Abstract
It has been shown that the adeno-associated virus (AAV) vector can deliver the VEGF gene efficiently into the ischemic mouse myocardium. However, the AAV genomes can be found in extracardiac organs after intramyocardial injection. To limit unwanted VEGF expression in organs other than the heart, we tested the use of the cardiac myosin light chain 2v (MLC-2v) promoter and the hypoxia-response element to mediate cardiac-specific and hypoxia-inducible VEGF expression. An AAV vector, MLCVEGF, with 250 bp of the MLC-2v promoter and nine copies of the hypoxia-response element driving VEGF expression, was constructed. Gene expression was studied in vitro by infection of rat cardiomyocytes, rat skeletal myocytes, and mouse fibroblasts with the vector and in vivo by direct injection of the vector into normal and ischemic mouse hearts. With MLCVEGF infection, VEGF expression was higher in cardiomyocytes than the other two cell lines and was hypoxiainducible. VEGF expression was also higher in ischemic hearts than in normal hearts. No VEGF expression was detectable in organs with detectable MLCVEGF vectors other than the heart. MLCVEGF-injected ischemic hearts had more capillaries and small vessels around the injection site, smaller infarct size, and better cardiac function than the negative controls. Hence, MLCVEGF can mediate cardiac-specific and hypoxia-inducible VEGF expression, neoangiogenesis, infarct-size reduction, and cardiac functional improvement.
Keywords: cardiac-specific gene expression, hypoxia induction, ischemic heart
Angiogenic factors have been investigated for the treatment of ischemic heart disease for more than a decade. Animal and clinical data indicate that angiogenic factors such as fibroblast growth factor (1-3), vascular endothelial growth factor (VEGF) (4-6), and angiopoietins (7) can induce angiogenesis in ischemic myocardium and improve cardiac function. However, a potential problem associated with prolonged and high-level expression of angiogenic factors, such as VEGF, is hemangioma formation in the heart (8, 9) and the limb (10). Hence, it is important to modulate the expression of the angiogenic proteins. For the treatment of ischemic heart disease, an ideal control is for the expression of angiogenic factors to respond to hypoxia. By using nine copies of the hypoxia-response element (HRE) isolated from the erythropoietin (Epo) gene enhancer, we showed previously that VEGF gene expression could be regulated to respond to hypoxia induction in vitro and in vivo (11). However, with intramyocardial delivery in the rat, the injected vectors have been found to infect extracardiac tissues (12). Unwanted angiogenesis may also occur in these tissues even with HRE-controlled angiogenic gene expression, because hypoxia may exist in these tissues as well because of vascular diseases, cardiac failure, or occult tumors.
In this study, we first investigated adeno-associated virus (AAV) vector distribution after intramyocardial injection in the mouse and found the vectors in several extracardiac organs. We then tested the use of a cardiac-specific promoter combined with the HRE to mediate both cardiac-specific and hypoxia-inducible gene expression. We chose the myosin light chain (MLC) 2v promoter because this contractile protein is abundant in slow-twitch skeletal and cardiac muscles (13). Although the MLC-2v promoter is 3 kb long, critical elements that mediate cardiac-specific gene expression are located within first 250 bp (14, 15). We constructed an AAV vector with the 250-bp MLC-2v promoter and nine copies of Epo HRE driving VEGF gene expression. VEGF expression mediated by this vector was studied in vitro and in vivo. Angiogenesis and functional improvement mediated by this vector in ischemic mouse hearts were also analyzed.
Materials and Methods
AAV Vector Construction and Production. A primer set, 5′-GGCCAGATCTTAGACAATGGCAGGACCCAG (sense) and 5′-GGCCAAGCTTGAATTCAAGGAGCCTGCTGG (antisense), was used to PCR-amplify the 250 bp of rat MLC-2v promoter. This promoter was cloned between nine copies of Epo HRE and human VEGF165 cDNA in an AAV vector to generate MLCVEGF (Fig. 1). CMVVEGF (AAV-VEGF) and CMVLacZ (AAV-LacZ) constructed for previous experiments (11, 16) were used as controls (Fig. 1). AAV serotype 2 vectors were prepared by using the three-plasmid cotransfection system (17).
Fig. 1.
AAV constructs. The name in parentheses was used in previous publications (16). ITR, inverted terminal repeat; 9HRE, nine copies of Epo HRE; MLCp, 250 bp of the MLC-2v promoter.
