Abstract
It has become apparent that chromatin modification plays a critical role in the regulation of cell-type-specific gene expression. Here, we show that an inhibitor of histone deacetylase, valproic acid (VPA), induced neuronal differentiation of adult hippocampal neural progenitors. In addition, VPA inhibited astrocyte and oligodendrocyte differentiation, even in conditions that favored lineage-specific differentiation. Among the VPA-up-regulated, neuron-specific genes, a neurogenic basic helix–loop–helix transcription factor, NeuroD, was identified. Overexpression of NeuroD resulted in the induction and suppression of neuronal and glial differentiation, respectively. These results suggest that VPA promotes neuronal fate and inhibits glial fate simultaneously through the induction of neurogenic transcription factors including NeuroD.
Keywords: cell fate specification, chromatin, neural stem cell, valproic acid
Multipotent neural progenitor cells can differentiate into neurons and glial cells (e.g., astrocytes and oligodendrocytes) in the mammalian CNS, but the molecular mechanisms that control their fate specification are not yet fully understood. It is becoming increasingly apparent that chromatin accessibility plays a key role in the transcriptional regulation of cell-type-specific gene expression. In eukaryotic cells, DNA is wrapped around core histones and forms nucleosomes that fold into higher-order chromatin structure (1). Modification of histone N-terminal tails through acetylation or deacetylation can alter the interaction between histones and DNA, serving as a mechanism to regulate gene expression (2–5). Histone acetylation has been established to correlate with gene activation (5). Transcriptional coactivators, such as CREB (cAMP-responsive element-binding protein)-binding protein/p300 and its associated factor, PCAF, have been shown to possess histone acetyltransferase activity (6). Conversely, gene repression is associated with histone deacetylation and transcriptional repressors, such as the nuclear receptor corepressor N-CoR and neuron-restrictive silencer factor/repressor element-1 silencing transcription factor (NRSF/REST), are found in complexes with histone deacetylases (HDACs) (7–9).
Although recent work suggests that HDAC recruitment at target promoters is important for lineage specification in a variety of nonneural cell types (10, 11), there is little understanding of the control of chromatin modification and histone acetylation at a global level in a multipotent neural progenitor cell. The identification of compounds that inhibit HDACs has provided useful tools for studying the connection between global chromatin effects and cell lineage specification.
Valproic acid (VPA; 2-propylpentanoic acid) is an established drug in the long-term treatment of epilepsy (12). Recent experiments in 293T, Neuro2A, and teratocarcinoma F9 cells have demonstrated that VPA can directly inhibit HDAC activity and cause hyperacetylation of histones in these cell lines (13, 14). Furthermore, the HDAC inhibition mediated by VPA has been shown to suppress the growth and increase the differentiation of many different tumor cell lines (12). In addition to its differentiation effects, VPA has been shown to mediate neuronal protection through the activation of signal transduction pathways, such as the extracellular signal-regulated kinase (ERK) pathway (15) and through the inhibition of proapoptotic factors (16). Here, we show that VPA induces neural progenitor cells to differentiate predominantly into neurons and that the effects of VPA are mediated, at least in part, by the neurogenic basic helix–loop–helix (bHLH) transcription factor NeuroD.
Materials and Methods
Cell Culture and in Vitro Differentiation Analysis. The hippocampal neural progenitor cells isolated from adult female Fisher 344 rats in this study have been characterized in refs. 17 and 18. Differentiation conditions for neurons, oligodendrocytes, and astrocytes were described in ref. 19. For VPA-induction experiments, cells were trypsinized and plated into N2 medium containing either 0.3 or 1 mM VPA (Sigma) for 4 days. In some cultures, 10 μM BrdUrd (Sigma) was added to label dividing cells, and 1 μg/ml propidium iodide or 1 μg/ml Hoechst 33342 (Sigma) were added to label dead cells or all cells, respectively. Trichostatin A (TSA; 100 nM) (Upstate Biotechnology, Lake Placid, NY) and 1 μM sodium butyrate (NaB; Sigma) were added in some cultures as an alternative HDAC inhibitor.
Immunocytochemistry and in Vitro Quantification. Cells were fixed with 4% paraformaldehyde, followed by immunocytochemical staining as described in ref. 20. Labeled cells were visualized by using a Nikon E800 upright microscope or a Nikon E600 inverted microscope and a Spot RT charge-coupled device camera (Diagnostic Instruments, Sterling Heights, MI). Quantification of cell phenotypes was with StereoInvestigator (MicroBrightfield, Williston, VT). The following primary antibodies were used: rabbit anti-Tuj1 (1:7,500; Covance, Princeton), mouse anti-microtubule-associated proteins 2a and 2b (MAP2ab; 1:250; Sigma); mouse anti-Rip (1:50; Hybridoma Bank, Iowa City, IA); guinea pig anti-glial fibrillary acidic protein (GFAP; 1:2,500; Advanced Immunochemical, Long Beach, CA); rat anti-BrdUrd (1:400; Accurate Chemicals). The detection of BrdUrd in cultured cells required treatment in 2 M HCL at 37°C for 30 min (20). All experiments were independently replicated at least three times.
