Summary
Clostridium difficile infection (CDI) is a challenging threat to human health. Infections occur after disruption of the normal microbiota, most commonly through the use of antibiotics. Current treatment for CDI largely relies on the broad‐spectrum antibiotics vancomycin and metronidazole that further disrupt the microbiota resulting in frequent recurrence, highlighting the need for C. difficile‐specific antimicrobials. The cell surface of C. difficile represents a promising target for the development of new drugs. C. difficile possesses a highly deacetylated peptidoglycan cell wall containing unique secondary cell wall polymers. Bound to the cell wall is an essential S‐layer, formed of SlpA and decorated with an additional 28 related proteins. In addition to the S‐layer, many other cell surface proteins have been identified, including several with roles in host colonization. This review aims to summarize our current understanding of these different C. difficile cell surface components and their viability as therapeutic targets.
Introduction
Clostridium difficile is a Gram positive, spore‐forming, anaerobic bacterium and is the leading cause of antibiotic‐associated nosocomial diarrhoea (Rupnik et al., 2009). Although the use of antibiotics has undoubtedly had an enormous positive impact on human health over the past seven decades, it is unfortunate that their use is also the main risk factor for C. difficile infection (CDI). Hospitalized patients are frequently treated with broad‐spectrum antibiotics, both as prophylactics and to treat infection, resulting in catastrophic damage to the gut microbiota (Dethlefsen et al., 2008). C. difficile can exploit the resulting dysbiosis to colonize and proliferate in the gut (Lawley et al., 2009).
C. difficile pathogenesis is a three‐step process that begins with disruption of the gut microbiota (Smits et al., 2016). This is followed by germination of indigenous or ingested spores, which starts the colonization phase – involving the attachment of the bacterium to the host intestinal epithelium and multiplication both at the surface and in the lumen. Colonization is required for the final phase of virulence, the release of toxins and the onset of disease symptoms. Most C. difficile clinical isolates produce two related toxins, TcdA and TcdB, that belong to the large clostridial cytotoxin family (Jank and Aktories, 2008). Both of these toxins act by glucosylating small GTPases including Rho, Rac and Cdc42. The action of these toxins is responsible for the clinical manifestations of disease, ranging from mild diarrhoea through to life‐threatening inflammatory complications such as pseudomembranous colitis and toxic megacolon (Smits et al., 2016). Thirty‐day mortality can exceed 30% in elderly populations (McGowan et al., 2011). Since 2001, the emergence of highly transmissible epidemic strains and advances in the availability of genetic tools has increased interest and rapidly advanced our understanding of this important pathogen (McDonald et al., 2005; Ng et al., 2013; Dembek et al., 2015). Despite these improvements, our understanding of C. difficile virulence is still in its infancy. It is clear that bacterial cell surface components will be crucial in the interaction between the bacterium and the host. However, the molecular details of these interactions are still largely uncharacterized.
Recent studies focussing on the C. difficile envelope have identified and characterized several cell wall polymers, as well as numerous surface proteins (Table 1). Many of these macromolecules are unique to C. difficile making the cell envelope a prime target for the development of species‐specific therapeutics. This review will describe our current understanding of C. difficile cell envelope architecture, highlighting the potential for novel drugs and vaccines to treat and prevent CDI.
Table 1.
Therapeutic potential of cell envelope components
| Target | Immunogenic | Vaccine formulation | In vitro characterization of mutants | In vivo characterization of mutants | Clostridium difficile‐specific | Other comments | Source |
|---|---|---|---|---|---|---|---|
| PS‐I | Y | Toxin B conjugate vaccine (untested) | N | N | Y | Only present in a minority of C. difficile strains | Ganeshapillai et al. (2008), Jiao et al. (2013) |
| PS‐II | Y | CRM197 and toxin fragment conjugate vaccines | N | N | Y | Found in all tested C. difficile strains. Anchors the essential protein SlpA to the cell surface | Oberli et al. (2011), Adamo et al. (2012), Romano et al. (2014) |
| PS‐III | Y | HAS and ExoA conjugate vaccines | N | N | PS‐III antibodies cross‐react with other members of the Clostridia | Cox et al. (2013), Martin et al. (2013) | |
| SlpA | Y | Various vaccination routes with various adjuvants | N | N | Y | Most abundant protein of the S‐layer. This protein is essential and may act as an important colonization factor. SlpA is therefore an interesting target for antimicrobial therapies, although vaccine studies have yielded inconclusive results | Calabi et al. (2002), O'Brien et al. (2005), Ni Eidhin et al. (2008), Ryan et al. (2011) |
| Cwp84 | Y | Subcutaneous, rectal and intragastric routes of vaccination with various adjuvants. Oral vaccine also tested | Mutants display poor growth in vitro | Mutant remains fully virulent in the hamster model of infection | Y | Although inactivation of Cwp84 does not reduce virulence, vaccination of hamsters with Cwp84 provides protection and results in greater survival rates when compared with control groups | Kirby et al. (2009), Pechine et al. (2011), Sandolo et al. (2011) |
| CbpA | Untested | N | Mutants display no significant decrease in adhesion to immobilized collagen or human fibroblasts | Mutants show no colonization fitness difference in a competitive mouse model | N | CbpA does not appear to be important for C. difficile virulence and is not C. difficile‐specific. For these reasons CbpA is unlikely to be a valid antimicrobial target | Janoir et al. (2013), Tulli et al. (2013) |
| FbpA | Y | N | Mutant displays no difference in adherence to Caco‐2 or HT29‐MTX cell lines | Mutant showed decreased caecal colonization in a monoxenic mouse model and was outcompeted in a dixenic mouse model | N | Fibronectin‐binding proteins are not C. difficile‐specific and the role of FbpA in intestinal adherence appears to be minor. Taken together this suggests that FbpA may not be a suitable target for anti‐C. difficile therapies | Pechine et al. (2005), Barketi‐Klai et al. (2011) |
| PPEP‐1 | Untested | N | PPEP‐1 mutants retain CD2831 on their cell surface, increasing binding to a collagen matrix | PPEP‐1 mutants display slightly lower virulence | Y | Due to the reduction in virulence in a PPEP‐1 mutant, PPEP‐1 remains a viable antimicrobial target | Hensbergen et al. (2015) |
| GroEL | Y | Intranasal immunization with recombinant GroEL | Co‐incubation of C. difficile with purified protein or anti‐GroEL antibodies reduce adherence to vero cells | N | N | Although vaccination of mice with GroEL reduces intestinal colonization by C. difficile, GroEL is a common bacterial protein and no in vivo experiments have been described. For these reasons GroEL does not, at the present, appear to be a good anti‐C. difficile target | Hennequin et al. (2001a,b), Pechine et al. (2013) |
| CD0873 | Untested | N | CD0873 mutants are unable to bind Caco‐2 cells | N | Y | CD0873 remains largely uncharacterized and no in vivo studies are available. Therefore, although CD0873 mutants show decreased adherence to Caco‐2 cells more research must be Performed before CD0873 can be characterized as a viable anti‐C. difficile target | Kovacs‐Simon et al. (2014) |
C. difficile cell wall
Peptidoglycan
Peptidoglycan (PG) is an essential component of the cell wall with pleiotropic functions, including maintenance of cell shape and integrity, and anchoring cell wall proteins (CWP). PG structure is largely conserved, consisting of long glycan polymers cross‐linked by short peptide chains. The polysaccharide backbone is composed of polymers of the β‐1→4 linked disaccharide N‐acetylglucosamine‐N‐acetylmuramic acid (GlcNAc‐MurNAc). A short peptide stem, which varies between bacterial species, is linked to the D‐lactoyl group of MurNAc [reviewed in (Vollmer et al., 2008)]. Muropeptide analysis of digested C. difficile PG identified the tetrapeptide stem: l‐Ala‐d‐Glu‐A2pm‐d‐Ala (A2pm: 2,6‐diaminopimelic acid) (Fig. 1) (Peltier et al., 2011). Cross‐linking of the glycan strands in bacteria most commonly occurs via 4‐3 cross‐links catalysed by d,d‐transpeptidases, the essential target of β‐lactam antibiotics (reviewed in Vollmer et al., 2008). However, C. difficile displays a very high abundance of 3‐3 peptide cross‐links generated by at least two l,d‐transpeptidases. The abundance of 3‐3 cross‐links in C. difficile PG is increased by the partial inhibition of d,d‐transpeptidases by ampicillin, suggesting that the l,d‐transpeptidases are insensitive to ampicillin. Despite this, C. difficile remains susceptible to ampicillin, suggesting that 4‐3 cross‐linking is essential for PG assembly (Peltier et al., 2011).
Figure 1.