Measurement of VEGF Production by Cultured Cells Infected with VEGF-Expressing Vectors. Rat cardiomyocytes (H9C2) (18), rat skeletal myocytes (L6), and mouse fibroblasts (NIH 3T3) (American Type Culture Collection) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FBS. H9C2 and L6 cells were cultured in 10% CO2 and NIH 3T3 in 5% CO2. Anoxia induction and measurement of VEGF expression were conducted by using the methods described in ref. 11.
Inoculation of Vectors into Ischemic Heart. Mice were housed in the animal care facility of the University of California, San Francisco (UCSF), and experiments were conducted according to guidelines for rodent surgery and a protocol approved by the Institutional Animal Care and Use Committee of UCSF. CD1 mice (male and female; Charles River Breeding Laboratories) were used. Left anterior descending coronary artery ligation and viral vector inoculation were performed as described (11, 16).
PCR and Real-Time RT-PCR Analyses. The TRIzol RNA isolation system (Invitrogen) was used to isolate RNA and genomic DNA. Genomic DNA (2 μg) was used for PCR amplification of AAV genomes. Total RNA was DNase-treated and purified by using the RNeasy Mini Kit (Qiagen, Valencia, CA). First-strand DNA was synthesized with SuperScript II reverse transcriptase (Invitrogen). A 2-μg aliquot of the treated total RNA was used for each sample. Five microliters of cDNA from each sample was used for PCR amplification. RNA without reverse transcriptase were also amplified and used as negative control to rule out possible genomic DNA contamination. Mouse hypoxanthine phosphoribosyl transferase (mHPRT) was used as an internal control. For real-time PCR, the cDNA was diluted 1:10, and 5 μl was added to each reaction. PCRs were performed by using TaqMan Master Mix and the TaqMan Real-Time PCR detection system (ABI PRISM 9700HT, Applied Biosystems). The primer sequences and the method used for real-time PCR data analysis are described in Supporting Text, which is published as supporting information on the PNAS web site.
Histological and Immunohistochemical Analyses. Serial frozen sections were made from the apex of the heart to the site of the ligation. Infarct size was reconstructed with serial trichrome-stained sections. CMVLacZ infected cells were detected by LacZ staining. Anti-platelet endothelial cell adhesion molecule 1 (Santa Cruz Biotechnology) and anti-smooth muscle α-actin (Sigma) antibodies were used to stain endothelial cells and vascular smooth muscles. Immunohistochemical staining was conducted by using the standard protocol of the Elite Vectastain ABC Kit (Vector Laboratories).
Echocardiogram. A transthoracic echocardiogram on conscious mice was performed with an Acuson Sequoia c256 (Sonora Medical Systems, Longmont, CO) by using a 15-MHz linear array transducer (15L8). After the anterior chest was shaved, the mouse was inserted into a plastic cone (Mouse Decapi-Cone, Braintree Scientific), in a prone position, to restrain its activity. The cone was fixed with adhesive tape, and warm ultrasound transmission gel (Aquasonic 100, Parker Laboratory, Fairfield, NJ) was used to fill the space between the chest and the cone. Care was taken to avoid excessive pressure on the thorax, which can induce bradycardia, and two-dimensional imaging was obtained.
Statistical Analysis. Student's t tests and ANOVA testing were used to comparing the differences among groups, with statistical significance considered if P ≤ 0.05. The data are presented as mean ± SD.
Results
Detection of AAV Genome in Multiple Extracardiac Organs After Intramyocardial Injection. AAV vector distribution in the mouse after intramyocardial injection of CMVVEGF was analyzed by PCR genomic DNA isolated from the brain, heart, lung, liver, kidney, spleen, gonad, and diaphragm 2 weeks (three mice) and 6 months (two mice) after the injections. Genomic DNAs isolated from a normal uninjected heart and a Hepes buffer-injected heart were used as negative controls. Vectors were detected from all three mouse hearts collected 2 weeks after vector inoculation and one of the two mouse hearts collected 6 months after vector injection (Fig. 2 Upper). No vector sequence was detected from the uninjected and Hepes buffer-injected hearts. The vector genome was also detected in the livers of all of the vector-injected mice and in the brain, kidney, spleen, gonad or diaphragm of some of the mice (Fig. 2 Lower). In the mouse in which we detected no vector sequence in the heart at the 6-month time point, vector sequences were detected in other organs. Most likely, the vector was injected into the chamber of left ventricle instead of the myocardium in this animal. No vector sequence was detected in the lung of any of the animals. These results suggest that the AAV vector had leaked out of the heart at or after intramyocardial injection.
Fig. 2.
