virus attachment protein stalk. The Journal of biological chemistry. 2012;287:38543–38551. doi: 10.1074/jbc.M112.410563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lou Z, Xu Y, Xiang K, Su N, Qin L, Li X, Gao GF, Bartlam M, Rao Z. Crystal structures of Nipah and Hendra virus fusion core proteins. Febs J. 2006;273:4538–4547. doi: 10.1111/j.1742-4658.2006.05459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Matthews JM, Young TF, Tucker SP, Mackay JP. The core of the respiratory syncytial virus fusion protein is a trimeric coiled coil. J Virol. 2000;74:5911–5920. doi: 10.1128/jvi.74.13.5911-5920.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Welch BD, Liu Y, Kors CA, Leser GP, Jardetzky TS, Lamb RA. Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc Natl Acad Sci U S A. 2012;109:166672–116677. doi: 10.1073/pnas.1213802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wong JJ, Paterson RG, Lamb RA, Jardetzky TS. Structure and stabilization of the Hendra virus F glycoprotein in its prefusion form. Proc Natl Acad Sci U S A. 2016;113:1056–1061. doi: 10.1073/pnas.1523303113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Xu K, Chan YP, Bradel-Tretheway B, Akyol-Ataman Z, Zhu Y, Dutta S, Yan L, Feng Y, Wang LF, Skiniotis G, Lee B, Zhou ZH, Broder CC, Aguilar HC, Nikolov DB. Crystal Structure of the Pre-fusion Nipah Virus Fusion Glycoprotein Reveals a Novel Hexamer-of-Trimers Assembly. PLoS Pathog. 2015;11:e1005322. doi: 10.1371/journal.ppat.1005322. This study showed that host-cell invasion may be a more coordinated task than previously anticipated. In prior publications, our understanding of membrane fusion has been quite two dimensional, and involved only one or two fusion protein trimers.
- 111.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bossart KN, Wang LF, Eaton BT, Broder CC. Functional expression and membrane fusion tropism of the envelope glycoproteins of Hendra virus. Virology. 2001;290:121–135. doi: 10.1006/viro.2001.1158. [DOI] [PubMed] [Google Scholar]
- 113.Krzyzaniak MA, Zumstein MT, Gerez JA, Picotti P, Helenius A. Host Cell Entry of Respiratory Syncytial Virus Involves Macropinocytosis Followed by Proteolytic Activation of the F Protein. Plos Pathogens. 2013:9. doi: 10.1371/journal.ppat.1003309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bolt G, Pederson LO, Birkeslund HH. Cleavage of the respiratory syncytial virus fusion protein is required for its surface expression: role of furin. Virus Research. 2000;68:25–33. doi: 10.1016/s0168-1702(00)00149-0. [DOI] [PubMed] [Google Scholar]
- 115.Sakaguchi T, Fujii Y, Kiyotani K, Yoshida T. Correlation of proteolytic cleavage of F protein precursors in paramyxoviruses with expression of the fur, PACE4, and PC6 genes in mammalian cells. Journal of General Virology. 1994;75:2821–2827. doi: 10.1099/0022-1317-75-10-2821. [DOI] [PubMed] [Google Scholar]
- 116.Xu Y, Gao S, Cole DK, Zhu J, Su N, Wang H, Gao GF, Rao Z. Basis for fusion inhibition by peptides: analysis of the heptad repeat regions of the fusion proteins from Nipah and Hendra viruses, newly emergent zoonotic paramyxoviruses. Biochem Biophys Res Commun. 2004;315:664–670. doi: 10.1016/j.bbrc.2004.01.115. [DOI] [PubMed] [Google Scholar]
- 117.Xu Y, Lou Z, Liu Y, Cole DK, Su N, Qin L, Li X, Bai Z, Rao Z, Gao GF. Crystallization and preliminary crystallographic analysis of the fusion core from two new zoonotic paramyxoviruses, Nipah virus and Hendra virus. Acta Crystallogr D Biol Crystallogr. 2004;60:1161–1164. doi: 10.1107/S0907444904009515. [DOI] [PubMed] [Google Scholar]
- 118.Lambert DM, Barney S, Lambert AL, Guthrie K, Medinas R, Davis DE, Bucy T, Erickson J, Merutka G, Petteway SR., Jr Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc Natl Acad Sci U S A. 1996;93:2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci U S A. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gardner AE, Dutch RE. A conserved region in the F(2) subunit of paramyxovirus fusion proteins is involved in fusion regulation. J Virol. 2007;81:8303–8314. doi: 10.1128/JVI.00366-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hashiguchi T, Kajikawa M, Maita N, Takeda M, Kuroki K, Sasaki K, Kohda D, Yanagi Y, Maenaka K. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc Natl Acad Sci U S A. 2007;104:19535–19540. doi: 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lawrence MC, Borg NA, Streltsov VA, Pilling PA, Epa VC, Varghese JN, McKimm-Breschkin JL, Colman PM. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J Mol Biol. 2004;335:1343–1357. doi: 10.1016/j.jmb.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 123.Hashiguchi T, Maenaka K, Yanagi Y. Measles virus hemagglutinin: structural insights into cell entry and measles vaccine. Frontiers in Microbiology. 2011:2. doi: 10.3389/fmicb.2011.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Plemper RK, Brindley MA, Iorio RM. Structural and mechanistic studies of measles virus illuminate paramyxovirus entry. PLoS Pathog. 2011;7:e1002058. doi: 10.1371/journal.ppat.1002058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Navaratnarajah CK, Oezguen N, Rupp L, Kay L, Leonard VH, Braun W, Cattaneo R. The heads of the measles virus attachment protein move to transmit the fusion-triggering signal. Nature structural & molecular biology. 2011;18:128–134. doi: 10.1038/nsmb.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gravel KA, Morrison TG. Interacting domains of the HN and F proteins of newcastle disease virus. J Virol. 2003;77:11040–11049. doi: 10.1128/JVI.77.20.11040-11049.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Aguilar HC, Ataman ZA, Aspericueta V, Fang AQ, Stroud M, Negrete OA, Kammerer RA, Lee B. A Novel Receptor-induced Activation Site in the Nipah Virus Attachment Glycoprotein (G) Involved in Triggering the Fusion Glycoprotein (F) J Biol Chem. 2009;284:1628–1635. doi: 10.1074/jbc.M807469200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lee JK, Prussi A, Paal T, White LK, Snyder JP, Plemper RK. Functional interaction between paramyxovirus fusion and attachment proteins. Journal of Biological Chemistry. 2008;283:16561–16572. doi: 10.1074/jbc.M801018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chen L, Gorman JJ, McKimm-Breschkin J, Lawrence LJ, Tulloch PA, Smith BJ, Colman PM, Lawrence MC. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure (Camb) 2001;9:255–266. doi: 10.1016/s0969-2126(01)00581-0. [DOI] [PubMed] [Google Scholar]
- 130.Kim YH, Donald JE, Grigoryan G, Leser GP, Fadeev AY, Lamb RA, DeGrado WF. Capture and imaging of a prehairpin fusion intermediate of the paramyxovirus PIV5. Proc Natl Acad Sci U S A. 2011;108:20992–20997. doi: 10.1073/pnas.1116034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.McLellan JS, Yang YP, Graham BS, Kwong PD. Structure of Respiratory Syncytial Virus Fusion Glycoprotein in the Postfusion Conformation Reveals Preservation of Neutralizing Epitopes. Journal of Virology. 2011;85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Swanson K, Wen XL, Leser GP, Paterson RG, Lamb RA, Jardetzky TS. Structure of the Newcastle disease virus F protein in the post-fusion conformation. Virology. 2010;402:372–379. doi: 10.1016/j.virol.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhao X, Singh M, Malashkevich VN, Kim PS. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc Natl Acad Sci U S A. 2000;97:14172–14177. doi: 10.1073/pnas.260499197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dutch RE. Entry and fusion of emerging paramyxoviruses. PLoS Pathog. 2010;6:e1000881. doi: 10.1371/journal.ppat.1000881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lamb RA, Jardetzky TS. Structural basis of viral invasion: lessons from paramyxovirus F. Curr Opin Struct Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Aguilar HC, Matreyek KA, Choi DY, Filone CM, Young S, Lee B. Polybasic KKR motif in the cytoplasmic tail of Nipah virus fusion protein modulates membrane fusion by inside-out signaling. J Virol. 2007;81:4520–4532. doi: 10.1128/JVI.02205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Aguilar HC, Matreyek KA, Filone CM, Hashimi ST, Levroney EL, Negrete OA, Bertolotti-Ciarlet A, Choi DY, McHardy I, Fulcher JA, Su SV, Wolf MC, Kohatsu L, Baum LG, Lee B. N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J Virol. 2006;80:4878–4889. doi: 10.1128/JVI.80.10.4878-4889.