The targeting of proteins to the endoplasmic reticulum (ER) is a topic of considerable interest since this organelle serves as an entry point for proteins destined for other organelles, as well as for the ER itself. A unique feature of plants is that they are able to store proteins in the ER in addition to other endomembrane compartments, and the deposition of such storage proteins provides important sources of both human and animal nutrition. Thus, increasing our knowledge of the mechanisms required for the targeting of storage proteins to this crucial organelle will ultimately allow the modification of critical steps, leading to improvements in plants as a protein source as well as in crop yield and productivity.
The entry of proteins into the endomembrane system is dependent on the presence of a transient N-terminal signal peptide. However, recent developments indicate that the localization of RNAs to specific ER subdomains may facilitate protein targeting within the endomembrane system. The aim of this Update is to introduce RNA localization as a means of targeting protein synthesis to specific intracellular locations with a focus on the localization of prolamine mRNA, and thus protein, to the ER-derived compartment known as the protein body. In addition to summarizing published research concerning prolamine mRNA localization, unpublished data showing the existence of multiple RNA localization pathways to specific ER subdomains and the role of mRNA targeting with respect to protein localization will be discussed.
RNA TARGETING: AN INTRODUCTION
In recent years, it has become clear that mRNA localization is a widespread and efficient means of targeting gene products to specific intracellular regions (for review, see Jansen, 2001; Kloc et al., 2002; Van de Bor and Davis, 2004; and for plants by Fedoroff, 2002; Okita and Choi, 2002). The localization of mRNA within the cytoplasm dictates cell polarity in both somatic cells and oocytes in addition to playing a central role in pattern formation and cell fate determination during embryonic development. This is often the result of site-specific translation of a protein that results in the formation of a morphogen gradient, typically transcription factors, that determines the overall body plan after fertilization (Ephrussi and St Johnston, 2004).
Historically, the most complete and best-characterized model systems for RNA localization are axis specification in the Drosophila oocyte and pole definition in the Xenopus embryo (Jansen, 2001; Kloc et al., 2002; Ephrussi and St Johnston, 2004). Other well-studied examples include the localization of mRNAs in polarized somatic cells such as fibroblasts and neurons (Jansen, 2001; Kloc et al., 2002). More recently, localization of Ash1 mRNA that encodes an inhibitor of mating-type switching has been found to be targeted to the budding tip of yeast daughter cells (Chartrand et al., 2001). The latter suggests that the process of RNA localization may be common to all eukaryotes. Despite the current lack of mature model systems from the plant kingdom, several examples of RNA localization in plants are known. These include the differential segregation of expansin mRNAs to the apical or basipetal end of xylem precursor cells, and the targeting of actin mRNA during the establishment of cell polarity and early cell divisions in embryos of the brown alga Fucus (for review, see Okita and Choi, 2002). The RNA-dependent localization of rice (Oryza sativa) seed storage protein mRNAs is well characterized and will be discussed in more depth below.
THE RNA LOCALIZATION PATHWAY
RNAs are localized by several different mechanisms, but the most common one, often called the RNA localization pathway (Wilhelm and Vale, 1993), initially involves the formation of a transport particle or granule resulting from the interaction of trans-acting RNA binding factors and accessory proteins with cis-acting localization elements (zipcodes) present in the targeted RNA (Jansen, 2001; Kloc et al., 2002). The final steps in the RNA localization pathway are transportion via the cytoskeleton and anchoring at the target site.
Although mRNA localization is a cytoplasmic event, it has become increasingly apparent that early events in the nucleus are important. It is now believed that proteins that determine the destinations of mRNAs are recruited on native transcripts in the nucleus to form a precursor transport particle (Farina and Singer, 2002). In addition, these nuclear proteins may be involved in many aspects of mRNA biogenesis and are likely to act in concert to provide specificity (Van de Bor and Davis, 2004). One such group of proteins are the heterogeneous nuclear ribonucleoprotein family (hnRNP), a member of which (hnRNP A2) is known to be involved in the localization of myelin basic protein in oligodendrocytes (Hoek et al., 1998). More recently, the Drosophila protein Hrp48 has been shown to be required for both the subcellular localization of RNAs (Huynh et al., 2004) and the subsequent regulation of their translation (Yano et al., 2004). Based on homology searches, several putative members of the hnRNP family exist in rice (GenBank accession nos. 32990960, 32969243, 32977743, and 32986113).