Western Blot Analysis. For Western blots detecting changes in histone acetylation, whole-cell lysates were prepared from neural progenitors cultured in undifferentiated conditions [fibroblast growth factor 2 (FGF-2)] or from differentiating conditions [retinoic acid plus forskolin for neurons, insulin-like growth factor 1 (IGF-1) for oligodendrocytes, and leukemia inhibitory factor (LIF) plus bone morphogenetic protein 2 (BMP-2) for astrocytes]. Antibodies recognizing acetylated and total histones H3 and H4 were all from Upstate Biotechnology: rabbit anti-acetyl H3 (1:10,000), rabbit anti-acetyl H4 (1:100), mouse anti-histone H3 (1:100), and rabbit anti-histone H4 (1:100). Immunoblotting of Akt, ERKs, and signal transducer and activator of transcription 3 (STAT3) were performed as described in ref. 21. The following antibodies from Cell Signaling Technology (Beverly, MA) were used: rabbit anti-phospho-Akt (1:100), rabbit anti-Akt (1:1000), rabbit anti-phospho-p44 and -p42 ERKs (1:1,000), rabbit anti-p44 and -p42 ERKs (1:1,000), rabbit anti-phosphotyrosine-STAT3 (1:500), and mouse anti-STAT3 (1:5000).
Expression Vectors and Neural Progenitor Electroporation. The mouse NeuroD cDNA was cloned into the pMY expression vector (22) containing internal ribosome entry site GFP. Electroporation (Amaxa Biosystems, Gaithersburg, MD) was performed according to the manufacturer's protocol, and cells were plated in N2 plus 20 ng/ml FGF-2 for 24 h. To induce differentiation, FGF-2 was withdrawn and replaced with fresh N2 media. FBS (0.5%) was included in all differentiation cultures to enhance survival of cells after electroporation. To induce oligodendrocyte-specific differentiation, electroporated cells were directly plated into insulin-free N2 medium plus 500 ng/ml IGF-1 for 4 days. To induce astrocyte-specific differentiation, 50 ng/ml LIF plus 50 ng/ml BMP-2 was added, and the cells were cultured for 4 days.
RT-PCR. Total RNA was isolated from cell cultures by using RNeasy columns (Qiagen, Valencia, CA). RT-PCR was performed essentially as described in ref. 19. Primer sequences are available upon request.
VPA Analysis in Vivo. Adult female Fisher 344 rats received two daily i.p. injections of 300 mg/kg VPA (experimental) or saline (control). VPA was also provided in the drinking water (12 g/liter) for the experimental group. All rats received one daily i.p. injection of 100 mg/kg BrdUrd for 6 consecutive days after the first day. After 14 days, animals were killed, perfused with 4% paraformaldehyde, and processed for BrdUrd immunohistochemical staining as described in refs. 17 and 23. The following antibodies were used: rabbit anti-Tuj1 (1:500), guinea pig anti-GFAP (1:1,000), and rat anti-BrdUrd (1:400). Images were acquired by using a Nikon E800 upright microscope equipped with a 10× objective lens and postprocessed in photoshop (Adobe Systems, San Jose, CA). All comparative analyses were focused in the subgranular zone (SGZ) of the dentate gyrus on the same side of matched planes of sections in the dorsal hippocampus. For quantification of BrdUrd-positive and BrdUrd/Tuj1-double positive cells, area counts of the SGZ were performed. In each section, four adjacent fields were sampled, starting from where the upper and lower blades meet.
Results
HDAC Inhibition Induces Neuronal Differentiation. Adult hippocampal neural progenitors were used in this study as a model system to elucidate molecular mechanisms that control fate determination. These cells fulfill the definition of multipotent neural progenitor cells: They self-renew when cultured with basic FGF-2 and can differentiate into neurons, oligodendrocytes, and astrocytes after stimulation with exogenous factors (retinoic acid plus forskolin for neurons, IGF-1 for oligodendrocytes, and LIF plus BMP-2 for astrocytes) (see Fig. 6, which is published as supporting information on the PNAS web site) (17–19).
To evaluate the functional significance of histone acetylation and deacetylation during multipotent neural progenitor proliferation and differentiation, cell behavior was monitored in the presence of the HDAC inhibitor VPA. Neural progenitors were plated in N2 media with 20 ng/ml FGF-2 for 24 h before switching to fresh N2 medium with or without 1 mM VPA for 4 days (Fig. 1). Lineage-specific differentiation was monitored with markers of neurons (Tuj1 or MAP2ab), oligodendrocytes (Rip), and astrocytes (GFAP). Surprisingly, instead of seeing nonspecific effects on gene expression leading to mixed differentiation of all lineages by VPA treatment, we saw a large increase in Tuj1- and MAP2ab-positive neurons in VPA-treated cultures (Fig. 1 A and D). The percentage of Tuj1- and MAP2ab-positive neurons was low (<1%) in control cultures (Fig. 1 A and D). The presence of oligodendrocytes and astrocytes was not detectable in control or VPA-treated cultures (data not shown). Neuronal differentiation by VPA appeared to be dose-dependent (Fig. 1D); however, because higher VPA concentrations (>3 mM) resulted in increased cell death, we performed our remaining studies by using concentrations that favored maximal differentiation and minimal toxicity (1 mM VPA; see Fig. 1D). Neural progenitors were also treated with 100 nM TSA and 1 μM NaB, two different HDAC inhibitors that also promoted neuronal differentiation (Fig. 1 A and D). Because of the higher toxicity of TSA and NaB in neural progenitor cultures, these experiments were carried out for only 2 days.
Fig. 1.
