Abstract
Binary expression systems are flexible and versatile genetic tools in Drosophila. The Q-system is a recently developed repressible binary expression system that offers new possibilities for transgene expression and genetic manipulations. In this review chapter, we focus on current state-of-the-art Q-system tools and reagents. We also discuss in vivo applications of the Q-system, together with GAL4/UAS and LexA/LexAop systems, for simultaneous expression of multiple effectors, intersectional labeling, and clonal analysis.
Keywords: Binary expression system, Q-system, QF, QF2, QF2w, LexAQF, GAL4QF, QUAS, QS, Quinic acid, Chimeric transactivators, Intersectional expression, Mitotic recombination, MARCM, Mosaic analysis, Neurospora crassa
1 Introduction
Spatial and temporal control of transgene expression is essential for studies of gene and cell function. Expression of GFP and other reporters uncover the anatomy of cellular circuits and the activity of cells [1 – 3]; gene knockdown by RNAi or gene overexpression emphasizes gene function [4, 5]; expression of neurotoxins, or expression of light- or temperature-sensitive channels, alters cell function by inhibiting synaptic transmission [6], or by hyperpolarizing or depolarizing the cell [7 – 9]; expression of cellular toxins or apoptotic genes [10, 11] eliminates defined cells to reveal their role in a circuit or complex tissue. Binary expression systems, such as GAL4/UAS [12], LexA/LexAop [13], tTA/TRE [14], and the Q-system [15], are designed to direct efficient transgene expression that can be fine-tuned to suit various experimental needs (reviewed in ref. [16)].
The Q-system is a binary expression system that offers an easy and flexible means to manipulate cell and circuit function [15, 17]. The Q-system consists of two core components, a “driver” and a “reporter”, and two additional components, a “repressor” and a drug that counteracts the repressor (Fig. 1). The driver components of the Q-system are transgenes that carry enhancer and promoter sequences specific to the cells of interest upstream of a transcription factor, QF. The reporter components are transgenes that carry the QF binding sequence, QUAS, upstream of genes coding for fluorescent proteins, toxins, ion channels, or other effectors. The repressor components are transgenes that carry the QS gene, placed downstream of UAS or another enhancer. Normally the “driver” and the “reporter” transgenes are kept in different fly stocks and are brought together by genetic crosses in variable combinations as determined by the purpose of a particular experiment.
Fig. 1.
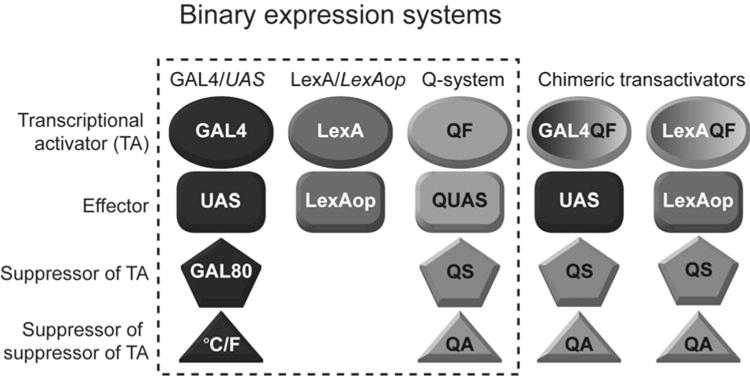
Binary expression systems. The GAL4/UAS and the Q-system consist of a transcriptional activator (TA, ovals), an effector (rectangles), a suppressor (pentagons), and a suppressor of the suppressor (triangles). The LexA/LexAop system consists only of a TA and an effector. The three systems function independently of each other, but reporter and suppressor activities can be swapped by using Chimeric transactivators (two right columns). GAL4QF binds to the UAS effector sequence (of the GAL4/UAS system), and is suppressed by the QS suppressor (of the Q-system). Similarly, LexAQF binds to the LexAop effector sequence (of the LexA/LexAop system), and is suppressed by QS
The design of the Q-system is similar to GAL4/UAS [12] and LexA/LexAop [13] systems (Fig. 1), and because the three systems do not cross-react, they can be used simultaneously for sophisticated genetic manipulations. The need for the Q-system came from two major limitations of the GAL4/UAS and LexA/LexAop systems. First, the transactivator LexA does not have its own repressor [13], thus it is not possible to repress GAL4 and LexA independently. Second, GAL80, the repressor of GAL4, cannot control temporal expression of transgenes independently of the ambient temperature [18, 19]. The Q-system offers a useful alternative to the GAL4/UAS system in experiments where the activity of repressor needs to be switched on or off at a certain point in time, but changes in ambient temperature need to be avoided, e.g., due to the strong behavioral preference Drosophila have toward temperatures around 24 °C [20]. GAL4/UAS and the Q systems can also be utilized to drive different reporters in overlapping subsets of cells, when independent temporal control of the reporters is required.
The Q-system also enables sophisticated intersectional and double-MARCM (Mosaic Analysis with a Repressible Cell Marker) experiments to be performed [15, 21 – 23]. It is often required to express transgenes in a small number of cells that have similar function, e.g., olfactory projection neurons that target a single glomerulus. There are several thousand existing GAL4 lines, but the use of them is often hampered by the fact that, in addition to the cells of interest (projection neurons from the previous example), GAL4 is also expressed in many other cells (e.g., other types of neurons, often with unknown function). Therefore, UAS-geneX effectors will alter the function of both cells in and outside of the target tissue. Intersectional approaches between GAL4/UAS and Q systems can “clean up” a line by restricting expression of reporters to a subset of cells where both GAL4 and QF are expressed [15, 17]. Conventional MARCM allows the expression of a reporter in one of the two cells that are produced by a mitotic cell division [21, 23]. Coupled MARCM utilizes GAL4/UAS and Q systems [15, 17] together, and allows the expression of two different reporters and/or effectors in the two clusters of cells that originated from one mitotic cell division. This feature is very useful in the studies of cell proliferation, cell fate, and other developmental processes (see also refs. [24 – 26)].
The applications of the Q-system reach beyond Drosophila genetics. Currently (June 2016), the Q-system has been successfully used in cultured mammalian cells [15], C. elegans [27] and zebrafish [28], and is under development in plants (Arabidopsis) and mosquitoes (Anopheles gambiae) (Riabinina et al, in review).
2 Neurospora qa Gene Cluster
The Q-system is based on the qa gene cluster of the bread fungus Neurospora crassa (Fig. 2a) [15, 29]. The genes of the qa cluster control the metabolism of quinic acid, which allows Neurospora to use quinic acid as an alternative carbon source in conditions of low glucose [29]. The qa cluster contains seven genes, two of which are regulatory (the transcriptional activator qa-1F and its repressor qa-1S), and five are enzymatic or structural. The QA-1F protein binds to a specific DNA sequence (5′- GGR TAA RγR γTT ATC C -3′, where R is A/G, Y is C/T) present in several copies upstream of the qa genes, initiating their transcription [30, 31]. In conditions of low quinic acid, the interaction between QA-1S and QA-1F proteins prevents QA-1F from binding transcriptional machinery required for activating the expression of the qa genes [32]. In conditions of high quinic acid, the quinic acid binds to QA-1S which disrupts its binding to QA-1F. QA-1F can now associate with the transcriptional machinery and activate transcription. Similarly to GAL4, QA-1F has been predicted to have a modular structure [31, 33 – 39], consisting of a Zn2/Cys6 zinc finger DNA binding domain (DBD), a middle domain (MD), and an acidic activation domain (AD) that binds QA-1S or molecular factors that initiate transcription. In contrast to GAL4 and QF, LexA consists of a DBD only, and thus needs to be paired with an activation domain (the commonly used ones are VP16 [21], p65 [40], GAL4 AD [21]) to initiate transcription. As such, LexA also does not have a specific independent suppressor.
Fig. 2.
