Abstract
The NB mitochondrial genome found in most fertile varieties of commercial maize (Zea mays subsp. mays) was sequenced. The 569,630-bp genome maps as a circle containing 58 identified genes encoding 33 known proteins, 3 ribosomal RNAs, and 21 tRNAs that recognize 14 amino acids. Among the 22 group II introns identified, 7 are trans-spliced. There are 121 open reading frames (ORFs) of at least 300 bp, only 3 of which exist in the mitochondrial genome of rice (Oryza sativa). In total, the identified mitochondrial genes, pseudogenes, ORFs, and cis-spliced introns extend over 127,555 bp (22.39%) of the genome. Integrated plastid DNA accounts for an additional 25,281 bp (4.44%) of the mitochondrial DNA, and phylogenetic analyses raise the possibility that copy correction with DNA from the plastid is an ongoing process. Although the genome contains six pairs of large repeats that cover 17.35% of the genome, small repeats (20–500 bp) account for only 5.59%, and transposable element sequences are extremely rare. MultiPip alignments show that maize mitochondrial DNA has little sequence similarity with other plant mitochondrial genomes, including that of rice, outside of the known functional genes. After eliminating genes, introns, ORFs, and plastid-derived DNA, nearly three-fourths of the maize NB mitochondrial genome is still of unknown origin and function.
Mitochondrial genomes have been sequenced from a large number of protists, algae, fungi, and animals, but from few plants (for review, see Burger et al., 2003). To date, complete mitochondrial DNA (mtDNA) sequences from only five plants have been published: one nonvascular plant (Marchantia polymorpha; Oda et al., 1992), three eudicots (Arabidopsis [Arabidopsis thaliana]: Unseld et al., 1997; sugar beet [Beta vulgaris]: Kubo et al., 2000; rapeseed [Brassica napus]: Handa, 2003), and one monocot (rice [Oryza sativa]; Notsu et al., 2002). Plant mitochondria have a number of distinctive features, including considerable variation in genome size and organization, which can occur even within a single species (Fauron et al., 1995).
Although angiosperm mitochondrial genomes are at least 10 times larger than those of mammals, the total number of known genes they encode is fewer than twice as many as their mammalian counterparts. The mtDNAs of both plants and animals include genes for ribosomal RNAs, tRNAs, and several subunits of the oxidative phosphorylation complexes. The greatest difference is that some of the ribosomal proteins and some of the proteins involved in the biogenesis of cytochrome c are coded for by mtDNA in plants, whereas they are coded for by nuclear DNA in animals. In several angiosperm genera, two subunits of the succinate dehydrogenase complex are also coded for by mtDNA (Adams et al., 2001). Unlike their animal counterparts, a few of the plant mitochondrial tRNAs are encoded in the nucleus and imported into the mitochondrion (for review, see Maréchal-Drouard et al., 1993). Furthermore, some of the transcribed and functional mitochondrial tRNAs are originally of plastid origin, present on fragments of chloroplast DNA (ctDNA) that have become incorporated into the mtDNA. This movement of DNA between cellular compartments is responsible for some of the variation in the known gene sets of different plants and appears to be an ongoing evolutionary process in plants (for review, see Palmer et al., 2000). An additional curiosity is that, although plant mitochondrial genes are translated according to the universal code, transcripts of many genes require editing in order for that to occur (for review, see Brennicke et al., 1999).
One inference from the small number of sequenced plant mitochondrial genomes is that their sizes vary independently of the number of functional genes. The mitochondrial genome of the liverwort, M. polymorpha, was reported to be 184 kb and to encode 66 identified genes, including ribosomal and tRNAs (Oda et al., 1992). The 367-kb Arabidopsis mitochondrial genome was reported to include only 59 identified genes (Unseld et al., 1997). Although 84 unidentified open reading frames (ORFs) larger than 100 codons were also reported, it is unknown if any of them are actually expressed (Marienfeld et al., 1999). The recently sequenced mitochondrial genome of rapeseed, a member of the same family as Arabidopsis, contains a nearly identical set of identified genes, despite being only 222 kb in length (Handa, 2003). Indeed, the only difference in protein-gene content between the Arabidopsis and rapeseed mitochondrial genomes is that rps14 is intact in rapeseed, whereas it is a pseudogene in Arabidopsis. The 369-kb sugar beet mitochondrial genome is similar in size to that of Arabidopsis and contains a similar set of genes (29 protein coding, 5 rRNA, and 25 tRNA; Kubo et al., 2000). Although the mitochondrial genome of rice is quite a bit larger at 491 kb, the number of functional genes is comparable (Notsu et al., 2002). The major variation among all plant mitochondrial genomes characterized so far is in the ribosomal protein and tRNA gene content.
Comparative analyses of mitochondrial genes have shown that, with rare exceptions (Palmer et al., 2000), the sequences of protein-coding genes are highly conserved. Indeed, the rates of nucleotide substitution in plant mitochondrial protein-coding genes are usually lower than those of chloroplast genes or of plant or animal nuclear genes (for review, see Palmer, 1990). However, comparisons among plant mitochondrial genomes show that the sequences that occur between genes can be highly variable. Although these intergenic regions can include retrotransposons of nuclear origin and integrated chloroplast sequences, approximately 50% of each of the previously sequenced angiosperm mtDNAs could not be found in the extant databases (Unseld et al., 1997; Marienfeld et al., 1999; Kubo et al., 2000; Handa, 2003). Furthermore, there are no obvious similarities among the mtDNA sequences of the eudicots (Arabidopsis, rapeseed, and sugar beet) in these “unknown” regions. Within cucurbits, where mitochondrial genome size variation is extreme, an expansion of short, dispersed repeats (SDRs) has been proposed to account for some of the size increase (Lilly and Havey, 2001).
It is not clear why plant mitochondrial genomes rearrange so readily, or how their genomes expand and contract over such short evolutionary times. Complete mitochondrial sequence data are needed for many more plants, including closely related taxa, to address the question of how rapid changes in their intergenic regions occur. Relationships among grasses have been extensively studied (e.g. Freeling, 2001), and the rice mitochondrial genome sequence has been published (Notsu et al., 2002). We now report the complete sequence and comparative analysis of a second monocot mitochondrial genome, the maize (Zea mays subsp. mays) NB cytotype, which is present in most commercial hybrid maize lines.
RESULTS AND DISCUSSION
Previous sequencing of plant mitochondrial genomes used cloned DNA: cosmids for M. polymorpha (Oda et al., 1992), rice (Notsu et al., 2002), and sugar beet (Kubo et al., 2000), bacterial artificial chromosomes for Arabidopsis (Unseld et al., 1997), and plasmids for rapeseed (Handa, 2003). Although a set of well-mapped cosmids exists for the maize NB mitochondrial genotype (Fauron and Havlik, 1988), a shotgun approach was used to sequence the genome to test the general applicability of this approach. Shotgun sequencing will be useful for plant mitochondrial genomes for which no prior mapping information is available. It is also a more cost-effective way by which to generate data in a short period of time.
Sequence Assembly and Genomic Architecture
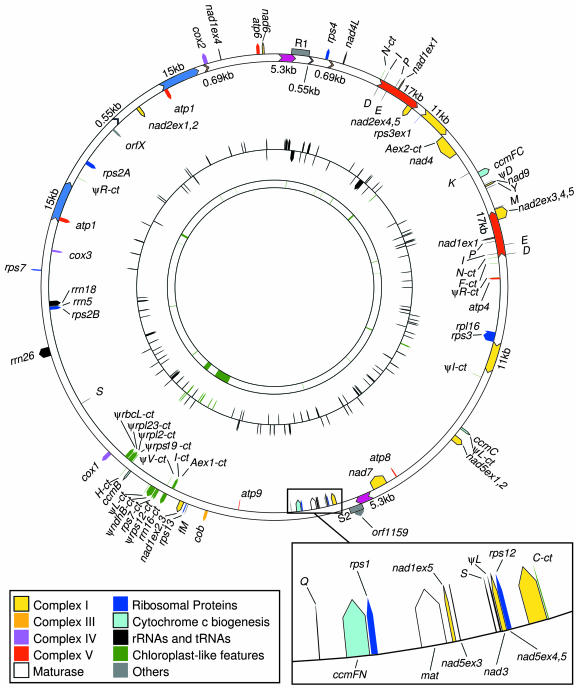
The final assembly of the maize NB mitochondrial sequences generated a single circular map of 569,630 bp (Fig. 1), larger than any of the previously sequenced plant mitochondrial genomes and remarkably similar to the estimates from early mapping studies (570 kb; Lonsdale et al., 1984; Fauron and Havlik, 1988). It should be noted that a circular map does not mean that the genome is actually composed of a circular molecule in vivo. Indeed, a number of studies suggest that there are alternative physical structures (Bendich, 1993; Backert et al., 1997; Senthilkumar and Narayanan, 1999). The NB mitochondrial genome has an average G + C content of 43.9%, which is comparable to other sequenced plant mitochondrial genomes (rice, 43.9%; sugar beet, 43.9%; Arabidopsis, 44.8%; and rapeseed, 45.2%).
Figure 1.
Circular map of the maize NB mitochondrial genome generated from sequence data. Known protein-coding, tRNA and rRNA genes, and gene fragments are shown on the outside circle. Colors indicate genes by function: Complex I (nad; yellow), Complex III (cob; orange), Complex IV (cox; purple), Complex V (atp; red), cytochrome assembly (ccm; light blue), ribosomal proteins (dark blue), maturase (white), other ORFs (gray), rRNA and tRNAs (black), and genes transferred from the chloroplast (green). Single-letter designations indicate tRNAs. Large repeats are color coded within the outer ring. Regions homologous to R1 and S2/R2 are indicated by gray blocks. The middle circle indicates positions of ORFs (≥99 amino acid predicted sizes). The inner ring shows regions of chloroplast homology with matches of at least 80% identity and lengths of at least 100 bp (green).