HDAC-inhibition-mediated neuronal differentiation. (A) Treatment of neural progenitors with 1 mM VPA for 4 days resulted in an increase in cells with MAP2ab (green) staining and neuronal morphology compared with control cultures, which lack neuronal differentiation. Similar results were observed with the addition of 100 nM TSA and 1 μM NaB for 2 days. Blue regions indicate 4′,6-diamidino-2-phenylindole-stained nuclei. (B) Staining of acetylated histone H3 (red) and MAP2ab (green) is higher in HDAC inhibitor-treated cultures. (C) Proliferation in untreated (control) and 1-day HDAC inhibitor-treated cultures as determined by BrdUrd (red) incorporation. (Scale bar, 50 μm.) (D and E) Quantifications of neuronal differentiation (D) and proliferation (E) in untreated and HDAC inhibitor-treated cultures. All data shown are from at least three experiments in parallel cultures with error bars representing standard deviations.
To verify that VPA, TSA, and NaB were acting as HDAC inhibitors in neural progenitors, we stained control and treated cultures with an antibody specific for acetylated histone H3. Neural progenitors treated with HDAC inhibitors showed increased levels of acetylated histone H3 in the nucleus, compared with untreated neural progenitors (control), suggesting that the increase in neuronal differentiation by VPA, TSA, and NaB was associated with a decrease in HDAC activity (Fig. 1B).
To evaluate the effect of HDAC inhibitors on neural progenitor proliferation, neural progenitors in the presence of N2 plus FGF-2 (20 ng/ml) were treated with either VPA, TSA, or NaB or left untreated for 24 h and labeled with 10 μM BrdUrd for 1 h (Fig. 1 C and E). We observed a dramatic decrease in the percentage of BrdUrd-positive cells in each HDAC inhibitor-treated condition. Approximately 50% of the cells in control cultures were BrdUrd-positive, compared to ≈20% of the cells being BrdUrd-positive in VPA-treated cultures (Fig. 1E). TSA and NaB also dramatically reduced neural progenitor proliferation (from 50% in control cultures to 8% in treated cultures).
Although the effects of HDAC inhibition in neural progenitors appears mainly to be through a reduction of proliferation and an increase in neuronal differentiation, HDAC inhibitors have also been shown to induce apoptosis (24, 25). We therefore evaluated the degree of cell death in neural progenitor cells by staining living cultures with 1 μg/ml propidium iodide, which stains dead cells, and 1 μg/ml Hoechst 33342, which stains live and dead cells. Staining of live (rather than fixed) cultures was used to avoid underestimating cell death because of the possible detachment of dying and/or dead cells from culture substrates. Neural progenitor cells treated with 1 mM VPA for 2 days resulted in 7.0 ± 0.1% dead cells, with 25.9% MAP2ab-positive neurons. Cells treated with a lower dose of VPA (0.3 mM) for 2 days resulted in 4.5 ± 1.6% dead cells, which is only slightly higher than the amount of cell death in untreated control cultures (2.0 ± 0.1% dead cells). Even with the lower dose of VPA when cell death was minimal, there was still evidence of neuronal differentiation (6.9% MAP2ab-positive cells), suggesting that a selective death of a subset of committed progenitors or nonneuronal cells does not appear to have a significant role in the increased neuronal differentiation with VPA treatment.
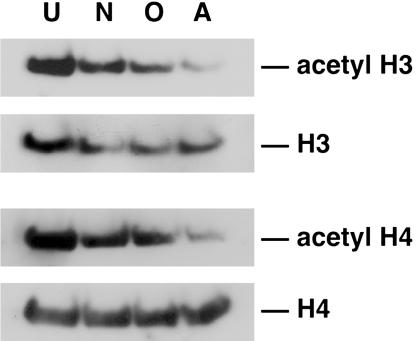
Neuronal Lineage Progression Is Associated with Maintenance of Histone Acetylation. To determine whether changes in histone acetylation occur during CNS lineage progression, we compared neural progenitors that had differentiated into neurons, oligodendrocytes, and astrocytes with undifferentiated cells (Fig. 2; see Fig. 6 for each differentiation condition) (19). To detect changes in histone acetylation, protein extracts were harvested from neural progenitors and analyzed by Western blot analysis with antibodies specific for acetylated histones H3 and H4. We detected an abundance of acetylated histones H3 and H4 in extracts of undifferentiated neural progenitors. Interestingly, the level of acetylated histone H3 and H4 was still relatively high in neuronal extracts as compared with undifferentiated extracts, suggesting that there is less deacetylation during neuronal lineage progression. However, there appeared to be less histone H3 and H4 acetylation in astrocyte and oligodendrocyte extracts, suggesting that there is more deacetylation during astrocyte and oligodendrocyte lineage progression compared with neuronal lineage progression (Fig. 2). To exclude the possibility that the changes in histone acetylation observed during CNS lineage progression were due to changes in overall histone levels, we also performed Western blot analysis with antibodies against total histones. As shown in Fig. 2, the steady-state levels of histones H3 and H4 did not change significantly under various conditions. The observations that VPA treatment increased neuronal differentiation and that the level of acetylated histones was higher in neurons compared with oligodendocytes and astrocytes suggested that histone acetylation is important for neuronal lineage progression of adult multipotent neural progenitor cells. The partial reduction of acetylated histones H3 and H4 in oligodendrocyte and astrocyte extracts suggested that the maintenance of histone acetylation is less important in nonneuronal cells; in fact, there is evidence that recruitment of HDAC activity by the transcriptional regulator NRSF/REST is important to repress the promoter of neuron-specific genes in nonneuronal cells (7, 8, 26). In addition, recent work suggests that histone deacetylase activity is important for oligodendrocyte lineage progression (27).