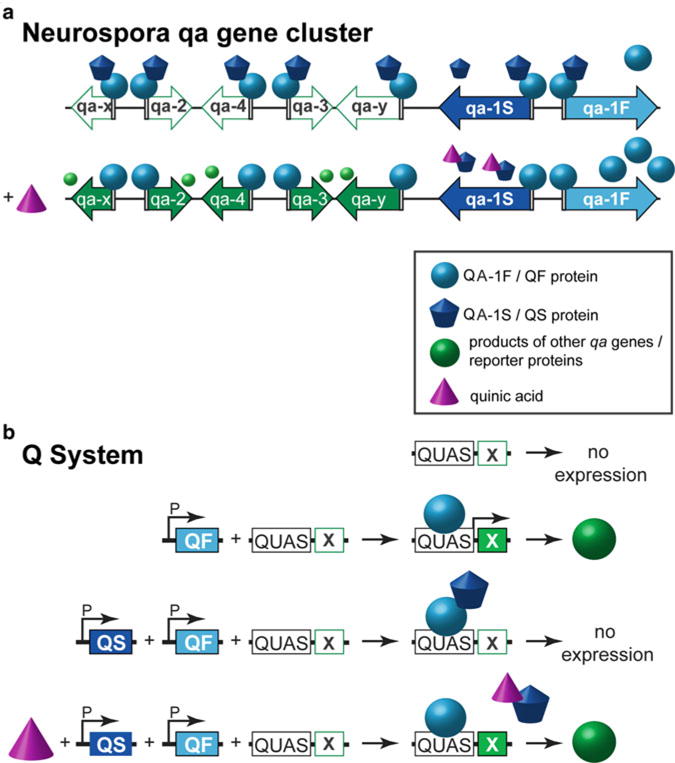
Activation and repression of transcription in the Neurospora qa gene cluster and in the Q-system. (a) Neurospora qa gene cluster. Top row: The qa gene cluster consists of five structural and enzymatic genes (white and green arrows) and two regulatory genes (blue arrows). The transactivator QA-1F (light blue sphere) binds to a 16bp sequence (white rectangle) upstream of the seven genes. The repressor QA-1S (dark blue cupcake) binds to QA-1F in conditions of low quinic acid. Bottom row: In conditions of high quinic acid (QA, purple cone), QA binds to QA-1S and prevents the interaction between QA-1F and QA-1S. QA-1F can now drive transcription of the genes involved in quinic acid catabolism (green arrows and green spheres), the QA-1S repressor, and also self-amplify. (b) The Q-system. First row: No expression of a reporter protein X is observed when only a QUAS-X construct is present in the genome. Second row: The expression of X is observed (green sphere) when a promoter-QF (P-QF) and a QUAS-X transgene are present in the genome. Third row: When QF and QS are expressed in the same cell (e.g., by the same promoter P), QS will suppress QF, and no reporter will be expressed. Fourth row: Feeding flies with quinic acid (QA) relieves QS-induced suppression, and the reporter X is expressed
3 Components of the Q-System
Regulatory genes qa-1F and qa-1S, together with the qa-1F binding sequence, were cloned out of Neurospora, and adapted for use in Drosophila as the Q-system [15]. For simplicity, QA-1F, QA-1S, and the qa-1F binding site were renamed to QF, QS, and QUAS, respectively. The components of the Q-system are analogous to those of GAL4/UAS and LexA/LexAop systems (Fig. 1): QF, GAL4, and LexA are transcription factors (TF) that can drive expression of transgenes by binding to their specific activation sequences (QF binds to QUAS, GAL4 binds to UAS, and LexA binds to LexAop). QS and GAL80 are repressors of TFs that bind to the activation domain of QF or GAL4 respectively and prevent initiation of transcription. The activity of the repressor can be silenced by a non-toxic drug, quinic acid, in the case of QS, or by temperature, in the case of a temperature-sensitive variant of GAL80 (Fig. 2b).
In the original version of the Q-system [15], the transactivator QF appeared to be toxic when broadly expressed. The cause of this toxicity was unknown. In addition, QF enhancer-trap lines were often mis-expressed in the trachea, presumably because a part of the QF DNA sequence acted as a cryptic tracheal enhancer in Drosophila. To remove the cryptic enhancer, second-generation versions of QF, named QF2 and QF2w, were re-codonized by manually choosing codons predicted to have average expression strength in Drosophila [41]. To find and eliminate the region of QF most responsible for general toxicity, the structural domains of QF (DBD, MD, and AD) were paired with those of GAL4 or LexA. Expression of these chimeric transactivators in vivo under the control of the strong neuronal promoter synaptobrevin indicated that the QF MD was the major source of lethal toxicity. In addition, the QF MD turned out to be dispensable for QF function. Therefore, the QF MD was removed from QF to yield QF2 (Fig. 3). Thus, in contrast to the original QF, QF2 has the QF DBD fused directly to QF AD. The resulting transactivator retains high-activity levels and is repressible by QS, similarly to the original QF [41].
Fig. 3.
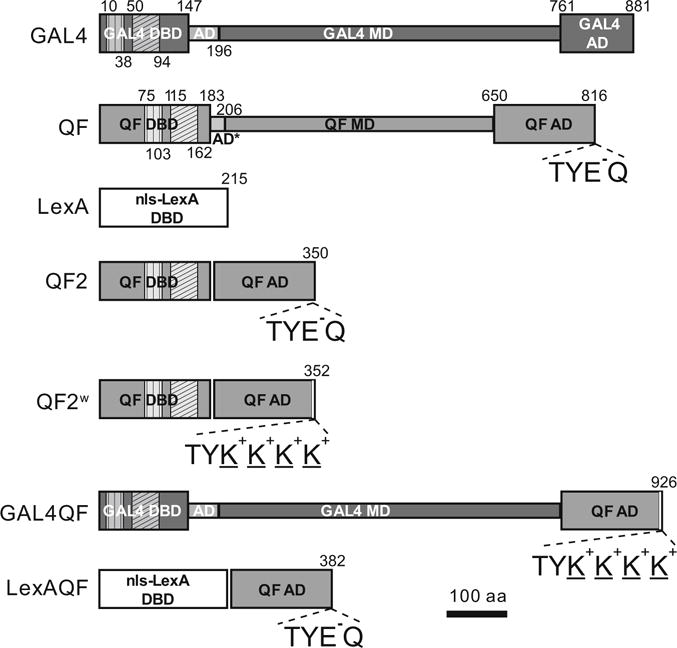
Transactivator schematics. Schematic representations of GAL4, original QF, LexA, QF2, QF2w, GAL4QF, and LexAQF. The transactivators consist of modular regions: DNA binding domain (DBD), middle domain (MD), and activation domain (AD). Vertical hatching indicates Zn 2/Cys 6 zinc finger motifs, diagonal hatchings mark dimerization domains. Numbers above and below schemes indicate amino acid position. Constructs are drawn to scale. C-terminal amino acids are indicated for transactivators with the QF AD to highlight differences between the ADs of QF/QF2/LexAQF and QF2w/GAL4QF
An alternative QF2 has also been generated: QF2w. There is only a slight difference between the coding sequences of QF2 and QF2w: the last two amino acids (glutamic acid and glutamine) on the C-terminus of QF2 were replaced by four lysines in QF2w (Fig. 3). This mutation changed the charge on the C-terminus from negative (E − Q) to positive (K+ K+ K+ K+), making QF2w a weaker transcriptional activator than QF2 (see also ref. 42), but also better repressible by QS. QF2 thus may be a preferred choice when very high transcriptional activity is required. We note, however, that having QF2 expressed ubiquitously or very widely may still compromise the health of fly stocks. A solution could be to have a tubulinP-QS transgene in the same stock (e.g., Bloomington Stock #51957), or to use the weaker QF2w transactivator for wide expression patterns. The fact that QS alleviates effects of QF2 indicates that the activation domain of QF2 may be sequestering nonspecific targets or the transcriptional machinery of a cell. This phenomenon is not unique to the QF2 activation domain, as it has been similarly observed that high levels of GAL4, driven by a strong promoter, compromise the health of a cell [43].
Expression levels of reporters, driven with QF2 and QF2w, can be fine-tuned in many ways. The expression levels can be dramatically increased by using 10, 15, 20, or 40 QUAS repeats for effector lines instead of the usual 5 [40, 44]; by including regulatory elements such as IVS, Syn21, and WPRE into effector constructs [45]; by creating multimeric reporter proteins [44]; by using site-directed φC31 integrase to place QF and QUAS transgenes into more highly expressing attP sites [40, 46]; or by using stronger terminator sequences such as SV40 or p10 instead of hsp70 in QF and QUAS constructs [45]. No clear temperature dependence of QF2/2w efficiency has been observed [41].
The Q-system can be used simultaneously with the GAL4/UAS system. To fully benefit from the availability of two repressible binary systems, they must function completely independently of each other. It has been verified in the adult brain and in the larval imaginal disks that QF fails to activate transcription downstream of UAS, and GAL4 fails to act on QUAS [15]. The cross-repression between the two systems is also absent: GAL80 does not reduce the activity levels of QF, and QS does not affect GAL4. Expression of GAL4 and QF2/QF2w simultaneously in the same cell does not lead to toxicity or downregulation of expression [41]. This mutual independence allows, among many other applications, to extend the classical MARCM analysis of mitotic clones with Coupled MARCM and Double MARCM (see below).
4 Temporal Control of Expression by Quinic Acid
All currently existing versions of transactivators with the QF AD (original QF, QF2, QF2w, GAL4QF, and LexAQF (see below)) are well repressed by QS, with the repression of QF2 by QS possibly more efficient at higher temperatures [41]. The QS-induced repression may be removed by feeding the larvae or adult flies food containing quinic acid (QA). QA is a naturally occurring non-toxic drug with anti-oxidative properties. It can be dissolved in water to concentrations of up to 6 % by weight, and added to the standard solidified fly medium by making holes in the food and pipetting the QA solution into them (0.3 ml of solution per 10 ml of food). This method works well for larva, but may lead to insufficient QA consumption in adult flies. QA consumption may be increased by keeping flies in vials with 1 % agarose gel, complemented with 1 % sucrose and 6 % QA [41]. To provide flies with a source of protein, the gel may be supplemented with fresh yeast paste made from dry yeast and 3 % QA solution, neutralized to pH 7 by NaOH. The effectiveness of QA to suppress QS does not appear to be altered when the QA solution is neutralized. Adding into the vial a small piece of tissue paper, moistened with the same neutralized QA solution, can provide extra moisture if necessary.
