Abstract
Previous studies indicated that the deficiency of starch-branching enzyme (SBE) Ia in the single mutant sbe1a∷Mu (sbe1a) has no impact on endosperm starch structure, whereas the deficiency of SBEIIb in the ae mutant is well known to reduce the branching of starch. We hypothesized that in maize (Zea mays) endosperm, the function of SBEIIb is predominant to that of SBEIa, and SBEIa would have an observable effect only on amylopectin structure in the absence of SBEIIb. To test this hypothesis, the mutant sbe1a was introgressed into lines containing either wx (lacking the granule-bound starch synthase GBSSI) or ae wx (lacking both SBEIIb and GBSSI) in the W64A background. Both western blotting and zymogram analysis confirmed the SBEIa deficiency in sbe1a wx and sbe1a ae wx, and the SBEIIb deficiency in ae wx and sbe1a ae wx. Using zymogram analysis, no pleiotropic effects of sbe1a genes on SBEIIa, starch synthase, or starch-debranching enzyme isoforms were observed. High-performance size exclusion chromatography analysis shows that the chain-length profiles of amylopectin as well as β-limit dextrin were indistinguishable between wx and sbe1a wx, whereas significant differences for both were observed between ae wx and sbe1a ae wx, suggesting an effect of SBEIa on amylopectin biosynthesis that is observable only in the absence of SBEIIb. The amylopectin branch density and the average number of branches per cluster were both higher in endosperm starch from sbe1a ae wx than from ae wx. These results indicate possible functional interactions between SBE isoforms that may involve enzymatic inhibition. Both the cluster repeat distance and the distance between branch points on the short intracluster chains were similar for all genotypes however, suggesting a similar pattern of individual SBE isoforms in cluster initiation and the determination of branch point location.
Starch biosynthesis involves the activities of four enzymes: ADP-Glc pyrophosphorylase, starch synthase (SS), starch-branching enzyme (SBE), and starch-debranching enzyme (DBE; Smith et al., 1997). The coordinated functions of these enzymes result in two forms of starch molecules: amylose, an essentially linear glucan chain consisting of α-1,4 linkages, and amylopectin, a highly branched glucan chain with multiple branch points formed by α-1,6 linkages.
SBE catalyzes the formation of branch points within glucan chains by cleaving an α-1,4 linkage and reattaching the chain to a glucan chain via α-1,6 bond. Three isoforms of SBE (SBEIa, SBEIIa, and SBEIIb) have been identified in maize (Zea mays; Boyer and Preiss, 1978; Dang and Boyer, 1988), and their expression patterns differ considerably (Gao et al., 1996). While the in vitro properties of SBEIa differ from those of SBEIIa and SBEIIb (Guan and Preiss, 1993; Takeda et al., 1993), the sbe1a or sbe2a single mutants obtained using Mu-mediated transposon mutagenesis indicated that the chain-length (CL) profiles of starch produced in endosperm in vivo were not affected by the deficiency of either SBEIa or SBEIIa (Blauth et al., 2001, 2002). Conversely, the ae mutant, which is deficient in SBEIIb, results in major changes in endosperm amylopectin structure (Yuan et al., 1993; Shi and Seib, 1995; Klucinec and Thompson, 2002). The in vivo functional behavior of these three single Sbe mutants led us to hypothesize that during amylopectin biosynthesis in maize endosperm, the function of SBEIIb is predominant to that of both SBEIa and SBEIIa, and therefore the sbe1a mutant might affect starch structure only in an ae background.
An appropriate description of SBE function in amylopectin biosynthesis requires a full appreciation of the structure of the constituent clusters of amylopectin (Hizukuri, 1986; Manners, 1989; Bertoft et al., 1999; Thompson, 2000). In previous in vivo studies, the amylopectin CL profile of a specific SBE mutation was usually interpreted as the outcome of SBE function in transferring chains with specific lengths (Nishi et al., 2001; Binderup et al., 2002; Nakamura, 2002; Satoh et al., 2003). However, we considered it unlikely that chains transferred by SBE in vivo would not undergo further elongation or branching, and any further action by SS, SBE, or DBE would eliminate the length information of the chains initially transferred by an SBE.
In order to evaluate the functional behavior of SBE isoforms, we developed additional parameters to describe the structure of amylopectin (Fig. 1). We believe that the understanding of the cluster model of amylopectin should go beyond the CL distribution (Hizukuri, 1986) and take into account the relationships among branch points as well (Bertoft et al., 1999; Thompson, 2000). This latter relationship may well be the outcome of in vivo SBE activity (Thompson, 2000) and was used to characterize the functional behavior of SBE isoforms. While amylopectin CL profiles provide a general description of all the constituent chains present, further analysis of the CL profile of amylopectin β-limit dextrin is necessary to understand the branching pattern of the clusters (Fuwa et al., 1987; Inouchi et al., 1987; Yuan et al., 1993; Yun and Matheson, 1993; Klucinec and Thompson, 2002). Using high-performance size-exclusion chromatography (HPSEC), Klucinec and Thompson (2002) identified a shorter chain population from intracluster B chains in several genotypes. This observation provides some insight concerning the distance between adjacent branch points.