Upon exiting the nucleus, the assembly of a large ribonucleoprotein particle or granule occurs as a result of the interaction of RNA-binding proteins with the targeted RNA, changing its conformation and thus triggering the binding of additional proteins. This RNA transport particle may contain multiple localized RNAs in addition to factors involved in targeting, anchoring, and translation so that protein synthesis can begin immediately once the target site is reached (Carson et al., 1998). To date, no general consensus sequences have been identified for RNA localization signals, and it is widely believed that the targeting process is likely to be more complex than first anticipated, with constant reorganization of the mRNA-protein complex en route to its final destination (for review, see Van de Bor and Davis, 2004).
The transport of RNA particles from the nucleus to the target site generally occurs via the cytoskeleton (other mechanisms exist but will not be discussed here), most frequently, but not always, using microtubules. These can form longer structures than actin microfilaments and are thus better suited for RNA localization in polarized, differentiated, vertebrate cells and oocytes (Kloc et al., 2002). The cytoskeleton provides a framework for the delivery of RNAs to their final destinations (de Heredia and Jansen, 2004) and acts as a scaffold for translating ribosomes or as a site enriched for translational factors (Hesketh, 1996). Indeed, there is evidence that the cytoskeleton reduces the lateral mobility of translocon complexes within the membrane of the rough ER (Nikonov and Kreibich, 2003), reaffirming the importance of the cytoskeleton for protein translation. That mRNAs are transported in a translationally silent state is of crucial importance, and RNA transport is inescapably coupled with translational arrest (discussed later in more detail), with encoded proteins being synthesized only once the final destination has been reached.
In summary, the RNA localization pathway contains a number of distinct components to identify and characterize: (1) cis-acting RNA signals, which determine the mRNA localization site; (2) trans-acting RNA-binding proteins (and cointeracting proteins) that recognize these signals; (3) motor proteins and cytoskeleton elements (either actin filaments or microtubules) responsible for directed transport of RNA particles from the nucleus to the target site; and finally (4) proteins involved in translational regulation and RNA anchoring. Significant progress has been made with respect to identifying signals and the cytoskeletal elements required for transport. Recent findings on these aspects in rice are described below together with additional questions that have been raised and currently remain unanswered.
RICE STORAGE PROTEINS: A UNIQUE SYSTEM
Unlike most plants, which preferentially accumulate a single major class of storage proteins, developing rice seeds synthesize large and essentially equimolar amounts of prolamines, the typical type accumulated by cereals, and glutelins, proteins homologous to the 11S globulins accumulated by legumes. In addition, the alcohol-soluble prolamines and salt-soluble globulin-like glutelins are stored within different compartments of the endomembrane system (Okita and Choi, 2002).
Over the last decade, our lab has been working to elucidate the mechanisms by which selective targeting of prolamine and glutelin to different compartments is accomplished. Considerable progress has been made in this regard, and it is now known that the targeting of the mRNAs encoding these proteins to different subdomains of the ER is responsible for their distinct protein localizations (Li et al., 1993; Choi et al., 2000).
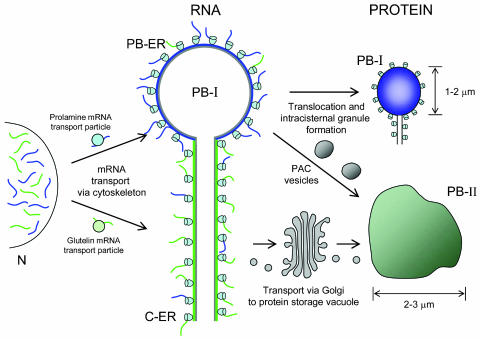
Figure 1 shows how both prolamine and glutelin mRNAs are targeted via separate RNA-based mechanisms from their site of transcription in the nucleus to distinct subdomains of the ER (Choi et al., 2000). Prolamine mRNAs are transported to the ER-derived protein bodies containing prolamines (PB-ER). Translation of this RNA results in the translocation of prolamine polypeptides into the ER lumen, which subsequently assemble to form intracisternal granules as a result of their hydrophobic nature. These spherical protein bodies are known as protein body type I (PB-I). In contrast to prolamine mRNAs, glutelin mRNAs are transported primarily to the cisternal ER (C-ER), whereupon the synthesized protein is sorted to the Golgi prior to its deposition in protein storage vacuoles (PSV or PB-II).