The effect of QA depends on the amount of QA in the food, and on the duration of exposure to QA. Larvae are particularly amenable to QA treatment [41]. In the case of adult flies, raised on normal fly medium and transferred after eclosion to a QA-containing vial, the maximum effect of QA was reached after about 3 days of exposure, although an increase in reporter expression may be seen in vivo already after 6 h [15].
The effect of QA can be seen only in cells that have absorbed QA from the environment or from fly hemolymph after the fly has fed on QA. For instance in the adult brain, olfactory and gustatory receptor neurons, neurons in the optic lobes and PI neurons appear to be most accessible to QA [41]. Other brain areas, however, are less affected by QA, presumably due to the glial “blood–brain barrier” [47] that prevents diffusion of QA into these cells.
5 Chimeric Transactivators
Chimeric transactivators GAL4QF and LexAQF were initially generated in an attempt to uncover the source of QF toxicity. GAL4QF was generated by fusing the GAL4 DBD and the GAL4 MD with the weakened QF AD (Fig. 3). LexAQF was generated by fusing the LexA DBD with the original QF AD (Fig. 3). Both transactivators drive strong expression in vivo of reporters placed under the control of UAS (for GAL4QF) or LexAop (for LexAQF), when examined in larval and adult neurons, larval imaginal disks and larval body wall muscles [41]. Both chimeric TAs are also suppressible by QS [41]. This feature is particularly useful for LexAQF as it allows for the suppression of LexAop reporter expression independently of GAL4/GAL80. This will allow for the use of LexAQF in MARCM experiments simultaneously with GAL4. GAL4QF also allows for the suppression of UAS reporter expression independently from GAL80. This may be used together with GAL4 in experiments where temporal control of expression by temperature-sensitive GAL80ts is desirable only for some cells of an expression pattern. In addition, the QS-induced repression of GAL4QF and LexAQF can be relieved by QA in much the same way as with QF, QF2, and QF2w [41].
6 Intersectional Expression with the Q-System
GAL4/UAS and Q systems are independent repressible binary expression systems, that, together with LexA/LexAop, chimeric transactivators and FLP/FRT [48] or other recombinases [49], enable the ability to achieve a variety of expression patterns. These modified expression patterns may be viewed as logic gates (Table 1). In the simplest possible case, the same effector or two different effectors may be expressed in cells that are covered by a GAL4 and a QF driver lines (Fig. 4). This approach has been used in a number of studies [50 – 60], e.g., to express different Ca2+ reporters in olfactory projection neurons and Kenyon cells of the Mushroom body [58], to express Channelrhodopsin in olfactory receptor neurons but a Ca2+ reporter in projection neurons [59, 61], or to label one subpopulation of projection neurons with GFP and another one with RFP to visualize their overlap [60].
Table 1.
Available Q-system reagents
| Stock number | Q-transgene | Purpose/function | Reference |
| Driver lines with original QF | |||
| 36347 | iav-QF | Expresses QF in chordotonal organs in the pattern of the iav (inactive) gene | [74] |
| 36346, 36349 | nompC-QF | Expresses QF in chordotonal organs vchA, vchB, lch5 and lch1, the class III sensory neurons vdaD, v’pda, ldaB, ddaA and ddaF and the dmd1 sensory neurons in the pattern of the nompC gene. | [74] |
| 36357 | Synj-QF | Expresses QF in the nervous system, imaginal disks, gut and salivary glands in the pattern of the synj (Synaptojanin) gene. | [74] |
| 36365 | Tbh-QF | Expresses QF in the larval nerve cord in the pattern of the Tyramine beta-hydroxylase gene, targets octopaminergic neurons. | [75] |
| 52244, 52245 | Tdc2-QF | Expresses QF under control of Tdc2 (Tyrosine decarboxylase 2) regulatory sequences, targets tyraminergic neurons. | [76] |
| 52250, 52251, 57359, 57362 | Trh-QF | Expresses QF under control of Trh (Tryptophan hydroxylase) regulatory sequences. | [76] |
| 36345, 36348 | TrpAl-QF | Expresses QF in class IV larval sensory neurons in the pattern of the TrpAl gene. | [74] |
| 36370 | Trpl-QF | Expresses QF in Bolwig’s organ and eye imaginal disks in the pattern of the trpl gene. | [74] |
| 30014, 30015, 30037, 30038, 30039 | GH146-QF | Expresses QF in subsets of projection neurons. | [15] |
| 41573 | Mz19-QF | Expresses QF in olfactory projection neurons. | [51] |
| 30016, 30043, 30042, 30019, 30018 | EnhancerTrap-QF | Enhancer trap lines generated by random insertion at various genomic loci | [15] |
| 30017, 30020 | SwappableEnhancer Trap-QF | Enhancer trap lines generated by random insertion at various genomic loci. These lines allow replacement of QF with another gene. | [15] |
| 57361, 57363, 57364 | Ddc-FRT-stop-FRT-QF | Expresses QF in the pattern of the Ddc (Dopa decarboxylase) gene upon FLP-mediated removal of a transcriptional stop cassette. | Contributed by Tom Hartl and Matt Scott |
| 57358, 57365 | Trh-FRT-stop-FRT-QF | Expresses QF in the pattern of the Trh (Tryptophan hydroxylase) gene upon FLP-mediated removal of a transcriptional stop cassette. | Contributed by Tom Hartl and Matt Scott |
| InSITE-QF | Enhancer-trap lines from InSITE collection | [64] | |
| IR76b-QF | Expresses QF in a subset of neurons in the pattern of chemosensory receptor IR76b | [77] | |
| Or67d-QF | Expresses QF in a subset of neurons in the pattern of olfactory receptor OR67d | [59] | |
| Driver lines with QF2 | |||
| 51957, 51958 | αTubulin-QF2 | Expresses QF2 ubiquitously under the control of αTubulin84B | [41] |
| 51955, 51956, 51959 | nsyb-QF2 | Expresses QF2 pan-neuronally under the control of n-syb. | [41] |
| 60320 | Chat-QF2 | Trojan-MiMIC line that expresses QF2 in the pattern of cholineacetyl transferase, targets cholinergic neurons. | [66] |
| 60315 | vGlut-QF2 | Trojan-MiMIC line that expresses QF2 in the pattern of vesicular glutamate transporter, targets glutamatergic neurons | [66] |
| 60323 | Gad1-QF2 | Trojan-MiMIC line that expresses QF2 in the pattern of glutamic acid decarboxylase 1, targets GABAergic neurons | [66] |
| Driver lines with QF2w(eak) | |||
| 51961 | Actin5C-QF2w | Expresses QF2w under the control of the Act5C promoter. | [41] |
| 51962, 51963 | αTubulin-QF2w | Expresses QF2w ubiquitously under the control of αTubulin84B | [41] |
| 51960, 51964, 51965 | nsyb-QF2w | Expresses QF2w pan-neuronally under the control of n-syb. | [41] |
| 59283 | pGMR-QF2w | Expresses QF2w under the control of the GMR promoter in the eye-antennal imaginal disk. | [41] |
| Driver lines with GAL4QF | |||
| 51945, 51946, 51947 | nsyb-GAL4QF | Expresses the GAL4 DNA-binding domain fused to the QF2w activation domain pan-neuronally under the control of n-syb. | [41] |
| 59284, 59285 | Actin5C-GAL4QF | Expresses GAL4QF under the control of the Act5C promoter. | [41] |
| Driver lines with LexAQF | |||
| 51953, 51954 | nsyb-LexAQF | Expresses the LexA DNA-binding domain fused to the QF activation domain pan-neuronally under the control of n-syb. | [41] |
| 62567, 62568 | Actin5C-LexAQFw | Expresses LexAQFw under the control of the Act5C promoter. | [41] |
| 60319 | Chat-LexAQF | Trojan-MiMIC line that expresses LexAQF in the pattern of cholineacetyl transferase, targets cholinergic neurons. | [66] |
| 60314 | vGlut-LexAQF | Trojan-MiMIC line that expresses LexAQF in the pattern of vesicular glutamate transporter, targets glutamatergic neurons | [66] |