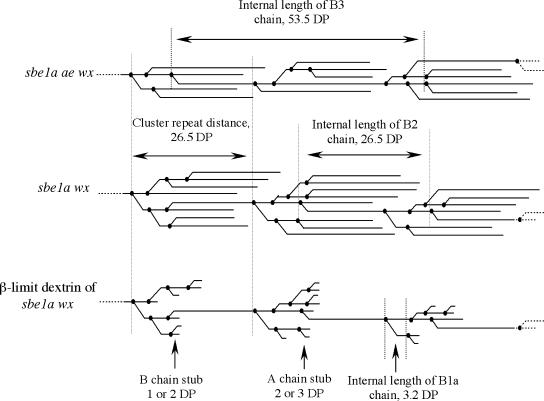
Figure 1.
Cluster diagrams of amylopectin to illustrate parameters employed. In a β-limit dextrin, the average length of B-chain stubs and A-chain stubs are DP 1.5 and DP 2.5, respectively (Yun and Matheson, 1993). The cluster repeat distance and the internal lengths of B1a, B2, and B3 chains are labeled in the figure.
In this study, we related the function of individual SBE isoforms to the branch density and branching pattern of amylopectin in SBE mutants. We hypothesized that in maize endosperm, the function of SBEIIb is predominant to that of SBEIa, and SBEIa would exhibit a demonstrable effect on amylopectin structure only in the absence of SBEIIb. To address this hypothesis, the multiple mutants sbe1a wx and sbe1a ae wx were isolated and used to investigate the in vivo functions of SBE isoforms by determining their effects on the branch density and branching pattern of endosperm amylopectin.
RESULTS
Verification of Mutant Genotypes
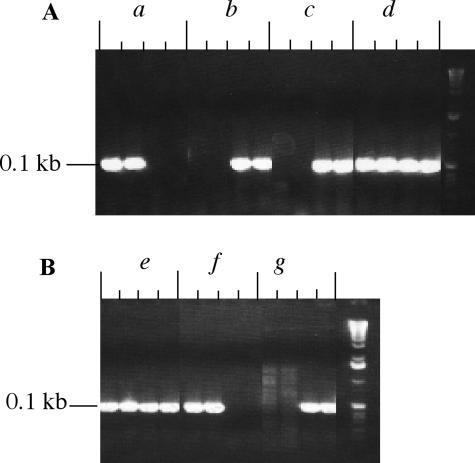
In order to study the role of SBE isoforms in starch biosynthesis, a series of mutant lines were generated. To verify the genotypes and the corresponding phenotypes of these lines, a complementary analysis by PCR, western blotting, and zymograms was performed. As shown in Figure 2, plants homozygous for sbe1a∷Mu allele were identified using PCR screening of the Mu transposon insertion in the Sbe1a gene. The plants homozygous for sbe1a were self-pollinated to produce ears with double (sbe1a wx) or triple (sbe1a ae wx) homozygous kernels. In order to confirm the presence or absence of SBE isoforms in the endosperm, protein extract from 20-d-after-pollination (DAP) endosperms of all genotypes were analyzed using starch zymograms (Fig. 3A) and western blotting (Fig. 4A) of native PAGE. The identities of SBE isoforms in native-PAGE gels were further verified using SDS-PAGE (SBEIa in Fig. 4B; SBEIIa and IIb, data not shown). The location of each SBE isoform was thus confirmed and labeled in Figures 3, A and B, and 4A.
Figure 2.
PCR genotyping of sbe1a∷Mu alleles in wx background (A) and ae wx background (B) using leaf DNA. For each sample, the first and second lanes were duplicate tests using primers 1A4 and 1A5 for the identification of Sbe1a allele, and the third and fourth lanes were duplicate tests using primers 1A4 and MuTIR9242 for the identification of sbe1a∷Mu allele. Samples a and f were identified as Sbe1a homozygous, or as having the genotype of wx and ae wx, respectively. Samples b, c, and g were identified as sbe1a homozygous, or as having the genotype of sbe1a wx (b and c) or sbe1a ae wx (g). Samples d and e were both heterozygous sbe1a/Sbe1a.
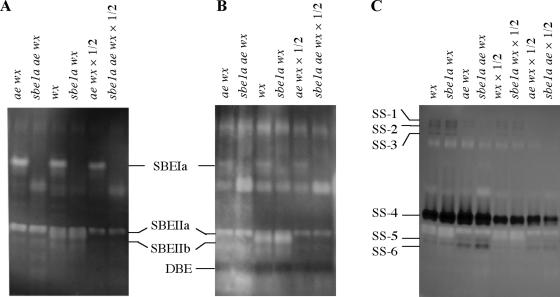
Figure 3.
Zymograms of crude protein extracts from 20-DAP endosperm using different starch substrates: potato starch (A), waxy-maize starch (B), and glycogen-ADP Glc (C), to detect the activities of SBE, DBE, and soluble SS, respectively. Activities of all known SBE isoforms can be identified in both A and B. The band identified as DBE produced a blue color indicating the activity of a major DBE, which was not affected by genotype.