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bishop KA, Stantchev TS, Hickey AC, Khetawat D, Bossart KN, Krasnoperov V, Gill P, Feng YR, Wang L, Eaton BT, Wang LF, Broder CC. Identification of Hendra virus G glycoprotein residues that are critical for receptor binding. J Virol. 2007;81:5893–5901. doi: 10.1128/JVI.02022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Corey EA, Iorio RM. Mutations in the stalk of the measles virus hemagglutinin protein decrease fusion but do not interfere with virus-specific interaction with the homologous fusion protein. J Virol. 2007;81:9900–9910. doi: 10.1128/JVI.00909-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Plemper RK, Hammond AL, Gerlier D, Fielding AK, Cattaneo R. Strength of envelope protein interaction modulates cytopathicity of measles virus. J Virol. 2002;76:5051–5061. doi: 10.1128/JVI.76.10.5051-5061.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Talekar A, Moscona A, Porotto M. Measles Virus Fusion Machinery Activated by Sialic Acid Binding Globular Domain. Journal of Virology. 2013;87:13619–13627. doi: 10.1128/JVI.02256-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bose S, Jardetzky TS, Lamb RA. Timing is everything: Fine-tuned molecular machines orchestrate paramyxovirus entry. Virology. 2015;479:518–531. doi: 10.1016/j.virol.2015.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Brindley MA, Chaudhury S, Plemper RK. Measles Virus Glycoprotein Complexes Preassemble Intracellularly and Relax during Transport to the Cell Surface in Preparation for Fusion. Journal of Virology. 2015;89:1230–1241. doi: 10.1128/JVI.02754-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhu Q, Biering SB, Mirza AM, Grasseschi BA, Mahon PJ, Lee B, Aguilar HC, Iorio RM. Individual N-glycans added at intervals along the stalk of the Nipah virus G protein prevent fusion but do not block the interaction with the homologous F protein. Journal of virology. 2013;87:3119–3129. doi: 10.1128/JVI.03084-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gui L, Jurgens EM, Ebner JL, Porotto M, Moscona A, Lee KK. Electron Tomography Imaging of Surface Glycoproteins on Human Parainfluenza Virus 3: Association of Receptor Binding and Fusion Proteins before Receptor Engagement. Mbio. 2015:6. doi: 10.1128/mBio.02393-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Smith EC, Popa A, Chang A, Masante C, Dutch RE. Viral entry mechanisms: the increasing diversity of paramyxovirus entry. Febs Journal. 2009;276:7217–7227. doi: 10.1111/j.1742-4658.2009.07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bissonnette MLZ, Connolly SA, Young DF, Randall RE, Paterson RG, Lamb RA. Analysis of the pH requirement for isolates of the paramyxovirus membrane fusion of different parainfluenza virus 5. Journal of Virology. 2006;80:3071–3077. doi: 10.1128/JVI.80.6.3071-3077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Srinivasakumar N, Ogra PL, Flanagan TD. Characteristics of Fusion of Respiratory Syncytial Virus with Hep-2 Cells as Measured by R18 Fluorescence Dequenching Assay. Journal of Virology. 1991;65:4063–4069. doi: 10.1128/jvi.65.8.4063-4069.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.San Roman K, Villar E, Munoz-Barroso I. Acidic pH enhancement of the fusion of Newcastle disease virus with cultured cells. Virology. 1999;260:329–341. doi: 10.1006/viro.1999.9841. [DOI] [PubMed] [Google Scholar]
- 150.Herfst S, Mas V, Ver LS, Wierda RJ, Osterhaus ADME, Fouchier RAM, Melero JA. Low-pH-induced membrane fusion mediated by human metapneumovirus F protein is a rare, strain-dependent phenomenon. Journal of Virology. 2008;82:8891–8895. doi: 10.1128/JVI.00472-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Seth S, Vincent A, Compans RW. Activation of fusion by the SER virus F protein: a low-pH-dependent paramyxovirus entry process. Journal of Virology. 2003;77:6520–6527. doi: 10.1128/JVI.77.11.6520-6527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pernet O, Pohl C, Ainouze M, Kweder H, Buckland R. Nipah virus entry can occur by macropinocytosis. Virology. 2009;395:298–311. doi: 10.1016/j.virol.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 153.Cantin C, Holguera J, Ferreira L, Villar E, Munoz-Barroso I. Newcastle disease virus may enter cells by caveolae-mediated endocytosis. Journal of General Virology. 2007;88:559–569. doi: 10.1099/vir.0.82150-0. [DOI] [PubMed] [Google Scholar]
