Unlike animals, plants are not able to escape from adverse circumstances. To cope with external stresses, plants modulate the endomembrane systems, especially the endoplasmic reticulum (ER), which is the most flexible and adaptable organelle (Staehelin, 1997). Alternatively, plants generate a specialized compartment from the ER to accumulate an enormous amount of proteins that are actively synthesized on the ER (Chrispeels and Herman, 2000). Active protein synthesis can be regarded as an internal stress. In response to the internal and external stresses, the ER differentiates into various types of compartments, each of which has its own responsibility.
The diversity of ER-derived compartments has been getting increased attention in plant cell biology. The ER networks of membranous tubules and cisternae are extended throughout the cytosol area surrounding the vacuoles. The ER membrane, which occupies nearly one-half the total area of membrane in cells, can be a source of a variety of compartments. Various ER-derived compartments with sizes ranging from 0.1 μm to 10 μm have been identified in plant cells (Larkins and Hurkman, 1978; Li et al., 1993; Hara-Nishimura et al., 1998; Schmid et al., 1998; Toyooka et al., 2000; Hayashi et al., 2001). All the compartments that have been characterized share three common features: (1) The ER-derived compartments accumulate a large amount of a single protein or only a few different proteins. (2) The protein components do not act within the compartments, even if they are functional proteins such as enzymes. (3) The compartments appear in a specific tissue at a particular time of the life cycle of plants. These features imply that ER-derived compartments have distinct responsibilities.
To understand a physiological role of an ER-derived compartment, it is necessary to identify the protein or proteins that accumulate in the compartment. The ER-derived compartments are classified into two types according to their contents: storage-protein type and hydrolytic-enzyme type. The former type compartments accumulate seed storage proteins in large quantities. The latter type compartments accumulate a hydrolytic enzyme, although they do not contain substrates for the respective enzyme. We are attempting to answer several questions. How are enormous amounts of storage proteins and hydrolytic enzymes accumulated in the compartments? How and where do the hydrolytic enzymes meet with their own substrate to fulfill their functions in the cells? How and when are the compartments formed from the ER? The aim of this Update is to try to give some answers to these questions. The mechanisms underlying the protein accumulation in the compartments and the formation of the compartments are discussed with the two types of ER-derived compartments.
PAC VESICLE: SELECTIVE UPTAKE OF STORAGE PROTEIN PRECURSORS
PAC (precursor-accumulating) vesicle is an ER-derived compartment that was found in maturing seeds of pumpkin (Cucurbita maxima) and castor bean (Ricinus communis; Hara-Nishimura et al., 1998). PAC vesicles accumulate precursors of storage proteins, 2S albumin and 11S globulin, to be transported to protein storage vacuoles. How do the vesicles accumulate the precursors? The precursor proteins form aggregates within the ER lumen, and the aggregates develop into PAC vesicles. PAC vesicles are responsible for an aggregate sorting of proteins to protein storage vacuoles in a Golgi-independent manner.
Recently, another way of Golgi-dependent uptake of the storage protein precursors into the PAC vesicles was discovered. A proteomic analysis of the isolated vesicles identified a type I integral membrane protein, PV72, on the membrane of the PAC vesicles (Shimada et al., 1997). PV72 was specifically and transiently accumulated in association with the synthesis of storage proteins, and has an ability to bind to the 2S albumin precursor via the C-terminal region including a vacuolar targeting signal (Shimada et al., 2002). PV72 has Ca2+-binding domains, and the binding of Ca2+ stabilizes the receptor-ligand complex even at pH 4.0. The association and dissociation of PV72 with the ligand is modulated by the Ca2+ concentration (EC50 = 40 μm) rather than the environmental pH (Watanabe et al., 2002). This indicates protein storage vacuoles that have an acidic interior pH and high Ca2+ concentrations are not an appropriate organelle to dissociate the precursor proteins from PV72. The questions are where PV72 traps the precursor molecules and where PV72 releases them. It has been shown that PV72 is localized not only on the PAC membrane but also on the Golgi complex (Shimada et al., 2002). PV72 might be recycled between PAC vesicles and the Golgi complex for recruiting the storage protein precursor molecules from the Golgi complex to the PAC vesicles.