| 60324 | Gad1-LexAQF | Trojan-MiMIC line that expresses LexAQF in the pattern of glutamic acid decarboxylase 1, targets GABAergic neurons | [66] |
| Available by request from C. Potter, JHU | αTubulin-FRT-stop-FRT-LexAQF | Expresses LexAQF ubiquitously under the control of αTubulin84B upon FLP-mediated removal of transcriptional stop cassette | |
| QUAS reporter lines | |||
| 52259, 52260 | 10xQUAS-Chr2.T159C-HA | A variant of Channelrhodopsin, excitatory blue light-activated sodium channel, tagged with HA. | [76] |
| 58401, 58402 | QUAS-FRT-stop-FRT-ChR2.TR-mCherry | A variant of Channelrhodopsin, excitatory blue light-activated sodium channel tagged with mCherry, that will be expressed upon excision of FRT-stop-FRT cassette. | [59] |
| 58400 | QUAS-ChR2.TR-mCherry | A variant of Channelrhodopsin, excitatory blue light-activated sodium channel, tagged with mCherry. | [59] |
| 36356, 36367 | QUAS-CHETA.YFP | A variant of Channelrhodopsin, excitatory blue light-activated sodium channel, tagged with YFP. | [74] |
| 55106, 55107, 55108, 55109 55110, 55115, 55116, 55120 | QUAS-X∷CycB | Cyclin B, tagged with RFP or Venus YFP, to mark cells in S, G2, and M phases of cell cycle. Reagent of the Fly-FUCCI system. | [78] |
| 55106, 55107, 55108, 55109 55115, 55116, 55119, 55120 | QUAS-X∷E2F1 | E2F1, tagged with CFP or GFP, to mark cells in S, G2, and M phases of cell cycle. Reagent of the Fly-FUCCI system. | [78] |
| 30008, 30126, 30127 | QUAS-FLP | FLP recombinase | [15] |
| 52226, 52227 | QUAS-R-GECO | RGECO calcium reporter | [58] |
| 30001, 30002, 30003 | QUAS-mCD8:GFP | Membrane-bound GFP | [15] |
| 52263, 52264 | 10xQUAS-6XGFP | Cytoplasmic hexameric GFP for extra-strong labeling. | [44] |
| 36351 | QUAS-nSyb:mCherry-HA | Cherry- and HA-tagged n-syb protein for labeling presynaptic terminals | [74] |
| 36358 | QUAS-nSyb:GFP-myc | GFP- and myc-tagged n-syb protein for labeling presynaptic terminals | [74] |
| 30134, 30135, 30136 | QUAS-FRT-stop-FRT-mCD8:GFP | GFP, fused to membrane tag CD8, expressed upon FLP-mediated removal of transcription stop | [15] |
| 36355, 36366 | QUAS-eNpHR | Halorhodopsin, inhibitory yellow light-activated chloride channel. | [74] |
| 30006, 30007 | QUAS-nuclacZ | LacZ (beta-galactosidase) for labeling nuclei. | [15] |
| 36352, 36353, 36363, 36364 | QUAS-Rab3 | Synaptic vesicle membrane-associated protein Rab3, tagged with GFP or mCherry. | [74, 76] |
| 52255 | 10xQUAS-2xHA-Rab3 | Synaptic vesicle membrane-associated protein Rab3, tagged with HA. | [74, 76] |
| 52269, 52270 | 10xQUAS-6XmCherry | Cytoplasmic hexameric red fluorescent protein mCherry for extra-strong labeling. | [44] |
| 30004, 30005, 30037, 30043 30118 | QUAS-mtdTomato-3xHA | Membrane-bound red fluorescent protein tdTomato. | [15] |
| 30128 | QUAS-FRT-stop-FRT-shibirets | Temperature-sensitive shibire protein, expressed upon FLP-mediated recombination that removes the transcriptional stop cassette. | [15] |
| 30010, 30011, 30012, 30013 | QUAS-shibirets | Temperature-sensitive shibire protein. Inhibits synaptic transmission at 29 °C and above. | [15] |
| 41565, 41566 | QUAS-Ten-a | Presynaptic teneurin | [51] |
| 41571, 41572 | QUAS-Ten-m | Postsynaptic teneurin | [51] |
| 61658, 61811 | QUAS-p65AD∷CaM | p65 activation domain, fused to Calmodulin. Component of activity-dependent labeling system TRIC. | [61] |
| 61659, 61660 | QUAS-FRT-stop-FRT-p65AD∷CaM | p65 activation domain, fused to Calmodulin. Expressed upon FLP-mediated recombination that removes the transcriptional stop cassette. Component of activity-dependent labeling system TRIC. | [61] |
| 52230 | QUAS-ChIEF-tdTomato | A variant of Channelrhodopsin, excitatory blue light-activated sodium channel, tagged with tdTomato. | Contributed by Kaiyu Wang |
| 30009 | QUAS-dm | Diminutive (Drosophila myc). | Contributed by Christopher Potter and Liqun Luo |
| 51948, 51949, 51950 | QUAS-GAL80 | GAL80, repressor of GAL4 | Contributed by Christopher Potter |
| 52231 | QUAS-GCaMP3 | GCaMP3 calcium reporter for live imaging | Contributed by Kaiyu Wang |
| QUAS-spH | Synapto-pHluorin, a pH-sensitive mutant of GFP that allows to monitor synaptic vesicle release. | [57] | |
| QUAS-Brp-Short-mStraw | Bruchpilot-Short protein, fused to mStrawberry, to label sites of presynaptic neurotransmitter release. | [52] | |
| QUAS-GFP11 | A GRASP version of GFP for labeling synaptic connections. | [79] | |
| QS repressor lines | |||
| 30021, 30022, 30024, 52112 | αTubulin-QS | Expresses QS, the repressor of TA’s with QF AD, under the control of αTubulin84B promoter | [15] |
| 30033 | UAS-QS | Expresses QS, the repressor of TA’s with QF AD, under the control of UAS | [15] |
Fig. 4.
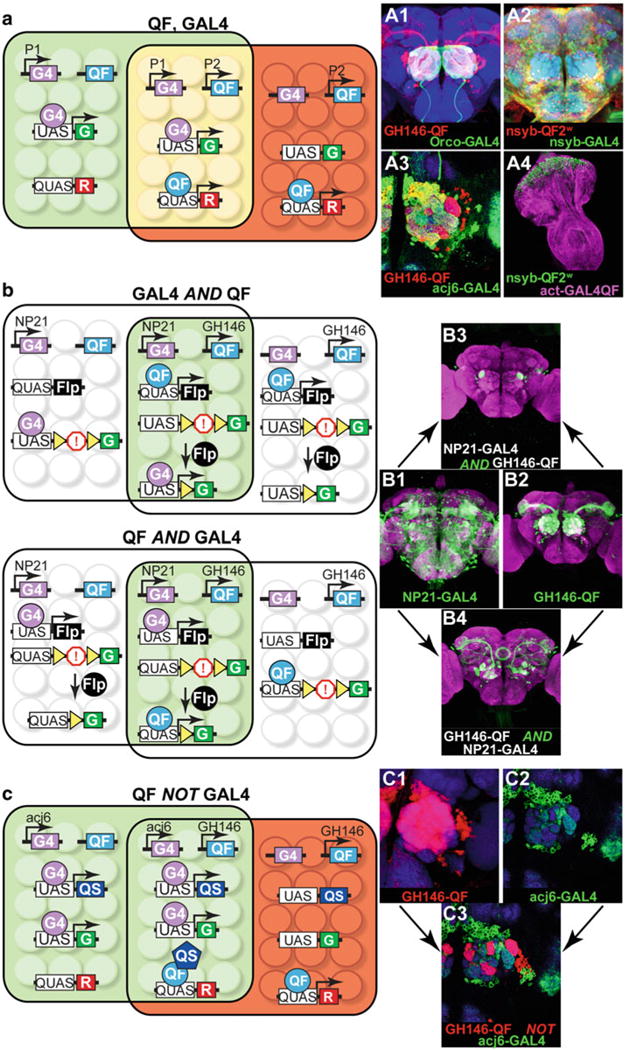
Intersectional labeling. (a) Simultaneous expression of two different reporters in overlapping subsets of cells with the GAL4/UAS and the Q-system. In the schematics on the left, the genotype of the fly is: P1-GAL4, P2-QF, UAS-GFP, QUAS-RFP. Enhancers P1 (green area) and P2 (red area) are active in partially overlapping subsets of cells (yellow area). A1: Expression in olfactory receptor neurons (green) and olfactory projection neurons (red) with GH146-QF, QUAS-mtdt-3xHA, Orco-GAL4, UAS -mCD8-GFP. A2: Pan-neuronal co-expression of nuclear β-galactosidase (red) and membrane-bound GFP (green). Genotype: nsyb-QF2w, QUAS-nucLacZ, nsyb-GAL4, UAS-mCD8-GFP. A3: Antennal lobe innervation of two partially overlapping projection neuron populations. Genotype: GH146-QF, QUAS-mdtd-3xHA, acj6-GAL4, UAS-mCD8-GFP. Antennae were removed to eliminate Acj6+ olfactory neuron innervations in the antennal lobe. A4: Ubiquitous (purple) and neuronal (green) expression in the eye-antennal imaginal disk of a third instar larva. Genotype: nsyb-QF2w, QUAS-mCD8-GFP, actin-GAL4QF, UAS-mtdt-3xHA. Brain regions (blue) depict anti-nc82 staining in A1 – A3. Schematic and image in (a) and A3 reprinted with permission from [15]. (b) Limiting expression to cells that express both transactivators. Two possible ways to achieve a GAL4 AND QF intersection are diagrammed. Genotype of top schematic and B3: NP21-GAL4, GH146-QF, QUAS-FLP, UAS-FRT-stop-FRT-GFP. Genotype of bottom schematic and B4: NP21-GAL4, GH146-QF, UAS-FLP, QUAS-FRT-stop-FRT-GFP. B1: Expression pattern of NP21-GAL4 line, as visualized by the UAS-mCD8-GFP reporter (green). B2: Expression pattern of GH146-QF lines as visualized by the QUAS-mCD8-GFP reporter (green). B3 and B4: Expression patterns of the AND intersections. The difference between B3 and B4 arises due to the developmental timing of FLPase expression. See main text for details. Purple color depicts anti-nc82 staining. Schematic and images in (b) reprinted with permission from ref. 15. (c) Using one transactivator to limit the expression pattern of the other transactivator: QF NOT GAL4 intersectional example. Genotype of the flies: acj6-GAL4, GH146-QF, UAS-QS, UAS-GFP, QUAS-RFP (schematic and C3). The acj6-GAL4 line drives the GFP reporter and the QS repressor, which silences QF in those cells where GAL4 and QF expression patterns overlap. QF is active in the cells where GAL4 is not expressed, resulting in the RFP labeling. C1: Expression pattern of GH146-QF, visualized with QUAS-mtdt-3xHA (red). C2: Expression pattern of acj6-GAL4, visualized with UAS-mCD8-GFP (green). C3: Expression pattern of QF NOT GAL4 intersection. The final expression pattern in red is limited to where QF, but not GAL4, is expressed. Blue color depicts anti-nc82 staining. Schematic and images in (c) reprinted with permission from ref. 15