PCR detection of AAV vector sequences in hearts and several extra-cardiac organs after intramyocardial injection of the vectors. (Upper) The PCR results with heart DNA. N, normal heart; 2 weeks, hearts collected 2 weeks after vector injection; Hepes, Hepes-injected heart; 6 months, hearts collected 6 months after vector injection. (Lower) An example of organs in one of the mice collected 2 weeks after intramyocardial injection. B, brain; H, heart; L, lung; Li, liver; K, kidney; S, spleen; O, ovary; D, diaphragm.
Enhancement of Anoxic Response in Cultured Cardiomyocytes by MLCVEGF. To limit the VEGF expression in the heart and to achieve hypoxia-inducible gene expression, an AAV vector (MLCVEGF, Fig. 1) with nine copies of Epo HRE and a 250-bp MLC-2v promoter driving VEGF was constructed. The gene expression mediated by the MLCVEGF vector was tested in vitro by using H9C2 (rat cardiac myocyte), L6 (rat skeletal myocyte), and NIH 3T3 (mouse fibroblast) cells. NIH 3T3 have been used in our previous study (11) and expressed LacZ 10 times higher in anoxic condition than normoxic condition after infection with an AAV vector carrying nine copies of Epo HRE and minimum SV40 promoter driving LacZ gene. CMVVEGF was also used to infect these cell lines as a control. The cells were cultured in normoxic and anoxic conditions after the infection. The ratios of VEGF expression between anoxic and normoxic conditions of all three cell lines were near 1 after CMVVEGF infection. In contrast, these ratios with MLCVEGF infection were 8.7 for H9C2 cells, 2.9 for L6 cells, and 3.7 for NIH 3T3 cells. The ratios of VEGF expression between H9C2 cells and NIH 3T3 cells were 1.8 (normoxic condition) and 4.2 (anoxic condition), and the ratios of VEGF expression between H9C2 cells and L6 cells were 6.2 (normoxia condition) and 8.7 (anoxic condition) (Table 1). Thus, the MLCVEGF vector mediated hypoxia-inducible VEGF expression predominantly in the cardiomyocyte (H9C2).
Table 1. VEGF expression by AAV vectors under normoxic and anoxic conditions (pg/ml per 16-h culture).
| MLCVEGF
|
CMVVEGF
|
|||||
|---|---|---|---|---|---|---|
| Cells | Anoxia | Normoxia | Anoxia/normoxia ratio | Anoxia | Normoxia | Anoxia/normoxia ratio |
| H9C2 | 156 ± 73 | 18 ± 2.1 | 8.7 | 119 ± 42 | 103 ± 24 | 1.2 |
| L6 | 18 ± 4.6 | 6.2 ± 0 | 2.9 | 110 ± 38 | 92 ± 6 | 1.2 |
| NIH 3T3 | 37 ± 5.5 | 10 ± 3 | 3.7 | 88 ± 15 | 105 ± 22 | 0.84 |
Induction of VEGF Gene Expression in Ischemic Mouse Hearts. To investigate whether the MLCVEGF could mediate hypoxiainducible VEGF expression in vivo, 5 × 1010 copies of MLCVEGF mixed with 10% (5 × 109 copies) of CMVLacZ were injected into normal and ischemic hearts. The CMVLacZ vector, in which the LacZ gene was driven by the CMV promoter, was used as an indicator of the location and the efficiency of vector inoculation. We also injected 5 × 1010 copies of CMVVEGF mixed with 10% CMVLacZ to normal and ischemic hearts as controls. Six mice were injected in each group. Gene expression was quantified by real-time RT-PCR 2 weeks after the vector inoculation. We observed, in our previous experiment, that the hematocrits of mice received intramyocardial injection of AAVEpo (serotype 2) began to rise after 2 weeks and reached a plateau at ≈5 weeks. A previous study with the brain showed that Hif-1α accumulated in tissue at the onset of hypoxia and decreased gradually to a normoxic level by 3 weeks despite the continuous low arterial oxygen tension (19). Thus, we chose to analyze the hypoxia responsive gene expression 2 weeks after the left anterior descending coronary artery ligation and vector injection. Compared with normal hearts, MLCVEGF-mediated VEGF expression increased by 74% in ischemic hearts and CMVVEGF remained unchanged (Fig. 3). The differences in gene expression between ischemic and normal hearts were significant (P = 0.01) with MLCVEGF inoculation.
Fig. 3.