The actual NB mitochondrial genome complexity is 520 kb, which is calculated by removing one copy of each of the large (>500 bp) repeats from the 570-kb circle. Either of these numbers is still larger than any previously sequenced plant mitochondrial genome. In comparison, the rice mitochondrial genome has been reported to have an overall size of 490.5 kb (Notsu et al., 2002), and its genome complexity is estimated to be 359 kb. Interestingly, the number of nucleotides with matches in GenBank is almost identical in both species: 88,560 in maize and 88,173 in rice.
Protein-Coding Genes
The maize NB main mitochondrial genome contains 58 identified genes, including 34 genes coding for 33 known proteins (Table I). They include 22 proteins of the electron transport chain—nine subunits of complex I: NADH dehydrogenase subunits 1, 2, 3, 4, 4L, 5, 6, 7, and 9) (NAD1, 2, 3, 4, 4L, 5, 6, 7, and 9); one subunit of complex III: apocytochrome b (cob); three subunits of complex IV: cytochrome c oxidase subunits 1, 2, and 3 (COX 1, 2, and 3); five subunits of complex V: ATP synthase F1 subunits 1, 4, 6, 8, and 9 (ATP 1, 4, 6, 8, and 9); and four subunits of a complex involved in the biogenesis of cytochrome c: subunits B, C, and F (CCMB, C, FN, and FC). Also present is a gene formerly called orfX, now renamed mttB, that codes for a transporter protein and is reported to be homologous to the Escherichia coli tatC gene (Bonnard and Grienenberger, 1995; Bogsch et al., 1998), as well as a maturase gene, mat-r, which lies within intron 4 of nad1 (Wahleithner et al., 1990). This is very similar to the protein-coding content of other sequenced angiosperm genomes (Table I). Exons account for only 6.22% of the total genome. The nucleotide coordinates of all the genes, introns, and ORFs are listed in Supplemental Table I (available at www.plantphysiol.org).
Table I.
Gene content of the maize NB genome compared with other plant mitochondrial genomes
| Gene | Maize NB | Rice | Sugar Beet | Arabidopsis | M. polymorpha | |
|---|---|---|---|---|---|---|
| Complex I | nad1a | +, 2-exon 1 | + | + | + | + |
| nad2a | +, 2-exons 4,5 | +, 2-exons 3–5 | + | + | + | |
| nad3 | + | + | + | + | + | |
| nad4 | + | + | + | + | + | |
| nad4L | + | + | + | + | + | |
| nad5a | + | +, 2-exon 1 | + | + | + | |
| nad6 | + | + | + | + | + | |
| nad7 | + | + | + | + | Ψ | |
| nad9 | + | + | + | + | − | |
| Complex II | sdh2 | − | − | − | − | + |
| sdh3 | − | − | − | − | + | |
| sdh4 | − | − | − | Ψ | + | |
| Complex III | cob | + | + | + | + | +, Ψ |
| Complex IV | cox1 | + | + | + | + | + |
| cox2 | + | + | + | + | + | |
| cox3 | + | + | + | + | + | |
| Complex V | atp1 | 2 copies | + | + | + | + |
| atp4 (orf25) | + | + | + | + | − | |
| atp6 | + | + | + | 2 copies | + | |
| atp8 (orfB) | + | + | + | + | − | |
| atp9 | + | + | + | + | + | |
| Cytochrome c biogenesis | ccmB | + | + | + | + | + |
| ccmC | + | + | Ψ | + | + | |
| ccmFN | + | + | + | − | + | |
| ccm-FN1 | − | − | − | + | − | |
| ccm-FN2 | − | − | − | + | − | |
| ccmFC | + | + | + | + | − | |
| ccm-FC1 | − | − | − | − | + | |
| ccm-FC2 | − | − | − | − | + | |
| Ribosomal proteins | rps1 | + | + | − | − | + |
| rps2A | + | + | − | − | + | |
| rps2B | + | − | − | − | − | |
| rps3 | +, 2-exon 1 | + | + | + | + | |
| rps4 | + | + | + | + | + | |
| rps7 | + | + | + | + | + | |
| rps8 | − | − | − | − | + | |
| rps10 | − | − | − | − | + | |
| rps11 | − | Ψ | − | − | + | |
| rps12 | + | + | + | + | + | |
| rps13 | + | + | + | − | + | |
| rps14 | − | Ψ | − | Ψ | + | |
| rps19 | − | + | − | Ψ | + | |
| rpl2 | − | + | − | + | + | |
| rpl5 | − | + | + | + | + | |
| rpl6 | − | − | − | − | + | |
| rpl16 | + | + | − | + | + | |
| Other proteins | mat-r | + | + | + | + | − |
| mttB (orfX) | + | + | + | + | − | |
| tRNA-Ala | trnA | Ψct | − | − | − | mt |
| Arg | trnR | Ψmt, Ψct | Ψct | − | − | 3-mt |
| Asn | trnN | 2-ct | ct | ct | ct | mt |
| Asp | trnD | 2-mt, Ψmt | mt | ct | ct | mt |
| Cys | trnC | ct | ct, Ψmt | 2-mt, Ψmt | mt | mt |
| Glu | trnE | 2-mt | 2-mt | mt | mt | mt |
| Gln | trnQ | mt | 2-mt | mt | mt | mt |
| Gly | trnG | − | − | mt | mt | 2-mt |
| His | trnH | ct | 2-ct | ct | ct | mt |
| Ile | trnI | 2-mt, ct, Ψct | mt, Ψct | mt | mt | mt |
| Lys | trnK | mt | mt | mt | 2-mt | mt |
| Leu | trnL | Ψmt, 2-Ψct | − | − | − | 3-mt |
| Met | trnM | ct | mt, 2-ct | ct | 2-ct | mt |
| f Met | trnfM | mt | mt | 4-mt | mt | 2-mt |
| Phe | trnF | ct | 2-ct | mt | − | mt |
| Pro | trnP | 2-mt, Ψctb | 2-mt, 2-Ψct | mt, Ψct | mt, | mt |
| Ser | trnS | 2-mt | 2-mt, ct, Ψct | 2-mt, ct | 4-mt, ct | 2-mt |
| Thr | trnT | − | − | − | − | mt |
| Trp | trnW | ctb | 2-ct | ct | ct | mt |
| Tyr | trnY | mt | 2-mt | mt | 2-mt | 2-mt |
| Val | trnV | Ψct | Ψct | Ψct | − | mt |
| rRNA | rrn5 | + | + | + | + | + |
| rrn18 | + | + | + | + | + | |
| rrn26 | + | + | 3 copies | + | + |
Trans-spliced in all except M. polymorpha.
Present on the 2.3-kb linear plasmid. For protein genes, extra copies of exons that are present are listed after the +. +, Present; −, absent; Ψ, pseudogene.
Despite the higher genome complexity of the maize NB mitochondrial genome, it actually encodes two fewer proteins than rice (33 in maize versus 35 in rice; Table I). As has been previously noted, when plant mitochondrial genomes differ in gene content, it is usually in the number of ribosomal proteins present (for review, see Palmer et al., 2000). In the maize NB mitochondrial genome, there are 9 apparently functional genes for ribosomal proteins, compared with 11 in rice, 6 in sugar beet, 7 in Arabidopsis, and 16 in Marchantia (Table I). Maize mtDNA lacks rpl5 and rpl2, both of which are present in the rice and Arabidopsis mitochondrial genomes. It also lacks rps19, which is found in rice mtDNA. Mitochondrial rps2 genes have not been found in eudicot mtDNAs, but they are present in Marchantia and in monocot mtDNAs (Perrotta et al., 2002). There are two copies in the maize NB mitochondrial genome. Both are transcribed, although rps2A has much higher levels of mRNA and is thought to be the major functional form of the gene (Perrotta et al., 2002). All known monocot mitochondrial rps2 genes encode proteins that have long C-terminal extensions relative to their bacterial counterparts. As was shown by Perrotta et al. (2002), these extensions are not conserved in sequence between the two genes in maize.
The functional copies of mitochondrial rps11 and rps14 lie in the nucleus in both rice and maize, but the rice mitochondrial genome retains pseudogenes of each (Notsu et al., 2002). Of the sequenced plant mitochondrial genomes, only M. polymorpha possesses functional rps11 and rps14 genes. In fact, M. polymorpha has the largest number of ribosomal protein-coding genes (Table I) despite having the smallest sequenced plant mitochondrial genome (186 kb; Oda et al., 1992).
Ribosomal RNAs and tRNAs
Maize NB mtDNA has 3 ribosomal RNA genes (5S, 18S, and 26S) and 32 tRNA genes (Sangaré et al., 1990; Table I). One functional tRNA gene, trnW, is located on a 2.3-kb linear plasmid (Leon et al., 1989) and is not included in our submitted sequence of the main mitochondrial genome. Ten of the tRNA sequences have been classified as pseudogenes (Kumar et al., 1996; L. Maréchal-Drouard, personal communication). The 22 presumably expressed tRNA genes recognize 15 amino acids. The tRNAs encoded within rice mtDNA recognize the same 15 amino acids.
Although plant mitochondria use the universal genetic code and require tRNAs for all 20 amino acids, which mitochondrial tRNAs are actually encoded by the mtDNAs of plants is quite variable (Table I). Functional tRNA genes for six amino acids—Ala, Arg, Gly, Leu, Thr, and Val—are missing from the NB mtDNA. Since all of them are required for protein synthesis in mitochondria, they are presumably encoded by the nuclear genome and imported from the cytosol into the mitochondria (see Maréchal-Drouard et al., 1993; Dietrich et al., 1996). In addition, some of the functional tRNA genes in plant mitochondrial genomes are of chloroplast origin. Overall, 10 of the 22 functional tRNA genes encoded within the maize NB mtDNA are of chloroplast origin, including the plasmid-localized trnW. In comparison, of the 25 functional tRNA genes in the rice mitochondrial genome (including 7 in duplications), 11 are of chloroplast origin (Notsu et al., 2002).
Twenty-one of the expressed tRNAs display a classic cloverleaf structure, whereas each of the two tRNA-Sers (tRNA-SerGCU and tRNA-SerUGA) fold into an unusual five-loop secondary structure. Five of the tRNA genes (trnD, trnN, trnI, trnQ, and trnP) are present in duplicate because they are located within a 17-kb repeated sequence. Posttranscriptional modification from C to U within the anticodon sequence is necessary to generate a functional tRNA-Ile, which is similar to the situation reported in potato mitochondria (Weber et al., 1990).