A practical application of the Q-system is to use it to narrow down expression patterns from many currently available GAL4 expression lines, such as the Janelia GAL4 enhancer collection [62], the MiMIC collection [63], or the InSITE collection [64]. It may be useful to express an effector only in the cells that belong to the overlapping expression patterns of two selected enhancer lines, e.g., a Janelia-GAL4 line and an InSITE-QF line. Alternatively, neurons of a GAL4 line may be selectively targeted based on their overlap (or lack of overlap) with the expression patterns of a neurotransmitter or other gene of interest, e.g., by utilizing such reagents as TßH-QF (octopaminergic neurons), Cha-QF2 (cholinergic neurons), GAD1-QF2 (GABAergic neurons), or nompC-QF (mechanosensory neurons) (Table 2). Limiting expression may be achieved in a variety of ways, conceptualized by AND, NOT, and XOR logic operations (Fig. 4, Table 2) [15, 65]. For many operations, an FRT-transcriptional stop-FRT cassette is essential. FLPase, when expressed in a cell, will permanently remove the transcriptional stop cassette from the cell and all its progeny. The optimal strategy depends on the availability of driver, reporter and repressor lines, and also on the possible off-target labeling that may arise at early developmental stages. For example, the two approaches to the AND intersection could give rise to different final readouts. These differences are based on two parameters: (1) which line (GAL4 or QF) drives FLPase expression; this can potentially label all cells that developmentally expressed the chosen transcription factor up to the timepoint of investigation; (2) which line (GAL4 or QF) drives final reporter expression; this potentially labels only those cells normally labeled at the chosen timepoint. For instance, the two ways to achieve the AND intersection between NP21-GAL4 and GH146-QF lines are shown in Fig. 4. NP21-GAL4 drives expression in many cells in the adult brain, including some of the olfactory projection neurons. GH146-QF drives expression only in the olfactory projection neurons in the adult. In the first case, NP21-GAL4 drives UAS-FLPase expression, while GH146-QF drives GFP expression from QUAS-FRT-transcriptional stop-FRT-GFP constructs that have excised their FRT-transcriptional stop-FRT cassette. The resulting expression pattern includes a subset of olfactory projection neurons targeting 5 glomeruli, normally labeled by GH146-QF in the adult fly, and also cells of the ellipsoid body, which are not normally labeled by GH146-QF in the adult. The labeled projection neurons are ones where NP21-GAL4 had been expressed at some point during development (which led to the removal of the FRT-transcriptional stop-FRT) and now visualized by GH146-QF activity. It is also possible that labeling is visible in the adult in cells where GFP had been produced at an earlier developmental stage, i.e., in the pupa, but is not produced in the adult. This perdurance of GFP is presumably the reason for the labeling in the ellipsoid body neurons. In the other possible AND intersectional approach, GH146-QF drives QUAS-FLPase expression, while NP21-GAL4 drives GFP expression from UAS-FRT-transcriptional stop-FRT-GFP constructs that have excised their FRT-transcriptional stop-FRT cassette. This genotype results in labeling of only olfactory projection neurons that target only one glomerulus since these are the only neurons that express NP21-GAL4 at the adult stage. Thus, the final readout from an AND intersection strongly depends on which line (GAL4 or QF) is used to report developmental versus final expression patterns. It is often informative to perform both approaches. In general, it is often advantageous to choose the line (GAL4 or QF) that expresses most strongly in the tissue of interest at the timepoint of interest for the final readout (visualized by the FRT-stop-FRT reporter).
Table 2.
Intersectional expression
| Logic operation | Description | Transgenes requireda “QF” stands for QF, QF2, or QF2w. “R” stands for Reporter. “>” stands for FRT site. |
|---|---|---|
| A OR B |
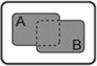
|
|
| B NOT A |
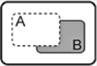
|
|
| A AND B |
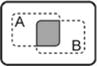
|
|
| NOT A |

|
|
| A→B |
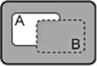
|
|
| A XOR B |
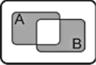
|
|
| A NOR B |
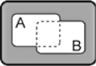
|
|
| A NAND B |
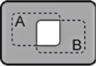
|
|
| A XNOR B |
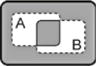
|
|
This list is not exhaustive; alternative genetic combinations are possible
Table 1 lists Q-system reagents that have been published or otherwise publicized, and will be good starting places for intersectional approaches. For example, InSITE [64], MiMIC [63], and Trojan-MiMIC [66] collections are useful tools for recapitulating interesting expression pattern with various driver constructs.
7 Mosaic Analyses with the Q-System
Genetic mosaics allow for the study of gene function in a small subpopulation of cells, to label one or a few cells out of a full expression pattern, or to investigate cells born at defined developmental timepoints. The GAL4/UAS system has been extensively used for these purposes [21, 23 – 25, 67 – 71]. The Q-system, used together with GAL4/UAS, allows for even more advanced mosaic labeling and gene manipulation. We discuss here two approaches for mosaic analyses: FLP-mediated removal of an FRT-transcriptional stop-FRT cassette [72, 73], and Mosaic Analysis with Repressible Cell Marker (MARCM) [15, 21].
The simplest form of mosaic analysis requires three transgenic components: (1) a driver line (e.g., GH146-QF) which drives expression of a reporter; (2) the reporter transgene (e.g., QUAS-FRT-stop-FRT-mtdT) where the transcriptional stop cassette (FRT-stop-FRT) can be removed by the FLP recombinase; and (3) a ubiquitous heatshock promoter driving FLPase expression (hsFLP). The FLP recombinase will be expressed when the flies are placed at temperatures above 29 °C. Longer heat shock times, or higher temperatures up to 38 °C, can induce more FLPase expression. This is a useful way to regulate the extent of the FRT-stop-FRT excision. FLP-mediated excision may occur during or shortly after the heatshock treatment, resulting in labeling in a random subset of cells where the driver line is expressing. By adjusting the duration of the heatshock and the strength of the hsFLP line, it is possible to label anywhere from only a few cells to most cells out of the driver line expression pattern [48]. In contrast to UAS-FLP/QUAS-FLP discussed above, the effects of hsFLP are much more random because the FLPase is expressed only during the heatshock treatment. This results in low amount of FLPase in a cell and thus low probability of successful DNA targeting. This method is particularly useful to study cell and circuit anatomy, and also to drive expression of effectors (e.g.,TrpA1, Channelrhodopsin, halorhodopsin, toxins, etc.) with the purpose of examining behavioral phenotypes of individual flies and relating them to the affected cells [73].
The Q FLP-out method, described above, may be used together with the analogous GAL4 FLP-out method, to independently label cells from two different expression patterns.