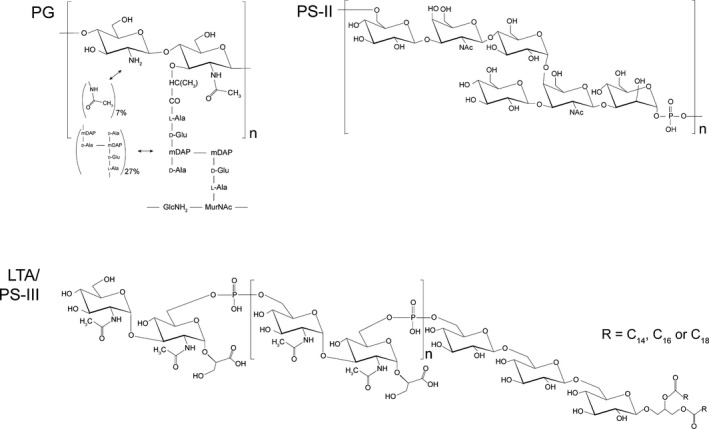
Structure of the conserved cell wall polymers of Clostridium difficile. PG: C. difficile produces a peptidoglycan characterized by a very high degree of N‐acetylglucosamine deacetylation (up to 93%), the stem peptide l‐Ala‐d‐Glu‐A2pm‐d‐Ala‐d‐Ala and an unusually high degree (73%) of 3‐3 cross‐links. PS‐II: a conserved cell wall polysaccharide polymer with a core hexasaccharide repeating unit of [→6)‐β‐d‐Glcp‐(1→3)‐β‐d‐Galp NAc‐(1→4)‐α‐d‐Glcp‐(1→4)‐[β‐d‐Glcp‐ (1→3]‐β‐d‐Galp NAc‐(1→3)‐α‐d‐Manp‐(1→P→]. PS‐III/LTA: a conserved lipid‐anchored cell wall polysaccharide in the extended lipoteichoic acid family with a core repeating unit of [→6)‐α‐d‐Glcp NAc‐(1→3)‐[→P‐6]‐α‐d‐Glcp NAc‐(1→2)‐d‐GroA]. This repeat unit is linked to →6)‐β‐d‐Glcp‐(1→6)‐β‐d‐Glcp‐(1→6)‐β‐d‐Glcp‐(1→1)‐Gro, with the terminal glycerol esterified with C14, C16, or C18 saturated or mono‐unsaturated fatty acids.
N‐deacetylation or O‐acetylation of either GlcNAc or MurNAc introduces further variation in PG structure between bacterial species. C. difficile displays very high levels of GlcNAc N‐deacetylation (89–93%), while all MurNAc residues remain fully acetylated. The proportion of N‐deacetylation observed is higher than that reported for other Gram positive bacteria (Peltier et al., 2011). In addition to this naturally high level of PG deacetylation, the percentage of acetylated GlcNAc residues decreases more than twofold in response to the introduction of lysozyme (Ho et al., 2014). Lysozyme is a key effector of the innate immune system that cleaves the PG backbone (Bevins and Salzman, 2011); however, PG deacetylation can confer resistance (Vollmer and Tomasz, 2000). Lysozyme induces expression of csfV, encoding an extracytoplasmic σ factor, which in turn upregulates expression of the polysaccharide deacetylase PdvA, resulting in further PG deacetylation. csfV mutants are severely attenuated in the hamster model of infection (Ho et al., 2014).
A notable observation is the presence of a functional vanG cd cluster in 85% of available C. difficile genomes. Vancomycin, an antibiotic of last resort used to treat severe or recurrent C. difficile infections, inhibits cell wall synthesis through interaction with the terminal d‐Ala‐d‐Ala of PG precursors (Reynolds, 1989; Johnson et al., 2014). Resistance to vancomycin can be conferred via modification of the terminal d‐Ala‐d‐Ala. For example, Enterococcus faecalis achieves low‐level resistance through synthesis of d‐Ala‐d‐Ser precursors by enzymes encoded in the vanG operon (Depardieu et al., 2003). Genes homologous to those in the vanG operon have been identified in the majority of C. difficile strains tested (Ammam et al., 2012; Peltier et al., 2013) and are able to synthesize d‐Ala‐d‐ser precursors in response to sublethal concentrations of vancomycin (Ammam et al., 2013) although this is debated (Peltier et al., 2013). Introduction of the vanG cd operon into a vancomycin‐sensitive strain of E. coli conferred low‐level resistance to vancomycin and a C. difficile mutant lacking this cluster displays a slightly lower vancomycin MIC relative to wild‐type (Ammam et al., 2013). The C. difficile genome also contains genes homologous to vanW and vanZ, although the function of these genes remains unknown (Ammam et al., 2012). Despite this, C. difficile remains susceptible to vancomycin treatment. This observation may be due to a number of factors including preferential incorporation of d‐Ala‐d‐Ala precursors by the cell wall synthesis machinery and weak activity of the resistance genes. However, it is possible that genomic alterations allowing vanG cd ‐mediated vancomycin resistance could emerge (Ammam et al., 2013). Another cell wall synthesis inhibitor, the lipoglycodepsipeptide Ramoplanin, displays efficacy against C. difficile in the hamster model and also reduces persistence of spores (Freeman et al., 2005). This drug has shown promising results in a phase II trial (Pullman et al., 2004) and was fast tracked for development by the US Food and Drug Administration. Ramoplanin was acquired by Nanotherapeutics in 2009 and is due to begin a new phase IIb clinical trial in 2016. An alternative approach to disrupting PG is the use of phage endolysins that degrade the cell wall resulting in cell lysis and death. The ΦCD27 endolysin, CD27L1‐179, can effectively lyse C. difficile cells (Mayer et al., 2011) by cleaving the bond between MurNAc and the first l‐Ala (Peltier et al., 2015). As PG architecture is more highly conserved than proteinaceous phage receptors, the endolysin is effective against diverse strains of C. difficile. However, the wider effect of such endolysins on the gut microbiota remains to be tested.
Secondary cell wall polysaccharides
To date, three anionic polymers have been identified on the cell surface of C. difficile. The first described polysaccharide (PS‐I) consists of a branched penta‐glycosylphosphate repeating unit, originally identified in a ribotype 027 strain (Ganeshapillai et al., 2008). PS‐I is only found in a minority of strains. The two other polysaccharides, a polymer of hexaglycosylphosphate repeat units (PS‐II) and a lipid bound glycosylphosphate polymer (PS‐III), are more widely distributed and have been found in all strains examined to date (Fig. 1) (Ganeshapillai et al., 2008; Reid et al., 2012). PS‐I and PS‐II have been described as teichoic acid‐like, although they differ significantly from the simple glycerol phosphate or ribitol phosphate classic teichoic acids (Ganeshapillai et al., 2008; Weidenmaier and Peschel, 2008). PS‐III is a member of the extended lipoteichoic acid family (Percy and Grundling, 2014). Although the biological significance of these polymers in C. difficile remains poorly understood, PS‐II has been identified as the cell wall ligand that anchors members of the CWP family to the cell surface (see below). Additionally, evidence suggests that secondary cell wall polymer synthesis may be an essential process linked to CWP secretion (Willing et al., 2015).
Due to the accessibility of these polymers in the cell wall, they have been investigated as potential vaccine targets. Glycans are T cell‐independent antigens, although when conjugated to a carrier protein, these molecules can elicit a T‐cell memory response (Berti and Adamo, 2013). Anti‐PS‐I antibodies can be detected in sera from healthy horses and chemically synthesized PS‐I has been fused to a subunit of C. difficile toxin B to form a potential dual conjugate vaccine (Jiao et al., 2013). These preliminary studies demonstrated the potential of cell wall polysaccharides as vaccine candidates. However, as PS‐I is not universal between disparate C. difficile strains, PS‐II and PS‐III may represent more promising vaccine targets. Following exposure to C. difficile, humans naturally produce anti‐PS‐II antibodies (Oberli et al., 2011) and elevated levels of anti‐PS‐II IgM antibodies can be detected in pigs in response to administration of non‐conjugate PS‐II (Bertolo et al., 2012). Several conjugate PS‐II vaccines have been developed, using carriers including CRM197, a non‐toxic mutant of the diphtheria toxin commonly used as a carrier protein in commercial vaccines (Adamo et al., 2012), and C. difficile toxin fragments (Romano et al., 2014). Both of these conjugate vaccines elicit a PS‐II specific IgG antibody response. Antibodies that react with synthetic PS‐III have also been detected in the blood of C. difficile‐infected patients (Martin et al., 2013) and conjugate PS‐III vaccines are able to elicit an immune response (Cox et al., 2013). However, anti‐PS‐III antibodies also cross‐react with other members of the Clostridium family suggesting that this lipoteichoic acid is not C. difficile‐specific.
S‐layer and cell wall proteins
C. difficile S‐layer
The bacterial surface layer (S‐layer) is a proteinaceous two‐dimensional para‐crystalline array coating the entire cell (Fagan and Fairweather, 2014). S‐layers are usually composed of one or more proteins, called S‐layer proteins (SLPs) that self‐assemble to form the array. SLPs are among the most abundant and metabolically expensive proteins in bacteria that produce them, suggesting that they play a critical role. For example, a typical C. difficile cell has a surface area of approximately 18.85 μm2 and an S‐layer unit cell of 64 nm2 containing two protein subunits (our unpublished data). The S‐layer therefore consists of approximately 590 000 S‐layer subunits, requiring synthesis, export and assembly of 164 subunits per second during exponential growth. C. difficile possesses an S‐layer with square‐ordered lattice (Kawata et al., 1984), consisting of two distinct proteins, a high‐molecular weight (HMW) SLP (42–50 kDa) and a low‐molecular weight (LMW) SLP (22–38 kDa) (Cerquetti et al., 2000). The two SLPs are generated by post‐translational cleavage of a pre‐protein encoded by slpA (Calabi et al., 2001). The S‐layer appears to be essential, as evidenced by an inability to generate transposon‐mediated insertional mutants within the slpA gene (Dembek et al., 2015). SlpA has three identifiable subdomains: an N‐terminal secretion signal, followed by the highly variable LMW region and finally the HMW region containing three tandem cell wall binding 2 motifs (CWB2, PF04122) (Fagan et al., 2011). The signal peptide directs translocation across the cell membrane via the accessory Sec system (Fagan and Fairweather, 2011), following which the pre‐protein is cleaved by the cell wall localized cysteine protease Cwp84 (Kirby et al., 2009; Dang et al., 2010), to generate the two SLPs. Following cleavage, the two SLPs form a stable heterodimeric complex (H/L complex; Fagan et al., 2009) that self‐assembles to form the mature S‐layer. Extended interaction domains stabilize the HMW and LMW SLP via non‐covalent interactions in the H/L complex (Fagan et al., 2009). The CWB2 motifs in the HMW SLP anchor the H/L complex to the cell wall via an interaction with PS‐II (Willing et al., 2015). The mechanism by which the H/L complex assembles to form the mature S‐layer is still not understood. It is believed that S‐layer self‐assembly is a thermodynamically driven process (Chung et al., 2010) and some SLPs possess a distinct crystallization domain that mediates lateral interactions in the array (Smit et al., 2002). However, no such domain has been identified in C. difficile SlpA.