Induction of VEGF expression in ischemic hearts by the MLCVEGF vector. Expression of human VEGF mRNA was shown as the relative threshold of VEGF and LacZ mRNA and was calculated based on the threshold cycle (CT). ΔCT, relative threshold cycle; Δ{ΔCT(VEGF-LacZ)} = ΔCTVEGF-ΔCTLacZ where ΔCTVEGF = CTVEGF-CTmHPRT; ΔCTLacZ = CTLacZ-CTmHPRT. The y axis indicates the relative threshold cycles. The larger the number, the more cycles were required to reach the threshold, indicating lower gene expression. The relative thresholds for CMVVEGF were similar for normal heart (1.50 ± 0.80) and ischemic hearts (1.51 ± 0.79), whereas the relative threshold for MLCVEGF-injected hearts decreased from 3.11 ± 0.38 (normal hearts) to 2.31 ± 0.34 (ischemic heart), indicating a 74% increase of VEGF expression in ischemic hearts compare with normal hearts.
Heart-Specific VEGF Gene Expression Mediated by MLCVEGF. Genomic DNA analysis showed that the liver is the most frequent organ that has detectable viral sequences, followed by the spleen and the diaphragm, after intramyocardial injection. We compared VEGF expression in the livers (by real-time RT-PCR) and the diaphragms (by RT-PCR) in which vector genomes were detected. Whereas VEGF expression was detected in the livers and diaphragms of CMVVEGF/CMVLacZ-injected mice, no expression was detected in the livers and diaphragms of mice infected with MLCVEGF/CMVLacZ, although the level of LacZ expression mediated by the coinjected CMVLacZ vector were about the same in CMVVEGF- and MLCVEGF-injected mice (Figs. 4 and 5). Thus, about the same amount of the AAV vector genomes was present in the livers and diaphragms of these animals. The lack of VEGF gene expression in the livers and diaphragms of MLCVEGF-injected mice was caused by the inactivity of the MLC-2v promoter in these organs.
Fig. 4.
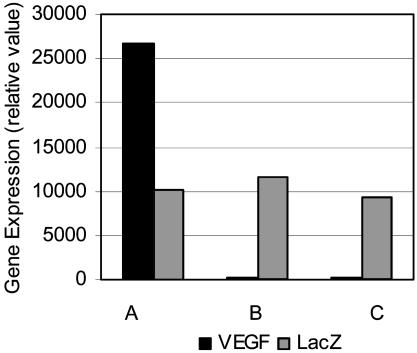
VEGF and LacZ expression in livers of mice that have AAV sequences after receiving intramyocardial injection of CMVVEGF/CMVLacZ (columns A) and MLCVEGF/CMVLacZ (columns B and C). Columns A and C, normal heart mice; columns B, ischemic heart mice. The y axis indicates the gene expression (relative value), calculated based on the threshold cycle (CT). Using the expression of mouse HPRT as the basis for comparison, the relative value of VEGF expression was calculated as 2-ΔCTVEGF where ΔCTVEGF = CTVEGF-CTmHPRT, and the relative value of LacZ expression was calculated as 2-ΔCTLacZ where ΔCTLacZ = CTLacZ-CTmHPRT. The relative value of VEGF expression in livers of CMVVEGF-injected normal heart mice was 26,615.9 ± 1.37. VEGF expression was not detectable in livers of MLCVEGF-injected mice with either normal or ischemic hearts. The relative values of LacZ expression in livers were 10,085.5 ± 2.03 (CMVVEGF/CMVLacZ group), 11,585.2 ± 1.53 (MLCVEGF/CMVLacZ group with ischemic hearts), and 9,410.1 ± 1.21 (MLCVEGF/CMVLacZ group with normal hearts).
Fig. 5.
PCR detection of AAV genomes and VEGF and LacZ gene expression in hearts and diaphragms 2 weeks after left anterior descending coronary artery ligation and intramyocardial vector injection. The templates used for the PCR were genomic DNA (DNA), cDNA (RT+) and RNA (RT-). H, heart; D, diaphragm.