Figure 1.
Schematic representation of RNA-dependent seed storage protein targeting in developing rice endosperm. Following transcription in the nucleus (N), prolamine and glutelin mRNAs are transported via the cytoskeleton in large transport particles to the cortical area of endosperm cells, where they segregate onto the PB-ER and C-ER, respectively. Prolamine polypeptides are translocated into the ER lumen where they form spherical intracisternal granules known as protein body type I (PB-I), while glutelin is exported via the Golgi and deposited in irregularly shaped protein storage vacuoles (PB-II). The existence of a precursor-accumulating (PAC) vesicle-dependent pathway leading from the PB-ER directly to the protein storage vacuole has been hypothesized. The approximate sizes of the protein storage compartments are indicated.
Both the PB-ER and C-ER subdomains are a part of the cortical ER in developing rice endosperm, a region closely associated with the cytoskeleton (Muench et al., 2000). The cortical ER is the predominant site of protein synthesis in rice endosperm and in the majority of plant cells. This region is also rich in actin microfilaments, which serve as an anchor for translating ribosomes as demonstrated by an abundance of EF1α, a protein synthesis factor that recruits aminoacyl-tRNAs to the acceptor site of ribosomes during peptide chain elongation (Clore et al., 1996).
The nature of RNA transport and the role of the cytoskeleton were determined by real-time observation of prolamine RNA transport in living rice seeds. This was accomplished by using a modified two-gene expression system where the green fluorescent protein (GFP) was employed as a fluorescent tag to monitor RNA transport (Hamada et al., 2003a). This methodology demonstrated that prolamine RNAs are transported as large particles that move in a stop-start fashion but in a generally consistent direction and with a typical average velocity of 0.3 to 0.4 μm/s. The addition of drugs, which disrupt actin filaments (but not microtubules) or act as inhibitors of myosin, strongly retarded particle movement, indicating that the transport process is mediated by the actomyosin system.
HOW ARE STORAGE PROTEIN RNAs TARGETED TO ER SUBDOMAINS?
Identification of cis-acting RNA localization signals indicates that there are multiple transport pathways to the cortical ER in rice. Initial studies demonstrated that prolamine peptide sequences were not essential for RNA localization, although initiation of translation is a prerequisite for correct prolamine mRNA localization to the PB-ER (Choi et al., 2000; Hamada et al., 2003b). This finding is in agreement with the fact that transport particles often contain ribosomes in addition to other factors required for translation (Krichevsky and Kosik, 2001). Moreover, it enabled the identification of cis-acting RNA localization signals within the prolamine gene by using a series of deletions of prolamine cDNA fused with a reporter RNA encoding GFP (Hamada et al., 2003b). Two cis-elements were identified: one located in the 5′ coding sequence distal to the signal peptide coding sequence and the second residing in the proximal half of the 3′untranslated region (UTR). Both regions are required for correct prolamine RNA targeting to the PB-ER, since deletion of either one of the signals leads to partial mislocalization of RNA to the cisternal ER. Deletion of both signals leads to RNA localization exclusively on the cisternal ER, supporting the existence of a default RNA localization pathway leading from the nucleus to the cisternal ER. This default pathway does not require the presence of a region encoding a signal peptide and has been observed for a variety of cytoplasmic proteins, including the reporter protein β-glucuronidase (Hamada et al., 2003b).
Interestingly, replacing the prolamine 3′UTR with that of glutelin results in prolamine RNAs being redirected to the cisternal ER (Choi et al., 2000; Hamada et al., 2003b). As the prolamine coding sequence has a single PB-ER targeting cis-element, the results support the presence of one or more cis-elements in the glutelin 3′UTR and the existence of a second regulated transport pathway involving glutelin RNAs. This result indicates that glutelin RNAs are directed to the cisternal ER by a third pathway, which may be dominant over the regulated prolamine RNA transport pathway.