A lack of isogenic slpA mutants has greatly hampered analysis of C. difficile S‐layer function. However, both the LMW and HMW SLPs have been implicated in binding to both cultured Hep‐2 cells and ex vivo human gastrointestinal tissue (Calabi et al., 2002). Isolated SLPs were also observed to inhibit attachment of C. difficile to Caco‐2BBE cells (Merrigan et al., 2013). In addition to a possible adhesive role, the S‐layer has also been implicated in host immune activation via TLR4 (Ryan et al., 2011). Taken together, these data suggest that the S‐layer plays a crucial role in interactions with the host but detailed analysis of the contribution of the S‐layer to pathogenesis will require isogenic mutants. As the S‐layer is the dominant surface structure on C. difficile cells and anti‐SLP antibody has been detected in sera from convalescent patients (Wright et al., 2008). Passive immunization with anti‐SLP antibodies also showed some promise in delaying death in the lethal hamster model (O'Brien et al., 2005). However, active immunization with purified SLPs alone, or SlpA in combination with cholera toxin, did not result in significant protection using the same model (Ni Eidhin et al., 2008; Bruxelle et al., 2016). SlpA is highly variable between C. difficile strains with at least 12 distinct sequence types known (Dingle et al., 2013). An effective therapeutic or vaccine targeting this protein would need to demonstrate protection against all types.
S‐layer locus
The C. difficile genome encodes 28 paralogues of SlpA (Fagan et al., 2011) that make up the CWP family (described below). slpA is encoded within a genomic locus including 11 of these paralogues (Fig. 2A) and adjacent to a cluster of polysaccharide synthesis genes thought to be responsible for the synthesis of PS‐II (Willing et al., 2015). Genome sequencing of a panel of 57 diverse C. difficile strains has identified a 10 kb cassette within the S‐layer locus that displays higher inter‐strain variability than the rest of the locus (Dingle et al., 2013). This core variable S‐layer cassette includes the slpA, secA2, cwp2 and cwp66 genes, and 12 distinct cassette sequence types have been identified to date. Interestingly, one of these S‐layer cassettes was found to contain a 24 kb polysaccharide synthesis gene cluster inserted in place of cwp2 (Fig. 2B). As the C. difficile S‐layer is generally not thought to be glycosylated (Qazi et al., 2009), it will be very interesting to determine if this SlpA type is glycosylated. The sequence diversity observed between cassette types suggests a strong selective pressure having shaped the antigenic types. This selective pressure can perhaps be attributed to both the host immune response and bacteriophage predation.
Figure 2.
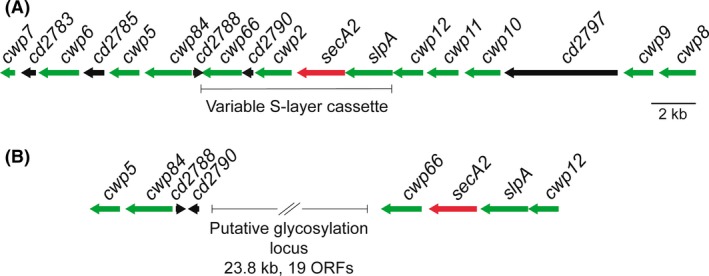
The S‐layer locus.
A. Clostridium difficile strain 630 encodes 29 cell wall proteins that use the CWB2 (PF04122) motif for non‐covalent anchoring to the cell wall. Twelve of these, including the S‐layer precursor SlpA, are encoded within a single genomic locus (green arrows) that also encodes the S‐layer secretion ATPase SecA2 (red arrow) and five unrelated proteins (black arrows). The core variable S‐layer cassette region is highlighted. An extensive glycan synthesis cluster is located immediately downstream of cwp7. It is believed that the proteins encoded in this cluster are responsible for the synthesis of PS‐II (Willing et al., 2015).
B. One of the 12 identified S‐layer cassettes (cassette type 11) has a 23.8 kb insertion that includes 19 putative ORFs (Dingle et al., 2013). Functional predictions of each of the encoded proteins identified all of the activities necessary for the synthesis of a complex glycan and transfer to a substrate. In cassette type 11, the cwp2 gene is missing and the order of cwp66 and cd2790 is reversed.
Cell wall protein family
The 28 members of the C. difficile CWP family all contain three tandem copies of the CWB2‐anchored surface proteins. Similar families of CWB2‐containing proteins have also been identified in C. botulinum and C. tetani (Bruggemann et al., 2003; Sebaihia et al., 2007). Bacillus anthracis also has an S‐layer and related family of CWP that share a common anchoring mechanism, the S‐layer homology (SLH) motif (Kern and Schneewind, 2008). The SLH motif is distinct to the CWB2 motif found in C. difficile surface proteins but is also found in three tandem copies. The B. anthracis SLH motifs adopt a pseudo‐trimeric arrangement forming a three‐pronged spindle that is required for non‐covalent binding to a pyruvylated secondary cell wall polysaccharide (Mesnage et al., 2000; Kern et al., 2011). Given the similarities to the arrangement of CWB2 motifs within SlpA and the CWPs, it is tempting to speculate that a similar mechanism may be responsible for anchoring these proteins to the anionic polymer PS‐II in the C. difficile cell wall (Willing et al., 2015). In addition to the CWB2‐anchoring domain, many of the CWPs include an additional domain that is believed to functionalize the S‐layer (Fagan and Fairweather, 2014) (Fig. 3). Only a small number of these CWPs have been characterized in any detail but several have been shown to play crucial roles in the interaction between C. difficile and the host.
Figure 3.
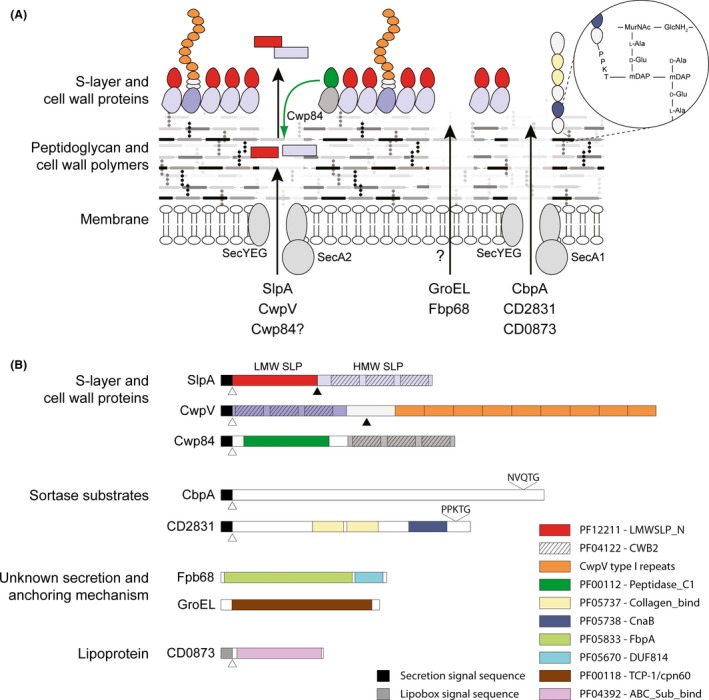
Organization of the Clostridium difficile cell envelope.
A. C. difficile has a normal Gram positive cell envelope with a surface exposed proteinaceous S‐layer on the outer surface. The S‐layer is decorated and functionalized by members of the CWP family; shown are the putative adhesin CwpV and cysteine protease Cwp84. Secretion of the S‐layer precursor SlpA and CwpV are dependent on the accessory ATPase SecA2. Following secretion, SlpA is cleaved by Cwp84 (green arrow), generating the LMW and HMW SLPs. These SLPs form a high‐affinity heterodimer that represents the basic subunit of the S‐layer. CwpV also undergoes post‐secretion processing via an enzyme‐independent auto‐proteolytic mechanism. In addition to the S‐layer and associated CWPs, C. difficile possesses numerous other cell surface proteins. The mechanism of secretion and cell wall anchoring of GroEL and Fbp68 (FbpA) is unclear but both can be detected on the cell surface. The lipoprotein CD0873 and sortase‐anchored proteins CbpA and CD2831 are likely secreted via the canonical Sec pathway. Following secretion, CD0873 is attached to the cell membrane via its lipid anchor and the sortase substrates are covalently linked to the peptidoglycan (Thr‐mDap) by the sortase enzyme CD2718.