Improved Cardiac Function in Ischemic Hearts Injected with MLCVEGF. To study the effect of MLCVEGF-mediated gene expression on angiogenesis, infarct size, and cardiac function, five groups of mice were injected into the myocardium (i) with 1010 copies of MLCVEGF mixed with 10% CMVLacZ; (ii) with CMVVEGF mixed with 10% CMVLacZ; (iii) with 1010 copies of CMVLacZ alone; (iv) with Hepes buffer; and (v) uninjected immediately after the left anterior descending coronary artery occlusion. The number of mice used for final analyses is shown in Table 2. Cardiac function was assessed 4 weeks after the surgery. Left ventricular end diastolic dimension (LVDd) and end systolic dimension (LVDs) were measured. The percentage of fractional shortening (FS%) was calculated as (LVDd-LVDs)/LVDd × 100. The FS% were 31.4 ± 6.4 for CMVVEGF group and 31.2 ± 9.9 for MLCVEGF group. The FS% for control groups were 26.6 ± 8.9 for the CMVLacZ group, 21.8 ± 4.6 for the Hepes buffer group, and 24.8 ± 5.6 for the uninjected group. The FS% for normal mouse was 53.8 ± 6.2 (Table 2). The differences in FS% between VEGF vector-injected groups and the Hepes buffer group are significant, at P ≤ 0.05. The LVDd and LVDs of VEGF vector-injected mice were significantly smaller than those of the Hepes buffer-injected mice (P ≤ 0.05) (Table 2 and Fig. 6). Thus, MLCVEGF-mediated VEGF expression improved cardiac function. The functional improvement was equivalent to that mediated by CMVVEGF.
Table 2. Infarct size and echocardiogram.
| Vectors (n) | Infarct size, % | LVDd, mm | LVDs, mm | FS(%) |
|---|---|---|---|---|
| CMVVEGF (5) | 37 ± 11 | 4.45 ± 0.19 | 3.06 ± 0.41 | 31.4 ± 6.4 |
| MLCVEGF (8) | 38 ± 7 | 4.42 ± 0.60 | 3.09 ± 0.87 | 31.2 ± 9.9 |
| CMVLacZ (6) | 46 ± 6 | 4.54 ± 0.20 | 3.34 ± 0.51 | 26.6 ± 8.9 |
| Hepes (5) | 46 ± 5 | 4.72 ± 0.17 | 3.69 ± 0.25 | 21.8 ± 4.6 |
| Uninjected (7) | 49 ± 8 | 4.57 ± 0.18 | 3.43 ± 0.28 | 24.8 ± 5.6 |
| Normal (4) | 0 | 3.60 ± 0.18 | 1.67 ± 0.29 | 53.8 ± 6.2 |
LVDd, left ventricular end diastolic dimension; LVDs, left ventricular end systolic dimension; FS(%), percentage of fractional shortening, calculated as (LVDd-LVDs)/LVDd × 100; n, number of animals used in each group. Infarct size was measured as a percentage of the left ventricular wall.
Fig. 6.
Echocardiogram of the left ventricle. A, anterior wall; P, posterior wall. Note the greater movement of the anterior wall of the MLCVEGF-injected heart compared with the CMVLacZ- and Hepes-injected hearts.
Induction of Neovasculature and Reduction of Infarct Size in Ischemic Hearts Injected with MLCVEGF. Hearts collected after echocardiography were sectioned and stained with anti-platelet endothelial cell adhesion molecule 1 and smooth muscle α-actin antibodies. Vessels were counted on six areas, three on the anterior wall and three on the posterior wall in cross sections of the left ventricle (Fig. 7a). Area 1 is made up entirely of muscle tissue, area 2 has both muscle and scar, and area 3 has scar only. Vectors were injected into area 2 at the anterior wall. Hence, comparison between the injected areas in the anterior and the corresponding uninjected posterior areas would indicate the effect of VEGF expression. Capillary density was expressed as the ratio of capillary to cardiac myocyte for area 1 and as the number of capillaries per mm2 for areas 2 and 3. The density of α-actin-positive vessels was expressed as the number of vessels per mm2 for all areas. In the two VEGF vector-injected groups, the capillaries and α-actin-positive vessels in all three areas of the anterior walls were increased compared with the posterior walls in the same hearts. The anterior walls in the hearts of these groups also had more capillaries and α-actin-positive vessels than the anterior walls of the three control groups (CMVLacZ, Hepes, and uninjected groups) (Fig. 7b). In anterior area 1, the differences in capillary:muscle ratios and density of α-actin-positive vessels were significant between the MLCVEGF group and all three control groups (P < 0.01). In anterior area 2, significant differences were seen between the MLCVEGF and Hepes groups for the α-actin-positive vessels and the uninjected group for capillary density (P ≤ 0.03). For anterior area 3, significant differences were shown between the MLCVEGF group and the LacZ or Hepes groups (P = 0.03) on the density of α-actin-positive vessels. The CMVVEGF group has similar densities of capillaries and α-actin-positive vessels as the MLCVEGF group.
Fig. 7.