The existence of these multiple RNA transport pathways is also supported by ongoing studies of rice storage protein mutants. Satoh and his colleagues (Ogawa et al., 1989, and references cited therein) have identified over 100 rice lines defective in storage protein synthesis, accumulation, and/or processing. Several classes of mutants have been identified by the abnormal accumulation of the 57-kD glutelin precursor, which is normally processed into acidic and basic subunits in the protein storage vacuole. Analysis of prolamine and glutelin mRNA localization by in situ reverse transcription-PCR in three mutants analyzed to date shows that they mislocalize either prolamine mRNA to the cisternal ER (glup2) or glutelin mRNA to the PB-ER (glup4 and glup6). The Glup2 gene is essential for RNA targeting to the PB-ER, and its genetic disruption leads to the displacement of prolamine RNAs to the cisternal ER, most likely via the default pathway. The partial localization of glutelin RNAs to PB-ER in glup4 and glup6 lines indicates that the glutelin and prolamine RNA transport pathways are not totally distinct processes but are related. Further characterization of these and other mutant lines is under way.
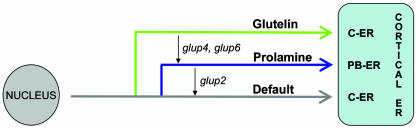
Based on the available evidence, we can devise a model of RNA transport in rice. Figure 2 shows the three separate (glutelin, prolamine, and default) RNA localization pathways from the nucleus to subdomains of the cortical ER, including branch points indicating their relative dominance and their relationship as well as the effect of the glup2, glup4, and glup6 mutations on RNA transport. The existence of a single shared pathway would presumably allow the use of common components to transport different RNAs. It remains to be resolved whether different RNA species are contained within the same transport particle or are present in different particles. In either case, additional components are necessary to confer specificity and to dictate which pathway should be followed by a particular RNA.
Figure 2.
Possible RNA localization pathways to cortical ER subdomains. Schematic representation of the distinct targeting pathways leading from the nucleus to the cortical ER. In addition to two regulated RNA pathways for prolamine and glutelin mRNA targeting, which require specific zipcode signals, there is evidence for the existence of a default pathway leading to the cisternal ER. Also depicted are the changes in RNA localization seen in the glup mutants. Based on the analysis of glup mutants, the three RNA pathways are not independent but are interrelated. This model does not distinguish the possibility that different RNAs reside in the same RNA transport particle and then segregate to PB-ER and cisternal ER or whether the three RNA transport pathways denote transport particles containing specific RNAs.
One question that remains unanswered is whether the same area of cisternal ER can serve as the target site for both glutelin and default RNA localization pathways. It is conceivable that the cisternal ER, often treated as a single entity, is in reality divided into regions with distinct functionalities. For example, RNAs that code for secretory proteins may be localized to an ER subdomain that is spatially isolated from that enriched for RNAs that code for proteins destined for the protein storage vacuole. This issue will undoubtedly be resolved by future research into ER function.
THE IMPORTANCE OF TRANSLATIONAL ARREST
A prerequisite for RNA localization is the transport of RNAs in a quiescent state; otherwise, translation during transit would litter the protein throughout the cell. Two examples that illustrate the demand for translational arrest during RNA transport are oskar (Macdonald, 2004), which codes for a posterior-end determinant in Drosophila oocytes, and Ash1 (Gu et al., 2004), a mating-type switch regulator in budding yeasts. When these RNAs are promiscuously translated during their transport to their normal destination, cell fate determination in these systems is disrupted, indicating that translational arrest is an essential step in this process. Although evidence denoting a similar requirement for RNA localization in rice has not been obtained, it is clear that the RNAs must be transported in a silenced state to facilitate protein localization (see below). For RNAs that encode proteins that enter the endomembrane system, translation arrest could potentially occur when the newly synthesized signal peptide is bound by the signal recognition particle. However, since translation commences when the signal recognition particle docks onto ER membranes, which are distributed throughout the cell, it is likely that more intricate and subtle processes will be employed to repress translation.