B. Domain organization of the proteins shown in A. N‐terminal secretion signals are shown as black boxes, the CD0873 lipobox is shown in grey and the (lipoprotein) signal peptidase cleavage sites are indicated with white arrows. Post‐secretion cleavage sites are indicated with black arrows. Functional domains demonstrated experimentally or identified using the Pfam database (Finn et al., 2016) are also highlighted. The sequence and location of sorting motifs are shown above CbpA and CD2831.
Cwp66 is a 66 kDa protein with N‐terminal CWB2 motifs. The C‐terminal domain contains an apparently surface‐exposed adhesin that can mediate adherence to Vero cells (Calabi et al., 2001; Waligora et al., 2001). cwp66 is transcribed only in early exponential phase as a polycistronic transcript (Savariau‐Lacomme et al., 2003).
Cwp84 has a papain class cysteine protease domain (Savariau‐Lacomme et al., 2003). Proteolytic enzymes are frequently involved in bacterial colonization process, serving to degrade host proteins including immunoglobulin, nutrient acquisition and processing bacterial proteins necessary in pathogenesis (Maeda, 1996). Purified Cwp84 exhibits proteolytic activity against fibronectin, laminin and type IV collagen, suggestive of a possible role in infection (Janoir et al., 2004, 2007). However, the relevance of these host targets is unclear and the activity observed may be opportunistic in nature rather than reflecting underlying biological function. Pull‐down experiments using an inhibitor of SlpA cleavage identified Cwp84 as the SlpA processing protease (Dang et al., 2010) and when a cwp84 insertional knockout strain was constructed, SlpA processing was completely abolished (Kirby et al., 2009). The cwp84 gene is also located close to slpA in the S‐layer cassette (Fig. 2A). Taken together, this suggests that SlpA is the principal target of Cwp84. Although potent inhibitors of Cwp84 have been developed (Dang et al., 2010; Tam Dang et al., 2012), a cwp84 mutant was fully virulent in the hamster model of acute infection, suggesting that this protease is not a viable antimicrobial target (Kirby et al., 2009). As Cwp84 is highly conserved between C. difficile strains, it has also been investigated as a possible vaccine candidate (Pechine et al., 2011; Sandolo et al., 2011). Immunization with Cwp84 induced a specific antibody response and increased survival in the lethal hamster model. However, complete protection was not achieved and further investigation will be required to determine if Cwp84 has potential as a component of an anti‐C. difficile vaccine.
A second cysteine protease, Cwp13 is a paralogue of Cwp84, displaying 63% amino acid identity (de la Riva et al., 2011). In the absence of Cwp84, Cwp13 can partially substitute in SlpA processing, however, it also displays proteolytic activity against a sequence in the HMW region of SlpA, distinct from that recognized by Cwp84 (de la Riva et al., 2011). It has been suggested that Cwp13 plays a role in the turnover of misfolded proteins on the cell surface (de la Riva et al., 2011).
CwpV is the largest member of the CWP family and is encoded outside the S‐layer cassette. In addition to N‐terminal CWB2 motifs, CwpV possesses a region of unknown function ending in a flexible serine‐glycine‐rich linker and a C‐terminal region containing 4–9 repeats of 79–120 amino acids (Reynolds et al., 2011). Expression of CwpV is phase‐variable, with only 5% of cells in a population expressing the protein in vitro (Emerson et al., 2009). However, when expressed, CwpV accounts for almost 15% of S‐layer associated protein. Expression of CwpV is controlled by a 195 bp invertible switch located immediately upstream of the gene. The switch is flanked by imperfect 21 bp inverted repeats that can be recombined by the site‐specific recombinase RecV, inverting the intervening DNA (Emerson et al., 2009; Reynolds et al., 2011). In the ‘OFF’ orientation, a stem loop terminator is formed that prevents transcriptional readthrough. In the ‘ON’ orientation, no stem loop is formed and the gene is expressed. CwpV secretion is mediated by SecA2 (Fagan and Fairweather, 2011), following which, it is cleaved into a ~42 kDa N‐terminal fragment and a 90–120 kDa C‐terminal fragment that form a non‐covalent heterodimeric complex on the cell surface. CwpV cleavage is via intra‐molecular, enzyme‐ and cofactor‐independent autoproteolysis (Dembek et al., 2012). The C‐terminal‐repeat region varies between strains and five distinct sequence types have been identified to date, types I–V (Reynolds et al., 2011). As with SlpA, antigenic variability of CwpV may be a result of host immune or bacteriophage selective pressure. Indeed, it has been observed that CwpV expression confers protection against bacteriophage infection using a novel mechanism that does not affect phage adsorption but rather prevents phage DNA replication (Sekulovic et al., 2015), similar to superinfection exclusion systems. CwpV expression also promotes auto‐aggregation of cells in solid and liquid media (Reynolds et al., 2011) similar to those reported in mouse models of colonization (Lawley et al., 2009). It is tempting to suggest that CwpV may play a role in host colonization, but further studies will be required to test this.
Sortase‐anchored proteins
In many Gram positive bacteria, surface proteins are covalently attached to the cell wall by the action of sortases. Staphylococcus aureus makes extensive use of this anchoring mechanism and the housekeeping sortase, SrtA, has been well characterized (Schneewind et al., 1992). SrtA recognizes a C‐terminal tripartite signal sequence containing a highly conserved pentapeptide cell wall sorting motif, LPxTG. Proteins are then anchored to the cell wall via the catalytic action of a conserved cysteine residue of the sortase which cleaves the LPxTG motif between the threonine and glycine residues and, subsequently, covalently attaches the substrate protein to PG precursors (Fig. 3A) (Perry et al., 2002). Six sortase families, with different functions within the cell, have been described, all of which recognize different substrate motifs (Spirig et al., 2011). Only one functional sortase gene has been identified in C. difficile, a second contains an internal stop codon and is considered a pseudogene (Sebaihia et al., 2006). cd2718 encodes a sortase that displays 32% identity to S. aureus SrtB (Donahue et al., 2014) and displays structural characteristics of class B sortases (Chambers et al., 2015). CD2718 acts on a sorting motif closely related to that of S. aureus SrtA, differing at only at the first position, (S/P)PxTG (Donahue et al., 2014; van Leeuwen et al., 2014). Sortases, although not usually essential for growth, are often required for virulence and are therefore considered targets for new anti‐infective compounds (Cascioferro et al., 2014). Small molecule protease inhibitors are able to inhibit the action of C. difficile sortase which may aid in the development of new CDI‐specific therapeutics (Donahue et al., 2014). However, the use of C. difficile sortase as a therapeutic target may prove ineffective as inactivation of cd2718 does not significantly reduce virulence in the hamster model of infection (Chambers et al., 2015).
Studies on C. difficile sortase substrates are somewhat limited. Although eight putative sortase substrates have been identified in strain 630 (Donahue et al., 2014), attachment to the cell wall has only been demonstrated for a few of these (Chambers et al., 2015; Peltier et al., 2015). Regulation of surface exposed adhesins is key to the switch between motile and sessile forms (Boyd and O'Toole, 2012). The collagen‐binding protein CD2831 and putative adhesin CD3246 both depend on sortase activity for attachment to the cell wall and are released through the activity of the highly specific and unique protease PPEP‐1 (Zmp1/CD2830) (Hensbergen et al., 2015). The bacterial second messenger cyclic‐di‐GMP (c‐di‐GMP) has been associated with the sessile to motile switch (Romling et al., 2013). c‐di‐GMP negatively regulates PPEP‐1 expression via a type I c‐di‐GMP riboswitch, and induces cd2831 via a type II riboswitch (Soutourina et al., 2013). Thus, low level of c‐di‐GMP reduces expression of CD2831, while also facilitating release of existing protein from the cell surface and perhaps facilitating the transition from sessile to motile forms (Peltier et al., 2015). A PPEP‐1 mutant shows significantly reduced virulence in the hamster model of infection, highlighting the importance of adhesin regulation in vivo and suggests that PPEP‐1 is a promising antimicrobial target (Hensbergen et al., 2015).
Collagen‐binding protein A (CbpA) has been identified as a putative sortase substrate due to the presence of an apparent SrtB sorting motif (NVQTG) (Tulli et al., 2013). Although sortase‐mediated anchoring has not been experimentally confirmed, CbpA is surface exposed. CbpA belongs to the MSCRAMM family (Patti et al., 1994), which includes proteins that interact with the host extracellular matrix, and displays high affinity for collagens I and V, the most common components of fibrils. Heterologous expression in Lactococcus lactis resulted in surface localization and an increased ability to adhere to both immobilized collagen V and human fibroblasts (Tulli et al., 2013). Despite these observations, a cbpA mutant displays no significant decrease in adherence to either immobilized collagen or human fibroblasts (Tulli et al., 2013). A cbpA mutant also showed no significant difference, compared with the parental strain, in colonization fitness in a competitive mouse model (Janoir et al., 2013), perhaps due to redundancy with CD2831 (Hensbergen et al., 2015) and other adhesion factors. CbpA is therefore unlikely to be an effective antimicrobial target.
Other cell surface proteins
In addition to the CWP family and sortase substrates, a number of other proteins are localized to the cell surface, through interaction with the cytoplasmic membrane or through uncharacterized mechanisms of cell surface association. Several of these proteins have been identified as important colonization factors during C. difficile infection, facilitating adherence to human tissue.