- 154.Kolokoltsov AA, Deniger D, Fleming EH, Roberts NJ, Karpilow JM, Davey RA. Small interfering RNA profiling reveals key role of clathrin-mediated endocytosis and early endosome formation for infection by respiratory syncytial virus. Journal of Virology. 2007;81:7786–7800. doi: 10.1128/JVI.02780-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Diederich S, Thiel L, Maisner A. Role of endocytosis and cathepsin-mediated activation in Nipah virus entry. Virology. 2008;375:391–400. doi: 10.1016/j.virol.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Andersson T, Wallen P, Grillner S, Norrby E, Kristensson K. A Calcium-Channel Antagonist Can Prevent Paramyxovirus-Induced Neurodegeneration. Neuroreport. 1991;2:145–148. doi: 10.1097/00001756-199103000-00009. [DOI] [PubMed] [Google Scholar]
- 157.Haywood AM, Boyer BP. Sendai Virus Membrane-Fusion - Time Course and Effect of Temperature, Ph, Calcium, and Receptor Concentration. Biochemistry. 1982;21:6041–6046. doi: 10.1021/bi00267a003. [DOI] [PubMed] [Google Scholar]
- 158.Donald JE, Zhang Y, Fiorin G, Carnevale V, Slochower DR, Gai F, Klein ML, DeGrado WF. Transmembrane orientation and possible role of the fusogenic peptide from parainfluenza virus 5 (PIV5) in promoting fusion. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3958–3963. doi: 10.1073/pnas.1019668108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bissonnette MLZ, Donald JE, DeGrado WF, Jardetzky TS, Lamb RA. Functional Analysis of the Transmembrane Domain in Paramyxovirus F Protein-Mediated Membrane Fusion. Journal of Molecular Biology. 2009;386:14–36. doi: 10.1016/j.jmb.2008.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bagai S, Lamb RA. Truncation of the COOH-terminal region of the paramyxovirus SV5 fusion protein leads to hemifusion but not complete fusion. J Cell Biol. 1996;135:73–84. doi: 10.1083/jcb.135.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dutch RE, Lamb RA. Deletion of the cytoplasmic tail of the fusion protein of the paramyxovirus simian virus 5 affects fusion pore enlargement. Journal of Virology. 2001;75:5363–5369. doi: 10.1128/JVI.75.11.5363-5369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Seth S, Vincent A, Compans RW. Mutations in the cytoplasmic domain of a paramyxovirus fusion glycoprotein rescue syncytium formation and eliminate the hemagglutinin-neuraminidase protein requirement for membrane fusion. J Virol. 2003;77:167–178. doi: 10.1128/JVI.77.1.167-178.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Joshi SB, Dutch RE, Lamb RA. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology. 1998;248:20–34. doi: 10.1006/viro.1998.9242. [DOI] [PubMed] [Google Scholar]
- 164.Hernandez LD, Hoffman LR, Wolfsberg TG, White JM. Virus-cell and cell-cell fusion. Annual Review of Cell and Developmental Biology. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 165.Wurth MA, Schowalter RM, Smith EC, Moncman CL, Dutch RE, McCann RO. The actin cytoskeleton inhibits pore expansion during PIV5 fusion protein-promoted cell-cell fusion. Virology. 2010;404:117–126. doi: 10.1016/j.virol.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gower TL, Peeples ME, Collins PL, Graham BS. RhoA is activated during respiratory syncytial virus infection. Virology. 2001;283:188–196. doi: 10.1006/viro.2001.0891. [DOI] [PubMed] [Google Scholar]
- 167.Schowalter RM, Wurth MA, Aguilar HC, Lee B, Moncman CL, McCann RO, Dutch RE. Rho GTPase activity modulates paramyxovirus fusion protein-mediated cell-cell fusion. Virology. 2006;350:323–334. doi: 10.1016/j.virol.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 168.Taylor MP, Koyuncu OO, Enquist LW. Subversion of the actin cytoskeleton during viral infection. Nature Reviews Microbiology. 2011;9:427–439. doi: 10.1038/nrmicro2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kallewaard NL, Boxena AL, Crowe JE. Cooperativity of actin and microtubule elements during replication of respiratory syncytial virus. Virology. 2005;331:73–81. doi: 10.1016/j.virol.2004.10.010. [DOI] [PubMed] [Google Scholar]