A variety of repressor proteins that bind to the 3′UTR of the mRNA and suppress translation have been identified. However, except in a few cases, the mechanisms by which these proteins repress translation remain largely unknown (Kloc et al., 2002; Huang and Richter, 2004). In addition to blocking initiation, protein synthesis can also be repressed at the elongation step as inferred by the association of translationally quiescent RNAs with sedimenting polysomes. The available evidence indicates that prolamine RNA transport requires a functional AUG start codon, suggesting that the RNAs in transit are loaded with ribosomes but are then prevented from elongating.
One process for which an increasing number of occurrences are being documented is repression of translation by microRNAs, which bind to the 3′UTR of mRNA. The most relevant example with reference to RNA localization is the Fragile X mental retardation protein (FMRP), which serves to modulate (mainly repress) translation as well as RNA transport in neurons (Siomi et al., 2004). This protein is capable of multivalent RNA interactions since it has two RNA-binding motifs, a pair of KH domains and an RGG domain. Although FMRP can bind directly to RNAs, it is likely that this interaction occurs through its binding to the microRNA BC1, which, in turn, anneals to the targeted RNA (Zalfa et al., 2003). The Drosophila homolog dFMR1 has been found to be a component of the RNAi-induced silencing complex (RISC), indicating a role for the RNAi machinery in translation control of transported RNAs (Murchison and Hannon, 2004; Siomi et al., 2004).
There is currently no direct evidence for the involvement of RNAi in RNA transport in rice, although available data does not exclude this possibility. In addition to an FMRP homolog, the RISC of Drosophila S2 cells contain TSN-1 (Murchison and Hannon, 2004), a nucleic acid-binding protein of which a rice homolog has been identified as a major RNA-binding protein in a cytoskeleton-enriched protein body fraction from developing seeds (Sami-Subbu et al., 2001). In addition to being tightly associated with microtubules, Rp120 has been detected in GFP-labeled prolamine RNA transport particles (C. Wang and T. Okita, unpublished data), indicating a role in RNA localization possibly as a RISC component. Colocalization studies with known RISC components should establish whether RNAi plays a role in RNA transport in rice.
THE RELATIONSHIP BETWEEN RNA AND PROTEIN LOCALIZATION
The central tenet behind the targeting of mRNAs encoding seed storage proteins is that it directly influences the localization of the encoded protein, permitting high localized concentrations to be generated (Kloc et al., 2002). This is clearly demonstrated by the segregation of prolamine and glutelin RNAs onto distinct ER subdomains and their consequently distinct protein localizations following export and deposition of glutelin in protein storage vacuoles. In addition, the resulting high concentrations of seed storage proteins such as prolamine made possible by the RNA targeting mechanism may be critical for the correct formation of intracisternal granules that develop into mature protein bodies (see Hara-Nishimura et al., 2004). Furthermore, the use of separate subdomains for the synthesis of prolamine and glutelin is likely to prevent nonproductive interactions between these proteins due to their disparate physical properties.
Examination of protein localization in glup2, glup4, and glup6 rice mutants has extended the evidence for tight coupling of RNA and protein localization. In these lines, mislocalization of prolamine RNAs to cisternal ER (glup2) or glutelin RNAs (glup4, glup6) to the PB-ER results in a corresponding relocalization of the resulting protein. For example, in glup4 and glup6 glutelin polypeptides were found not only in storage vacuole PB-II but also within the prolamine-containing PB-I (M. Ogawa, T. Kumamaru, and H. Satoh, unpublished data). This indicates that RNA localization is a crucial mechanism for ensuring correct mRNA and thus protein targeting.
This new data does, however, raise a number of questions about protein transport and diffusivity within the ER lumen. First, how can redirecting prolamine mRNA to the cisternal ER cause that protein to interfere with glutelin deposition instead of being secreted? Second, why is it that glutelin encoded by an RNA that has been redirected to the PB-ER does not diffuse into the cisternal ER from where it might exit, but is instead trapped within this ER subdomain? It is possible that the answers to these questions lie in the physical properties of storage proteins and the high throughput of the actively synthesizing ER. A prolamine polypeptide that finds itself in the cisternal ER may become assembled with glutelins (with which it would not usually be in close proximity) and thus be escorted via the Golgi to protein storage vacuoles. The trapping of glutelin within prolamine protein bodies could be explained by limited lateral diffusion within the ER lumen. This would prevent such a protein from moving laterally into the cisternal ER. Irrespective of the mechanisms at work, it seems possible that interfering with RNA localization may trigger nonphysiological events, which, in the active cortical ER of endosperm tissue, contrive to result in aberrant protein localization.