Introns
A total of 22 identifiable group II introns are present within 8 of the protein-coding genes, including 7 trans-spliced introns that are part of nad1, nad2, and nad5. Fifteen cis-spliced introns are located in cox2, nad1, nad2, nad4, nad5, nad7, ccmFC, and rps3. The numbers and locations of introns are almost identical to those in the other sequenced angiosperm mitochondrial genomes. The only differences found are that sugar beet lacks the rps3 intron and the second intron of nad4 (Kubo et al., 2000). No group I introns have been identified in the NB mitochondrial genome.
The functional mitochondrial rRNA and tRNA genes of the sequenced angiosperms lack introns, but insertions are found within four of the tRNA pseudogenes in the maize NB mtDNA. The mitochondrial ψ-tRNA-Leu gene contains an intron (relative to the functional tRNA-Leu of liverwort mtDNA). Two plastid-derived ψ-tRNA-Leu genes also appear to contain small insertions adjacent to the anticodon site. A plastid-derived ψ-tRNA-Ala has two exons located 250 kb apart in the maize NB genome (Aex1 and Aex2 in Fig. 1). Presumably, they were transferred together with their intron and were later separated by mitochondrial recombination processes because the plastid sequences flanking the exons are also present. If this gene were to be expressed, trans-splicing would have to occur.
ORFs
An ORF was defined as an in-frame sequence 300 bp or longer that is bounded by a start and a stop codon, with no match to a coding sequence in the public databases. This definition excludes smaller proteins and does not indicate whether the sequence is expressed. There are 121 mitochondrial ORFs, most of which are unique to maize, and 7 plastid-derived ORFs (Supplemental Table I). None of the maize mtDNA ORFs are maintained among all sequenced higher-plant mtDNAs; only one (orf140-b) is found in Arabidopsis and three (orf99-a, orf140-b, and orf146-a) are found in rice. Compared with maize, orf99-a is the same size in rice, orf146-a is slightly shorter in rice, and orf140-b is shorter in rice and slightly longer in Arabidopsis. Ten of the maize ORFs are found within cis-spliced introns, nine of them within nad2. A well-known intron-located gene in plants is the maturase-related mat-r gene (Wahleithner et al., 1990), which resides within an intron of nad1 (Fig. 1; Table I).
Only two ORFs (orf186 and orf127) have been found to be truly chimeric in the NB mitochondrial genome of maize. A short segment (79 nucleotides) derived from cox2 resides within orf186, whereas orf127 contains 209 bp from rps12. Some other genes or portions of genes are duplicated because they are present within repeated DNA. The atp1 gene is present within the 15-kb repeat; nad1-exon1, a trans-spliced exon, lies within the 17-kb repeat (Fig. 1). In addition, exons from rps3 (exon 1) and nad2 (exons 4 and 5) are duplicated because their adjacent introns span the borders of the 11-kb and 17-kb repeats, respectively (Fig. 1).
In the NB mitochondrial genome, there are many more ORFs (≥300 nucleotides) than would be expected by chance (128 versus 1–7 in ten 600-kb, randomly generated genomes [see “Materials and Methods”]). However, for the vast majority of these ORFs, there is no evidence that they are expressed (Meyer, 2004).
Plastid DNA Insertions
The NB mitochondrial genome contains two large insertions of ctDNA, one of 12.6 kb (Stern and Lonsdale, 1982) and one of 4.1 kb (Lonsdale et al., 1983). There are multiple, smaller inserts, bringing the total amount of transferred ctDNA to about 23.9 kb (Table II). These sequences are derived primarily from the ctDNA inverted repeats (IR; Fig. 2A). Although some of the transferred tRNAs are functional, there is no evidence for the expression of the transferred protein-coding genes. Because some of the transferred segments are located in mtDNA repeats, the total ctDNA in the mitochondrial genome is about 25.3 kb, which represents 4.44% of the NB mitochondrial genome. The identified plastid-derived pieces range in size from 12,592 bp (the 12.6 kb segment) to 28 bp (Table II).
Table II.
Plastid-derived sequences found in the maize NB mitochondrial genome
| mtDNA Coordinates | Size | Plastid Coordinates | Features/Plastid Genes |
|---|---|---|---|
| bp | |||
| 10,079–10,513 | 435 | 42–486 | psbA] |
| 36,156–36,183 | 28 | 53–26 | Intergenic |
| 36,337–36,478 | 142 | 84,865–84,716 IR | Intergenic |
| 41,350–41,637 | 288 | 71,922–71,527 | [psbB] |
| 42,656–42,806 also 130,364–130,214 | 151 | 103,144–102,970 IR | trnN |
| 44,517–44,573 also 128,503–128,447 | 57 | 25,537–25,593 | [rpoC1] |
| 52,845–53,259 also 120,175–119,761 | 415 | 112,170–112,600 | Intergenic, ndhE] |
| 74,856–76,086 | 1,231 | 99,909–98,669 IR | [23S, trnA exon 2 |
| 83,346–83,391 | 46 | 93,134–93,177 IR | Intergenic |
| 92,531–92,591 | 61 | 39,340–39,392 | [psaB] |
| 95,560–95,633 | 74 | 78,787–78,721 | [rps8] |
| 100,986–101,341 | 356 | 51,666–52,094 | [ndhK, ndhC] |
| 105,417–105,478 | 62 | 97,981–98,047 IR | Intergenic |
| 132,426–133,125 | 700 | 50,129–49,391 | trnF, trnL exon 2 |
| 133,112–133,145 | 34 | 47,175–47,140 | [rps4 |
| 138,500–138,596 | 97 | 102,801–102,699 IR | [Pseudo-trnR |
| 144,818–144,888 | 71 | 36,200–36,268 | [atpF exon 2] |
| 179,605–180,470 | 866 | 84,868–85,750 IR | Pseudo-trnI, ORF241] |
| 201,498–201-584 | 87 | 84,956–84,880 IR | trnI |
| 212,531–212,674 | 144 | 81,992–82,141 | [rpl22] |
| 259,180–259,358 | 179 | 20,022–20,199 | trnC |
| 267,293–267,339 | 47 | 93,134–93,177 IR | Intergenic |
| 326,021–338,612 | 12,592 | 98,651–86,092 IR | trnA exon1, trnI, 16S, trnV, rps12 exons 2 & 3, rps7, ndhB, trnL |
| 346,550–348,769* | 2,220 | 82,476–84,710* IR | rps19, trnH, rpl2, rpl23 |
| 348,510–350,668* | 2,159 | 58,820–56,669* | Pseudo rpl23, rbcL |
| 364,096–364-573 | 478 | 24,989–25,432 | [rpoC1] |
| 374,462–374,512 | 51 | 103,944–103,994 IR | Intergenic |
| 410,255–410,386 | 132 | 70,562–70,701 | [psbB] |
| 430,342–430,381 | 40 | 99,255–99,216 IR | [23S] |
| 462,498–462,578 also 531,435–531,515 | 81 | 56,246–56,327 | Intergenic |
| 466,326–466,411 also 535,263–535,348 | 86 | 39,781–39,700 | [psaB] |
| 473,375–474,825 | 1,451 | 102,900–101,365 IR | trnR, 5S, 4.5S, 23S] |
| 520,792–520,961 | 170 | 92,546–92,713 | Intergenic |
| 521,721–521,786 | 66 | 58,666–58,605 or 84,607–84,668** | Pseudo-rpl23 or rpl23 |
| 564,791–564,825 | 35 | 102,767–102,801 | [trnR] |
Maize plastid sequences found in maize NB mitochondrial DNA. Sequences present in two locations in the mtDNA are parts of large repeats. IR indicates plastid inverted repeat sequence; coordinates are given only for the first IR. Brackets indicate truncation at the 5′ or 3′ end of genes. *, Part of 4.1-kb chimeric region in mtDNA (see text). **, Plastid sequences are identical, so the exact progenitor is uncertain.
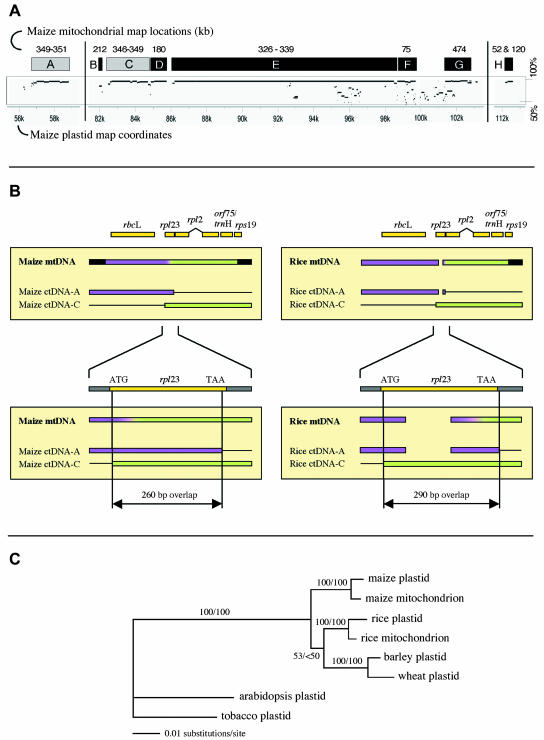
Figure 2.
Three regions of plastid DNA found in the mitochondrial genome. A, Locations of plastid sequences in the plastid and mitochondrial genomes. Lower axis is plastid genome coordinates; the plastid IR extends from 82 kb to 105 kb. Boxes indicate regions of ctDNA present in the NB mtDNA; numbers above them are their coordinates (kb) in the mitochondrial genome. Lighter boxes are the components of the 4.1-kb region. Below the boxes are the MultiPip representations of sequence similarity between the maize plastid genome and the maize mitochondrial genome. Vertical scale is 50% to 100%. B, Alignment of sequences relevant to the 4.1-kb segment of ctDNA present in the maize and rice mitochondrial genomes. Purple and green colored boxes indicate regions A and C in section A that recombined to form the 4.1-kb segment. Lower alignment is an enlargement of the region of overlap between the A and C regions, showing their relationship to rpl23. Purple-to-green gradients indicate areas where recombination occurred, but where definitive assignment to A or C is not possible. C, Parsimony phylogenetic tree for the 4.1-kb plastid sequence in the mitochondrion. Depicted branch lengths for Arabidopsis and tobacco are substantially shorter than their actual lengths because highly divergent and thus unalignable sequences were deleted from the data set. Parsimony/maximum likelihood bootstrap values for 100 replicates are indicated on branches.