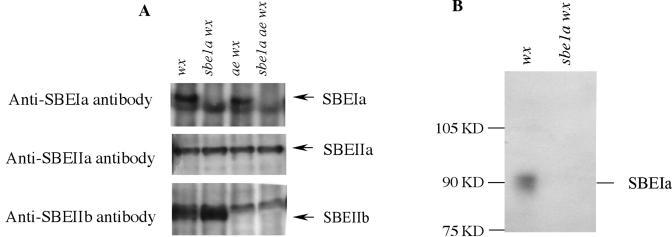
Figure 4.
A, Western blotting after native PAGE of crude protein extracts from 20-DAP endosperm of wx, sbe1a wx, ae wx, and sbe1a ae wx. The gel after native PAGE parallel with the zymogram test (Fig. 3) was blotted to a PVDF membrane (see “Materials and Methods”). The membrane was separated into three slices and incubated with antibodies to SBEIa, SBEIIa, and SBEIIb, respectively. The locations of SBEIa, SBEIIa, and SBEIIb in the native PAGE thus predicted are consistent with the genotyping result via PCR. B, Western blotting after SDS-PAGE of isolated gel slices at the predicted location of SBEIa from the native PAGE. The gel after SDS-PAGE was blotted to a PVDF membrane and incubated with antibody to SBEIa (see “Materials and Methods”). The molecular mass of protein was determined to be 90 kD, in consistence with that reported by Blauth et al. (2001, 2002). Therefore, the protein in the isolated gel slice was confirmed to be SBEIa.
Figure 3, A and B, indicated the deficiency of SBEIa in sbe1a wx and sbe1a ae wx genotypes, and the deficiency of SBEIIb in ae wx and sbe1a ae wx. SBEIIa activity was evident in the endosperm of all genotypes used in this study. The absence of SBEIa or SBEIIb has no detectable pleiotropic effect on the activity of SBEIIa.
The sbe1a Mutant Does Not Affect the Levels of DBE and SS Isoforms
Figure 3B indicates that both the ae and the sbe1a mutations did not have pleiotropic effects on the activities of the observable DBE isoform. Figure 3C indicates that the ae mutant did not affect the activity of the major SS isoform, whereas the sbe1a mutant did not affect the activities of either the major or the minor SS isoforms. The labels for SSs in Figure 3C (SS-1 to SS-6) were used only to indicate individual activity bands in the zymogram of SSs, not to indicate specific identities of SS isoforms.
Starch Granule Size, Kernel Phenotype, and Average Weight
Microscopy revealed that, while the granules of ae wx and sbe1a ae wx are smaller than those of wx and sbe1a wx, no differences can be observed between wx and sbe1a wx, and between ae wx and sbe1a ae wx (Fig. 5). Phenotypes of dried kernels were recorded by photographing mature ears, whereas endosperm starch granules were purified and imaged by light microscopy. As shown in Figure 5, while the kernel phenotype of sbe1a wx is indistinguishable from wx, the kernels of both ae wx and sbe1a ae wx appear shrunken compared to those of wx and sbe1a wx. Additionally, the kernels of sbe1a ae wx appear darker than those of the other three genotypes. The kernel weight for each genotype was 0.217 ± 0.012, 0.217 ± 0.013, 0.141 ± 0.007, and 0.162 ± 0.008 g/kernel for wx, sbe1a wx, ae wx, and sbe1a ae wx, respectively.
Figure 5.
A, Mature kernel pictures of wx, sbe1a wx, ae wx, and sbe1a ae wx. Kernels of sbe1a ae wx give a darker appearance than wx, sbe1a wx, and ae wx, while both ae wx and sbe1a ae wx appear shrunken compared to wx and sbe1a wx. The kernel phenotypes of wx and sbe1a wx are essentially the same. B, Endosperm starch granules from mature kernels of wx, sbe1a wx, ae wx, and sbe1a ae wx stained with iodine. No evident size distribution difference can be observed between wx and sbe1a wx, or between ae wx and sbe1a ae wx. A much larger population of small-size granules exists for both ae wx and sbe1a ae wx compared to wx and sbe1a wx.
Amylopectin Fine Structure
To gain insight into the in vivo roles of the SBEIa isoform on starch biosynthesis, the endosperm starch isolated from mature kernels of the mutant genotypes was structurally examined. A group of structural parameters were used to describe the branch density and branching pattern of amylopectin. The branch density was calculated from the average CL of amylopectin, whereas the branching patterns were expressed in terms of the average number of branches per cluster (ANBPC), the modal cluster repeat distance, and the modal distance between adjacent branch points in the cluster (Fig. 1).
CL Profile, Average CL, and Branch Density of Amylopectin Are Affected by sbe1a Mutant in the ae Background
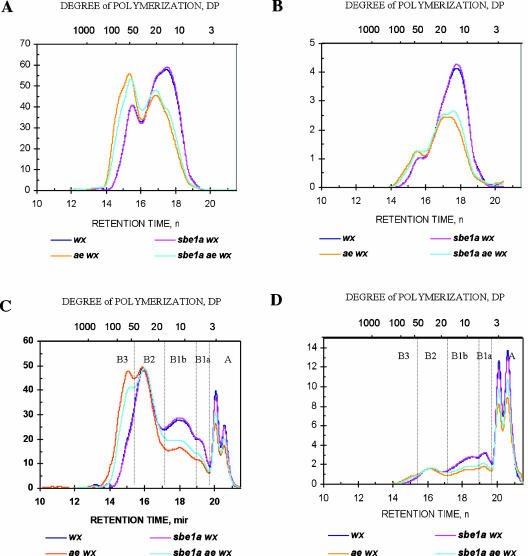
The normalized CL profile of amylopectins from wx, sbe1a wx, ae wx, and sbe1a ae wx genotypes are shown in Figure 6, A and B. Comparison among the four genotypes revealed that the profile for sbe1a wx is essentially the same as for wx. Consistent with previous studies, the starch of ae wx comprises considerably more long chains and fewer short chains than wx and sbe1a wx (Yuan et al., 1993; Klucinec and Thompson, 2002; Yao et al., 2002). The CL profile of sbe1a ae wx differs from that of ae wx exhibiting fewer long chains and more short chains.