Induction of neovasculature formation in ischemic hearts by VEGF-expressing vector. (a) A diagram of a cross section of the left ventricle. The areas used for analyses of vessel density are marked. (b) Blood vessel densities in different areas. MLC, MLCVEGF-injected group; CMV, CMVVEGF-injected group; LacZ, CMVLacZ-injected group; Hepes, Hepes-injected group; Uninj, uninjected group.*, the differences between the MLCVEGF group and control groups (CMVLacZ, Hepes, and uninjected groups) are significant.**, the differences between the CMVVEGF group and control groups are significant.
Infarct size was reconstructed on trichrome-stained serial sections and expressed as a percentage of the left ventricular wall. The CMVVEGF- and MLCVEGF-injected groups had smaller infarct sizes compared with the three control groups (Table 2). The differences between the VEGF-injected groups and all three control groups were significant (P ≤ 0.05 for all). Thus, MLCVEGF mediated new blood vessel formation around injection sites and resulted in smaller infarcts in treated mice than in control mice.
Discussion
Ischemic heart disease remains the leading cause of morbidity and mortality in the Western world. Current therapeutic approaches aim to relieve symptoms and cardiac events by reducing myocardial oxygen demand with medical therapy or by restoring blood flow by coronary angioplasty or bypass surgery. However, the ability to accomplish revascularization in some patients with diffuse disease is limited. Therapeutic angiogenesis, using angiogenic growth factors or cytokines to stimulate collateral blood vessel formation, is being tested as an alternative treatment (20, 21).
Gene therapy has the potential advantage of maintaining expression of the angiogenic factors at a sufficient concentration over a long period from a single administration. We have used the AAV vector to deliver the VEGF gene into the ischemic myocardium of mice, and we were able to induce neovasculature formation in ischemic hearts (16). However, uncontrolled exogenous VEGF expression has been shown to cause tumor-like vessels and hemangiomas (8-10, 22-24). To limit unregulated VEGF expression, we used the Epo HRE to regulate VEGF expression to respond to hypoxia induction (11). It is possible that injected vectors could enter the bloodstream and become distributed to extracardiac organs, where they might cause unwanted angiogenesis. In this study, we demonstrated (by PCR) vector sequences in genomic DNA isolated from brains, hearts, livers, kidneys, spleens, gonads, and diaphragms of mice that had received intramyocardial injection of AAV vectors. The liver was the most frequent extracardiac organ that showed positive vector genome. It has been shown that AAV delivered four copies of Epo HRE, and a minimum SV40 promoter driving transgene expression was detected in ischemic liver and skeletal muscle (25). Thus, even with HRE-controlled VEGF expression, angiogenesis at an unwanted site cannot be completely prevented, because hypoxia might occur in organs other than the heart because of vascular disease affecting other organs or because of congestive heart failure. The finding of the AAV vector in extracardiac organs may be caused by the leakage of the vector into the left ventricular cavity during the injection, because the wall of the mouse ventricle is thin. This finding may explain the absence of vector DNA in the lung. Perhaps leakage would be less likely in ventricles that have thicker walls. Nevertheless, for safe angiogenic therapy for ischemic hearts, it will be desirable to have the gene expression both cardiac-specific and hypoxia-responsive.
In this study, we used the 250-bp MLC-2v promoter and nine copies of Epo HRE to drive VEGF gene expression specifically in the heart in response to hypoxia induction. The MLC-2v promoter is the best characterized promoter for ventricular-specific gene expression in embryonic and adult hearts (26). Although the MLC-2v promoter is 3 kb long, transgenic mouse studies have identified the first 250-bp sequence of the promoter to be adequate for ventricular-specific gene expression in embryonic and adult hearts (13, 27). The key element for ventricular-specific gene expression is the 28-bp HF-1 (14, 15, 28). The endogenous cardiac MLC-2v gene is expressed at high levels in both mouse cardiac and slow-twitch skeletal muscles. However, a high level of luciferase activity was detected only in cardiac muscle of transgenic mice harboring the 250-bp rat cardiac MLC-2v promoter-driving luciferase. Luciferase was not expressed at detectable levels in slow-twitch skeletal muscle of these mice (13). When inserted into an adenoviral vector or an AAV vector, this promoter mediated cardiac-specific reporter gene expression in both neonatal rat and adult mouse after i.v. and intraventricular cavity injection (29-31). In our study, this promoter was used to mediate VEGF expression in the acute ischemic adult mouse heart. We demonstrated that the 250-bp MLC-2v promoter plus nine copies of Epo HRE increased VEGF expression by 74% in ischemic hearts compared with normal hearts. Because RNA was isolated from whole heart in our study, the hypoxia induction of gene expression may be underestimated. Although the MLCVEGF DNA sequence was detected in livers and diaphragms of MLCVEGF-injected mice, no VEGF expression was detected. In contrast, LacZ expression mediated by coinjected CMVLacZ vector was detected in these organs, and VEGF expression was found in the organs of CMVVEGF-injected mice that have detectable vector sequences. With cardiac-specific gene expression, the densities of capillaries and small vessels were increased in the myocardium and in the scar around the MLCVEGF injection site. Also, the MLCVEGF-injected mice, compared with non-VEGF-injected controls, had smaller infarct sizes and better cardiac functions that were comparable to the CMVVEGF-injected group.