Fig. 2.
Changes in histone acetylation during neural progenitor lineage progression. Western blot analysis of neural progenitor extracts from undifferentiated (U) cultures grown in N2 media plus 20 ng/ml FGF-2 or differentiated with 1 μM retinoic acid plus 5 μM forskolin for 4 days (neuron, N), 500 ng/ml IGF-1 for 4 days (oligodendrocyte, O), or 50 ng/ml LIF plus 50 ng/ml BMP-2 for 6 days (astrocyte, A). Immunoblotting was performed by using antibodies against histones H3 and H4 and acetylated histones H3 and H4. Data shown are from at least three independent experiments.
VPA Actively Suppresses Oligodendrocyte and Astrocyte Differentiation While Promoting Neuronal Differentiation. The decreased histone acetylation in oligodendrocytes and astrocytes suggests that histone deacetylation is important for oligodendrocyte and astrocyte differentiation. To directly test this hypothesis, we induced oligodendrocyte and astrocyte differentiation of neural progenitors in the presence or absence of VPA. IGF-1 (500 ng/ml) induced neural progenitors to differentiate into Rip-positive oligodendrocytes with characteristic web-like processes (Fig. 3) (19). Addition of VPA to IGF-1-treated cultures dramatically reduced the percentage of Rip-positive cells. Intriguingly, the percentage of Tuj1-positive neurons increased in IGF-1-treated cultures when VPA was added. As was seen with neural progenitors that differentiated into astrocytes with 50 ng/ml LIF plus 50 ng/ml BMP-2 (28), addition of VPA reduced the number of GFAP-positive astrocytes and increased the number of Tuj1-positive neurons (Fig. 3). Taken together, these results suggest that histone deacetylation is important for both oligodendrocyte and astrocyte lineage progression, and increased histone acetylation is associated with neuronal lineage progression.
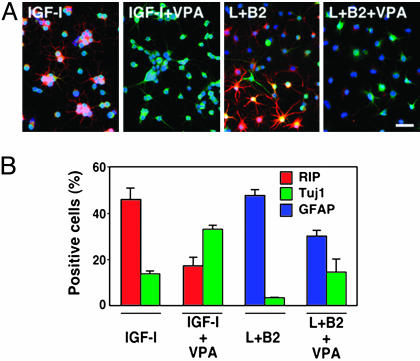
Fig. 3.
VPA-mediated suppression of glial differentiation. (A) Neural progenitors induced to differentiate into Rip-positive oligodendrocytes (red) with 500 ng/ml IGF-1 or GFAP-positive astrocytes (red) with 50 ng/ml LIF (L) plus 50 ng/ml BMP-2 (B2). Treatment of cultures with 1 mM VPA for 4 days reduced the number of Rip-positive cells (red) in IGF-1-treated cultures and GFAP-positive cells (red) in LIFplusBMP-2-treated cultures, while increasing the number of Tuj1-positive cells (green) in each case. (Scale bar, 50 μm.) (B) Quantification of neurons, oligodendrocytes, and astrocytes in various differentiation conditions in the presence or absence of 1 mM VPA. All data shown are from at least three experiments in parallel cultures with error bars representing standard deviations.
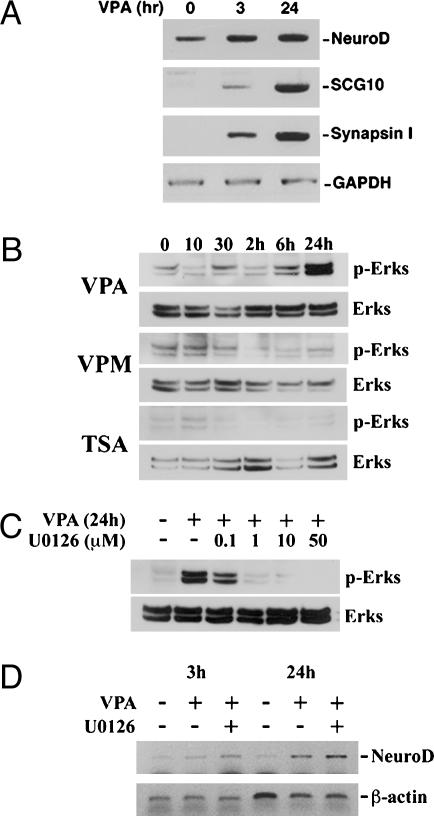
Up-Regulation of HDAC-Dependent, Neuron-Specific Genes After VPA Treatment. The protein NRSF/REST binds to the neuron-restrictive silencer element sequence to mediate transcriptional repression of target genes by the recruitment and association at its N terminus of the mSin3A/HDAC1,2 complex (8, 26). Recent work examined the role of HDAC-dependent repression of neuron-restrictive silencer element-containing, neuron-specific genes and showed that treatment of Rat-1 fibroblasts with the HDAC inhibitor TSA resulted in the ectopic activation of a number of neuron-specific genes (29). We therefore performed RT-PCR analysis of HDAC-dependent, neuron-specific genes after VPA treatment (Fig. 4A). We could detect an up-regulation of NeuroD, SCG10, and Synapsin I as early as 3 h and at 24 h after VPA treatment. GAPDH levels did not change after VPA treatment and was used as an internal control.