Figure 6.
HPSEC chromatograms of debranched amylopectin (A and B) and β-limit dextrin (C and D) of wx, sbe1a wx, ae wx, and sbe1a ae wx genotypes. The y axis was normalized to the total area (mass amount) of the profile (A and C) and then transformed into molar amount (B and D). A chains (external chains attaching to other chains via α-1,6-glucosidic linkages) and each population of B chains (bearing one or more chains while attaching to another chain) are labeled in C and D. The B chains are further defined by their functions in constituting the amylopectin clusters as B1 chains (intracluster chains), B2 chains (intercluster chains connecting two clusters), and B3 chains (intercluster chains connecting three clusters; Hizukuri, 1986). B1 chains are divided into B1a (short B1) and B1b (long B1) chains.
As shown in Table I, the CL values of both ae-containing genotypes (24.7 and 23.1 for ae wx and sbe1a ae wx, respectively) were significantly (P < 0.01) higher than the CL of non-ae genotypes (18.2 and 18.1 for wx and sbe1a wx, respectively). The CL of sbe1a ae wx was lower than that of ae wx (P < 0.05). The branch density varied in the order ae wx < sbe1a ae wx ≪ sbe1a wx = wx.
Table I.
Parameters describing the structure of amylopectin and its clusters
| Genotype (SBE Isoforms Present) | CLa (DP) | A : B1a : B1b : B2 : B3b | ANBPCc |
|---|---|---|---|
| wx (SBEIa, SBEIIa, SBEIIb) | 18.3 ± 0.1 | 56 : 11 : 21 : 10 : 2 | 7.0 ± 0.1 |
| sbe1a wx (SBEIIa, SBEIIb) | 18.1 ± 0.2 | 55 : 11 : 21 : 10 : 2 | 7.1 ± 0.1 |
| ae wx (SBE1a, SBEIIa) | 24.7 ± 0.2 | 56 : 9 : 16 : 14 : 6 | 4.0 ± 0.1 |
| sbe1a ae wx (SBEIIa) | 23.1 ± 0.3 | 55 : 9 : 17 : 14 : 4 | 4.5 ± 0.0 |
Number-based average CL of amylopectin, expressed as mean ± sd (n = 2).
Mean value of number ratio among A, B1a, B1b, B2, and B3 chains.
Average number of branches per cluster, expressed as mean ± sd (n = 2).
CL Profiles of β-Limit Dextrin and the Ratio among Chain Populations Are Affected by sbe1a Mutant in the ae Background
The CL profile of β-limit dextrin allows us to investigate the length distribution of individual chain populations. Figure 6, C and D, shows the normalized profiles of debranched β-limit dextrins. The profiles can be categorized into two general groups: the non-ae genotypes wx and sbe1a wx, which express SBEIIb in the endosperm, and the ae genotypes ae wx and sbe1a ae wx, which lack SBEIIb, reflecting the large impact of SBEIIb on amylopectin structure. It is also evident that the profiles of wx and sbe1a wx are nearly identical, whereas an appreciable difference can be observed between ae wx and sbe1a ae wx.
The molar ratios among chain populations are shown in Table I. For all genotypes, the A/B chain ratios are similar, around 55/45. However, starch from the ae mutant exhibits increased proportion of intercluster chains (B2 + B3 chains) from 12% for wx and sbe1a wx to 18% for sbe1a ae wx and 20% for ae wx. In the ae background, sbe1a slightly decreased the intercluster/intracluster B chains ratio (from 20/25 for ae wx to 18/26 for sbe1a ae wx), and it also decreased the portion of B3 chains (from 6% for ae wx to 4% for sbe1a ae wx).
Effect of sbe1a on the Branching Pattern of Amylopectin
As indicated in Table I, while the ANBPC of sbe1a wx (7.1) and wx (7.0) are similar, the ANBPC value of sbe1a ae wx (4.5) is higher than for ae wx (4.0; P < 0.01), and both are much lower than those of wx and sbe1a wx. Therefore, ANBPC is increased by sbe1a in the ae background.
The modal lengths of B2 (degree of polymerization [DP] 29) and B3 (DP 56) chains after limit β-amylolysis were determined from Figure 6, C and D. The cluster repeat distance thus calculated was DP 26.5 for all four genotypes studied (Fig. 1). Therefore, cluster repeat distance is unaffected by these genotypes.
As shown in Figure 6D, the modal CL of B1a chains in the β-limit dextrins is similar for all genotypes studied (DP 5.7). The modal internal length of B1a chains is calculated to be DP 3.2 as described above. As depicted in Figure 1, the internal length of B1a chains reflects the distance between two adjacent branch points. Therefore, the distance between adjacent branch points in clusters appears to be unaffected by the Sbe mutations.