Detailed analyses revealed that the 12.6-kb segment appears to be part of a much larger portion of the IR and adjacent DNA that has come to reside in the mtDNA (Fig. 2A). The transferred region would have extended approximately 21 kb from plastid coordinate 82 to 103 kb, with the additional sequences located on either side of the segment originally reported by Stern and Lonsdale (1982). None of the segments derived from this 21-kb region overlap. A single transfer is suspected because (1) essentially the entire region is present, and (2) when the homologous segments from this region that are located throughout the mitochondrial genome are aligned with their plastid counterparts, there are only small gaps (5–300 bp) between them (Fig. 2A). Had multiple transfers occurred, it is highly unlikely that several nonoverlapping but closely adjoined segments would have been transferred. The fragmentation of the transferred DNA presumably took place subsequent to its incorporation into mtDNA via the same mechanisms that are responsible for other mtDNA rearrangements.
The transferred ctDNA segments are present as single copies, except for those that occur in the NB mtDNA large repeats. Of the 18 segments larger than 100 bp, only 3 are also present in rice. One of them, a 550-bp portion of rpoC1 (RNA polymerase subunit C1 from maize plastid coordinate 25 kb), is part of a 7-kb plastid segment in the rice mitochondrial genome. The other two fragments together constitute a single 4.1-kb region in the mitochondrial DNA, as described below.
The 4.1-kb (4,098 bp) region of plastid origin is located between maize mitochondrial coordinates 346.5 kb and 350.5 kb, and is a composite of two separate regions of ctDNA. This segment includes homologs of the plastid genes rbcL, rpl23, rpl2, orf75, trnH, and rps19 (Fig. 2B). In the plastid genome, rpl23 through rps19 are located in the IR (segment C in Fig. 2A), whereas rbcL and an rpl23 pseudogene are located outside of the IR in the large single-copy region (segment A in Fig. 2A). The joining of these two sequences appears to have been the result of recombination between the 260 bp that is common to both the rpl23 pseudogene and the IR rpl23.
The rice mitochondrial genome contains a similar composite region at a sequence homologous to the maize insertion site (Fig. 2B, top). The right end of the plastid insertion ends at the same point within rps19 in both maize and rice. At the other (rbcL) end, the rice plastid-derived sequence extends an additional 3.3 kb upstream. Rice and maize also have different points of recombination within the overlapping region in rpl23 (Fig. 2B, bottom). The fact that the same terminus and the same insertion point, along with the unusual organization of the 4.1-kb region, occur in both maize and rice mtDNA suggests that this transfer of plastid DNA occurred before the divergence of rice and maize. However, if this hypothesis were correct, the plastid-derived insert in maize mtDNA would be expected to be more similar to the plastid-derived insert in rice mtDNA than to its own plastid sequence. In fact, the opposite is observed; the plastid-derived insert in maize mtDNA is more similar to maize plastid DNA, as is shown by phylogenetic analyses of the concatenated 4.1-kb region (Fig. 2C). The finding that the maize 4.1-kb plastid-derived mitochondrial insert groups with maize ctDNA, and that the similar rice plastid-derived mitochondrial insert groups with rice ctDNA, held true for distance, maximum parsimony, and maximum likelihood methods, as well as in analyses using components of each sequence separately, e.g. the rbcL or the IR segment and individual genes (data not shown). Similar results were obtained for the mtDNA copy of rbcL by Cummings et al. (2003). This result is not consistent with an ancient origin and implies that the plastid-derived mitochondrial sequences in both species were transferred after the divergence of rice and maize.
If the plastid DNA transfer did indeed occur after the divergence of maize and rice, one must account for the facts that (1) the IR plastid segment has the same right end breakpoint and insertion point, and (2) both segments of the insertion appear to be roughly contemporary in both species. This would require that, when the IR segments from the plastid genomes of maize and rice inserted into the maize and rice mitochondrial genomes, recombination occurred using the same plastid and mitochondrial sequences, yielding identical products at the rps19 end. This would have been followed in each species by recombination with the rbcL-containing segment within their regions of homology within rpl23. If both events in both species occurred well after the divergence of the maize and rice lineages, the sequences would yield the phylogenetic results obtained here and by Cummings et al. (2003).
An alternative explanation for the phylogenetic results is that the chimeric, plastid-derived region was generated within the mitochondrial genome prior to the divergence of rice and maize, but that copy correction is occurring; i.e. there is an ongoing process in which DNA from the plastid recombines with homologous, plastid-derived DNA in the mitochondrion. Thus, the presence and organization of the 4.1-kb region would be old, but the plastid sequences comprising it would be recently acquired. In the copy-correction scenario, the transfer of plastid DNA into the mitochondrion could be a relatively common event in which the transferred DNA integrates only rarely, but more freely participates in recombination with existing mtDNA sequences of plastid origin. The recombinogenic nature of plant mitochondrial genomes would assist in this process, as would the proclivity of plant mitochondria to take up exogenous DNA.
The six largest plastid-derived regions within maize NB mtDNA are found to be at least 96% identical to their plastid counterparts, which provides further evidence for recent uptake of plastid sequences by the mitochondrial genome. Indeed, the 12.6-kb region is 99.8% identical over the nucleotides common to the mitochondrial and plastid genomes. Although this type of data has been considered evidence of their recent acquisition (Cummings et al., 2003), we suggest that dating their acquisition by sequence similarity is fraught with uncertainty, especially if copy-correction is occurring.
Repeated Sequences
A variety of repetitive DNA motifs occur in the maize NB mitochondrial genome. There are five pairs of large directly repeated sequences, with repeat lengths of 14,936; 11,092; 5,270; 719; and 543 bp, as well as one large IR of 16,870 bp (Fig. 1). Overall, the large repeats account for 17.35% of the genome. The 0.7-kb and 0.5-kb repeats are small enough that our sequencing strategy allowed an individual determination of each repeat sequence. One copy of the 0.7-kb repeat is slightly larger (725 bp) due to a 6-bp insertion.
Studies with cucurbit mtDNAs (Lilly and Havey, 2001) and Chlamydomonas reinhardtii ctDNA (Maul et al., 2002) suggested that SDRs could partially account for organelle genome expansions. Therefore, searches for any smaller sequences that were duplicated within the NB mitochondrial intergenic regions were performed. Using BLAST we located any sequence between 20 bp and 500 bp that was present more than once, beyond that already accounted for in the larger repeats. Using a 90% sequence similarity criterion, 5.59% of the NB genome was identified as smaller repeats (20–500 bp; Table III). As the minimum similarity requirement was decreased, the amount of the genome attributable to SDRs increased slightly, but even at 70% similarity the proportion of the genome that could be accounted for by smaller repeats was only 30% greater than at 90% similarity (Table IV). RepeatFinder (http://ser-loopp.tc.cornell.edu/cbsu/repeatfinder.htm), which does not allow user specification of similarity criteria, yielded results similar to our 90% similarity data.
Table III.
Repeated DNA sequences found in maize and rice
| Repeat Length
|
Maize
|
Rice
|
||||
|---|---|---|---|---|---|---|
| Number | Bases | Percent | Number | Bases | Percent | |
| 20–49 | 917 | 19,794 | 3.47 | 476 | 10,855 | 2.21 |
| 50–99 | 96 | 6,118 | 1.07 | 95 | 6,109 | 2.15 |
| 100–199 | 34 | 4,959 | 0.87 | 23 | 2,992 | 0.61 |
| 200–499 | 4 | 1,045 | 0.18 | 9 | 2,497 | 0.51 |
| 500–999 | 4 | 2,530 | 0.44 | 0 | 0 | 0.00 |
| 1,000–1,999 | 0 | 0 | 0.00 | 0 | 0 | 0.00 |
| 2,000–4,999 | 0 | 0 | 0.00 | 7 | 27,964 | 5.70 |
| 5,000–9,999 | 2 | 10,540 | 1.85 | 0 | 0 | 0.00 |
| 10,000–19,999 | 6 | 85,796 | 15.06 | 0 | 0 | 0.00 |
| 20,000+ | 0 | 0 | 0.00 | 6 | 231,332 | 47.16 |
| Total | 659 | 130,782 | 22.94 | 616 | 281,749 | 58.34 |
The numbers of repeats and the numbers of bases in each size class of repeats, in roughly log-incremental size classes. All copies of repeats are included. Percents of genomes are calculated from total genome sizes. Sequence-identity criterion is 90%.
Table IV.
Total nucleotides contained in SDRs at four minimum-similarity criteria
| Minimum Similarity
|
Maize
|
Rice
|
||
|---|---|---|---|---|
| Bases | Percent | Bases | Percent | |
| 100% | 11,624 | 2.04 | 7,804 | 1.59 |
| 90% | 31,843 | 5.59 | 19,519 | 3.98 |
| 80% | 39,306 | 6.90 | 22,748 | 4.64 |
| 70% | 41,371 | 7.26 | 24,155 | 4.92 |
Total nucleotides within the maize and rice genomes that are parts of SDRs (20–499 bp). Nucleotides that are parts of overlapping repeats are counted only once; thus the maize 90% number is slightly less than the sum of the maize components in Table III (31,916). Percent is percentage of total genome.