Fig. 4.
VPA up-regulates neuron-specific genes, including the bHLH transcription factor NeuroD.(A) RT-PCR of NeuroD, SCG10, and Synapsin I in neural progenitors at time 0 or treated with 1 mM VPA for 3 h and 24 h. GAPDH was used as a normalization control. (B) Western blot time-course analysis of ERK activation after 1 mM VPA, 100 nM TSA, and 1 mM valpromide (VPM) treatment. (C) ERK activation with 1 mM VPA treatment with and without addition of U0126 (different doses are indicated). (D) RT-PCR analysis of VPA-mediated up-regulation of NeuroD (3 and 24 h) with and without U0126 (1 μM).
The molecular basis of the reduced proliferation in VPA-treated neural progenitor cells was also examined. Cell cycle regulators, such as cyclin-dependent kinase (CDK) inhibitors, have been shown to be important for the antiproliferative effects of HDAC inhibitors (30, 31). Consistent with this finding, we found that there was increased expression of CDK inhibitors p21WAF1/CIP1 and p27KIP1 in neural progenitors after VPA treatment (data not shown). These results suggest that neuronal differentiation mediated by VPA involves an up-regulation of both neuron-specific genes and CDK inhibitors to reduce cell proliferation and promote neuronal fate commitment.
It was recently reported that VPA could phosphorylate and activate ERKs 1 and 2, resulting in the differentiation of E18 cortical neurons (32). To determine whether VPA-mediated neuronal differentiation in adult hippocampal neural progenitors was directly caused by the activation of the ERK pathway, we performed a Western blot time-course analysis. Although we did not see ERK activation by VPA after 1 h (Fig. 7 and Supporting Text, which are published as supporting information on the PNAS web site), we decided to carry out the experiment for longer time points. Neural progenitor cells were treated with 1 mM VPA, 1 mM valpromide (a VPA analog that is not a HDAC inhibitor, but is also an antiepileptic), and 100 nM TSA for the indicated times (0, 10, and 30 min and 2, 6, and 24 h) and extracts were collected for Western blots (Fig. 4B). Although neural progenitors treated with VPA did not show an increase in ERK activation at the earlier time points, there was ERK activation by 24 h. Interestingly, we did not observe ERK activation with TSA or valpromide. To determine whether ERK activation by VPA at 24 h was necessary to trigger neuronal fate commitment, we decided to block ERK activation with an inhibitor of mitogen-activated protein kinase kinase 1/2 (U0126), which is the upstream kinase that phosphorylates ERKs. Cells were pretreated with control (DMEM/F12 media) or 0.1–50 μM U0126 for 2 h and treated with or without 1 mM VPA for 24 h (Fig. 4C). We found that 1 μM U0126 was sufficient to block the ERK activation by VPA at 24 h. We next examined whether neuronal fate commitment mediated by VPA, as evidenced by an up-regulation of the bHLH transcription factor NeuroD, could still occur, even when ERK activation was blocked. Cells were pretreated with 1 μM U0126 for 2 h before adding 1 mM VPA for 3 or 24 h (Fig. 4D). RT-PCR analysis confirmed that there was up-regulation of NeuroD after VPA treatment at 3 h, with even higher levels at 24 h (compared with untreated cells). More importantly, blocking ERK activation did not appear to inhibit the VPA-mediated up-regulation of NeuroD, suggesting that ERK activation was not necessary to trigger neuronal differentiation.
The Mechanism of VPA-Mediated Neuronal Differentiation Is Associated with the Activation of NeuroD. Among the HDAC-dependent neuron-specific genes that became up-regulated with VPA treatment, NeuroD was the best candidate for triggering neuronal fate commitment and promoting differentiation. Strikingly, mice deficient for NeuroD completely lack the dentate gyrus granule cell layer, supporting the role of NeuroD in the proliferation, differentiation, and survival of granule cells in the hippocampus (33, 34). We therefore examined whether forced expression of NeuroD alone could have effects on neuronal differentiation (Fig. 8, which is published as supporting information on the PNAS web site). We found that neural progenitors overexpressing NeuroD differentiated into >75% Tuj1-positive and MAP2ab-positive neurons, respectively, and reduced their ability to differentiate into oligodendrocytes and astrocytes under appropriate conditions (Fig. 8). These results suggest that NeuroD by itself can recapitulate many of the effects of VPA and is sufficient to induce neuronal differentiation of adult multipotent neural progenitor cells.
VPA-Mediated Neuronal Differentiation in Vivo. Our findings raise the question of whether the effects of VPA on adult hippocampal neural progenitors in vitro are related to the clinical indications of VPA as an antiepileptic in vivo. Pilocarpine-induced status epilepticus increases cell proliferation in the SGZ of the adult dentate gyrus (35). Prolonged seizure activity has also been shown to markedly increase dentate granule cell neurogenesis (35). To examine the potential effects of VPA on dentate granule cell proliferation and neurogenesis, we injected adult rats with VPA for a total of 14 days, while including a 6-day BrdUrd regimen to label dividing cells (Fig. 5A). Consistent with previous studies (35), BrdUrd immunohistochemistry in saline-injected rats (controls, n = 4) labeled mitotically active cells mostly located in the SGZ of the dentate gyrus (Fig. 5B, white arrows). In contrast, VPA-injected rats (n = 6) showed a marked reduction in BrdUrd incorporation in the SGZ (Fig. 5B, white arrows). No obvious differences in BrdUrd immunostaining were observed in the hilar region or the molecular layer of the dentate gyrus. Quantitative analysis of BrdUrd labeling within the SGZ revealed a significant decrease (≈2-fold) (P < 0.001, t test) in the number of dividing cells in VPA-injected rats compared with controls (Fig. 5C). The VPA-mediated decrease in granule cell proliferation within the SGZ is consistent with the effects of VPA in decreasing proliferation of neural progenitors in vitro.