The above biochemical and cell biological analyses suggest the PV72 acts as a vacuolar sorting receptor. For an in vivo demonstration, a reverse-genetic approach was used with an Arabidopsis (Arabidopsis thaliana) mutant lacking the homologous gene to PV72. Arabidopsis has seven homologs of PV72, which are designated AtVSR1 to AtVSR7 (Arabidopsis thaliana vacuolar sorting receptor). Only the atvsr1 mutant seeds abnormally accumulate the precursors of storage proteins, 2S albumin and 12S globulin, together with the mature forms of these proteins (Shimada et al., 2003). The atvsr1 mutant missorts storage proteins by secreting them from cells, resulting in an enlarged and electron-dense extracellular space in the seeds. The in planta analysis demonstrated that AtVSR1/PV72 is a sorting receptor that sorts storage proteins for protein storage vacuoles.
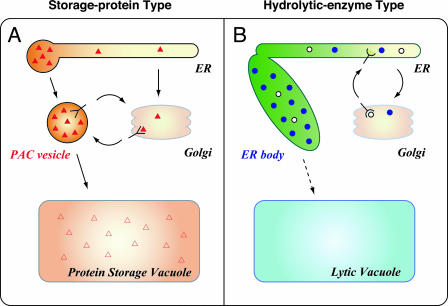
How does the sorting receptor function? Figure 1A shows a hypothetical model for the selective uptake of the storage protein precursors into the PAC vesicles. Most of the storage protein precursor molecules synthesized on the ER form an aggregate within the ER. However, inevitably, some molecules are not incorporated into the aggregates and remain free. The free molecules leave the ER for the Golgi complex, where they are trapped by the vacuolar sorting receptor and recruited to the PAC vesicles. After releasing the precursor molecules, the receptor is recycled to the Golgi complex.
Figure 1.
Schematic drawing of two types of ER-derived compartments in plant cells. Plant cells develop ER-derived compartments, which are classified into two types according to their contents: storage protein-type compartments (A) and hydrolytic enzyme-type compartments (B). PAC vesicles accumulate a large amount of storage protein precursors (solid triangles), while ER bodies accumulate a KDEL-tailed β-glycosidase, PYK10 (blue circles). Recycling receptors, vacuolar sorting receptor and KDEL receptor, are involved in the accumulation of the lumenal components in PAC vesicles and ER bodies, respectively. Open triangles, Mature form of storage proteins; open circles, ER resident proteins.
Selective uptake of the storage protein precursors into the PAC vesicles is conducted in two ways: one is aggregate sorting and the other is receptor-dependent sorting. The former sorting mechanism is advantageous to maturing seeds that actively synthesized a large quantity of storage proteins. The latter ensures proper delivery of the proteins by avoiding the missorting of the escaped molecules. This mechanism can be comparable to that for the recruitment of the escaped ER resident proteins by the KDEL receptor that is localized in the Golgi complex.
ER BODY: ACCUMULATION OF A KDEL-TAILED HYDROLYTIC ENZYME
Green fluorescent protein (GFP) from luminescent jellyfish allows us to visualize various organelles in living cells and in real time. A large ER-derived compartment, the ER body, is easily observed as bright fluorescing spindle-shaped bodies in Arabidopsis cotyledon cells that express an ER-targeted GFP, which is composed of a signal peptide and GFP followed by a tetrapeptide sequence, K(H)DEL (Lys/His-Asp-Glu-Leu) (Haseloff et al., 1997; Ridge et al., 1999; Hawes et al., 2001; Hayashi et al., 2001).