An SDR family was defined as sequences having at least 90% similarity and identical length. Similar families were grouped into superfamilies, as shown in Figure 3. For example, all four SDR families within superfamily 13 (Fig. 3A) have 22 bp in common, but the individual families, with two to four members each, have larger or smaller terminal extensions. Superfamily 2 (Fig. 3B) is an example of a simple sequence repeat superfamily, containing families that each have the sequence TACTA tandemly repeated four to five times. It is a large superfamily, having 13 families with as many as 17 members each and containing 91 members overall. It accounts for 2,011 bp of the NB genome. The largest group is superfamily 1 (Fig. 3C), with 30 SDR families that include 117 sequences and account for 3,797 bp of the NB genome. The families constitute a sliding sample across a “consensus” region (e.g. SDR274 in Fig. 3C), such that the repeats at the top of the alignment have no region of overlap with the sequences at the bottom of the alignment.
Figure 3.
Superfamilies of SDR elements. A, Elements in Superfamily 13 common to all 4 members of the superfamily are bold and underlined. Each line in Superfamily 13 is a family containing from 2 to 4 members (column 3) of length 25 to 36 bp (column 2). In the entire superfamily there are a total of 12 repeats covering 349 bp. B, The SDR families of Superfamily 2 are composed of members that themselves contain tandemly arranged pentanucleotide simple sequence repeat sequences, here alternately highlighted by bold underlining. C, Superfamily 1 possesses a sliding consensus, ultimately joining 30 families, and is the largest superfamily in the maize NB genome. Σ bp is the number of bp included in all copies of the repeats in each SDR family. All members of a given family are at least 90% identical in sequence.
There are 197 SDR families, of which 143 are grouped into 31 superfamilies. The remaining 54 families each contain 2 to 5 members. In both groups, 99% of the SDRs are 20 to 50 bp long, and 57% are 20 to 30 bp long (Table III). In a genome of the size of the maize NB mitochondrion, most of the observed repeats of less than 30 bp are probably the result of chance. Analyses of a set of 10 computer-generated 600-kb genomes (see “Materials and Methods”) having the same base composition as the maize NB genome showed that, of a total of 2,783 repeats, 2,321 (83.4%) were 20 to 24 bp and 405 (14.6%) were 25 to 29 bp. There were no repeats in the randomly generated genomes larger than 45 bp, and an average of only one of 40 to 44 bp and two of 35 to 39 bp. Thus, repeats longer than 40 bp within the NB mitochondrial genome are probably not the result of chance. Overall, the maize NB mitochondrial genome has an under-representation of SDRs of 20 to 50 bp, and an over-representation of larger repeats. Nonetheless, the total contribution of repeats of less than 500 bp in length to the NB genome is only 31,843 bp, or 5.59% (Table III).
Searches for transposable element sequences in the NB mitochondrial genome showed little evidence for this type of repetitive DNA. Using the Institute for Genomic Research (TIGR) grass transposable element database as a reference (http://www.tigr.org/tdb/e2k1/plant.repeats/index.shtml) and with a minimum match of 50 bp, only four small fragments (50–277 bp) with similarity to known retrotransposons were found (Supplemental Table II). Searching for IRs of 11 to 14 bp separated by 100 to 700 bp with FindMITE (Tu 2001; http://jaketu.biochem.vt.edu/) yielded 120 families of potential elements, none of which matched any of the known miniature IR transposable elements in grasses.
mtDNA within the Maize Nuclear Genome
An in silico search for the presence of NB mtDNA within the nuclear genome of the B73 inbred line was performed. The Assembled Zea mays Database (AZM; Whitelaw et al., 2003), consisting of assemblies of methyl-filtered and high-Cot sequences, was compared to the mitochondrial NB sequence, using methodologies to screen out possible mitochondrial contamination in the libraries (see “Materials and Methods”). Keeping in mind that the maize nuclear genome sequence is incomplete and the assembled contigs are relatively short, a rough estimate of the minimum amount of mitochondrial sequence that could be present in the nuclear genome was made. Alignment summaries categorized all of the matching sequences identified by BLAST searches either as probable contaminants or probable noncontaminants. There were 543 alignments, of which 72 aligned in a manner suggestive of being noncontaminants. These were further reduced to 64 robust alignments between the AZM nuclear contigs and the NB mitochondrial genome that could not be attributed to contamination. This set included 35,240 bp of the mitochondrial genome in a total of approximately 93 Mb assayed. The portion of the 2,500-Mb maize nuclear genome represented in the AZM contigs (which exclude most of the methylated or highly repetitive DNA) is estimated to be 413 Mb (Whitelaw et al., 2003). Assuming that the sampling was representative, there are approximately 155,000 bp of maize mitochondrial sequence within the portion of the maize nuclear genome captured by the methylation filtration and high-Cot methods.
To estimate the amount of mtDNA present in the total maize nuclear genome, the extent of mitochondrial sequence within a random, unfiltered B73 genome shotgun library (Whitelaw et al., 2003) was determined. Repeating the BLAST exercises described above on 32,955 assemblies from the shotgun sequences, 8 were found to contain mitochondrial sequences that are likely to reside within the nuclear genome. There were 2,758 bp of noncontaminant mitochondrial sequence within the 24 Mb of total DNA sampled, yielding an estimate of approximately 287,000 bp of mtDNA within the 2,500-Mb B73 maize nuclear genome. The copy number of the mitochondrial sequences was not determined.
The amount of mtDNA in the nuclear genome of maize inbred B73 appears to be at least 290 kb, which is a rough estimate. There may be mitochondrial sequences within the nuclear genome that are closely associated with repetitive sequences that would be excluded during high Cot selection, or methylated sequences that would be excluded from methyl-filtered libraries. In addition, the less than 1% random sampling of B73 sequences may not be representative of the nuclear genome as a whole.
The transfer of individual organelle genes to the nucleus is well documented (e.g. Palmer et al., 2000), as well as transfer of large stretches of organelle genomic DNA (see Martin, 2003). The tendency for nuclei to take up and integrate mtDNA (for review, see Richly and Leister, 2004) and for mitochondria to take up and to integrate ctDNA into their genomes, contrasts with the lack of evidence for incorporation of nuclear DNA into the maize mitochondrial genome.
Comparative Analyses of Angiosperm Mitochondrial Genomes
Comparisons of the angiosperm mitochondrial genomes were conducted using MultiPipMaker (Schwartz et al., 2000, 2003) with the maize NB mtDNA and its genes as the “reference” genome. The software computes alignments of similar regions in two or more DNA sequences and summarizes them as a percent identity plot. MultiPipMaker retains the linear order of the reference genome and aligns the other genomes with it, irrespective of the order that the sequences occur in those genomes. The percent identity (50%–100%) of each of the other genomes to the reference genome is indicated.
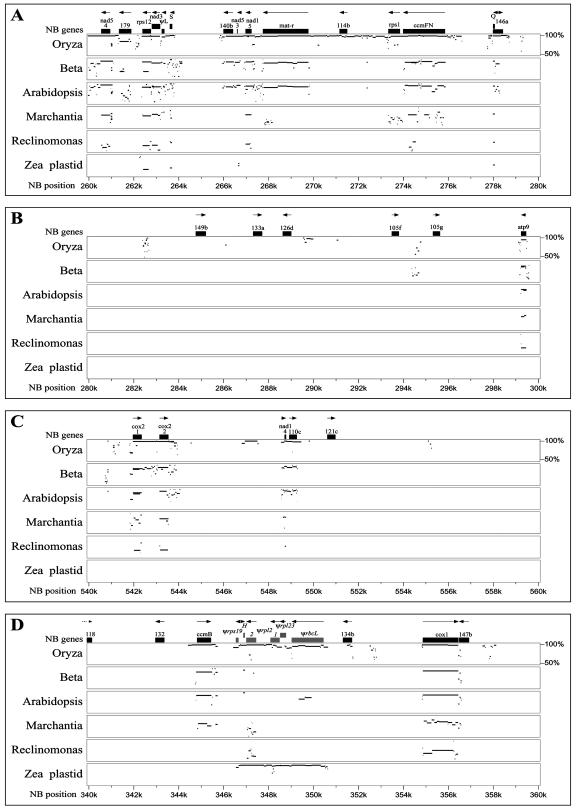
Although genes generally are not clustered in the NB mtDNA, there are regions relatively richer or poorer in known genes (Fig. 4, A versus B). The MultiPip alignments show that the similarities among the mtDNAs are associated mainly with known coding regions (Fig. 4). Indeed, coding-region conservation can be high even in comparisons with M. polymorpha and Reclinomonas americana, a protist containing the most bacteria-like mtDNA (Lang et al., 1997). The greater similarity of maize and rice mtDNAs can be seen readily in Figure 4. Intron sequences tend to be well conserved between maize and rice (e.g. cox2 intron; Fig. 4C), although these similarities disappear with increasing phylogenetic distances.
Figure 4.
MultiPipMaker analysis of several sequenced mitochondrial genomes. Maize NB is the reference genome to which the mitochondrial genomes of rice, sugar beet, Arabidopsis, M. polymorpha, and R. americana, and the plastid genome of maize (Maier et al., 1995) are compared. At the top of the figure, positions of NB genes are marked by black boxes, and their orientations are shown by arrows. A number directly below the gene designation indicates the exon number. ORFs are shown as numbers (for their predicted sizes), and tRNAs are indicated by their single-letter designations. The full MultiPipMaker analysis is included in supplemental materials (Supplemental Fig. 1). A, A relatively gene-rich region of the NB genome, showing conservation of mitochondrial genes and a 10-kb region of sequence similarity between maize NB and rice that includes noncoding DNA. B, A gene-poor region of NB mtDNA, showing very little similarity with other mitochondrial genomes. C, The intron and 3′ region of cox2 is conserved with rice but becomes less so with increasing phylogenetic distance. A region at 547 kb with no obvious feature is highly conserved between maize NB and rice. D, 4.1 kb of plastid-derived DNA present in both maize NB and rice mtDNAs; plastid-derived genes are italicized.