DISCUSSION
Structural Characterization of Amylopectin Cluster Is Fundamental to Revealing the Function of SBEs
A better understanding of starch fine structure may help refine the cluster model or guide evaluation of modifications to it, and is fundamental to understanding the molecular mechanism of starch biosynthesis as well. The definition of the cluster employed in the present report is in the tradition of Hizukuri (1986) but with emphasis on the clustering of branch points rather that the periodicity of CLs. Bertoft's laboratory has used a different approach to analyze clusters operationally defined by the proximity among branch points (Bertoft et al., 1999; Gerard et al., 2000). Although Bertoft has noted that the definition of a cluster is somewhat arbitrary, the calculation of ANBPC can be understood as describing a general tendency in the nature of clustering of branch points.
It has been suggested that during the development of amylopectin clusters, SBE and DBE determine the number and location of branch points, whereas SSs elongate the glucan chains (Ball et al., 1996; Myers et al., 2000). The CL profiles that appear to be related to altered branching enzyme activities (Martin and Smith, 1995) may be actually an indirect effect of an altered distribution of branches. We suggest that if branching enzyme activities are altered in starch biosynthesis, then one would look first for a change in the distribution of branches, rather than a change in the CL profile that would be secondary to an altered relationship among branches (Thompson, 2000). In this study, the distribution of branches in amylopectin is characterized using both the branch density and aspects of the branching pattern of amylopectin.
Maize SBE Isoforms Function Similarly in Determining the Cluster Repeat Distance and the Distance between Adjacent Branch Points
We consider that the cluster repeat distance and the distance between adjacent branch points are two important parameters showing the pattern of SBE isoforms in determining the positioning of branches. Assuming that B1a chains have only a single branch point, the modal internal length of B1a chains (DP 3.2) reflects the modal distance between adjacent branch points in a cluster for a subpopulation of the B chains (Thompson, 2000). Our data indicate that the modal internal length of B1a chain is not distinguishable for the SBE combinations studied. Based on the assumption that the distance for this subpopulation reflects the distance between branch points throughout the cluster, we propose that SBE isoforms behave similarly in determining the distance between branch points in a cluster.
Different SBE isoform combinations do not affect the cluster repeat distance either, indicating that the mechanism governing the initiation of a new cluster is unresolved by our research. Such a mechanism might involve collaborative actions of SBE, DBE, and SS, or it may involve a tendency for physical association of chains once a certain length is reached. Others have suggested that physical forces may be responsible for a similar cluster repeat length (Waigh et al., 2000). Our observation that the cluster repeat distance is indistinguishable for different genotypes is consistent with the apparent conservation of what Jenkins et al. (1993) termed size of an amylopectin cluster. By small-angle x-ray scattering, they observed a repeat value of 9 nm for amylopectins from a wide variety of sources throughout the plant kingdom (Jenkins et al., 1993).
In the Absence of SBEIIb, the Deficiency of SBEIa Increases the Branching of Amylopectin
While the branch density is a general description of the functional intensity of SBE during amylopectin branching, ANBPC reflects the functional intensity of SBE at the level of individual clusters. It is evident that both the branch densities and ANBPC values display the same orders for genotypes studied: ae wx < sbe1a ae wx ≪ sbe1a wx = wx. For sbe1a wx, both the branch density and ANBPC are the same as for wx. In contrast, the sbe1a mutant causes an altered branching in the ae background. These observations indicate that in maize endosperm, the function of SBEIIb is predominant to that of SBEIa, and SBEIa has an observable effect only on amylopectin structure in the absence of SBEIIb.
This study is consistent with that of Satoh et al. (2003) in that mutation of the sbe1 gene may result in increased branching. Endosperm starch isolated from a rice (Oryza sativa) mutant deficient in the endosperm BEI was characterized by marked increase in short chains, a significant decrease in long chains, and a slight increase in intermediate chains. Based on this data, Satoh et al. suggested that BEI specifically creates B chains.
At face value, the biochemical activity of the SBE enzymes might lead us to speculate that the deficiency of any SBE isoform should correlate with decreased amylopectin branching, as in the case of ae-containing mutants. However, as our data indicate, perhaps surprisingly, such a prediction is not correct. There is no evidence indicating that the increased branching in the sbe1a ae wx was caused by altered expressions of other starch biosynthetic enzymes, including SBEIIa, SS isoforms, and debranching enzymes. One possible explanation for this observation is that the increased branching observed in sbe1a ae wx (containing SBEIIa alone) compared to ae wx (containing both SBEI and SBEIIa) implies that SBEIa may inhibit SBEIIa in vivo. Such an inhibition may be caused by either substrate competition or protein-protein interaction. Recently, Tetlow et al. (2004) reported that protein phosphorylation in amyloplasts regulates SBE activity and protein-protein interactions in wheat (Triticum aestivum). They showed that SBEIIb and starch phosphorylase each coimmunoprecipitated with SBEI in a phosphorylation-dependent manner, suggesting that these enzymes may form protein complexes within the amyloplast in vivo.