Temporally refined mosaic analysis is possible using the MARCM technique [21]. This technique induces mosaic labeling based on birthdates of the labeled cells. Here the transgene expression (e.g., driven by GH146-QF) is suppressed throughout the animal (e.g., by ubiquitous QS from tubulinP-QS), but may be relieved in cells that, due to hsFLP -mediated recombination of homologous chromosomes during mitotic cell division, have lost the tubP-QS repressor-coding gene (Fig. 5). By experimentally selecting the developmental stage of the animals to be heatshocked, it is possible to reproducibly label specific cells of interest born at particular developmental times. In the example shown in Fig. 5a, two of the homologous chromosomes carry the FRT sequence at the same genomic position, with one of the chromosomes carrying the ubiquitous repressor transgene tubP-QS distal to the FRT sequence. Ubiquitous expression of QS prevents QF activity in all cells. The same animal also carries P1-QF (e.g., GH146-QF), QUAS-GFP, and hsFLP transgenes that can be located elsewhere in the genome apart from the chromosome arms distal to the FRT sites. Upon 37 °C heatshock, the FLP recombinase will be expressed, and can cause mitotic recombination at the FRT sites in a random subset of cells born at or shortly after the heatshock treatment. Only one of the cells produced from the cell division will be positively labeled by the marker (e.g., GFP). All progeny (if any) of the labeled cell will also be labeled, resulting in clones of labeled cells (Fig. 5 A1–A4). If mitotic recombination had not occurred, the two daughter cells would remain unlabeled.
Fig. 5.
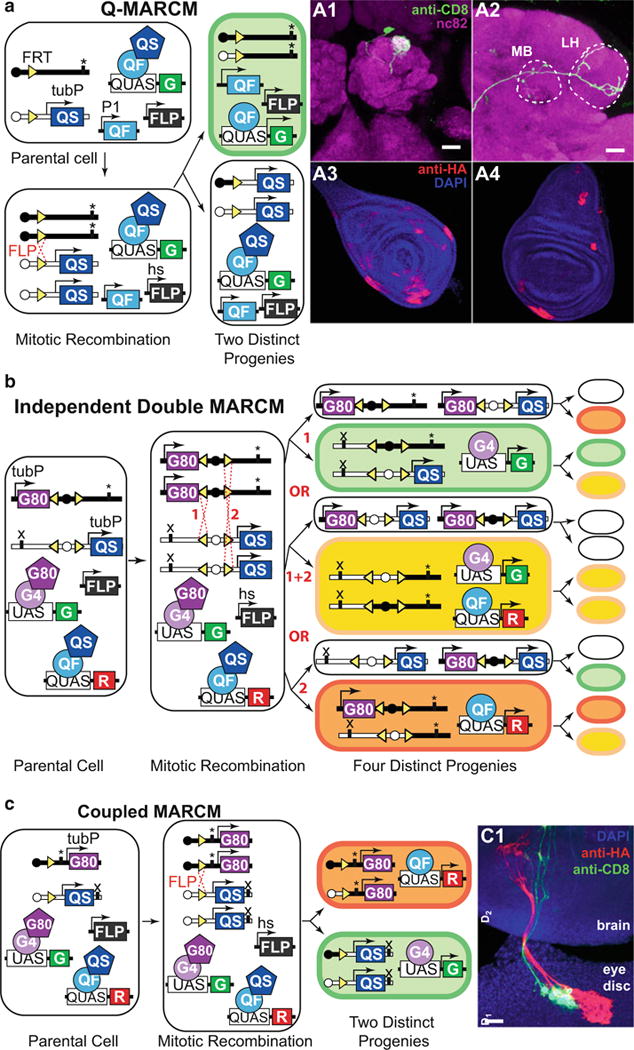
MARCM analysis. (a) Q-MARCM. The genotype of the parental cell is: hsFLP, FRT site (homozygous) recombined with tubulin-QS (heterozygous), P1-QF, QUAS-GFP. “*” marks the location of a recessive mutation that may be studied in the labeled cells. Upon FLPase-mediated mitotic recombination, one of the two postmitotic cells will lose the tubulin-QS transgene and start expressing GFP (top). The other postmitotic cell will remain unlabeled (bottom). A1 and A2: Q-MARCM labeling of a single olfactory projection neuron, visualized in the antennal lobe (A1), the mushroom body (MB, A2), and the lateral horn (LH, A2). Genotype: hsFLP, UAS-mCD8-GFP (X); GH146-QF#53, 82BFRT, tub-QS/82BFRT (III). A3 and A4: Q-MARCM labeled clones in the leg (A3) and wing (A4) imaginal disks of a third instar larva. Genotype: hsFLP1, QUAS-mtdT-3xHA (X); ET40-QF (II); 82BFRT, tubP-QS/82BFRT (III). Schematic and brain images reprinted with permission from ref. 15. (b) Independent double MARCM. The genotype of the parental cell is: hsFLP (also present in all progeny cells), P1-QF, P2-GAL4, UAS-GFP, QUAS-RFP, FRT site (homozygous) recombined with tubulin-GAL80 (heterozygous), a different FRT site (homozygous) recombined with tubulin-QS (heterozygous). “*” and “x” mark independent recessive mutations that may be studied in postmitotic cells. There are three possible outcomes of a heatshock-induced mitotic recombination at the FRT sites (1 or 2 or 1 + 2). The progenitors for each event are schematized with each generating a labeled and an unlabeled cell. Upon a second heatshock, mitotic recombination may happen again, altering the expression profiles of the progeny. See main text for details. Schematic modified with permission from ref. 15. (c) Coupled MARCM. The genotype of the parental cell is: hsFLP (also present in all progeny cells), P1-GAL4, P1-QF, UAS-GFP, QUAS-RFP, FRT site (homozygous) recombined with tubulin-GAL80 (heterozygous) or with tubulin-QS (heterozygous). “*” and “x” mark independent recessive mutations that may be studied in postmitotic cells. FLP-mediated recombination during mitosis at the FRT site followed by chromosome segregation result in all progeny being labeled either with GFP or with RFP. C1: Coupled MARCM clones in the eye-antennal imaginal disk send processes that innervate the brain of a third-instar larva. Genotype: hsFLP1, QUAS-mtdT-3xHA, UAS-mCD8-GFP (X); ET40-QF (II); 82BFRT tubP-QS/tubP-GAL4 82BFRT tubP-GAL80 (III). Schematic and image reprinted with permission from ref. 15
GAL4 - MARCM [21] may be used simultaneously with Q-MARCM [15] or LexAQF-MARCM [41]. The two MARCM events will drive two different reporters (e.g., UAS-GFP and QUAS-RFP) and, depending on the genomic arrangement of transgenes, can be either independent of each other (independent double MARCM for mosaic labeling of overlapping or non-overlapping subsets of cells), or they can be coupled to label both progenies of a single cell division (coupled MARCM). Independent double MARCM might be utilized to simultaneously label (and manipulate) cells not marked by the same GAL4 reporter (e.g., glia labeled by GAL4 and neurons labeled by QF; or two different neuronal populations that innervate a common target). Independent double MARCM could also be used to label cells born at the same developmental timepoint but which are not marked by the same GAL4 line. In addition, independent and coupled MARCM enables clones of labeled cells to be generated that also carry homozygous mutant alleles of genes of interest. By examining morphology, number and distribution of the labeled cells it is possible to study the effects of gene mutations on development or anatomy. Coupled MARCM also allows to examine the fate of two progenies born in one cell division, which can be used for mapping cell lineages and division patterns [15]. Figure 5b provides the schematics of independent double MARCM labeling for partially overlapping GAL4 and QF driver lines, driving GFP and RFP, respectively. The repressor transgenes GAL80 and QS can be located on different arms of two homologous chromosomes (as shown on Fig. 5b) or on non-homologous chromosomes. The driver and reporter transgenes must be located on chromosome arms non-homologous to those with repressor transgenes. Upon FLP-mediated recombination, some GAL4-expressing cells may lose the GAL80 transgene and express GFP (case 1). If a heatshock is repeated at a later stage, another FLP-mediated recombination may occur, leading to the loss of the QS repressor and expression of both GFP and RFP, thus labeling the cells yellow. Simultaneous RFP and GFP expression is possible only in the cells that belong to the expression patterns of both the GAL4 and the QF drivers. Interestingly, progeny of the unlabeled twin, produced after the first heatshock, may produce RFP-labeled cells upon a second heatshock, provided that both GAL4 and QF drivers are active in these cells. Subsequent heatshocks will not change the expression profile of the cells that already underwent two FLP-mediated recombinations during mitotic cell divisions, but may affect the cells that underwent zero or one FLP-recombination prior to the heatshock. Another possible option (case 2) is that upon the first heatshock a cell will lose both GAL80 and QS repressor genes, thus being labeled yellow. Subsequent heatshocks will not alter the expression profile of this cell’s or its twin’s progeny. The last option (case 3) is analogous to case 1, but results in the loss of the QS repressor in QF-expressing cells. The outcome of independent double MARCM will be labeling that includes red, green, and yellow cells, with yellow being dominant when the expression patterns of the GAL4 and QF lines are identical. Some of the cells that underwent FLPase-mediated recombination will remain unlabeled. Independent double MARCM thus allows for the investigation of cells born at two distinct timepoints. However, due to the random nature of FLPase-mediated recombination, the number of cells that undergo two FLP recombination events may be low.