Of the 569,630-bp maize NB mitochondrial genome, 27.9% is at least 90% identical to rice mtDNA (minimum match of 20 contiguous bp). Conversely, 39.5% of the rice mitochondrial sequence is similar to maize NB mtDNA. Although the numbers of nucleotides in common are the same, the percentage is larger because the rice genome is smaller. When the stringency is decreased from 90% to 80%, the proportion of NB present in rice increases minimally, from 27.8% to 29.0% of the genome, and the proportion of rice present in NB increases from 39.5% to 41.0%. Decreasing the stringency from 80% to 70% does not increase the proportions further. Only 9.5% of the maize NB mtDNA is shared with Arabidopsis mtDNA and 13.8% of the Arabidopsis mitochondrial genome is shared with maize. Thus, even between two grasses, most of the mitochondrial sequences do not seem to have an identifiably common ancestry, and between a monocot and a dicot, all that the mitochondrial genomes seem to have in common are genes (Fig. 4).
Most of the regions identified in maize mtDNA as containing ORFs are not conserved among the other taxa (Fig. 4B). Still, some of the mtDNA without known genes, but encompassing several maize ORFs or intergenic regions, was found to be shared between maize and rice mtDNAs (Fig. 4A). However, except for orf99a, orf140b, and orf146, the sequences that were ORFs in maize were not ORFs in rice (e.g. Fig. 4B). A striking feature of the maize NB mitochondrial genome is that most of the intergenic regions are not conserved with mtDNA even from another grass, and, in fact, they showed no sequence similarity to any other known sequences.
The maize NB and rice mitochondrial genomes have approximately the same number of SDRs that are at least 50 bp long (Table III), repeats of a length that are unlikely to be present due to chance. This is despite the fact that the rice genome is 15% smaller than the maize NB genome and has a complexity that is only 77% that of NB.
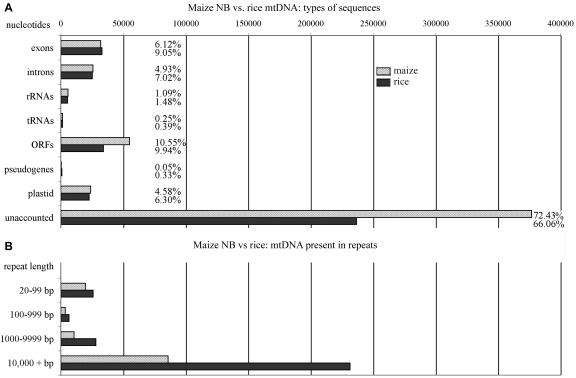
Because maize and rice are both grasses, separated by only about 50 million years of evolution (see Gaut, 2002), one might have expected a relatively high level of genome conservation between them. However, except for the known coding regions and a few small intergenic stretches, the rice mitochondrial sequences appear to be highly divergent from the maize sequences. This is similar to the comparison between the crucifers, Arabidopsis and rapeseed (Handa, 2003), which are separated by approximately 20 million years (Koch et al., 2001). The nonrepeated proportion that has no matches in the databases is similar in the maize and rice mitochondrial genomes (Fig. 5). Based on genome complexities (i.e. subtracting the repeats greater than 500 bp) and excluding known genes, pseudogenes, and plastid DNA but including ORFs, the enigmatic DNAs represent 83.0% and 76.0% of the maize and rice mitochondrial genomes, respectively.
Figure 5.
Composition of maize NB and rice mitochondrial genomes. A, Comparison of types of sequences. For each type (e.g. exons), the number of nucleotides accounted for is based upon genome complexity (520 kb for maize and 363 kb for rice). Percentage of each genome is also indicated. B, Amount of DNA present in repeats (90% identity criterion). The amount present in the total genome is shown for each species (570 kb for maize and 491 kb for rice).
Sequence Conservation between Two Fertile Genotypes of Maize
The other major mitochondrial genotype in male-fertile North American cultivated maize is termed NA. It was identified in the A188 inbred line, which is the cytoplasm present in most of the lines used to transform maize, and it has a larger, rearranged genome relative to NB (Fauron and Casper, 1994). The first 40 kb of the maize NA sequence was compared with the complete NB mtDNA sequence to make a preliminary estimate of the similarities between these two cytotypes.
NA was used as the reference genome in the MultiPip analysis shown in Figure 6. Of this 40 kb, only 26,080 bp are also present as long, homologous regions in NB, and they are found in four locations in the NB mtDNA. By design, the sequences for NA and NB begin at the same point, and the first 6,834 bp are shared (I in Fig. 6). There is a single nucleotide substitution and a single 10-bp gap in NB relative to NA. Following a 465-bp gap in NB relative to NA, segment II (6,065 bp) is present in an inverted orientation and includes 4 nucleotide substitutions. Segment II is composed of sequences homologous to the maize R1 plasmid (found as a free plasmid in two Latin American lines of maize; Weissinger et al., 1982). The next 12 kb of the NA mitochondrial genome is not present in the NB mtDNA (Fig. 6). The following homologous sequences, segments III and IV, are present at distant locations in the NB genome. Segment III (5,321 bp long) starts at coordinate 180,210 bp in the NB mtDNA. Segment IV (7,823 bp) starts at coordinate 326,030 bp in NB and is inverted relative to NA. It is only near the end of segment IV that mitochondrial genes are located: trnfM, rps13, and exon 3 of nad1.
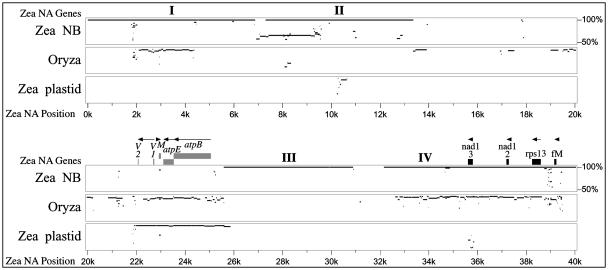
Figure 6.
MultiPip comparison of the first 40 kb of the maize NA mitochondrial genome with the complete mitochondrial genomes of maize NB and rice, as well as the maize plastid genome. Representations are as for Figure 4. I to IV identify large regions of homology between NA and NB.
In addition to the large segments of shared sequence, there are eight small regions of similarity (86, 69, 53, 40, 36, 30, 29, 25, and 24 bp) that are found at dispersed locations in the NB genome. The smallest are not depicted in Figure 6. An additional 36 small sequences (25–182 bp long, with at least 78% identity) that are present in segments I to IV are also repeated elsewhere in NB mtDNA.
The first 40 kb of the NA genome also contains three insertions of plastid DNA. The smallest, starting at 10,351 bp, is identical to that at the same position in NB. The second smallest constitutes the first 260 bp of segment III. The third plastid-derived segment is a 3.7-kb region that contains four genes: trnV, trnM, atpE, and atpB. Interestingly, this ctDNA insertion is not present in the NB mtDNA.
The rice mitochondrial genome also contains the 3.7-kb ctDNA insertion. Other than the plastid-derived sequence, approximately one-fourth of the rest of the first 40 kb of the NA mitochondrial genome is present in rice mtDNA. Segments II and III and much of segment I are absent from the rice mitochondrial genome. Most of segment IV, which contains three mitochondrial genes, is shared among NA, NB, and rice mtDNAs. Further comparisons are ongoing and will be reported elsewhere.
CONCLUSION
Although the maize mitochondrial genome is the largest that has been sequenced to date, the known gene space in maize is not larger than those of other sequenced plant mitochondrial genomes. In the 570-kb maize NB mitochondrial genome, identifiable mitochondrial exons comprise only 35.4 kb (6.22%) of the genome, and the cis introns account for 25.7 kb (4.51%). It is not yet known how much more of the genome is accounted for by trans-spliced introns, as well as by upstream and downstream regulatory regions, but it is unlikely to be a major fraction. Plastid-derived sequences comprise only 4.44%, and SDRs (20–49 bp) only 3.47% of the NB genome. Thus, compared to other sequenced plant mitochondrial genomes, the maize mitochondrial genome has even more DNA of undetermined origin.
The mechanisms for rapid mitochondrial genome rearrangement and expansion in plants remain enigmatic. Plant mitochondrial DNAs could be very interesting for evolutionary studies, because they are composed of two components: (1) slowly evolving DNA that includes coding regions and (2) noncoding DNA of obscure origin. Sequencing and comparing mtDNAs from a large number of closely related taxa should assist in understanding the evolution of plant mitochondrial genomes.
MATERIALS AND METHODS
mtDNA Preparation
Seeds from normal, male-fertile maize (Zea mays) inbred B37N were obtained from Pioneer-Hi-Bred (Des Moines, IA). Mitochondria were prepared by differential centrifugation, and mtDNA was purified on CsCl gradients (Fauron et al., 1987).
Library Construction
To generate random fragments, the mtDNA was processed in a French press (Schriefer et al., 1990) at 150 psi, achieved by placing the cell at 56 cm with 4.573 kg of weight applied at 92 cm on the lever arm to fractionate the DNA. Linkers (Invitrogen, Carlsbad, CA; BstXI/XhoI) were added to each end of the repaired fragments according to manufacturer's directions. Using gel electrophoresis, two fractions were selected, one of 3.5 to 4.5 kb and one of 4.5 to 6.5 kb. Successive gel electrophoresis removed excess linkers. The fragments were retrieved from the matrix by degrading the agar, followed by phenol extraction. The DNA was concentrated using ethanol precipitation.
The DNA fragments were cloned into plasmid vector pOTMI (Lander et al., 2001). The vector was linearized with BstXI and the 1.7-kb vector fragment was gel purified to eliminate the SacB stuffer fragment. The vector fragment was dephosphorylated and purified by preparative gel electrophoresis. Ligation was performed according to manufacturer's directions (New England Biolabs, Beverly, MA), and the Escherichia coli host, DH10B-T1 phage-resistant, was transformed with the chimeric plasmid DNA. Transformed cells were plated for production using chloramphenicol and isopropylthio-β-galactoside selection.