Fig. 5.
VPA decreases granule cell proliferation and increases neuronal differentiation in the dentate gyrus of adult rats. (A) Schematic of VPA and BrdUrd injection paradigm. (B) Representative images of brain sections focusing on the dentate gyrus (DG) of animals injected with saline (control) or VPA. Sections were processed for BrdUrd staining. White arrow indicates the dentate gyrus. Hi, Hilus. (Scale bar, 200 μm.) (C) The average number of BrdUrd-positive cells (in four adjacent fields) per section (control animals, n = 4; VPA-treated animals, n = 6) is plotted. The asterisk indicates that values are significantly different between control and VPA-treated animals (P < 0.001, t test). (D) The percentage of Tuj1-/BrdUrd-double positive cells (in four adjacent fields) per section (control animals, n = 4; VPA-treated animals, n = 6) is plotted. Asterisks indicate statistical significance (P < 0.05, t test). Error bars represent standard deviations.
We next examined the fate of dividing cells in the SGZ by assessing colocalization of BrdUrd with lineage-specific markers (Tuj1 for neurons and GFAP for astrocytes) (Fig. 5D). In control animals, ≈60% of the BrdUrd-positive cells were also Tuj1-positive. There was a significant increase (from 60% to 80%) in the percentage of BrdUrd-positive cells that were Tuj1-positive in VPA-treated animals (P < 0.05, t test), suggesting that VPA can mediate an increase in neuronal differentiation in vivo as well. The percentage of BrdUrd-positive cells that were also GFAP-positive was relatively low (≈1%) in control animals and did not significantly change compared with VPA-treated animals (data not shown). These results support the ability of VPA to decrease neural progenitor proliferation and increase neuronal differentiation in vivo as well as in vitro.
Discussion
In this series of experiments, we used a well established model system of adult multipotent neural progenitor cells to investigate the connection between global histone acetylation and neural cell fate specification. When cells are cultured in the presence of HDAC inhibitors, such as VPA, TSA, or NaB, neural progenitors reduced their proliferation and largely differentiated into neurons. These findings were also confirmed in our in vivo experiments. When neural progenitors were treated with conditions that promoted astrocyte and oligodendrocyte differentiation, VPA actively suppressed differentiation into astrocytes and oligodendrocytes while promoting differentiation into neurons. Analysis of the mechanism of VPA-mediated neuronal differentiation revealed an up-regulation of the neurogenic bHLH transcription factor NeuroD in VPA-treated cultures. Furthermore, overexpression of NeuroD alone in neural progenitors could recapitulate the effects of VPA.
We and others have previously shown that IGF-1 induces oligodendrocyte differentiation (19) and LIF induces astrocyte differentiation (36, 37). The VPA-mediated suppression of oligodendrocyte and astrocyte differentiation could be due to an inhibition of IGF-1 and LIF signal transduction pathways. Two of the major downstream mediators of IGF-1 signaling are the phosphoinositide 3-kinase/Akt and Ras/ERK pathways (38, 39), and the major downstream mediator of LIF signaling is the Janus tyrosine kinase/STAT pathway (36, 37). First, we did not detect an activation of Akt/ERKs or STAT3 with VPA treatment alone, at least within 1 h (see Fig. 7). Second, the activation status of Akt/ERKs or STAT3 after IGF-1 or LIF stimulation did not change in VPA-treated cultures compared with control cultures. This finding suggests that the VPA-mediated neuronal differentiation and suppression of glial differentiation was not due to a direct activation or inhibition, respectively, of these signaling molecules and point to another mechanism of VPA action.
Recently, Hao et al. (32) described the effects of VPA on the activation of the ERK pathway in E18 cortical neurons. In their assay system, VPA promoted neurite growth and cell reemergence in an ERK pathway-dependent manner. Although we were unable to detect an activation of ERKs within 1 h, we did observe ERK activation by VPA after 24 h. However, when we blocked ERK activation with U0126, we were still able to detect an up-regulation of NeuroD (at 3 and 24 h), suggesting that neuronal fate commitment triggered by VPA is not directly due to an activation of the ERK pathway. Furthermore, the observation that TSA was not able to activate ERKs but was able to up-regulate NeuroD (data not shown) and promote neuronal differentiation further confirmed that direct activation of the ERK pathway is not solely responsible for neuronal differentiation of adult neural progenitor cells.