The shape and size of ER bodies (approximately 1 μm diameter × approximately 10 μm long) are completely different from two other hydrolytic enzyme-type compartments: KDEL vesicles (Φ = 0.2–0.5 μm) in Vigna mungo cotyledons (Toyooka et al., 2000) and ricinosomes (Φ = 0.2–0.5 μm) in castor bean endosperm (Schmid et al., 1998). Despite these differences, ER bodies, KDEL vesicles, and ricinosomes share an accumulation of a hydrolytic enzyme with an ER-retention signal (KDEL) at the C terminus. ER bodies accumulate a KDEL-tailed β-glucosidase (PYK10) as a major component (Matsushima et al., 2003), while both KDEL vesicles and ricinosomes accumulate a KDEL-tailed Cys proteinase (Schmid et al., 1998; Toyooka et al., 2000).
The question raised is whether the KDEL-tailed protein PYK10 is selectively transported and accumulated in ER bodies. KDEL and HDEL are typical ER retention signals that are found at the C terminus of the ER resident proteins. K(H)DEL-tailed protein molecules that leave the ER for the Golgi complex are recycled to the ER by the action of a KDEL receptor (Fig. 1B). In a similar manner, GFP-HDEL is also accumulated in the ER, resulting in the formation of bright fluorescent ER bodies in the transgenic Arabidopsis seedlings. Is PYK10 also accumulated in the ER bodies by this mechanism? Comparison of the subcellular distributions of GFP-HDEL and PYK10 shows that PYK10 is more concentrated in ER bodies than the ER, while GFP-HDEL is less concentrated in the ER bodies than in the ER (Matsushima et al., 2003).
What is the mechanism responsible for the selective concentration of PYK10 in the ER bodies? One possibility is that PYK10 mRNA is segregated on the ER membrane in a similar manner as discussed by Okita and colleagues in this Focus Issue (Crofts et al., 2004). This is supported by the presence of ribosomes on the membrane of ER bodies. Another possibility is that PYK10, which is abundant in the seedlings, contributes to the formation of the ER bodies by forming aggregations in the ER cisternae. β-Glucosidase aggregations in plant cells have been reported (Nisius, 1988; Fieldes and Gerhardt, 1994; Kim et al., 2000). The ER bodies also contain two stress-inducible proteinases, RD21 and γVPE, which have neither KDEL nor HDEL at their C termini. These proteinases may be trapped into the ER bodies during the aggregate formation of the abundant β-glucosidase. A similar phenomenon is also found in the storage protein-type compartment. Vacuolar processing enzyme is found in the PAC vesicles in developing seeds of castor bean (Hiraiwa et al., 1993).
INDUCTION OF DIVERSE ER-DERIVED COMPARTMENTS
ER bodies have a limited distribution in Arabidopsis seedlings. They appear in the epidermal cells of cotyledons and hypocotyls after seed germination, and disappear during senescence of the tissues. On the other hand, the rosette leaves have no ER bodies at all under normal conditions. However, once rosette leaves are wounded, many ER bodies are induced around the wound site of the leaves (Matsushima et al., 2002). The induction of ER bodies in response to an environmental stress demonstrates the flexibility of the ER of plants.
The induction of ER bodies is under hormonal control (Matsushima et al., 2002). ER bodies are induced throughout the epidermal cells of the rosette leaves treated with methyl jasmonate (MeJA), which mediates plant defenses against mechanical wounding and chewing by insects. On the contrary, ethylene suppresses the effect of MeJA on the induction of ER bodies in rosette leaves. The induction of ER bodies is strictly coupled with the signal transduction of MeJA and ethylene, both of which have an antagonistic effect on it.
What mechanism is involved in differentiation of the ER of plants? Recently, another factor that regulates the formation of the ER bodies was identified by a genetic approach. We isolated an Arabidopsis mutant, nai1, whose seedlings hardly have any ER bodies. The NAI1 gene encodes a transcription factor that has a basic-helix-loop-helix domain, and transient expression of NAI1 induced ER bodies in the nai1-1 mutant (Matsushima et al., 2004). Proteomic analysis showed that nai1 has no PYK10. The absence of ER bodies in nai1 mutants is due to the loss of a transcription factor (NAI1) that functions upstream of a major component (PYK10) of the compartment. The transcription factor regulates the formation of the ER bodies. A similar transcription factor responsible for the formation of ER-derived compartments has been identified in maize (Zea mays). Opaque-2 (O2), a basic Leu zipper-type transcription factor, regulates the expression of α-zein, a major storage protein of maize, which is accumulated in protein bodies derived from the ER (Hartings et al., 1989). The protein bodies of the o2 mutant seeds are much smaller (Φ = 0.1–0.3 μm) than normal protein bodies (Φ = 1–2 μm). This implies that both types of ER-derived compartments share a similar mechanism for their formation.