Template Preparation
DNA template was purified on a Packard Bioscience DNATrak (CCS Packard, Torrence, CA) robot using a magnetic bead preparation (Hawkins et al., 1997). Briefly, paramagnetic carboxylated microspheres (Seradyn, Indianapolis) were prepared manually by washing four times in buffer, using a Dynal (Oslo) MPC-1 magnet to collect the beads before the wash buffer was decanted. The washed beads were resuspended in dilution buffer and kept on ice for immediate use, or stored at 4°C for up to 4 d. The thawed E. coli cells in 96-well flat-bottom inoculation plates were manually resuspended in buffer, and the plates were vortexed on a platform shaker for at least 12 min to resuspend the cell pellets. The plates of resuspended cells were placed on the DNATrak robot, and all further steps through the washes were accomplished robotically. Fully resuspended beads in dilution buffer were added to a cocktail of detergent, lysis buffer, and polyethylene glycol and were dispensed into plates containing the resuspended bacterial pellets. The plates were vortexed for 10 min and moved to magnet plates for 12 min. Supernatant from the lysis plates was transferred to round-bottom microtiter plates (purify plates), with care being taken to avoid transferring any beads. Clean beads were added to the purify plates, and the plates were placed on ring magnets for 10 min. The beads were washed four times using a wash solution dispensed by the robot. Any excess wash solution was decanted, and the plates were dried in a plate dryer produced at the Genome Sequencing Center (GSC) as a modification of a Hydra 96 robot (Robbins Scientific, Sunnyvale, CA) for 30 s at 110°C. When completely dry, the plates were stored at room temperature for up to 4 weeks.
DNA was eluted from the paramagnetic beads with double-distilled water. The plates were placed on ring magnets during DNA transfer. Plates were sealed with foil tape and stored at −20°C.
Sequencing Reactions
ABI DyeTerminator (Sunnyvale, CA) reactions were used for sequencing. The reactions were performed in a 384-well format and were assembled using a BiomekFX robot. Thermocycling reaction times were as previously reported (Lander et al., 2001).
Production Work Flow
After the library was constructed, an initial batch of 96 clones was purified and sequenced to assess library quality, after which other clones from the library were placed in the sequencing queue. From all templates processed, 80% were used for assembly, after low quality reads had been removed by the ASP script in use at the GSC. Chromatograms were processed using Phred (Ewing and Green, 1998; Ewing et al., 1998). The Staden Package vector-clipping program (Staden, 1996; Wendl et al., 1998) was used to screen out the vector.
Finishing
The maize NB mtDNA was sequenced by the whole genome shotgun method to a coverage depth of 22×. Following sequence assembly by Phrap, the database was viewed in Consed (Gordon et al., 1998). No sequencing reactions were required to close gaps or resolve low consensus quality. However, the project was misassembled because of the many exact duplications throughout the genome. Only a single copy of each duplication was represented in the initial Phrap assembly, since cross-matching was unable to distinguish reads from individual copies. Duplicated regions in the Phrap assembly usually had twice the average number of reads, and every region with above-average read depth was inspected as a potential region of duplication. Plasmid forward and reverse primer pairs were also used to identify misassemblies indicated by incorrect pair orientation. Duplications were detected by identifying contig ends in Miropeats (Parsons, 1995) with similarities to contiguous regions in the assembly. Two opposing contig ends with similarities to a contiguous region might be spanned by duplication of the intervening sequence. Replication of the intervening sequence to join contigs was not done unless corroborated by read depth, plasmid pairs, and Miropeats information. Assemblies across duplicated regions were further validated by amplification across duplications using PCR. Finally, in silico digests of the finished assembly were compared to the gene map of Fauron et al. (1995) to verify that the two assemblies were congruent.
Annotation
The primary database used for annotation was AceDB (http://www.acedb.org/). ORFs were initially identified using Artemis (Rutherford et al., 2000; http://www.sanger.ac.uk/Software/Artemis). The ORFs were used to query nonredundant databases using BLAST similarity searches, applying a cutoff of 70% sequence identity over at least 80% of the ORF length. To find putative orthologs in other completed genomes, predicted proteins were searched for in the COG database (Tatusov et al., 2001), in the protein family database Pfam (Bateman et al., 2002; http://pfam.wustl.edu), and in the integrated view of the signature databases Interpro (http://www.ebi.ac.uk/interpro). Additionally, to help define genes, ORF-Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), BLASTN, BLASTX (Altschul et al., 1990), and tRNAscan-SE (Lowe and Eddy, 1997; http://www.genetics.wustl.edu/eddy/tRNAscan-SE/) were used. To improve the accuracy of the identifications, the NB mtDNA annotation was compared with other plant mitochondrial genome annotations, and all differences in coding predictions were reassessed, based upon choice of start codon, length of conservation in plant mtDNA, and presence of identifiable motifs. In addition to known genes, ORFs of at least 300 nt (≥99 amino acids) were included.
Data Representation
The circular representation of the genome was drawn using DOCIRCLE (M. Gibson and C. Fauron, unpublished data), a program that draws circular DNA maps with genome features taken from text files. It is written in C++ and uses Embedded JavaScript (SpiderMonkey, http://www.mozilla.org/js/spidermonkey/) configuration files to specify how the figure is drawn.
Alignments were generated using MultiPipMaker, a web-based tool for genomic sequence alignments (Schwartz et al., 2000, 2003; http://bio.cse.psu.edu/pipmaker). The annotated maize NB mtDNA genomic sequence was used as the reference genome and compared with mtDNA sequences from rice (AB076665, AB076666), sugar beet (Beta vulgaris; NC002511), Arabidopsis (Arabidopsis thaliana; NC001284), Marchantia polymorpha (NC001660), and Reclinomonas americana (NC001823), as well as maize ctDNA (NC001666).
Phylogenetic Analyses
Sequences were initially aligned using Pileup (GCG), ClustalW (Thompson et al., 1994), or both, and then manually optimized. Phylogenetic analyses were performed with PAUP 4.0b10 (Swofford, 2002) using the default settings, except that in maximum likelihood analyses empirical base frequencies were used. Bootstraps used 100 replicates.
Analysis of Repeats
To search for transposable elements, the TIGR transposable element database was used in a BLASTN search of the NB genome. A minimum length of 15 bp and 90% sequence identity was required for assignment of a match. MITEs were searched for using the program FindMITE (Tu, 2001; http://jaketu.biochem.vt.edu). The requirements were 10 to 14 bp IRs with specific trinucleotide termini flanking 100 to 700 bp of nonspecific intervening DNA.
SDRs were defined as sequences of at least 20 bp but less than 500 bp that are present more than once in the NB genome, are at least 90% identical in sequence, and are of exactly the same length. The NB genome was searched against itself using National Center for Biotechnology Information (NCBI) BLASTALL. Using RepeatExtractor (http://zeamtdna.missouri.edu/nlsap-gui.htm), an in-house script, all self-matches were removed (i.e. the query against its exact self), as were the 12 matches of the 6 known repeats of greater than 500 bp. BLAST lists reverse complements as separate elements, so all found repeats were compared, and reverse complement matches were combined if they were unique, or one of them was eliminated if they were redundant. BLAST sometimes erroneously finds small repeats only among large repeats, so large repeats were masked. However, repeats that are present exclusively inside a large repeat were counted, as well as repeats that are present both outside and inside a large repeat. For example, a repeat with one copy outside a large repeat and one inside would be counted three times. Other parameters were as noted in the text. The SDRs were then grouped into superfamilies using an in-house script based on BLASTN and aligned with Pileup (GCG).
Ten random genomes were generated whose lengths were approximately the same as that of maize NB (600 kb) and whose overall nucleotide composition matched NB. The pseudo-genomes were strings of nucleotides generated using probabilities weighted to the proportions of the nucleotides in the NB genome, and verified in the resulting product. These genomes were subjected to the same SDR analyses as the NB genome itself.
Search for mtDNA within the Maize Nuclear Genome
B73 nuclear sequences available from TIGR as assemblies of methyl-filtered and high-Cot sequences (AZM release 2.0; http://www.tigr.org/tdb/tgi/maize/; Whitelaw et al., 2003) were used. To rule out the presence of mitochondrial sequence contamination in the both libraries, three sets of data were produced. First, a set of BLAST alignment summaries was made where at least one good high-scoring pair (HSP) existed for the AZM versus the mitochondrial sequence. All AZM contigs that had at least one alignment to a mitochondrial sequence that extended across the length of the AZM contig were treated as potential contaminants. Alignments that contained both AZM-contig and mtDNA sequences were treated as noncontaminants. Second, to account for instances where two HSPs were very close to each another, e.g. separated by a few bases and thereby degrading the alignment sufficiently to miss a call of a single, long alignment, a final manual inspection was performed. All such HSPs were removed, because these could have been falsely identified as noncontaminants. The remaining assemblies contained HSPs indicating robust alignments between a portion of the AZM contig and the mitochondrial sequence. Most of these are likely to represent cases of mitochondrial sequence within the nuclear genome. False results could still be generated where there is a misassembly of the maize nuclear genome reads. For example, an assembly of reads that represent mitochondrial contamination along with reads that are truly from the nuclear genome cannot be distinguished from true instances of mtDNA present within the nuclear genome.
This is likely to yield an underestimate because there may be mitochondrial sequences within the nuclear genome that are closely associated with repetitive sequences that would be excluded during high-Cot selection, or methylated sequences that would be excluded from methyl-filtered libraries.
The complete sequence described in this study, Zea mays ssp. mays cytotype NB mtDNA, is GenBank accession number AY506529. The partial sequence described in this study, Zea mays ssp. mays cytotype NA mtDNA, is GenBank accession number AY705912.
Supplementary Material
Acknowledgments
We thank John Spieth, Brandi Chiapelli, Warren Gish, William Nash, Jill Cifrese, and Leah Westgate for their contributions to this project. We thank Laurence Maréchal-Drouard, Jeffrey Palmer, David Stern, and Pankaj Jaiswal for helpful comments on the data and Makedonka Mitreva for help with final formatting and submission. We are grateful to our educational partners at Truman State University, Diane Janick-Buckner, Brent Buckner, and their students, particularly Anup Parikh, for input to the project.
This work was supported by the National Science Foundation Plant Genome Research Program (grant no. DBI–0110168).
The online version of this article contains Web-only data.