To label both progeny from a single cell division, Q-MARCM and GAL4 -MARCM can be coupled by placing repressor transgenes on homologous chromosome arms (Fig. 5c). This way, after a FLPase-mediated homologous recombination event, one twin will lose the GAL80 repressor but retain QS, while the other will retain GAL80 but lose QS. The first twin will start expressing GAL4-driven GFP, and the second twin will start expressing QF-driven RFP. Subsequent heatshocks will not alter the labeling pattern.
MARCM analysis is not limited to the labeling of cells, but also allows mosaic expression of effector transgenes (e.g., RNAi, toxin, Channelrhodopsin, etc.). It can also be used to generate positively labeled homozygous mutant cells if a mutant gene allele is located on the same homologous FRT chromosome arm as the FRT repressor transgenes [21]. Wildtype and mutant cells will be labeled by different markers, and their morphology and number may be easily examined [15, 21]. In addition, Q-MARCM and GAL4-MARCM can be utilized alongside standard GAL4 and Q-system-mediated transgenic expression. For example, Q-MARCM could be used to label single neurons, while GAL4 utilized to express RNAi in varying target tissues. This could reveal how genetic disruption of one tissue population affects the neuronal targeting of a different population.
8 Future Directions
The Q-system represents a versatile set of genetic tools that can be used in situations in which a single binary expression system has proven to be experimentally insufficient. Many QF, QUAS, and QS fly stocks are already publicly available from the Bloomington Drosophila Stock Center (http://flystocks.bio.indiana.edu/Browse/Qsystem/Qintro.htm). Table 1 summarizes all currently published or available Q-system lines. With the recent development of QF2, QF2w, GAL4QF, and LexAQF, we expect many more transgenic lines to be generated in the near future by our and other labs. As additional creative uses for the Q-system are introduced, the utility of the Q-system will extend beyond what is described here.
A number of useful modifications could be developed in the future for the Drosophila Q-system: split-QF for additional inter-sectional expression control; QF2/QF2w enhancer trap lines or QF2 expression collections to increase the number of available Q-system expression patterns; improved methods to deliver QA across the glia to the neurons to increase QA effects on central brain neurons; the development of QS variants with altered affinities for QA; and the development of temperature-sensitive QF or QS variants for temporal control of Q-system activity.
A genetic technique allowing GAL4 lines to be easily converted to QF2 lines was reported. This work includes many new useful QF2 driver lines. Lin C-C, Potter CJ (2016) Editing transgenic DNA components by inducible gene replacement in Drosophila melanogaster. Genetics In press.
Acknowledgments
We thank Chun-Chieh Lin, Qili Liu, Sha Liu, Darya Task and Olga Markova for their helpful comments on the manuscript.
References
- 1.Ramaekers A, Quan X-J, Hassan BA. The making and un-making of neuronal circuits in Drosophila. Humana Press; Totowa, NJ: 2012. Genetically encoded markers for drosophila neuroanatomy; pp. 49–59. [Google Scholar]
- 2.Silbering AF, Bell R, Galizia CG, Benton R. Calcium imaging of odor-evoked responses in the Drosophila antennal lobe. J Vis Exp. 2012:e2976. doi: 10.3791/2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simpson JH. Mapping and manipulating neural circuits in the fly brain, Advances in genetics. Academic; New York: 2009. pp. 79–143. [DOI] [PubMed] [Google Scholar]
- 4.Dietzl G, Chen D, Schnorrer F, Su K-C, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome- wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 5.Duffy JB. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- 6.Sweeney ST, Broadie K, Keane J, Niemann H, O’Kane CJ. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron. 1995;14:341–351. doi: 10.1016/0896-6273(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 7.Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, Garrity PA. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454:217–220. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lima SQ, Miesenbböck G. Remote control of behavior through genetically targeted photostimulation of neurons. Cell. 2005;121:141–152. doi: 10.1016/j.cell.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Paradis S, Sweeney ST, Davis GW. Homeostatic control of presynaptic release is triggered by postsynaptic membrane depolarization. Neuron. 2001;30:737–749. doi: 10.1016/s0896-6273(01)00326-9. [DOI] [PubMed] [Google Scholar]
- 10.Hay BA, Wassarman DA, Rubin GM. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- 11.Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- 12.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 13.Lai S-L, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- 14.Bello B, Resendez-Perez D, Gehring WJ. Spatial and temporal targeting of gene expression in Drosophila by means of a tetracycline-dependent transactivator system. Development. 1998;125:2193–2202. doi: 10.1242/dev.125.12.2193. [DOI] [PubMed] [Google Scholar]
- 15.Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venken KJT, Simpson JH, Bellen HJ. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron. 2011;72:202–230. doi: 10.1016/j.neuron.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potter CJ, Luo L. Using the Q system in Drosophila melanogaster. Nat Protoc. 2011;6:1105–1120. doi: 10.1038/nprot.2011.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science (New York, NY) 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 19.Lue NF, Chasman DI, Buchman AR, Kornberg RD. Interaction of GAL4 and GAL80 gene regulatory proteins in vitro. Mol Cell Biol. 1987;7:3446–3451. doi: 10.1128/mcb.7.10.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sayeed O, Benzer S. Behavioral genetics of thermosensation and hygrosensation in Drosophila. Proc Natl Acad Sci. 1996;93:6079–6084. doi: 10.1073/pnas.93.12.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 22.Luo L, Lee T, Nardine T, Null B, Reuter J. Using the MARCM system to positively mark mosaic clones in Drosophila. Dros Inf Serv. 1999;82:102–105. [Google Scholar]
- 23.Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- 24.del Valle Rodriguez A, Didiano D, Desplan C. Power tools for gene expression and clonal analysis in Drosophila. Nat Methods. 2012;9:47–55. doi: 10.1038/nmeth.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffin R, Binari R, Perrimon N. Genetic odyssey to generate marked clones in Drosophila mosaics. Proc Natl Acad Sci. 2014;111:4756–4763. doi: 10.1073/pnas.1403218111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.St Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet. 2002;3:176–188. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- 27.Wei X, Potter CJ, Luo L, Shen K. Controlling gene expression with the Q repressible binary expression system in Caenorhabditis elegans. Nat Methods. 2012;9:391–395. doi: 10.1038/nmeth.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subedi A, Macurak M, Gee ST, Monge E, Goll MG, Potter CJ, Parsons MJ, Halpern ME. Adoption of the Q transcriptional regulatory system for zebrafish transgenesis. Methods (San Diego, Calif) 2013;66(3):433–440. doi: 10.1016/j.ymeth.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giles NH, Geever RF, Asch DK, Avalos J, Case ME. The Wilhelmine E. Key 1989 invitational lecture. Organization and regulation of the qa (quinic acid) genes in Neurospora crassa and other fungi. J Hered. 1991;82:1–7. doi: 10.1093/jhered/82.1.1. [DOI] [PubMed] [Google Scholar]
- 30.Patel VB, Schweizer M, Dykstra CC, Kushner SR, Giles NH. Genetic organization and transcriptional regulation in the qa gene cluster of Neurospora crassa. Proc Natl Acad Sci. 1981;78:5783–5787. doi: 10.1073/pnas.78.9.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baum JA, Geever R, Giles NH. Expression of qa-1F activator protein: identification of upstream binding sites in the qa gene cluster and localization of the DNA-binding domain. Mol Cell Biol. 1987;7:1256–1266. doi: 10.1128/mcb.7.3.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huiet L, Giles NH. The qa repressor gene of Neurospora crassa: wild-type and mutant nucleotide sequences. Proc Natl Acad Sci. 1986;83:3381–3385. doi: 10.1073/pnas.83.10.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuji G, Kenmochi Y, Takano Y, Sweigard J, Farrall L, Furusawa I, Horino O, Kubo Y. Novel fungal transcriptional activators, Cmr1p of Colletotrichum lagenarium and Pig1p of Magnaporthe grisea, contain Cys2His2 zinc finger and Zn(II)2Cys6 binuclear cluster DNA-binding motifs and regulate transcription of melanin biosynthesis genes in a developmentally specific manner. Mol Microbiol. 2000;38:940–954. doi: 10.1046/j.1365-2958.2000.02181.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhang L, Bermingham-McDonogh O, Turcotte B, Guarente L. Antibody-promoted dimerization bypasses the regulation of DNA binding by the heme domain of the yeast transcriptional activator HAP1. Proc Natl Acad Sci. 1993;90:2851–2855. doi: 10.1073/pnas.90.7.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hidalgo P, Ansari AZ, Schmidt P, Hare B, Simkovich N, Farrell S, Shin EJ, Ptashne M, Wagner G. Recruitment of the transcriptional machinery through GAL11P: structure and interactions of the GAL4 dimerization domain. Genes Dev. 2001;15:1007–1020. doi: 10.1101/gad.873901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walters KJ, Dayie KT, Reece RJ, Ptashne M, Wagner G. Structure and mobility of the PUT3 dimer. Nat Struct Mol Biol. 1997;4:744–750. doi: 10.1038/nsb0997-744. [DOI] [PubMed] [Google Scholar]