An alternative mechanism of the neuron-promoting effect of VPA could be the direct activation of neurogenic transcription factors. Our work indicates that VPA treatment up-regulates NeuroD in neural progenitor cells, suggesting that VPA could, in fact, induce neuronal differentiation through the transcriptional activation of neurogenic transcription factors. Lunyak et al. (29) showed that the expression of NeuroD and of other neuron-specific genes is regulated by the HDAC-dependent transcriptional repressor NRSF/REST. Further examination of other neuron-specific genes, such as Synapsin I and SCG10, showed that each had increased gene expression after VPA treatment, consistent with previous reports (29). In addition to up-regulating NRSF/REST-regulated neuron-specific genes, VPA up-regulates neuronal markers, such as Tuj1 and MAP2ab, which are not direct targets of NRSF/REST. This finding suggests that the mechanism of VPA-mediated neuronal differentiation is not solely due to a direct release of NRSF/REST repression of neuron-specific genes and that VPA must activate factors important for neuronal lineage progression. The observation that forced expression of NeuroD alone can promote neuronal differentiation indicates that VPA may exert its effects through the activation of NeuroD. More studies are needed to determine whether the up-regulation of NeuroD after VPA treatment is merely correlative or whether there is a causal relationship between the VPA-mediated neuronal differentiation and neurogenic bHLH transcription factors.
Adult multipotent neural progenitor cells need to choose between self-renewal and differentiation into neurons, oligodendrocytes, or astrocytes. We hypothesized that chromatin modification plays an important role in these stem cell decisions because of its ability to control the simultaneous expression of many genes. The observation that VPA can have a profound effect on promoting neuronal differentiation and suppressing glial differentiation suggests that the maintenance of the acetylated state, revealed by HDAC inhibition, may have global and dominant effects on neuronal lineage progression.
Examination of total acetylated histones H3 and H4 in neural progenitor extracts under various conditions revealed that histone acetylation levels were highest in the undifferentiated state. Interestingly, among the differentiation conditions examined, acetylated histone levels were higher in neuronal extracts relative to astrocytic extracts. The level of acetylated histones in oligodendrocytic extracts was always intermediate to that of neurons and astrocytes, possibly because of a partial requirement of histone acetylation for oligodendrocyte lineage progression or the presence of neurons in oligodendrocyte cultures. In fact, a previous study showed that treatment of neural progenitors with IGF-1 (the oligodendrocyte-promoting condition) can also result in a low percentage of Tuj1-positive neurons (Fig. 6) (19). These results support the idea that there are global changes in chromatin states underlying differences in adult multipotent neural progenitor differentiation. Does acetylation of chromatin selectively activate specific master control genes, such as NeuroD, that go on to trigger downstream genes important for neuronal differentiation, or does NeuroD itself possess the ability to regulate global chromatin-based events, such as the maintenance of histone acetylation? Future studies are needed to determine the precise order and relationship of chromatin modification and the regulation of gene expression in the control of adult multipotent neural progenitor cell fate specification.
Supplementary Material
Acknowledgments
We thank M. L. Gage for editorial comments. This work was supported in part by grants from the National Institutes of Health, the George E. Hewitt Foundation for Medical Research, Project A.L.S., the Christopher Reeve Paralysis Foundation, the Pritzker Neurogenesis Research Consortium, and a Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad.
Abbreviations: HDAC, histone deacetylase; VPA, valproic acid; TSA, trichostatin A; NRSF/REST, neuron-restrictive silencer factor/repressor element-1 silencing transcription factor; ERK, extracellular signal-regulated kinase; bHLH, basic helix–loop–helix; NaB, sodium butyrate; MAP2ab, microtubule-associated proteins 2a and 2b; GFAP, glial fibrillary acidic protein; FGF-2, fibroblast growth factor 2; IGF-1, insulin-like growth factor 1; STAT3, signal transducer and activator of transcription 3; BMP-2, bone morphogenetic protein 2; LIF, leukemia inhibitory factor; SGZ, subgranular zone.
References
- 1.Georgopoulos, K. (2002) Nat. Rev. Immunol. 2, 162–174. [DOI] [PubMed] [Google Scholar]
- 2.Grunstein, M. (1997) Nature 389, 349–352. [DOI] [PubMed] [Google Scholar]
- 3.Kuo, M. H. & Allis, C. D. (1998) BioEssays 20, 615–626. [DOI] [PubMed] [Google Scholar]
- 4.Struhl, K. (1998) Genes Dev. 12, 599–606. [DOI] [PubMed] [Google Scholar]
- 5.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 6.Chan, H. M. & La Thangue, N. B. (2001) J. Cell Sci. 114, 2363–2373. [DOI] [PubMed] [Google Scholar]