The ER bodies are inducible in nai1-1 rosette leaves that have been treated with MeJA, although they exhibit irregular shapes (Matsushima et al., 2004). MeJA and ethylene have an antagonistic effect on their induction in the mutant leaves. MeJA treatment induces the expression of NAI1 and PYK10 genes, and this induction is suppressed in the presence of ethylene. The expression pattern of NAI1 and PYK10 genes are parallel to the induction pattern of the ER bodies. MeJA-induced NAI1 is needed for the proper formation of ER bodies.
The question is whether the nature of the protein components determines the shapes of the ER-derived compartments, such as the large spindle shape of ER bodies and the small globe shape of KDEL vesicles. A recent study of the overexpression of KDEL-tailed proteinase answers this question. As described above, a hydrolytic enzyme-type compartment, the KDEL vesicle, accumulates a KDEL-tailed Cys proteinase in V. mungo (Toyooka et al., 2000). Overexpression of the proteinase in Arabidopsis plants induces the formation of an ER-derived compartment, which is a spindle-shaped structure with a characteristic fibrous pattern inside (Okamoto et al., 2003). Interestingly, the induced compartments are more similar to the ER bodies than to KDEL vesicles. This indicates that a different protein has an ability to form ER body-like architectures in Arabidopsis plants. There should be some relationship between the ER body and the other hydrolytic-type compartment.
The storage protein-type compartment is also inducible in transgenic Arabidopsis plants by overexpression of a chimeric protein of 2S albumin subunit and a selectable marker enzyme, phosphinothricin acetyltransferase (Hayashi et al., 1999). Despite the limited distribution of PAC vesicles in maturing seeds, the transgenic Arabidopsis plants induce PAC vesicles in the vegetative organs, rosette leaves, hypocotyls, and roots. Aggregates of the expressed proteins may trigger the formation of PAC vesicles.
PHYSIOLOGICAL FUNCTION OF ER BODY
ER-derived compartments have their own specialized function. The physiological function of each compartment can be estimated by identifying the protein components and the temporal and spatial development. The ER bodies are abundant in the epidermal cells of cotyledons and hypocotyls of seedlings and root cells of Arabidopsis. These cells are most sensitive to environmental stresses. This implies that the development of ER bodies is linked to environmental stresses. The induction of ER bodies in the rosette leaves might also have a defense function against chewing insects and may be related in some way to other wound stresses. The development and induction of ER bodies is a plant strategy that assists the cells under stressed conditions.
To clarify the ER body-mediated defense system at the molecular level, it is necessary to identify the natural substrate of β-glucosidase PYK10. β-Glucosidases acts against pathogens by producing toxic compounds from the substrates. PYK10 and the substrate should be stored separately within the cells. PYK10 in the ER bodies will come into contact with its natural substrates to produce the toxic compounds, when plant tissues are injured or stressed.
The physiological function of β-glucosidases depends on the nature of the aglycon moiety of its natural substrates, such as glucosinolates for thioglucosidases and cyanoglucosides for cyanogenic glucosidases. For identification of the substrate of PYK10, the nai1 mutant can be an ideal tool. The natural substrates of PYK10 will be identified by comparing the chemical components in wild-type plants with those in the nai1 plants. Further analysis of the susceptibility of nai1 to herbivore insects or pathogens will provide new insight into the ER body-mediated plant defense system.
Why do ER bodies develop in seedlings but not in rosette leaves? The reason may be related to the economy of costs of nutrients and energy. ER bodies are not essential for the fundamental process of plant life, considering the normal growth of nai1 plants. Therefore, putting off the synthesis of PYK10 and the formation of ER bodies in rosette leaves until the need arises may save costs. On the other hand, cotyledons, which are most sensitive to wounding and chewing by insects, may pay the costs for the defense until rosette leaves develop.