- 37.Kraulis PJ, Raine ARC, Gadhavi PL, Laue ED. Structure of the DNA-binding domain of zinc GAL4. Nature. 1992;356:448–450. doi: 10.1038/356448a0. [DOI] [PubMed] [Google Scholar]
- 38.Marmorstein R, Carey M, Ptashne M, Harrison SC. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- 39.Ma J, Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- 40.Pfeiffer BD, Ngo T-TB, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riabinina O, Luginbuhl D, Marr E, Liu S, Wu MN, Luo L, Potter CJ. Improved and expanded Q-system reagents for genetic manipulations. Nat Methods. 2015;12:219–222. doi: 10.1038/nmeth.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gill G, Ptashne M. Mutants of GAL4 protein altered in an activation function. Cell. 1987;51:121–126. doi: 10.1016/0092-8674(87)90016-x. [DOI] [PubMed] [Google Scholar]
- 43.Kramer JM, Staveley BE. GAL4 causes developmental defects and apoptosis when expressed in the developing eye of Drosophila melanogaster. Genet Mol Res. 2003;2:43–47. [PubMed] [Google Scholar]
- 44.Shearin HK, Macdonald IS, Spector LP, Stowers RS. Hexameric GFP and mCherry reporters for the Drosophila GAL4, Q, and LexA transcription systems. Genetics. 2014;196:951–960. doi: 10.1534/genetics.113.161141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfeiffer BD, Truman JW, Rubin GM. Using translational enhancers to increase transgene expression in Drosophila. Proc Natl Acad Sci U S A. 2012;109:6626–6631. doi: 10.1073/pnas.1204520109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edwards TN, Meinertzhagen IA. The functional organisation of glia in the adult brain of Drosophila and other insects. Prog Neurobiol. 2010;90:471–497. doi: 10.1016/j.pneurobio.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- 49.Bischof J, Basler K. Drosophila. Humana Press; Totowa, NJ: 2008. Recombinases and their use in gene activation, gene inactivation, and transgenesis; pp. 175–195. [DOI] [PubMed] [Google Scholar]
- 50.Pitman JL, Huetteroth W, Burke CJ, Krashes MJ, Lai S-L, Lee T, Waddell S. A pair of inhibitory neurons are required to sustain labile memory in the Drosophila mushroom body. Curr Biol. 2011;21:855–861. doi: 10.1016/j.cub.2011.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong W, Mosca TJ, Luo L. Teneurins instruct synaptic partner matching in an olfactory map. Nature. 2012;484:201–207. doi: 10.1038/nature10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mosca TJ, Luo L. Synaptic organization of the Drosophila antennal lobe and its regulation by the Teneurins. eLife. 2014;3:03726. doi: 10.7554/eLife.03726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prieto-Godino LL, Diegelmann S, Bate M. Embryonic origin of olfactory circuitry in Drosophila: contact and activity-mediated interactions pattern connectivity in the antennal lobe. PLoS Biol. 2012;10:e1001400. doi: 10.1371/journal.pbio.1001400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang K, Gong J, Wang Q, Li H, Cheng Q, Liu Y, Zeng S, Wang Z. Parallel pathways convey olfactory information with opposite polarities in Drosophila. Proc Natl Acad Sci. 2014;111:3164–3169. doi: 10.1073/pnas.1317911111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herrera SC, Martín R, Morata G. Tissue homeostasis in the wing disc of Drosophila melanogaster: immediate response to massive damage during development. PLoS Genet. 2013;9:e1003446. doi: 10.1371/journal.pgen.1003446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silies M, Gohl DM, Fisher YE, Freifeld L, Clark DA, Clandinin TR. Modular use of peripheral input channels tunes motion-detecting circuitry. Neuron. 2013;79:111–127. doi: 10.1016/j.neuron.2013.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parnas M, Lin AC, Huetteroth W, Miesenböck G. Odor discrimination in Drosophila: from neural population codes to behavior. Neuron. 2013;79:932–944. doi: 10.1016/j.neuron.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Li Y, Lei Z, Wang K, Guo A. Transformation of odor selectivity from projection neurons to single mushroom body neurons mapped with dual-color calcium imaging. Proc Natl Acad Sci. 2013;110:12084–12089. doi: 10.1073/pnas.1305857110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang L, Li Y, Potter CJ, Yizhar O, Deisseroth K, Tsien RW, Luo L. GABAergic projection neurons route selective olfactory inputs to specific higher-order neurons. Neuron. 2013;79:917–931. doi: 10.1016/j.neuron.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strutz A, Soelter J, Baschwitz A, Farhan A, Grabe V, Rybak J, Knaden M, Schmuker M, Hansson BS, Sachse S. Decoding odor quality and intensity in the Drosophila brain. eLife. 2014;3:e04147. doi: 10.7554/eLife.04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao XJ, Riabinina O, Li J, Potter CJ, Clandinin TR, Luo L. A transcriptional reporter of intracellular Ca2+ in Drosophila. Nat Neurosci. 2015;18:917–925. doi: 10.1038/nn.4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pfeiffer BD, Jenett A, Hammonds AS, Ngo T-TB, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci U S A. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Venken KJT, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW, Levis RW, Spradling AC, Hoskins RA, et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods. 2011;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gohl DM, Silies MA, Gao XJ, Bhalerao S, Luongo FJ, Lin C-C, Potter CJ, Clandinin TR. A versatile in vivo system for directed dissection of gene expression patterns. Nat Methods. 2011;8:231–237. doi: 10.1038/nmeth.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pérez-Garijo A, Fuchs Y, Steller H. Apoptotic cells can induce non-autonomous apoptosis through the TNF pathway. eLife. 2013;2:01004. doi: 10.7554/eLife.01004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Diao F, Ironfield H, Diao F, Luan H, Shropshire W, Ewer J, Marr E, Potter CJ, Landgraf M, White BH. Plug-and-play genetic access to Drosophila cell types using exchangeable exon cassettes. Cell Rep. 2015;10:1410–1421. doi: 10.1016/j.celrep.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Griffin R, Sustar A, Bonvin M, Binari R, del Valle Rodriguez A, Hohl AM, Bateman JR, Villalta C, Heffern E, Grunwald D, et al. The twin spot generator for differential Drosophila lineage analysis. Nat Methods. 2009;6:600–602. doi: 10.1038/nmeth.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu H-H, Chen C-H, Shi L, Huang Y, Lee T. Twin-spot MARCM to reveal the developmental origin and identity of neurons. Nat Neurosci. 2009;12:947–953. doi: 10.1038/nn.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Evans CJ, Olson JM, Ngo KT, Kim E, Lee NE, Kuoy E, Patananan AN, Sitz D, Tran P, Do M-T, et al. G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat Methods. 2009;6:603–605. doi: 10.1038/nmeth.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hadjieconomou D, Rotkopf S, Alexandre C, Bell DM, Dickson BJ, Salecker I. Flybow: genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster. Nat Methods. 2011;8:260–266. doi: 10.1038/nmeth.1567. [DOI] [PubMed] [Google Scholar]
- 71.Hampel S, Chung P, McKellar CE, Hall D, Looger LL, Simpson JH. Drosophila Brainbow: a recombinase-based fluorescence labeling technique to subdivide neural expression patterns. Nat Methods. 2011;8:253–259. doi: 10.1038/nmeth.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kosman D, Small S. Concentration-dependent patterning by an ectopic expression domain of the Drosophila gap gene knirps. Development. 1997;124:1343–1354. doi: 10.1242/dev.124.7.1343. [DOI] [PubMed] [Google Scholar]
- 73.Stockinger P, Kvitsiani D, Rotkopf S, Tirián L, Dickson BJ. Neural circuitry that governs Drosophila male courtship behavior. Cell. 2005;121:795–807. doi: 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 74.Petersen LK, Stowers RS. A gateway MultiSite recombination cloning toolkit. PLoS One. 2011;6:e24531. doi: 10.1371/journal.pone.0024531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stowers RS. An efficient method for recombineering GAL4 and QF drivers. Fly. 2011;5:371–378. doi: 10.4161/fly.5.4.17560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shearin HK, Dvarishkis AR, Kozeluh CD, Stowers RS. Expansion of the gateway multisite recombination cloning toolkit. PLoS One. 2013;8:e77724. doi: 10.1371/journal.pone.0077724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang YV, Ni J, Montell C. The molecular basis for attractive salt-taste coding in Drosophila. Science. 2013;340:1334–1338. doi: 10.1126/science.1234133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zielke N, Korzelius J, van Straaten M, Bender K, Schuhknecht GFP, Dutta D, Xiang J, Edgar BA. Fly-FUCCI: a versatile tool for studying cell proliferation in complex tissues. Cell Rep. 2014;7:588–598. doi: 10.1016/j.celrep.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 79.Cavanaugh DJ, Geratowski JD, Wooltorton JRA, Spaethling JM, Hector CE, Zheng X, Johnson EC, Eberwine JH, Sehgal A. Identification of a circadian output circuit for rest:activity rhythms in Drosophila. Cell. 2014;157:689–701. doi: 10.1016/j.cell.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]