CONCLUSION
To study the function of SBEIa in vivo, the genotype sbe1a ae wx was constructed, and its effects on the activities of other SBEs, on SS, and on DBE isoforms were investigated. The branch density and branching pattern of amylopectin were used to describe the functional outcome of combinations of SBE isoforms. A contribution to functional activity by SBEIa was evident only in the absence of SBEIIb, consistent with the hypothesis that the function of SBEIIb is predominant to that of SBEIa in the formation of branches. In the absence of SBEIIb, the deficiency of SBEIa led to increased branching, suggesting in vivo inhibition of SBEIIa by SBEIa. The cluster repeat distance and the modal distance between adjacent branch points in a cluster appeared to be similar for all genotypes studied, suggesting that SBE isoforms behave similarly in initiating a cluster and in determining the location of branches within a cluster.
MATERIALS AND METHODS
Plant Growth and Breeding
Plants were grown in a greenhouse with supplemental light (14 h/d, 400 W high-pressure sodium light), 60% humidity, and 20°C to 22°C in dark period, 22°C to 24°C in light period. The wx and ae wx lines were isogenic in the W64A background (Penn State University maize genotype program). The mutant line segregating for a Mutator transposon within Sbe1a was identified in a reverse-genetics PCR screen (Blauth et al., 2002) and backcrossed into W64A-inbred line six times to obtain an isogenic mutant line. The sbe1a gene was then introgressed into wx and ae wx mutant lines by reciprocal crossing and then selfing. The progeny kernels displaying typical wx or ae wx visual phenotypes were screened for sbe1a∷Mu by PCR (see below). The identification of wx kernels using visual phenotype proved reliable since 100% of kernels tested showed a total lack of amylose by HPSEC analysis.
For genotyping of sbe1a mutants, DNA was extracted from 200 mg of leaf tissue of 10-d-after-emergence seedlings as described by Dellaporta (1994) and detected using PCR as described by Blauth et al. (2002).
Endosperm Protein Extraction
For each of wx, sbe1a wx, ae wx, and sbe1a ae wx genotypes, total endosperm proteins were extracted from 20-DAP endosperm tissues flash frozen in liquid nitrogen. Five grams of endosperm tissue were ground using mortar and pestle under liquid nitrogen. To the powder, 10 mL of native protein extraction buffer (0.05 m sodium acetate, 0.02 m DTT, pH 6.0) was added followed by further grinding in a chilled mortar. The homogenates were centrifuged (6,000g, 10 min, 4°C) and supernatants stored at −80°C.
Starch Isolation from Mature Kernels
For each of wx, sbe1a wx, ae wx, and sbe1a ae wx genotypes, 20 dry kernels from mature ears were randomly selected and starches were isolated as described by Yao et al. (2002).
Zymograms of Native PAGE
Crude protein extracts (10–20 μg) were separated on native polyacrylamide gels as described by Dinges et al. (2001). For activity detection of SBE and DBE, separating gels were electroblotted to polyacrylamide gels of the same size containing 7% (w/v) acrylamide, 0.3% (w/v) potato starch (Sigma-Aldrich, St. Louis) or waxy-maize starch (Sigma-Aldrich), and 375 mm Tris-HCl, pH 8.8. The transfer was performed overnight at 68 mA in the electrode buffer at 22°C. SBE and DBE activities were observed by staining the potato starch- and waxy-maize starch-containing gel, respectively, with iodine solution (I2 0.02% [w/v], KI 0.2% [w/v]) for 10 min at 22°C, after which the gel was photographed immediately.
For the detection of soluble SS activity, separating gels were electroblotted to 7% (w/v) polyacrylamide gels containing 0.3% (w/v) rabbit liver glycogen (Sigma-Aldrich) and 375 mm Tris-HCl, pH 8.8 (activity gel), for 3 h at 158 mA at 22°C. The activity gels were then incubated for 36 h in 50 mm glycylglycine/NaOH, pH 9.0, with 100 mm (NH4)2SO4, 15 mm 2-mercaptoethanol, 5 mm MgCl2, 0.5 mg/mL BSA, and 4 mm ADP-Glc (Sigma-Aldrich) at 22°C, then stained with iodine solution (I2 0.02% w/v, KI 0.2% w/v) for 10 min at 22°C and photographed immediately.
Western Blotting of Native PAGE
Western blotting was used to locate the individual SBE isoforms in native gels. Protein extracts were separated using native PAGE as used in zymograms, blotted onto Immunobulin polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA) using a HEP-1 semi-dry electroblotting device (Owl Scientific, Portsmouth, NH) and Towbin's buffer (Towbin et al., 1979). Immuno-detection of SBEIa, SBEIIa, and SBEIIb was performed as described by Blauth et al. (2001, 2002) using anti-SBE antibodies at 1:500 dilutions.
SBEIa, SBEIIa, and SBEIIb Confirmed Using SDS-PAGE
Gel slices corresponding to putative SBEIa, IIa, and IIb isoforms separated by native PAGE were excised and applied to the wells of SDS-PAGE gels. After SDS-PAGE electrophoresis and blotting to PVDF membranes, anti-SBE antibodies were used to verify the identities of particular gel fractions as described above.