Fibronectin‐binding protein (Fbp68/FbpA) is another member of the MSCRAMM family and is surface associated in C. difficile, despite lacking obvious mechanisms of cell surface association or a secretion signal (Hennequin et al., 2003). Fbp68 is a manganese‐dependent fibronectin‐binding protein, capable of binding to immobilized fibronectin and cultured vero cells (Hennequin et al., 2003; Lin et al., 2011). An fbp68 mutant displayed no significant defect in adherence to either Caco‐2 or HT29‐MTX cells but did show a significant decrease in caecal colonization in a monoxenic mouse model and was outcompeted by the parental strain in a dixenic mouse model (Barketi‐Klai et al., 2011). Anti‐Fbp68 antibodies have been found in CDI patient sera suggesting that Fbp68 may perhaps be a useful component of a C. difficile vaccine (Pechine et al., 2005).
Heat shock proteins have been shown to be important for survival in the host for many pathogenic bacteria (Zügel and Kaufmann, 1999), and C. difficile adherence to tissue cultures can be increased through varying stresses including heat shock, acidic pH and low iron levels (Eveillard et al., 1993; Waligora et al., 1999). GroEL is a member of the Hsp60 chaperonin family and its expression is upregulated in response to all of these stresses (Hennequin et al., 2001a). Co‐incubation of C. difficile with anti‐GroEL antibodies or purified GroEL significantly decreased adherence of C. difficile cells to cultured vero cells, suggesting that GroEL acts as an adhesin (Hennequin et al., 2001b). GroEL is also associated with the cell surface, although it lacks a signal sequence or obvious mechanism of cell surface association (Hennequin et al., 2001a,b). GroEL is an immunogenic protein in cell wall extracts of C. difficile and immunization of mice with recombinant protein reduces intestinal colonization by C. difficile (Pechine et al., 2013).
CD0873 is a lipoprotein with 21% sequence identity to the PsaA protein of Streptococcus pneumoniae, a multifunctional lipoprotein component of an ABC‐type transporter involved in adhesion to the host cell (Rajam et al., 2008; Kovacs‐Simon et al., 2014). Immunofluorescence microscopy revealed that CD0873 is surface exposed, and likely anchored to the membrane through attached acyl moieties. Although no in vivo experiments have been reported, a mutant strain incapable of producing CD0873 is unable to bind Caco‐2 cells, suggesting a role in adhesion to enteric cells. CD0873 is widely conserved (Kovacs‐Simon et al., 2014) and therefore represents an interesting antimicrobial target and vaccine candidate.
The cell surface as a target of phage therapy
Bacteriophage and phage‐like particles have great potential as anti‐C. difficile therapeutics (Hargreaves and Clokie, 2014). Although no C. difficile phage receptors have been identified to date, the S‐layer, CWPs and cell wall polysaccharides are all likely candidates. Indeed, it is possible that the evolution of S‐layer sequence diversity is driven by phage predation, at least in part. The long evolutionary history of C. difficile–phage interactions is apparent in the genome, with numerous prophage (Sebaihia et al., 2006), an extensive CRISPR system (Hargreaves et al., 2014) and an unusual resistance system based on the phase‐variable protein CwpV (Sekulovic et al., 2015). Although no strictly lytic phage have been identified to date, a number of studies have demonstrated the utility of phage therapy against C. difficile (Ramesh et al., 1999; Meader et al., 2010, 2013; Nale et al., 2015). ΦCD140 has been successfully used to treat CDI in hamsters, with 14 of 18 hamsters surviving a lethal challenge model (Ramesh et al., 1999). However, although the phage therapy was successful, it failed to prevent re‐infection when surviving hamsters were re‐challenged 2 weeks later. Two recent studies have also demonstrated the potential of phage therapy in vitro. ΦCD27 dramatically reduced the number of viable C. difficile and reduced production of the toxins, TcdA and TcdB, in both batch fermentation (Meader et al., 2010) and an in vitro gut model (Meader et al., 2013), with no apparent effect on the microbiota. However, these studies also highlight the therapeutic limitations of lysogenic phage. In one replicate, ΦCD27 failed to prevent C. difficile proliferation in the in vitro gut model and this was attributed to early lysogeny (Meader et al., 2013). It has also been reported that lysogeny with another phage, ΦCD38‐2, can lead to an increase in toxin production by an epidemic ribotype 027 strain (Sekulovic et al., 2011). One possible solution to this potential problem is the use of a varied phage cocktail rather than a single phage species. In one recent study, a panel of seven distinct phage species caused significant lysis of C. difficile and prevented appearance of resistant, lysogenic clones (Nale et al., 2015). This same phage cocktail delayed onset of symptoms by 33 h in the hamster model of acute disease.
Phage tail‐like particles are also an interesting alternative to traditional phage therapy. Some C. difficile strains produce R‐type bacteriocins that display antibacterial activity against other strains of C. difficile (Gebhart et al., 2012). These bacteriocins have similar structure to the tail filaments of Myoviridae phages of C. difficile, including ΦCD119 and ΦCD2, and presumably kill C. difficile by puncturing the cell envelope and dissipating the membrane potential. These naturally occurring bacteriocins have been modified to increase stability and have demonstrated impressive activity in vivo in a mouse model of infection. Importantly, killing is highly specific and these bacteriocins do not perturb the gut microbiota (Gebhart et al., 2015).
Conclusion
Clostridium difficile is a major cause of morbidity and mortality worldwide and is the leading cause of antibiotic‐associated diarrhoea. Dysbiosis, normally as a result of antibiotic treatment, is a prerequisite for CDI. Although it is encouraging to note the resurgence in research aimed at the discovery and development of new broad‐spectrum antibiotics, it is clear that we must take a more targeted approach to the treatment and prevention of CDI. The structure and function of the cell envelope is critical to our understanding of bacterial pathogenesis and also in the search for novel therapeutic and vaccine candidates. The recent surge in interest in C. difficile pathogenesis and the development of a genetic toolbox for the precise manipulation of the Clostridia has greatly improved our understanding of cell envelope architecture and function. Several components of the envelope show promise as potential drug or vaccine targets, including an unusual PG, at least two conserved secondary wall polymers and a large number of conserved surface proteins. Further study of the cell envelope in years to come will hopefully lead to development of new C. difficile‐specific treatments.
Conflict of interest
R.P.F. received a research grant from AvidBiotics Corp. (South San Francisco, USA) related to the development of C. difficile therapeutics.
Microbial Biotechnology (2017) 10(1), 76–90
Funding Information
The authors gratefully acknowledge the financial support provided by grants from the UK Medical Research Council (MR/N000900/1), the Society for Applied Microbiology and AvidBiotics Corp. O.B. is supported by a PhD studentship from the Imagine: Imaging Life initiative at the University of Sheffield. J.A.K. is supported by a PhD studentship from the University of Sheffield.