FUTURE PERSPECTIVES
The ER is a subcellular factory producing almost all the proteins and lipids for the ER itself, the Golgi complex, endosomes, prevacuolar compartments, vacuoles, lysosomes, secretory vesicles, and the plasma membrane, which are connected by both forward and backward vesicle traffic to constitute the endomembrane system. The molecular mechanisms underlying the endomembrane system in plants, animals, and yeast share common components. ER-derived compartments are closely related to the endomembrane system. However, most of the compartments are induced in specific organs in response to the internal and external signals. The induction of ER-derived compartments is controlled in a sophisticated way by the conditions under which plants grow. Thus, the regulation mechanism should be unique to plants and directly concerned with the life processes of higher plants.
Although ER-derived compartments vary in their structures and functions, some molecular mechanisms underlying their development and formation might be shared. Proteomic analyses of isolated compartments should identify key factors playing in the plant system. Isolation of mutants deficient in the formation of the compartments is another useful approach. For example, transgenic Arabidopsis that induces PAC vesicles by overexpressing a chimeric protein of the 2S albumin subunit followed by a selectable marker enzyme (described above) are still sensitive to the respective antibiotics. A mutant resistant to the antibiotics should have a defect in the formation of PAC vesicles. DNA array analysis with the nai1 mutant should also identify downstream genes of NAI1. Identification of such key factors will provide a valuable insight into the induction of the ER-derived compartments and the strategy that higher plants have evolved.
This work was supported by CREST of the Japan Science and Technology Corporation, and Grants-in-Aid for Scientific Research (no. 16085203) and for 21st Century COE Research Kyoto University (A14) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Chrispeels MJ, Herman EM (2000) Endoplasmic reticulum-derived compartments function in storage and as mediators of vacuolar remodeling via a new type of organelle, precursor protease vesicles. Plant Physiol 123: 1227–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofts AJ, Washida H, Okita TW, Ogawa M, Kumamaru T, Satoh H (2004) Targeting of proteins to endoplasmic reticulum-derived compartments in plants. The importance of RNA localization. Plant Physiol 136: 3414–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fieldes MA, Gerhardt KE (1994) An examination of the β-glucosidase (linamarase) banding pattern in flax seedlings using Ferguson plots and sodium dodecyl sulphate-polyacrylamide gel electrophoresis. Electrophoresis 15: 654–661 [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I, Shimada T, Hatano K, Takeuchi Y, Nishimura M (1998) Transport of storage proteins to protein-storage vacuoles is mediated by large precursor-accumulating vesicles. Plant Cell 10: 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartings H, Maddaloni M, Lazzaroni N, Di Fonzo N, Motto M, Salamini F, Thompson R (1989) The O2 gene which regulates zein deposition in maize endosperm encodes a protein with structural homologies to transcriptional activators. EMBO J 8: 2795–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes C, Saint-Jore C, Martin B, Zheng H-Q (2001) ER confirmed as the location of mystery organelles in Arabidopsis plants expressing GFP. Trends Plant Sci 6: 245–246 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Hara-Nishimura I, Nishimura M (1999) Accumulation of a fusion protein containing 2S albumin induces novel vesicles in vegetative cells of Arabidopsis. Plant Cell Physiol 40: 263–272 [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Yamada K, Shimada T, Matsushima R, Nishizawa NK, Nishimura M, Hara-Nishimura I (2001) A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol 42: 894–899 [DOI] [PubMed] [Google Scholar]