Microscopy of Starch Granules
Starch samples dispersed in deionized water were diluted with equal volumes of iodine solution (I2/KI = 0.04%/0.4%). The dispersions were viewed using a light microscope (BX50; Olympus, Melville, NY) and imaged with a digital camera (SPOT II; Bioscan, Warrendale, PA).
Kernel Phenotype and Weight
Images of dried kernels on mature ears of genotypes wx, sbe1a wx, ae wx, and sbe1a ae wx were captured by digital photography. Average weights of kernels were determined by weighing 20 kernels.
Preparation of Debranched Amylopectin and Debranched β-Limit Dextrin
The dispersions of debranched amylopectin and debranched β-limit dextrin were prepared as described by Klucinec and Thompson (1998, 2002). After debranching, the dimethyl sulfoxide/water dispersions were concentrated in a vacuum evaporator (SpeedVac SC100; Savant Instruments, Holbrook, NY).
HPSEC Analysis
Concentrated debranched starch dispersions were diluted 30 times (v/v) with 90% dimethyl sulfoxide, and a 50-μL aliquot was analyzed using HPSEC as described by Klucinec and Thompson (1998, 2002). Differential refractive index data (a mass response) were collected using Millennium software (Waters, Milford, MA), exported to Excel, and normalized on the basis of total area.
Branch Density of Amylopectin
From the CL profile of amylopectin, the average CL was calculated as described by Yuan et al. (1993). The branch density was described as the inverse of CL.
Parameters Describing the Branching Pattern of Amylopectin
Three parameters were used to describe the branching pattern of amylopectin: the ANBPC, the cluster repeat distance, and the distance between adjacent branch points in the cluster.
For calculating these parameters, the y axes of CL profiles of β-limit dextrin were transformed into molar amount as described by Klucinec and Thompson (2002). Based on the minima or inflections observed, the profiles were divided into regions of A chains, intracluster B chains (B1), and intercluster B chains (Hizukuri, 1986). Intracluster chains were further divided into what we now term B1a (a portion of B chains likely to have only one branch) and B1b (a portion of chains likely to have more than one branch), and the intercluster B chains were further divided into B2 chains (assumed to connect two clusters) and B3 chains (assumed to connect three clusters). The molar ratios of chain populations were calculated from the areas of corresponding regions.
Based on the molar ratios of chain populations, the ANBPC was calculated as the total number of chains divided by the total number of clusters. Since one B2 chain is assumed to initiate one cluster (Fig. 1) and one B3 chain to initiate two clusters, the total number of clusters was estimated as the number of B2 chains + the number of B3 chains × 2.
The cluster repeat distance was defined as the distance between two initiation points of two consecutive clusters (Fig. 1). The average cluster repeat distance will equal the average internal length of a B2 chain or half of the average internal length of a B3 chain (Fig. 1). We considered the modal (peak) value in the β-limit dextrin chromatograms to be a reasonable estimate of the average internal length of a population of B2 or B3 chains. Therefore, the internal length of B2 or B3 chains was estimated by subtracting DP 2.5 (Yun and Matheson, 1993) from the modal length of B2 or B3 chains of the debranched β-limit dextrin.
In this study, the distance between adjacent branch points in the cluster was estimated using the distance between branch points on B1a chain, assuming that the pattern of SBE isoforms in positioning a branch is constant during the development of a whole cluster. Since it is likely that only one chain attaches to each B1a chain, the distance between branch points on a B1a chain equals its internal length, which can be obtained by subtracting DP 2.5 (Fig. 1) from the modal length of B1a chains determined from the CL profile peak for this fraction of debranched β-limit dextrin.
This work was supported by a grant from the U.S. Department of Energy Bioscience Program (grant no. DE–FG02–96ER20234 to M.J.G. and D.B.T.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.043315.
References
- Ball SG, Guan HP, James MG, Myers AM, Keeling P, Mouille G, Buleon A, Colonna P, Preiss J (1996) From glycogen to amylopectin: a model for the biogenesis of the plant starch granule. Cell 86: 349–352 [DOI] [PubMed] [Google Scholar]
- Bertoft E, Zhu Q, Andtfolk H, Jungner M (1999) Structural heterogeneity in waxy-rice starch. Carbohydr Polym 38: 349–359 [Google Scholar]
- Binderup K, Mikkelsen R, Preiss J (2002) Truncation of the amino terminus of branching enzyme changes its chain transfer pattern. Arch Biochem Biophys 397: 279–285 [DOI] [PubMed] [Google Scholar]
- Blauth SL, Kim K, Klucinec JD, Shannon JC, Thompson DB, Guiltinan MJ (2002) Identification of Mutator insertional mutants of starch-branching enzyme 1 (sbe1) in Zea mays L. Plant Mol Biol 48: 287–297 [DOI] [PubMed] [Google Scholar]
- Blauth SL, Yao Y, Klucinec JD, Shannon JC, Thompson DB, Guiltinan MJ (2001) Identification of Mutator insertional mutants of starch-branching enzyme 2a in corn. Plant Physiol 125: 1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer C, Preiss J (1978) Multiple forms of (1,4)-α-D-glucan-6-glucosyl transferase from developing Zea Mays L. kernels Carbohydr Res 61: 321–334 [Google Scholar]