References
- Adamo, R. , Romano, M.R. , Berti, F. , Leuzzi, R. , Tontini, M. , Danieli, E. , et al (2012) Phosphorylation of the synthetic hexasaccharide repeating unit is essential for the induction of antibodies to Clostridium difficile PSII cell wall polysaccharide. ACS Chem Biol 7: 1420–1428. [DOI] [PubMed] [Google Scholar]
- Ammam, F. , Marvaud, J.C. , and Lambert, T. (2012) Distribution of the vanG‐like gene cluster in Clostridium difficile clinical isolates. Can J Microbiol 58: 547–551. [DOI] [PubMed] [Google Scholar]
- Ammam, F. , Meziane‐Cherif, D. , Mengin‐Lecreulx, D. , Blanot, D. , Patin, D. , Boneca, I.G. , et al (2013) The functional vanGCd cluster of Clostridium difficile does not confer vancomycin resistance. Mol Microbiol 89: 612–625. [DOI] [PubMed] [Google Scholar]
- Barketi‐Klai, A. , Hoys, S. , Lambert‐Bordes, S. , Collignon, A. , and Kansau, I. (2011) Role of fibronectin‐binding protein A in Clostridium difficile intestinal colonization. J Med Microbiol 60: 1155–1161. [DOI] [PubMed] [Google Scholar]
- Berti, F. , and Adamo, R. (2013) Recent mechanistic insights on glycoconjugate vaccines and future perspectives. ACS Chem Biol 8: 1653–1663. [DOI] [PubMed] [Google Scholar]
- Bertolo, L. , Boncheff, A.G. , Ma, Z. , Chen, Y.H. , Wakeford, T. , Friendship, R.M. , et al (2012) Clostridium difficile carbohydrates: glucan in spores, PSII common antigen in cells, immunogenicity of PSII in swine and synthesis of a dual C. difficile‐ETEC conjugate vaccine. Carbohydr Res 354: 79–86. [DOI] [PubMed] [Google Scholar]
- Bevins, C.L. , and Salzman, N.H. (2011) Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol 9: 356–368. [DOI] [PubMed] [Google Scholar]
- Boyd, C.D. , and O'Toole, G.A. (2012) Second messenger regulation of biofilm formation: breakthroughs in understanding c‐di‐GMP effector systems. Annu Rev Cell Dev Biol 28: 439–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggemann, H. , Baumer, S. , Fricke, W.F. , Wiezer, A. , Liesegang, H. , Decker, I. , et al (2003) The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc Natl Acad Sci U S A 100: 1316–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruxelle, J.F. , Mizrahi, A. , Hoys, S. , Collignon, A. , Janoir, C. , and Pechine, S. (2016) Immunogenic properties of the surface layer precursor of Clostridium difficile and vaccination assays in animal models. Anaerobe 37: 78–84. [DOI] [PubMed] [Google Scholar]
- Calabi, E. , Ward, S. , Wren, B. , Paxton, T. , Panico, M. , Morris, H. , et al (2001) Molecular characterization of the surface layer proteins from Clostridium difficile . Mol Microbiol 40: 1187–1199. [DOI] [PubMed] [Google Scholar]
- Calabi, E. , Calabi, F. , Phillips, A.D. , and Fairweather, N.F. (2002) Binding of Clostridium difficile surface layer proteins to gastrointestinal tissues. Infect Immun 70: 5770–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascioferro, S. , Totsika, M. , and Schillaci, D. (2014) Sortase A: an ideal target for anti‐virulence drug development. Microb Pathog 77: 105–112. [DOI] [PubMed] [Google Scholar]
- Cerquetti, M. , Molinari, A. , Sebastianelli, A. , Diociaiuti, M. , Petruzzelli, R. , Capo, C. , and Mastrantonio, P. (2000) Characterization of surface layer proteins from different Clostridium difficile clinical isolates. Microb Pathog 28: 363–372. [DOI] [PubMed] [Google Scholar]
- Chambers, C.J. , Roberts, A.K. , Shone, C.C. , and Acharya, K.R. (2015) Structure and function of a Clostridium difficile sortase enzyme. Sci Rep 5: 9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, S. , Shin, S.H. , Bertozzi, C.R. , and De Yoreo, J.J. (2010) Self‐catalyzed growth of S layers via an amorphous‐to‐crystalline transition limited by folding kinetics. Proc Natl Acad Sci U S A 107: 16536–16541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, A.D. , St Michael, F. , Aubry, A. , Cairns, C.M. , Strong, P.C. , Hayes, A.C. , and Logan, S.M. (2013) Investigating the candidacy of a lipoteichoic acid‐based glycoconjugate as a vaccine to combat Clostridium difficile infection. Glycoconj J 30: 843–855. [DOI] [PubMed] [Google Scholar]
- Dang, T.H. , de la Riva, L. , Fagan, R.P. , Storck, E.M. , Heal, W.P. , Janoir, C. , et al (2010) Chemical probes of surface layer biogenesis in Clostridium difficile . ACS Chem Biol 5: 279–285. [DOI] [PubMed] [Google Scholar]
- Dembek, M. , Reynolds, C.B. , and Fairweather, N.F. (2012) Clostridium difficile cell wall protein CwpV undergoes enzyme‐independent intramolecular autoproteolysis. J Biol Chem 287: 1538–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembek, M. , Barquist, L. , Boinett, C.J. , Cain, A.K. , Mayho, M. , Lawley, T.D. , et al (2015) High‐throughput analysis of gene essentiality and sporulation in Clostridium difficile . MBio 6: e02383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depardieu, F. , Bonora, M.G. , Reynolds, P.E. , and Courvalin, P. (2003) The vanG glycopeptide resistance operon from Enterococcus faecalis revisited. Mol Microbiol 50: 931–948. [DOI] [PubMed] [Google Scholar]
- Dethlefsen, L. , Huse, S. , Sogin, M.L. , and Relman, D.A. (2008) The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6: e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle, K.E. , Didelot, X. , Ansari, M.A. , Eyre, D.W. , Vaughan, A. , Griffiths, D. , et al (2013) Recombinational switching of the Clostridium difficile S‐layer and a novel glycosylation gene cluster revealed by large‐scale whole‐genome sequencing. J Infect Dis 207: 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue, E.H. , Dawson, L.F. , Valiente, E. , Firth‐Clark, S. , Major, M.R. , Littler, E. , et al (2014) Clostridium difficile has a single sortase, SrtB, that can be inhibited by small‐molecule inhibitors. BMC Microbiol 14: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson, J.E. , Reynolds, C.B. , Fagan, R.P. , Shaw, H.A. , Goulding, D. , and Fairweather, N.F. (2009) A novel genetic switch controls phase variable expression of CwpV, a Clostridium difficile cell wall protein. Mol Microbiol 74: 541–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eveillard, M. , Fourel, V. , Barc, M.C. , Kerneis, S. , Coconnier, M.H. , Karjalainen, T. , et al (1993) Identification and characterization of adhesive factors of Clostridium difficile involved in adhesion to human colonic enterocyte‐like Caco‐2 and mucus‐secreting HT29 cells in culture. Mol Microbiol 7: 371–381. [DOI] [PubMed] [Google Scholar]
- Fagan, R.P. , and Fairweather, N.F. (2011) Clostridium difficile has two parallel and essential Sec secretion systems. J Biol Chem 286: 27483–27493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan, R.P. , and Fairweather, N.F. (2014) Biogenesis and functions of bacterial S‐layers. Nat Rev Microbiol 12: 211–222. [DOI] [PubMed] [Google Scholar]
- Fagan, R.P. , Albesa‐Jove, D. , Qazi, O. , Svergun, D.I. , Brown, K.A. , and Fairweather, N.F. (2009) Structural insights into the molecular organization of the S‐layer from Clostridium difficile . Mol Microbiol 71: 1308–1322. [DOI] [PubMed] [Google Scholar]
- Fagan, R.P. , Janoir, C. , Collignon, A. , Mastrantonio, P. , Poxton, I.R. , and Fairweather, N.F. (2011) A proposed nomenclature for cell wall proteins of Clostridium difficile . J Med Microbiol 60: 1225–1228. [DOI] [PubMed] [Google Scholar]
- Finn, R.D. , Coggill, P. , Eberhardt, R.Y. , Eddy, S.R. , Mistry, J. , Mitchell, A.L. , et al (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44: D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman, J. , Baines, S.D. , Jabes, D. , and Wilcox, M.H. (2005) Comparison of the efficacy of ramoplanin and vancomycin in both in vitro and in vivo models of clindamycin‐induced Clostridium difficile infection. J Antimicrob Chemother 56: 717–725. [DOI] [PubMed] [Google Scholar]
- Ganeshapillai, J. , Vinogradov, E. , Rousseau, J. , Weese, J.S. , and Monteiro, M.A. (2008) Clostridium difficile cell‐surface polysaccharides composed of pentaglycosyl and hexaglycosyl phosphate repeating units. Carbohydr Res 343: 703–710. [DOI] [PubMed] [Google Scholar]
- Gebhart, D. , Williams, S.R. , Bishop‐Lilly, K.A. , Govoni, G.R. , Willner, K.M. , Butani, A. , et al (2012) Novel high‐molecular‐weight, R‐type bacteriocins of Clostridium difficile . J Bacteriol 194: 6240–6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhart, D. , Lok, S. , Clare, S. , Tomas, M. , Stares, M. , Scholl, D. , et al (2015) A modified R‐type bacteriocin specifically targeting Clostridium difficile prevents colonization of mice without affecting gut microbiota diversity. MBio 6: e02368–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves, K.R. , and Clokie, M.R. (2014) Clostridium difficile phages: still difficult? Front Microbiol 5: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves, K.R. , Flores, C.O. , Lawley, T.D. , and Clokie, M.R. (2014) Abundant and diverse clustered regularly interspaced short palindromic repeat spacers in Clostridium difficile strains and prophages target multiple phage types within this pathogen. MBio 5: e01045–01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennequin, C. , Collignon, A. , and Karjalainen, T. (2001a) Analysis of expression of GroEL (Hsp60) of Clostridium difficile in response to stress. Microb Pathog 31: 255–260. [DOI] [PubMed] [Google Scholar]