- 7.Huang, Y., Myers, S. J. & Dingledine, R. (1999) Nat. Neurosci. 2, 867–872. [DOI] [PubMed] [Google Scholar]
- 8.Jepsen, K., Hermanson, O., Onami, T. M., Gleiberman, A. S., Lunyak, V., McEvilly, R. J., Kurokawa, R., Kumar, V., Liu, F., Seto, E., et al. (2000) Cell 102, 753–763. [DOI] [PubMed] [Google Scholar]
- 9.Kao, H. Y., Downes, M., Ordentlich, P. & Evans, R. M. (2000) Genes Dev. 14, 55–66. [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang, C. L., McKinsey, T. A., Chang, S., Antos, C. L., Hill, J. A. & Olson, E. N. (2002) Cell 110, 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koipally, J., Renold, A., Kim, J. & Georgopoulos, K. (1999) EMBO J. 18, 3090–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blaheta, R. A. & Cinatl, J., Jr. (2002) Med. Res. Rev. 22, 492–511. [DOI] [PubMed] [Google Scholar]
- 13.Gottlicher, M., Minucci, S., Zhu, P., Kramer, O. H., Schimpf, A., Giavara, S., Sleeman, J. P., Lo Coco, F., Nervi, C., Pelicci, P. G. & Heinzel, T. (2001) EMBO J. 20, 6969–6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phiel, C. J., Zhang, F., Huang, E. Y., Guenther, M. G., Lazar, M. A. & Klein, P. S. (2001) J. Biol. Chem. 276, 36734–36741. [DOI] [PubMed] [Google Scholar]
- 15.Coyle, J. T. & Duman, R. S. (2003) Neuron 38, 157–160. [DOI] [PubMed] [Google Scholar]
- 16.Perez, M., Rojo, A. I., Wandosell, F., Diaz-Nido, J. & Avila, J. (2003) Biochem. J. 372, 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gage, F. H., Coates, P. W., Palmer, T. D., Kuhn, H. G., Fisher, L. J., Suhonen, J. O., Peterson, D. A., Suhr, S. T. & Ray, J. (1995) Proc. Natl. Acad. Sci. USA 92, 11879–11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer, T. D., Takahashi, J. & Gage, F. H. (1997) Mol. Cell Neurosci. 8, 389–404. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh, J., Aimone, J. B., Kaspar, B. K., Kuwabara, T., Nakashima, K. & Gage, F. H. (2004) J. Cell Biol. 164, 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer, T. D., Markakis, E. A., Willhoite, A. R., Safar, F. & Gage, F. H. (1999) J. Neurosci. 19, 8487–8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakashima, K., Narazaki, M. & Taga, T. (1997) FEBS Lett. 401, 49–52. [DOI] [PubMed] [Google Scholar]
- 22.Misawa, K., Nosaka, T., Morita, S., Kaneko, A., Nakahata, T., Asano, S. & Kitamura, T. (2000) Proc. Natl. Acad. Sci. USA 97, 3062–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kempermann, G., Kuhn, H. G. & Gage, F. H. (1997) Nature 386, 493–495. [DOI] [PubMed] [Google Scholar]
- 24.Tang, X. X., Robinson, M. E., Riceberg, J. S., Kim, D. Y., Kung, B., Titus, T. B., Hayashi, S., Flake, A. W., Carpentieri, D. & Ikegaki, N. (2004) Clin. Cancer Res. 10, 5837–5844. [DOI] [PubMed] [Google Scholar]
- 25.Phillips, A., Bullock, T. & Plant, N. (2003) Toxicology 192, 219–227. [DOI] [PubMed] [Google Scholar]
- 26.Naruse, Y., Aoki, T., Kojima, T. & Mori, N. (1999) Proc. Natl. Acad. Sci. USA 96, 13691–13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marin-Husstege, M., Muggironi, M., Liu, A. & Casaccia-Bonnefil, P. (2002) J. Neurosci. 22, 10333–10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakashima, K., Yanagisawa, M., Arakawa, H., Kimura, N., Hisatsune, T., Kawabata, M., Miyazono, K. & Taga, T. (1999) Science 284, 479–482. [DOI] [PubMed] [Google Scholar]
- 29.Lunyak, V. V., Burgess, R., Prefontaine, G. G., Nelson, C., Sze, S. H., Chenoweth, J., Schwartz, P., Pevzner, P. A., Glass, C., Mandel, G. & Rosenfeld, M. G. (2002) Science 298, 1747–1752. [DOI] [PubMed] [Google Scholar]
- 30.Archer, S. Y., Meng, S., Shei, A. & Hodin, R. A. (1998) Proc. Natl. Acad. Sci. USA 95, 6791–6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaziri, C., Stice, L. & Faller, D. V. (1998) Cell Growth Differ. 9, 465–474. [PubMed] [Google Scholar]
- 32.Hao, Y., Creson, T., Zhang, L., Li, P., Du, F., Yuan, P., Gould, T., Manji, H. & Chen, G. (2004) J. Neurosci. 24, 6590–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyata, T., Maeda, T. & Lee, J. E. (1999) Genes Dev. 13, 1647–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, M., Pleasure, S. J., Collins, A. E., Noebels, J. L., Naya, F. J., Tsai, M. J. & Lowenstein, D. H. (2000) Proc. Natl. Acad. Sci. USA 97, 865–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parent, J. M., Yu, T. W., Leibowitz, R. T., Geschwind, D. H., Sloviter, R. S. & Lowenstein, D. H. (1997) J. Neurosci. 17, 3727–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonni, A., Sun, Y., Nadal-Vicens, M., Bhatt, A., Frank, D. A., Rozovsky, I., Stahl, N., Yancopoulos, G. D. & Greenberg, M. E. (1997) Science 278, 477–483. [DOI] [PubMed] [Google Scholar]
- 37.Takizawa, T., Nakashima, K., Namihira, M., Ochiai, W., Uemura, A., Yanagisawa, M., Fujita, N., Nakao, M. & Taga, T. (2001) Dev. Cell 1, 749–758. [DOI] [PubMed] [Google Scholar]
- 38.LeRoith, D., Werner, H., Beitner-Johnson, D. & Roberts, C. T., Jr. (1995) Endocr. Rev. 16, 143–163. [DOI] [PubMed] [Google Scholar]
- 39.Kulik, G., Klippel, A. & Weber, M. J. (1997) Mol. Cell. Biol. 17, 1595–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.