In conclusion, both cardiac-specific and hypoxia-inducible VEGF expression were achieved with the 250-bp MLC-2v promoter and nine copies of Epo HRE.
Intramyocardial injection of this AAV vector resulted in neovasculature formation, reduction of infarct size, and improvement of cardiac function in ischemic hearts. This approach may lead to a safe and effective angiogenic gene therapy for ischemic heart disease in potential clinical application.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant HL67969 (to Y.W.K.) and in part by a grant from the Wayne and Gladys Valley Foundation (to W.G.).
Author contributions: H.S. and Y.W.K. designed research; H.S., S.J., Y.H., A.B., J.A.-H. performed research; H.S., W.G., and Y.W.K. analyzed data; H.S. wrote the paper; S.J. contributed new reagents/analytic tools; W.G. provided consultation; W.G. designed the cardiac functional study; and Y.W.K. supervised the research.
Abbreviations: AAV, adeno-associated virus; VEGF, vascular endothelial growth factor; HRE, hypoxia-response element; Epo, erythropoietin; MLC, myosin light chain; mHPRT, mouse hypoxanthine phosphoribosyl transferase.
References
- 1.Schumacher, B., Pecher, P., von Specht, B. U. & Stegmann, T. (1998) Circulation 97, 645-650. [DOI] [PubMed] [Google Scholar]
- 2.Giordano, F. J., Ping, P., McKirnan, M. D., Nozaki, S., DeMaria, A. N., Dillmann, W. H., Mathieu-Costello, O. & Hammond, H. K. (1996) Nat. Med. 2, 534-539. [DOI] [PubMed] [Google Scholar]
- 3.Harada, K., Grossman, W., Friedman, M., Edelman, E. R., Prasad, P. V., Keighley, C. S., Manning, W. J., Sellke, F. W. & Simons, M. (1994) J. Clin. Invest. 94, 623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearlman, J. D., Hibberd, M. G., Chuang, M. L., Harada, K., Lopez, J. J., Gladstone, S. R., Friedman, M., Sellke, F. W. & Simons, M. (1995) Nat. Med. 1, 1085-1089. [DOI] [PubMed] [Google Scholar]
- 5.Losordo, D. W., Vale, P. R., Symes, J. F., Dunnington, C. H., Esakof, D. D., Maysky, M., Ashare, A. B., Lathi, K. & Isner, J. M. (1998) Circulation 98, 2800-2804. [DOI] [PubMed] [Google Scholar]
- 6.Takeshita, S., Pu, L. Q., Stein, L. A., Sniderman, A. D., Bunting, S., Ferrara, N., Isner, J. M. & Symes, J. F. (1994) Circulation 90, II228-II234. [PubMed] [Google Scholar]
- 7.Shyu, K. G., Manor, O., Magner, M., Yancopoulos, G. D. & Isner, J. M. (1998) Circulation 98, 2081-2087. [DOI] [PubMed] [Google Scholar]
- 8.Schwarz, E. R., Speakman, M. T., Patterson, M., Hale, S. S., Isner, J. M., Kedes, L. H. & Kloner, R. A. (2000) J. Am. Coll. Cardiol. 35, 1323-1330. [DOI] [PubMed] [Google Scholar]
- 9.Lee, R. J., Springer, M. L., Blanco-Bose, W. E., Shaw, R., Ursell, P. C. & Blau, H. M. (2000) Circulation 102, 898-901. [DOI] [PubMed] [Google Scholar]
- 10.Springer, M. L., Chen, A. S., Kraft, P. E., Bednarski, M. & Blau, H. M. (1998) Mol. Cell 2, 549-558. [DOI] [PubMed] [Google Scholar]
- 11.Su, H., Arakawa-Hoyt, J. & Kan, Y. W. (2002) Proc. Natl. Acad. Sci. USA 99, 9480-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pachori, A. S., Melo, L. G., Zhang, L., Loda, M., Pratt, R. E. & Dzau, V. J. (2004) Biochem. Biophys. Res. Commun. 313, 528-533. [DOI] [PubMed] [Google Scholar]