- Dang PL, Boyer CD (1988) Maize leaf and kernel starch synthases and starch branching enzymes. Phytochemistry 27: 1255–1259 [Google Scholar]
- Dellaporta S (1994) Plant DNA miniprep and microprep: versions 2.1–2.3. In M Freeling, V Walbot, eds, The Maize Handbook. Springer-Verlag, New York, pp 522–525
- Dinges JR, Colleoni C, Myers AM, James MG (2001) Molecular structure of three mutations at the maize sugary1 locus and their allele-specific phenotypic effects. Plant Physiol 125: 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuwa H, Glover DV, Miyaura K, Inouchi N, Konishi Y, Sugimoto Y (1987) Chain-length distribution of amylopectins of double-mutants and triple-mutants containing the waxy gene in the inbred oh43 maize background. Starch-Starke 39: 295–298 [Google Scholar]
- Gao M, Fisher DK, Kim KN, Shannon JC, Guiltinan MJ (1996) Evolutionary conservation and expression patterns of maize starch branching enzyme I and IIb genes suggests isoform specialization. Plant Mol Biol 30: 1223–1232 [DOI] [PubMed] [Google Scholar]
- Gerard C, Planchot V, Colonna P, Bertoft E (2000) Relationship between branching density and crystalline structure of A- and B-type maize mutant starches. Carbohydr Res 326: 130–144 [DOI] [PubMed] [Google Scholar]
- Guan HP, Preiss J (1993) Differentiation of the properties of the branching isozymes from maize (Zea mays L.). Plant Physiol 102: 1269–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizukuri S (1986) Polymodal distribution of the chain lengths of amylopectins, and its significance. Carbohydr Res 147: 342–347 [Google Scholar]
- Inouchi N, Glover DV, Fuwa H (1987) Chain-length distribution of amylopectins of several single mutants and the normal counterpart, and sugary-1 phytoglycogen in maize (zea-mays-l). Starch-Starke 39: 259–266 [Google Scholar]
- Jenkins JPJ, Cameron RE, Donald AM (1993) A universal feature in the structure of starch granules from different botanical sources. Starch-Starke 45: 417–420 [Google Scholar]
- Klucinec JD, Thompson DB (1998) Fractionation of high-amylose maize starches by differential alcohol precipitation and chromatography of the fractions. Cereal Chem 75: 887–896 [Google Scholar]
- Klucinec JD, Thompson DB (2002) Structure of amylopectins from ae-containing maize starches. Cereal Chem 79: 19–23 [Google Scholar]
- Manners DJ (1989) Recent developments in our understanding of amylopectin structure. Carbohydr Polym 11: 87–112 [Google Scholar]
- Martin C, Smith AM (1995) Starch biosynthesis. Plant Cell 7: 971–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AM, Morell MK, James MG, Ball SG (2000) Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol 122: 989–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y (2002) Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: rice endosperm as a model tissue. Plant Cell Physiol 43: 718–725 [DOI] [PubMed] [Google Scholar]
- Nishi A, Nakamura Y, Tanaka N, Satoh H (2001) Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol 127: 459–472 [PMC free article] [PubMed] [Google Scholar]
- Satoh H, Nishi A, Yamashita K, Takemoto Y, Tanaka Y, Hosaka Y, Sakurai A, Fujita N, Nakamura Y (2003) Starch-branching enzyme I-deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiol 133: 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YC, Seib PA (1995) Fine-structure of maize starches from 4 wx-containing genotypes of the w64a inbred line in relation to gelatinization and retrogradation. Carbohydr Polym 26: 141–147 [Google Scholar]
- Smith AM, Denyer K, Martin C (1997) The synthesis of the starch granule. Annu Rev Plant Physiol Plant Mol Biol 48: 65–87 [DOI] [PubMed] [Google Scholar]
- Takeda Y, Guan HP, Preiss J (1993) Branching of amylose by the branching isoenzymes of maize endosperm. Carbohydr Res 240: 253–263 [Google Scholar]
- Tetlow IJ, Wait R, Lu Z, Akkasaeng R, Bowsher CG, Esposito S, Kosar-Hashemi B, Morell MK, Emesa MJ (2004) Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein-protein interactions. Plant Cell 16: 694–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DB (2000) On the non-random nature of amylopectin branching. Carbohydr Polym 43: 223–239 [Google Scholar]
- Towbin H, Staehelin T, Gordon J (1979) Electrophoetic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waigh TA, Kato KL, Donald AM, Gidley MJ, Clarke CJ, Riekel C (2000) Side-chain liquid-crystalline model for starch. Starch-Starke 52: 450–460 [Google Scholar]
- Yao Y, Guiltinan MJ, Shannon JC, Thompson DB (2002) Single kernel sampling method for maize starch analysis while maintaining kernel vitality. Cereal Chem 79: 757–762 [Google Scholar]
- Yuan RC, Thompson DB, Boyer CD (1993) Fine-structure of amylopectin in relation to gelatinization and retrogradation behavior of maize starches from 3 wx-containing genotypes in 2 inbred lines. Cereal Chem 70: 81–89 [Google Scholar]
- Yun SH, Matheson NK (1993) Structures of the amylopectins of waxy, normal, amylose-extender, and wx-ae genotypes and of the phytoglycogen of maize. Carbohydr Res 243: 307–321 [DOI] [PubMed] [Google Scholar]