FUTURE PROSPECTS: IDENTIFYING THE MAJOR PLAYERS
Following the identification of zipcode sequences for prolamine mRNA targeting and characterization of the mRNA transport mechanism, the obvious next step is to identify and characterize trans-acting RNA-binding proteins and other cointeracting proteins. One strategy is to isolate RNA-binding proteins from cytoskeleton-rich developing rice seed extracts using biotinylated bait RNA with sequences corresponding to prolamine RNA localization signals. Map-based cloning of the glup2, glup4, and glup6 mutants is also under way to identify the genes that result in storage protein RNA mislocalization.
The available information on RNA localization in plants raises a number of interesting questions that will be the subject of future research. Are there similar mRNA-targeting mechanisms that direct storage proteins to ER-derived protein bodies in other plant species, such as maize, or to storage vacuoles, as in legumes? What transport pathway is undertaken by RNAs that code for proteins packaged in precursor-accumulating vesicles? Are nonstorage protein RNAs targeted to specific regions of the ER according to their function or final subcellular destination? What is the role of RNA localization in the synthesis of cytoplasmic proteins? In the longer term, it is hoped that new model systems for RNA localization within the plant kingdom, such as embryogenesis and polar cell growth (root tip hairs, pollen tubes), will address the general significance of RNA localization in plant biology and lead to the identification of a whole crop of new protein targets to characterize.
Acknowledgments
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers 32990960, 32969243, 32977743, and 32986113.
This work was supported by the National Science Foundation (grant no. 0235140), by the U.S. Department of Agriculture (grant no. USDA–NRICRG 2003–00934), by the Agriculture Research Center, Washington State University (Project 0590), and by the Ministry of Education, Science and Culture of Japan (grant nos. 10660009 and 1213826).
References
- Carson JH, Kwon S, Barbarese E (1998) RNA trafficking in myelinating cells. Curr Opin Neurobiol 8: 607–612 [DOI] [PubMed] [Google Scholar]
- Chartrand P, Singer RH, Long RM (2001) RNP localization and transport in yeast. Annu Rev Cell Dev Biol 17: 297–310 [DOI] [PubMed] [Google Scholar]
- Choi S-B, Wang C, Muench DG, Ozawa K, Franceschi VR, Wu Y, Okita T (2000) Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature 407: 765–767 [DOI] [PubMed] [Google Scholar]
- Clore AM, Dannenhoffer JM, Larkins BA (1996) eEF1A is associated with the cytoskeletal network surrounding protein bodies in maize endosperm cells. Plant Cell 8: 2003–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Heredia ML, Jansen R-P (2004) mRNA localization and the cytoskeleton. Curr Opin Cell Biol 16: 80–85 [DOI] [PubMed] [Google Scholar]
- Ephrussi A, St Johnston D (2004) Seeing is believing: the bicoid morphogen gradient matures. Cell 116: 143–152 [DOI] [PubMed] [Google Scholar]
- Farina KL, Singer RH (2002) The nuclear connection in RNA transport and localization. Trends Cell Biol 12: 466–472 [DOI] [PubMed] [Google Scholar]
- Fedoroff NV (2002) RNA-binding proteins in plants: the tip of an iceberg? Curr Opin Plant Biol 5: 452–459 [DOI] [PubMed] [Google Scholar]