- Hennequin, C. , Porcheray, F. , Waligora‐Dupriet, A. , Collignon, A. , Barc, M. , Bourlioux, P. , and Karjalainen, T. (2001b) GroEL (Hsp60) of Clostridium difficile is involved in cell adherence. Microbiology 147: 87–96. [DOI] [PubMed] [Google Scholar]
- Hennequin, C. , Janoir, C. , Barc, M.C. , Collignon, A. , and Karjalainen, T. (2003) Identification and characterization of a fibronectin‐binding protein from Clostridium difficile . Microbiology 149: 2779–2787. [DOI] [PubMed] [Google Scholar]
- Hensbergen, P.J. , Klychnikov, O.I. , Bakker, D. , Dragan, I. , Kelly, M.L. , Minton, N.P. , et al (2015) Clostridium difficile secreted Pro‐Pro endopeptidase PPEP‐1 (ZMP1/CD2830) modulates adhesion through cleavage of the collagen binding protein CD2831. FEBS Lett 589: 3952–3958. [DOI] [PubMed] [Google Scholar]
- Ho, T.D. , Williams, K.B. , Chen, Y. , Helm, R.F. , Popham, D.L. , and Ellermeier, C.D. (2014) Clostridium difficile extracytoplasmic function sigma factor sigmaV regulates lysozyme resistance and is necessary for pathogenesis in the hamster model of infection. Infect Immun 82: 2345–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jank, T. , and Aktories, K. (2008) Structure and mode of action of clostridial glucosylating toxins: the ABCD model. Trends Microbiol 16: 222–229. [DOI] [PubMed] [Google Scholar]
- Janoir, C. , Grenery, J. , Savariau‐Lacomme, M.P. , and Collignon, A. (2004) Characterization of an extracellular protease from Clostridium difficile . Pathol Biol (Paris) 52: 444–449. [DOI] [PubMed] [Google Scholar]
- Janoir, C. , Pechine, S. , Grosdidier, C. , and Collignon, A. (2007) Cwp84, a surface‐associated protein of Clostridium difficile, is a cysteine protease with degrading activity on extracellular matrix proteins. J Bacteriol 189: 7174–7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoir, C. , Deneve, C. , Bouttier, S. , Barbut, F. , Hoys, S. , Caleechum, L. , et al (2013) Adaptive strategies and pathogenesis of Clostridium difficile from in vivo transcriptomics. Infect Immun 81: 3757–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, Y. , Ma, Z. , Hodgins, D. , Pequegnat, B. , Bertolo, L. , Arroyo, L. , and Monteiro, M.A. (2013) Clostridium difficile PSI polysaccharide: synthesis of pentasaccharide repeating block, conjugation to exotoxin B subunit, and detection of natural anti‐PSI IgG antibodies in horse serum. Carbohydr Res 378: 15–25. [DOI] [PubMed] [Google Scholar]
- Johnson, S. , Louie, T.J. , Gerding, D.N. , Cornely, O.A. , Chasan‐Taber, S. , Fitts, D. , et al (2014) Vancomycin, metronidazole, or tolevamer for Clostridium difficile infection: results from two multinational, randomized, controlled trials. Clin Infect Dis 59: 345–354. [DOI] [PubMed] [Google Scholar]
- Kawata, T. , Takeoka, A. , Takumi, K. , and Masuda, K. (1984) Demonstration and preliminary characterization of a regular array in the cell wall of Clostridium difficile . FEMS Microbiol Lett 24: 323–328. [Google Scholar]
- Kern, J.W. , and Schneewind, O. (2008) BslA, a pXO1‐encoded adhesin of Bacillus anthracis . Mol Microbiol 68: 504–515. [DOI] [PubMed] [Google Scholar]
- Kern, J. , Wilton, R. , Zhang, R. , Binkowski, T.A. , Joachimiak, A. , and Schneewind, O. (2011) Structure of surface layer homology (SLH) domains from Bacillus anthracis surface array protein. J Biol Chem 286: 26042–26049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby, J.M. , Ahern, H. , Roberts, A.K. , Kumar, V. , Freeman, Z. , Acharya, K.R. , and Shone, C.C. (2009) Cwp84, a surface‐associated cysteine protease, plays a role in the maturation of the surface layer of Clostridium difficile . J Biol Chem 284: 34666–34673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs‐Simon, A. , Leuzzi, R. , Kasendra, M. , Minton, N. , Titball, R.W. , and Michell, S.L. (2014) Lipoprotein CD0873 is a novel adhesin of Clostridium difficile . J Infect Dis 210: 274–284. [DOI] [PubMed] [Google Scholar]
- Lawley, T.D. , Clare, S. , Walker, A.W. , Goulding, D. , Stabler, R.A. , Croucher, N. , et al (2009) Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore‐mediated transmission, and severe disease in immunocompromised hosts. Infect Immun 77: 3661–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen, H.C. , Klychnikov, O.I. , Menks, M.A. , Kuijper, E.J. , Drijfhout, J.W. , and Hensbergen, P.J. (2014) Clostridium difficile sortase recognizes a (S/P)PXTG sequence motif and can accommodate diaminopimelic acid as a substrate for transpeptidation. FEBS Lett 588: 4325–4333. [DOI] [PubMed] [Google Scholar]
- Lin, Y.P. , Kuo, C.J. , Koleci, X. , McDonough, S.P. , and Chang, Y.F. (2011) Manganese binds to Clostridium difficile Fbp68 and is essential for fibronectin binding. J Biol Chem 286: 3957–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, H. (1996) Role of microbial proteases in pathogenesis. Microbiol Immunol 40: 685–699. [DOI] [PubMed] [Google Scholar]
- Martin, C.E. , Broecker, F. , Oberli, M.A. , Komor, J. , Mattner, J. , Anish, C. , and Seeberger, P.H. (2013) Immunological evaluation of a synthetic Clostridium difficile oligosaccharide conjugate vaccine candidate and identification of a minimal epitope. J Am Chem Soc 135: 9713–9722. [DOI] [PubMed] [Google Scholar]
- Mayer, M.J. , Garefalaki, V. , Spoerl, R. , Narbad, A. , and Meijers, R. (2011) Structure‐based modification of a Clostridium difficile‐targeting endolysin affects activity and host range. J Bacteriol 193: 5477–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, L.C. , Killgore, G.E. , Thompson, A. , Owens, R.C. Jr , Kazakova, S.V. , Sambol, S.P. , et al (2005) An epidemic, toxin gene‐variant strain of Clostridium difficile . N Engl J Med 353: 2433–2441. [DOI] [PubMed] [Google Scholar]
- McGowan, A.P. , Lalayiannis, L.C. , Sarma, J.B. , Marshall, B. , Martin, K.E. , and Welfare, M.R. (2011) Thirty‐day mortality of Clostridium difficile infection in a UK National Health Service Foundation Trust between 2002 and 2008. J Hosp Infect 77: 11–15. [DOI] [PubMed] [Google Scholar]
- Meader, E. , Mayer, M.J. , Gasson, M.J. , Steverding, D. , Carding, S.R. , and Narbad, A. (2010) Bacteriophage treatment significantly reduces viable Clostridium difficile and prevents toxin production in an in vitro model system. Anaerobe 16: 549–554. [DOI] [PubMed] [Google Scholar]
- Meader, E. , Mayer, M.J. , Steverding, D. , Carding, S.R. , and Narbad, A. (2013) Evaluation of bacteriophage therapy to control Clostridium difficile and toxin production in an in vitro human colon model system. Anaerobe 22: 25–30. [DOI] [PubMed] [Google Scholar]
- Merrigan, M.M. , Venugopal, A. , Roxas, J.L. , Anwar, F. , Mallozzi, M.J. , Roxas, B.A. , et al (2013) Surface‐layer protein A (SlpA) is a major contributor to host‐cell adherence of Clostridium difficile . PLoS ONE 8: e78404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage, S. , Fontaine, T. , Mignot, T. , Delepierre, M. , Mock, M. , and Fouet, A. (2000) Bacterial SLH domain proteins are non‐covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J 19: 4473–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nale, J.Y. , Spencer, J. , Hargreaves, K.R. , Buckley, A.M. , Trzepinski, P. , Douce, G.R. , and Clokie, M.R. (2015) Bacteriophage combinations significantly reduce Clostridium difficile growth in vitro and proliferation in vivo. Antimicrob Agents Chemother 60: 968–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, Y.K. , Ehsaan, M. , Philip, S. , Collery, M.M. , Janoir, C. , Collignon, A. , et al (2013) Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyrE alleles. PLoS ONE 8: e56051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Eidhin, D.B. , O'Brien, J.B. , McCabe, M.S. , Athie‐Morales, V. , and Kelleher, D.P. (2008) Active immunization of hamsters against Clostridium difficile infection using surface‐layer protein. FEMS Immunol Med Microbiol 52: 207–218. [DOI] [PubMed] [Google Scholar]
- Oberli, M.A. , Hecht, M.L. , Bindschadler, P. , Adibekian, A. , Adam, T. , and Seeberger, P.H. (2011) A possible oligosaccharide‐conjugate vaccine candidate for Clostridium difficile is antigenic and immunogenic. Chem Biol 18: 580–588. [DOI] [PubMed] [Google Scholar]
- O'Brien, J.B. , McCabe, M.S. , Athie‐Morales, V. , McDonald, G.S. , Ni Eidhin, D.B. , and Kelleher, D.P. (2005) Passive immunisation of hamsters against Clostridium difficile infection using antibodies to surface layer proteins. FEMS Microbiol Lett 246: 199–205. [DOI] [PubMed] [Google Scholar]
- Patti, J.M. , Allen, B.L. , McGavin, M.J. , and Hook, M. (1994) MSCRAMM‐mediated adherence of microorganisms to host tissues. Annu Rev Microbiol 48: 585–617. [DOI] [PubMed] [Google Scholar]
- Pechine, S. , Gleizes, A. , Janoir, C. , Gorges‐Kergot, R. , Barc, M.C. , Delmee, M. , and Collignon, A. (2005) Immunological properties of surface proteins of Clostridium difficile . J Med Microbiol 54: 193–196. [DOI] [PubMed] [Google Scholar]
- Pechine, S. , Deneve, C. , Le Monnier, A. , Hoys, S. , Janoir, C. , and Collignon, A. (2011) Immunization of hamsters against Clostridium difficile infection using the Cwp84 protease as an antigen. FEMS Immunol Med Microbiol 63: 73–81. [DOI] [PubMed] [Google Scholar]