- Hiraiwa N, Takeuchi Y, Nishimura M, Hara-Nishimura I (1993) A vacuolar processing enzyme in maturing and germinating seeds: its distribution and associated changes during development. Plant Cell Physiol 34: 1197–1204 [Google Scholar]
- Kim Y-W, Kang K-S, Kim S-Y, Kim I-S (2000) Formation of fibrillar multimers of oat β-glucosidase isoenzymes is mediated by the As-Glu1 monomer. J Mol Biol 303: 831–842 [DOI] [PubMed] [Google Scholar]
- Larkins BA, Hurkman WJ (1978) Synthesis and deposition of protein bodies of maize endosperm. Plant Physiol 62: 256–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wu Y, Zhang D-Z, Gillikin JW, Boston RS, Franceschi VR, Okita TW (1993) Rice prolamine protein body biogenesis: a BiP-mediated process. Science 262: 1054–1056 [DOI] [PubMed] [Google Scholar]
- Matsushima R, Fukao Y, Nishimura M, Hara-Nishimura I (2004) NAI1 gene that encodes a basic-helix-loop-helix-type putative transcription factor that regulates the formation of a novel ER-derived structure, the ER body. Plant Cell 16: 1536–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R, Hayashi Y, Kondo M, Shimada T, Nishimura M, Hara-Nishimura I (2002) An endoplasmic reticulum-derived structure that is induced under stress conditions in Arabidopsis. Plant Physiol 130: 1807–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R, Kondo M, Nishimura M, Hara-Nishimura I (2003) A novel ER-derived compartment, the ER body, selectively accumulates a β-glucosidase with an ER retention signal in Arabidopsis. Plant J 33: 493–502 [DOI] [PubMed] [Google Scholar]
- Nisius A (1988) The stromacentre in Avena plastids: an aggregation of β-glucosidase responsible for the activation of oat-leaf saponins. Planta 173: 474–481 [DOI] [PubMed] [Google Scholar]
- Okamoto T, Shimada T, Hara-Nishimura I, Nishimura M, Minamikawa T (2003) C-terminal KDEL sequence of a KDEL-tailed cysteine proteinase (sulfhydryl-endopeptidase) is involved in formation of KDEL vesicle and in efficient vacuolar transport of sulfhydryl-endopeptidase. Plant Physiol 132: 1892–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge RW, Uozumi Y, Plazinski J, Hurley UA, Williamson RE (1999) Developmental transitions and dynamics of the cortical ER of Arabidopsis cells seen with green fluorescent protein. Plant Cell Physiol 40: 1253–1261 [DOI] [PubMed] [Google Scholar]
- Schmid M, Simpson D, Kalousek F, Gietl C (1998) A cysteine endopeptidase with a C-terminal KDEL motif isolated from castor bean endosperm is a marker enzyme for the ricinosome, a putative lytic compartment. Planta 206: 466–475 [DOI] [PubMed] [Google Scholar]
- Shimada T, Fuji K, Tamura K, Kondo M, Nishimura M, Hara-Nishimura I (2003) Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 16095–16100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Kuroyanagi M, Nishimura M, Hara-Nishimura I (1997) A pumpkin 72-kDa membrane protein of precursor accumulating vesicles has characteristics of a vacuolar sorting receptor. Plant Cell Physiol 38: 1414–1420 [DOI] [PubMed] [Google Scholar]
- Shimada T, Watanabe E, Tamura K, Hayashi Y, Nishimura M, Hara-Nishimura I (2002) A vacuolar sorting receptor PV72 on the membrane of vesicles that accumulate precursors of seed storage proteins (PAC vesicles). Plant Cell Physiol 43: 1086–1095 [DOI] [PubMed] [Google Scholar]
- Staehelin LA (1997) The plant ER: a dynamic organelle composed of a large number of discrete functional domains. Plant J 11: 1151–1165 [DOI] [PubMed] [Google Scholar]
- Toyooka K, Okamoto T, Minamikawa T (2000) Mass transport of proform of a KDEL-tailed cysteine proteinase (SH-EP) to protein storage vacuoles by endoplasmic reticulum-derived vesicle is involved in protein mobilization in germinating seeds. J Cell Biol 148: 453–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe E, Shimada T, Kuroyanagi M, Nishimura M, Hara-Nishimura I (2002) Calcium-mediated association of a putative vacuolar sorting receptor PV72 with a propeptide of 2S albumin. J Biol Chem 277: 8708–8715 [DOI] [PubMed] [Google Scholar]