References
- Adams KL, Rosenbleuth M, Qiu YL, Palmer JD (2001) Multiple losses and transfers to the nucleus of two mitochondrial succinate dehydrogenase genes during angiosperm evolution. Genetics 158: 1289–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Backert S, Nielsen BL, Börner T (1997) The mystery of the rings: structure and replication of mitochondrial genomes from higher plants. Trends Plant Sci 2: 477–484 [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL (2002) The Pfam protein families database. Nucleic Acids Res 30: 276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendich AJ (1993) Reaching for the ring: the study of mitochondrial genome structure. Curr Genet 24: 279–290 [DOI] [PubMed] [Google Scholar]
- Bogsch EG, Sargent F, Stanley NR, Berks BC, Robinson C, Palmer T (1998) An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J Biol Chem 273: 18003–18006 [DOI] [PubMed] [Google Scholar]
- Bonnard G, Grienenberger JM (1995) A gene proposed to encode a transmembrane domain of an ABC transporter is expressed in wheat mitochondria. Mol Gen Genet 246: 91–99 [DOI] [PubMed] [Google Scholar]
- Brennicke A, Zabaleta E, Dombrowski S, Hoffmann M, Binder S (1999) Transcription signals of mitochondrial and nuclear genes for mitochondrial proteins in dicot plants. J Hered 90: 345–350 [DOI] [PubMed] [Google Scholar]
- Burger G, Gray MW, Lang BF (2003) Mitochondrial genomes: anything goes. Trends Genet 19: 709–716 [DOI] [PubMed] [Google Scholar]
- Cummings MP, Nugent JM, Olmstead RG, Palmer JD (2003) Phylogenetic analysis reveals five independent transfers of the chloroplast gene rbcL to the mitochondrial genome in angiosperms. Curr Genet 43: 131–138 [DOI] [PubMed] [Google Scholar]
- Dietrich A, Small I, Cosset A, Weil JH, Maréchal-Drouard L (1996) Editing and import: strategies for providing plant mitochondria with a complete set of functional transfer RNAs. Biochimie 78: 518–529 [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8: 186–194 [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8: 175–185 [DOI] [PubMed] [Google Scholar]
- Fauron C, Casper M, Gao Y, Moore B (1995) The maize mitochondrial genome: dynamic, yet functional. Trends Genet 11: 228–235 [DOI] [PubMed] [Google Scholar]
- Fauron CM, Casper M (1994) A second type of normal maize mitochondrial genome: an evolutionary link. Genetics 137: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauron CM-R, Abbott AG, Brettell RIS, Gesteland RF (1987) Maize mitochondrial DNA rearrangements between the normal type, the Texas male sterile cytoplasm, and a fertile revertant CMS-T regenerated plant. Curr Genet 11: 339–346 [Google Scholar]
- Fauron CM-R, Havlik M (1988) The BamHI/XhoI, SmaI restriction maps of the normal maize mitochondrial genotype B37. Nucleic Acids Res 16: 10395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M (2001) Grasses as a single genetic system: reassessment 2001. Plant Physiol 125: 1191–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS (2002) Evolutionary dynamics of grass genomes. New Phytol 154: 15–28 [Google Scholar]
- Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8: 195–202 [DOI] [PubMed] [Google Scholar]
- Handa H (2003) The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res 31: 5907–5916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins TL, McKernan KJ, Jacotot LB, MacKenzie JB, Richardson PM, Lander ES (1997) A magnetic attraction to high-throughput genomics. Science 276: 1887–1889 [DOI] [PubMed] [Google Scholar]
- Koch M, Haubold B, Mitchell-Olds T (2001) Molecular systematics of the Brassicaceae: evidence from coding plastidic matK and nuclear Chs sequences. Am J Bot 88: 534–544 [PubMed] [Google Scholar]
- Kubo T, Nishizawa S, Sugawara A, Itchoda N, Estiati A, Mikami T (2000) The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA(Cys)(GCA). Nucleic Acids Res 28: 2571–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Marechal-Drouard L, Akama K, Small I (1996) Striking differences in mitochondrial tRNA import between different plant species. Mol Gen Genet 252: 404–411 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Lang BF, Burger G, O'Kelly CJ, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Gray MW (1997) An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 387: 493–497 [DOI] [PubMed] [Google Scholar]
- Leon P, Walbot V, Bedinger P (1989) Molecular analysis of the linear 2.3 kb plasmid of maize mitochondria: apparent capture of tRNA genes. Nucleic Acids Res 17: 4089–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly JW, Havey MJ (2001) Small, repetitive DNAs contribute significantly to the expanded mitochondrial genome of cucumber. Genetics 159: 317–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale DM, Hodge TP, Fauron CM (1984) The physical map and organisation of the mitochondrial genome from the fertile cytoplasm of maize. Nucleic Acids Res 12: 9249–9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale DM, Hodge TP, Howe CJ, Stern DB (1983) Maize mitochondrial DNA contains a sequence homologous to the ribulose-1,5-bisphosphate carboxylase large subunit gene of chloroplast DNA. Cell 34: 1007–1014 [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier RM, Neckermann K, Igloi GL, Kössel H (1995) Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Biol 251: 614–628 [DOI] [PubMed] [Google Scholar]
- Maréchal-Drouard L, Weil JH, Dietrich A (1993) Transfer RNAs and transfer RNA genes in plants. Annu Rev Plant Physiol Plant Mol Biol 44: 13–32 [Google Scholar]
- Marienfeld JR, Unseld M, Brennicke A (1999) The mitochondrial genome of Arabidopsis is composed of both native and immigrant information. Trends Plant Sci 4: 495–502 [DOI] [PubMed] [Google Scholar]
- Martin W (2003) Gene transfer from organelles to the nucleus: frequent and in big chunks. Proc Natl Acad Sci USA 100: 8612–8614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul JE, Lilly JW, Cui L, dePamphilis CW, Miller W, Harris EH, Stern DB (2002) The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell 14: 2659–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer LJ (2004) Tissue-specific ORF and gene expression analysis in maize mitochondria. MS thesis. University of Missouri, Columbia, MO
- Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, Nakazono M, Hirai A, Kadowaki K (2002) The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet Genomics 268: 434–445 [DOI] [PubMed] [Google Scholar]
- Oda K, Yamato K, Ohta E, Nakamura Y, Takemura M, Nozato N, Akashi K, Kanegae T, Ogura Y, Kohchi T, et al (1992) Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA. A primitive form of plant mitochondrial genome. J Mol Biol 223: 1–7 [DOI] [PubMed] [Google Scholar]
- Palmer JD (1990) Contrasting modes and tempos of genome evolution in land plant organelles. Trends Genet 6: 115–120 [DOI] [PubMed] [Google Scholar]
- Palmer JD, Adams KL, Cho Y, Parkinson CL, Qiu YL, Song K (2000) Dynamic evolution of plant mitochondrial genomes: mobile genes and introns and highly variable mutation rates. Proc Natl Acad Sci USA 97: 6960–6966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JD (1995) Miropeats: graphical DNA sequence comparisons. Comput Appl Biosci 11: 615–619 [DOI] [PubMed] [Google Scholar]
- Perrotta G, Grienenberger JM, Gualberto JM (2002) Plant mitochondrial rps2 genes code for proteins with a C-terminal extension that is processed. Plant Mol Biol 50: 523–533 [DOI] [PubMed] [Google Scholar]
- Richly E, Leister D (2004) NUMTs in sequenced eukaryotic genomes. Mol Biol Evol 21: 1081–1084 [DOI] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B (2000) Artemis: sequence visualization and annotation. Bioinformatics 16: 944–945 [DOI] [PubMed] [Google Scholar]
- Sangaré A, Weil JH, Grienenberger JM, Fauron C, Lonsdale D (1990) Localization and organization of tRNA genes on the mitochondrial genomes of fertile and male sterile lines of maize. Mol Gen Genet 223: 224–232 [DOI] [PubMed] [Google Scholar]
- Schriefer LA, Gebauer BK, Qui LQ, Waterston RH, Wilson RK (1990) Low pressure DNA shearing: a method for random DNA sequence analysis. Nucleic Acids Res 18: 7455–7456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Elnitski L, Li M, Weirauch M, Riemer C, Smit A, Green ED, Hardison RC, Miller W (2003) MultiPipMaker and supporting tools: alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res 31: 3518–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W (2000) PipMaker: a web server for aligning two genomic DNA sequences. Genome Res 10: 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthilkumar P, Narayanan KK (1999) Analysis of rice mitochondrial genome organization using pulsed-field gel electrophoresis. J Biosci 24: 215–222 [Google Scholar]
- Staden R (1996) The Staden sequence analysis package. Mol Biotechnol 5: 233–241 [DOI] [PubMed] [Google Scholar]
- Stern DB, Lonsdale DM (1982) Mitochondrial and chloroplast genomes of maize have a 12-kilobase DNA sequence in common. Nature 299: 698–702 [DOI] [PubMed] [Google Scholar]
- Swofford D (2002) Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sinauer Associates, Sunderland, MA
- Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV (2001) The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 29: 22–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z (2001) Eight novel families of miniature inverted repeat transposable elements in the African malaria mosquito, Anopheles gambiae. Proc Natl Acad Sci USA 98: 1699–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unseld M, Marienfeld JR, Brandt P, Brennicke A (1997) The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet 15: 57–61 [DOI] [PubMed] [Google Scholar]
- Wahleithner JA, MacFarlane JL, Wolstenholme DR (1990) A sequence encoding a maturase-related protein in a group II intron of a plant mitochondrial nad1 gene. Proc Natl Acad Sci USA 87: 548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Dietrich A, Weil JH, Maréchal-Drouard L (1990) A potato mitochondrial isoleucine tRNA is coded for by a mitochondrial gene possessing a methionine anticodon. Nucleic Acids Res 18: 5027–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissinger AK, Timothy DH, Levings CS, Hu WWL, Goodman MM (1982) Unique plasmid-like mitochondrial DNAs from indigenous maize races of Latin America. Proc Natl Acad Sci USA 79: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendl MC, Dear S, Hodgson D, Hillier L (1998) Automated sequence preprocessing in a large-scale sequencing environment. Genome Res 8: 975–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw CA, Barbazuk WB, Pertea G, Chan AP, Cheung F, Lee Y, Zheng L, van Heeringen S, Karamycheva S, Bennetzen JL, et al (2003) Enrichment of gene-coding sequences in maize by genome filtration. Science 302: 2118–2120 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.