- 13.Lee, K. J., Ross, R. S., Rockman, H. A., Harris, A. N., O'Brien, T. X., van Bilsen, M., Shubeita, H. E., Kandolf, R., Brem, G., Price, J. et al. (1992) J. Biol. Chem. 267, 15875-15885. [PubMed] [Google Scholar]
- 14.Lee, K. J., Hickey, R., Zhu, H. & Chien, K. R. (1994) Mol. Cell. Biol. 14, 1220-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu, H., Garcia, A. V., Ross, R. S., Evans, S. M. & Chien, K. R. (1991) Mol. Cell. Biol. 11, 2273-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su, H., Lu, R. & Kan, Y. W. (2000) Proc. Natl. Acad. Sci. USA 97, 13801-13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsushita, T., Elliger, S., Elliger, C., Podsakoff, G., Villarreal, L., Kurtzman, G. J., Iwaki, Y. & Colosi, P. (1998) Gene Ther. 5, 938-945. [DOI] [PubMed] [Google Scholar]
- 18.Kimes, B. W. & Brandt, B. L. (1976) Exp. Cell Res. 98, 367-381. [DOI] [PubMed] [Google Scholar]
- 19.Chavez, J. C., Agani, F., Pichiule, P. & LaManna, J. C. (2000) J. Appl. Physiol. 89, 1937-1942. [DOI] [PubMed] [Google Scholar]
- 20.Simons, M., Annex, B. H., Laham, R. J., Kleiman, N., Henry, T., Dauerman, H., Udelson, J. E., Gervino, E. V., Pike, M., Whitehouse, M. J., et al. (2002) Circulation 105, 788-793. [DOI] [PubMed] [Google Scholar]
- 21.Laham, R. J., Chronos, N. A., Pike, M., Leimbach, M. E., Udelson, J. E., Pearlman, J. D., Pettigrew, R. I., Whitehouse, M. J., Yoshizawa, C. & Simons, M. (2000) J. Am. Coll. Cardiol. 36, 2132-2139. [DOI] [PubMed] [Google Scholar]
- 22.Isner, J. M., Pieczek, A., Schainfeld, R., Blair, R., Haley, L., Asahara, T., Rosenfield, K., Razvi, S., Walsh, K. & Symes, J. F. (1996) Lancet 348, 370-374. [DOI] [PubMed] [Google Scholar]
- 23.Pettersson, A., Nagy, J. A., Brown, L. F., Sundberg, C., Morgan, E., Jungles, S., Carter, R., Krieger, J. E., Manseau, E. J., Harvey, V. S., et al. (2000) Lab. Invest. 80, 99-115. [DOI] [PubMed] [Google Scholar]
- 24.Sundberg, C., Nagy, J. A., Brown, L. F., Feng, D., Eckelhoefer, I. A., Manseau, E. J., Dvorak, A. M. & Dvorak, H. F. (2001) Am. J. Pathol. 158, 1145-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pachori, A. S., Melo, L. G., Hart, M. L., Noiseux, N., Zhang, L., Morello, F., Solomon, S. D., Stahl, G. L., Pratt, R. E. & Dzau, V. J. (2004) Proc. Natl. Acad. Sci. USA 101, 12282-12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Small, E. M. & Krieg, P. A. (2004) Trends Cardiovasc. Med. 14, 13-18. [DOI] [PubMed] [Google Scholar]
- 27.O'Brien, T. X., Lee, K. J. & Chien, K. R. (1993) Proc. Natl. Acad. Sci. USA 90, 5157-5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross, R. S., Navankasattusas, S., Harvey, R. P. & Chien, K. R. (1996) Development 122, 1799-1809. [DOI] [PubMed] [Google Scholar]
- 29.Griscelli, F., Gilardi-Hebenstreit, P., Hanania, N., Franz, W. M., Opolon, P., Perricaudet, M. & Ragot, T. (1998) Hum. Gene Ther. 9, 1919-1928. [DOI] [PubMed] [Google Scholar]
- 30.Franz, W. M., Rothmann, T., Frey, N. & Katus, H. A. (1997) Cardiovasc. Res. 35, 560-566. [DOI] [PubMed] [Google Scholar]
- 31.Phillips, M. I., Tang, Y., Schmidt-Ott, K., Qian, K. & Kagiyama, S. (2002) Hypertension 39, 651-655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.