- Gu W, Deng Y, Zenklusen D, Singer RH (2004) A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev 18: 1452–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Ishiyama K, Choi SB, Wang C, Singh S, Kawai N, Franceschi VR, Okita TW (2003. a) The transport of prolamine RNAs to prolamine protein bodies in living rice endosperm cells. Plant Cell 15: 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Ishiyama K, Sakulsingharoj C, Choi SB, Wu Y, Wang C, Singh S, Kawai N, Messing J, Okita TW (2003. b) Dual regulated RNA transport pathways to the cortical region in developing rice endosperm. Plant Cell 15: 2265–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura I, Matsushima R, Shimada T, Nishimura M (2004) Diversity and formation of endoplasmic reticulum-derived compartments in plants. Are these compartments specific to plant cells? Plant Physiol 136: 3435–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh JE (1996) Intracellular sorting of macromolecules. Biochem Soc Trans 24: 521–528 [DOI] [PubMed] [Google Scholar]
- Hoek KS, Kidd GJ, Carson JH, Smith R (1998) hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry 37: 7021–7029 [DOI] [PubMed] [Google Scholar]
- Huang YS, Richter JD (2004) Regulation of local mRNA translation. Curr Opin Cell Biol 16: 308–313 [DOI] [PubMed] [Google Scholar]
- Huynh JR, Munro TP, Smith-Litiere K, Lepesant JA, Johnston DS (2004) The Drosophila hnRNPA/B homolog, Hrp48, is specifically required for a distinct step in osk mRNA localization. Dev Cell 6: 625–635 [DOI] [PubMed] [Google Scholar]
- Jansen RP (2001) mRNA localization: message on the move. Nat Rev Mol Cell Biol 2: 247–256 [DOI] [PubMed] [Google Scholar]
- Kloc M, Zearfoss NR, Etkin LD (2002) Mechanisms of subcellular mRNA localization. Cell 108: 533–544 [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS (2001) Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron 32: 683–696 [DOI] [PubMed] [Google Scholar]
- Li X, Franceschi VR, Okita TW (1993) Segregation of storage protein mRNAs on the rough endoplasmic reticulum membranes of rice endosperm cells. Cell 72: 869–879 [DOI] [PubMed] [Google Scholar]
- Macdonald PM (2004) Translational control: a cup half full. Curr Biol 14: R282–R283 [DOI] [PubMed] [Google Scholar]
- Muench DG, Chuong SDX, Franceschi VR, Okita TW (2000) Developing prolamine protein bodies are associated with the cortical cytoskeleton in rice endosperm cells. Planta 211: 227–238 [DOI] [PubMed] [Google Scholar]
- Murchison EP, Hannon GJ (2004) miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr Opin Cell Biol 16: 223–229 [DOI] [PubMed] [Google Scholar]
- Nikonov AV, Kreibich G (2003) Organization of translocon complexes in ER membranes. Biochem Soc Trans 31: 1253–1256 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Kumamaru T, Satoh H, Omura T, Park T, Shintaku K, Baba K (1989) Mutants for rice storage proteins. 2. Isolation and characterization of protein bodies from rice mutants. Theor Appl Genet 78: 305–310 [DOI] [PubMed] [Google Scholar]
- Okita TW, Choi S-B (2002) mRNA localization in plants: targeting to the cell's cortical region and beyond. Curr Opin Plant Biol 5: 553–559 [DOI] [PubMed] [Google Scholar]
- Sami-Subbu R, Choi S-B, Wu Y, Wang C, Okita TW (2001) Identification of a cytoskeleton-associated 120 kDa RNA binding protein in developing seeds. Plant Mol Biol 46: 79–88 [DOI] [PubMed] [Google Scholar]
- Siomi H, Ishizuka A, Siomi MC (2004) RNA interference: a new mechanism by which FMRP acts in the normal brain? What can Drosophila teach us? Ment Retard Dev Disabil Res Rev 10: 68–74 [DOI] [PubMed] [Google Scholar]
- Van de Bor V, Davis I (2004) mRNA localisation gets more complex. Curr Opin Cell Biol 16: 300–307 [DOI] [PubMed] [Google Scholar]
- Wilhelm JE, Vale RD (1993) RNA on the move: the mRNA localization pathway. J Cell Biol 123: 269–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, de Quinto SL, Matsui Y, Shevchenko A, Ephrussi A (2004) Hrp48, a Drosophila hnRNPA/B homolog, binds and regulates translation of oskar mRNA. Dev Cell 6: 637–648 [DOI] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C (2003) The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell 112: 317–327 [DOI] [PubMed] [Google Scholar]