- Pechine, S. , Hennequin, C. , Boursier, C. , Hoys, S. , and Collignon, A. (2013) Immunization using GroEL decreases Clostridium difficile intestinal colonization. PLoS ONE 8: e81112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier, J. , Courtin, P. , El Meouche, I. , Lemee, L. , Chapot‐Chartier, M.P. , and Pons, J.L. (2011) Clostridium difficile has an original peptidoglycan structure with a high level of N‐acetylglucosamine deacetylation and mainly 3‐3 cross‐links. J Biol Chem 286: 29053–29062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier, J. , Courtin, P. , El Meouche, I. , Catel‐Ferreira, M. , Chapot‐Chartier, M.P. , Lemee, L. , and Pons, J.L. (2013) Genomic and expression analysis of the vanG‐like gene cluster of Clostridium difficile . Microbiology 159: 1510–1520. [DOI] [PubMed] [Google Scholar]
- Peltier, J. , Shaw, H.A. , Couchman, E.C. , Dawson, L.F. , Yu, L. , Choudhary, J.S. , et al (2015) Cyclic diGMP regulates production of sortase substrates of Clostridium difficile and their surface exposure through ZmpI protease‐mediated cleavage. J Biol Chem 290: 24453–24469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percy, M.G. , and Grundling, A. (2014) Lipoteichoic acid synthesis and function in gram‐positive bacteria. Annu Rev Microbiol 68: 81–100. [DOI] [PubMed] [Google Scholar]
- Perry, A.M. , Ton‐That, H. , Mazmanian, S.K. , and Schneewind, O. (2002) Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase‐catalyzed surface protein anchoring. J Biol Chem 277: 16241–16248. [DOI] [PubMed] [Google Scholar]
- Pullman, J. , Prieto, J. and Leach, T.S. (2004) Ramoplanin vs vancomycin in the treatment of Clostridium difficile diarrhea: a phase II study In 44th Interscience Conference Antimicrob Agents Chemother, abstr K‐985a. American Society for Microbiology, Washington, DC, USA. [Google Scholar]
- Qazi, O. , Hitchen, P. , Tissot, B. , Panico, M. , Morris, H.R. , Dell, A. , and Fairweather, N. (2009) Mass spectrometric analysis of the S‐layer proteins from Clostridium difficile demonstrates the absence of glycosylation. J Mass Spectrom 44: 368–374. [DOI] [PubMed] [Google Scholar]
- Rajam, G. , Anderton, J.M. , Carlone, G.M. , Sampson, J.S. , and Ades, E.W. (2008) Pneumococcal surface adhesin A (PsaA): a review. Crit Rev Microbiol 34: 131–142. [DOI] [PubMed] [Google Scholar]
- Ramesh, V. , Fralick, J.A. , and Rolfe, R.D. (1999) Prevention of Clostridium difficile‐induced ileocecitis with Bacteriophage. Anaerobe 5: 69–78. [Google Scholar]
- Reid, C.W. , Vinogradov, E. , Li, J. , Jarrell, H.C. , Logan, S.M. , and Brisson, J.R. (2012) Structural characterization of surface glycans from Clostridium difficile . Carbohydr Res 354: 65–73. [DOI] [PubMed] [Google Scholar]
- Reynolds, P.E. (1989) Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur J Clin Microbiol Infect Dis 8: 943–950. [DOI] [PubMed] [Google Scholar]
- Reynolds, C.B. , Emerson, J.E. , de la Riva, L. , Fagan, R.P. , and Fairweather, N.F. (2011) The Clostridium difficile cell wall protein CwpV is antigenically variable between strains, but exhibits conserved aggregation‐promoting function. PLoS Pathog 7: e1002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Riva, L. , Willing, S.E. , Tate, E.W. , and Fairweather, N.F. (2011) Roles of cysteine proteases Cwp84 and Cwp13 in biogenesis of the cell wall of Clostridium difficile . J Bacteriol 193: 3276–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano, M.R. , Leuzzi, R. , Cappelletti, E. , Tontini, M. , Nilo, A. , Proietti, D. , et al (2014) Recombinant Clostridium difficile toxin fragments as carrier protein for PSII surface polysaccharide preserve their neutralizing activity. Toxins (Basel) 6: 1385–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling, U. , Galperin, M.Y. , and Gomelsky, M. (2013) Cyclic di‐GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77: 1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupnik, M. , Wilcox, M.H. , and Gerding, D.N. (2009) Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol 7: 526–536. [DOI] [PubMed] [Google Scholar]
- Ryan, A. , Lynch, M. , Smith, S.M. , Amu, S. , Nel, H.J. , McCoy, C.E. , et al (2011) A role for TLR4 in Clostridium difficile infection and the recognition of surface layer proteins. PLoS Pathog 7: e1002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandolo, C. , Pechine, S. , Le Monnier, A. , Hoys, S. , Janoir, C. , Coviello, T. , et al (2011) Encapsulation of Cwp84 into pectin beads for oral vaccination against Clostridium difficile . Eur J Pharm Biopharm 79: 566–573. [DOI] [PubMed] [Google Scholar]
- Savariau‐Lacomme, M.P. , Lebarbier, C. , Karjalainen, T. , Collignon, A. , and Janoir, C. (2003) Transcription and analysis of polymorphism in a cluster of genes encoding surface‐associated proteins of Clostridium difficile . J Bacteriol 185: 4461–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneewind, O. , Model, P. , and Fischetti, V.A. (1992) Sorting of protein A to the staphylococcal cell wall. Cell 70: 267–281. [DOI] [PubMed] [Google Scholar]
- Sebaihia, M. , Wren, B.W. , Mullany, P. , Fairweather, N.F. , Minton, N. , Stabler, R. , et al (2006) The multidrug‐resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38: 779–786. [DOI] [PubMed] [Google Scholar]
- Sebaihia, M. , Peck, M.W. , Minton, N.P. , Thomson, N.R. , Holden, M.T. , Mitchell, W.J. , et al (2007) Genome sequence of a proteolytic (Group I) Clostridium botulinum strain Hall A and comparative analysis of the clostridial genomes. Genome Res 17: 1082–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulovic, O. , Meessen‐Pinard, M. , and Fortier, L.C. (2011) Prophage‐stimulated toxin production in Clostridium difficile NAP1/027 lysogens. J Bacteriol 193: 2726–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulovic, O. , Ospina Bedoya, M. , Fivian‐Hughes, A.S. , Fairweather, N.F. , and Fortier, L.C. (2015) The Clostridium difficile cell wall protein CwpV confers phase‐variable phage resistance. Mol Microbiol 98: 329–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, E. , Jager, D. , Martinez, B. , Tielen, F.J. , and Pouwels, P.H. (2002) Structural and functional analysis of the S‐layer protein crystallisation domain of Lactobacillus acidophilus ATCC 4356: evidence for protein‐protein interaction of two subdomains. J Mol Biol 324: 953–964. [DOI] [PubMed] [Google Scholar]
- Smits, W.K. , Lyras, D. , Lacy, D.B. , Wilcox, M.H. , and Kuijper, E.J. (2016) Clostridium difficile infection. Nat Rev Dis Primers 2: 16020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina, O.A. , Monot, M. , Boudry, P. , Saujet, L. , Pichon, C. , Sismeiro, O. , et al (2013) Genome‐wide identification of regulatory RNAs in the human pathogen Clostridium difficile . PLoS Genet 9: e1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirig, T. , Weiner, E.M. , and Clubb, R.T. (2011) Sortase enzymes in Gram‐positive bacteria. Mol Microbiol 82: 1044–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam Dang, T.H. , Fagan, R.P. , Fairweather, N.F. , and Tate, E.W. (2012) Novel inhibitors of surface layer processing in Clostridium difficile . Bioorg Med Chem 20: 614–621. [DOI] [PubMed] [Google Scholar]
- Tulli, L. , Marchi, S. , Petracca, R. , Shaw, H.A. , Fairweather, N.F. , Scarselli, M. , et al (2013) CbpA: a novel surface exposed adhesin of Clostridium difficile targeting human collagen. Cell Microbiol 15: 1674–1687. [DOI] [PubMed] [Google Scholar]
- Vollmer, W. , and Tomasz, A. (2000) The pgdA gene encodes for a peptidoglycan N‐acetylglucosamine deacetylase in Streptococcus pneumoniae . J Biol Chem 275: 20496–20501. [DOI] [PubMed] [Google Scholar]
- Vollmer, W. , Blanot, D. , and de Pedro, M.A. (2008) Peptidoglycan structure and architecture. FEMS Microbiol Rev 32: 149–167. [DOI] [PubMed] [Google Scholar]
- Waligora, A.J. , Barc, M.C. , Bourlioux, P. , Collignon, A. , and Karjalainen, T. (1999) Clostridium difficile cell attachment is modified by environmental factors. Appl Environ Microbiol 65: 4234–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waligora, A.J. , Hennequin, C. , Mullany, P. , Bourlioux, P. , Collignon, A. , and Karjalainen, T. (2001) Characterization of a cell surface protein of Clostridium difficile with adhesive properties. Infect Immun 69: 2144–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidenmaier, C. , and Peschel, A. (2008) Teichoic acids and related cell‐wall glycopolymers in Gram‐positive physiology and host interactions. Nat Rev Microbiol 6: 276–287. [DOI] [PubMed] [Google Scholar]
- Willing, S.E. , Candela, T. , Shaw, H.A. , Seager, Z. , Mesnage, S. , Fagan, R.P. , and Fairweather, N.F. (2015) Clostridium difficile surface proteins are anchored to the cell wall using CWB2 motifs that recognise the anionic polymer PSII. Mol Microbiol 96: 596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, A. , Drudy, D. , Kyne, L. , Brown, K. , and Fairweather, N.F. (2008) Immunoreactive cell wall proteins of Clostridium difficile identified by human sera. J Med Microbiol 57: 750–756. [DOI] [PubMed] [Google Scholar]
- Zügel, U. , and Kaufmann, S.H. (1999) Role of heat shock proteins in protection from and pathogenesis of infectious diseases. Clin Microbiol Rev 12: 19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]


