Abstract
Arabidopsis (Arabidopsis thaliana) possesses two isoforms of plastidic ATP/ADP transporters (AtNTT1 and AtNTT2) exhibiting similar biochemical properties. To analyze the function of both isoforms on the molecular level, we examined the expression pattern of both genes by northern-blot analysis and promoter-β-glucuronidase fusions. AtNTT1 represents a sugar-induced gene mainly expressed in stem and roots, whereas AtNTT2 is expressed in several Arabidopsis tissues with highest accumulation in developing roots and young cotyledons. Developing lipid-storing seeds hardly contained AtNTT1 or -2 transcripts. The absence of a functional AtNTT1 gene affected plant development only slightly, whereas AtNTT2∷T-DNA, AtNTT1-2∷T-DNA, and RNA interference (RNAi) plants showed retarded plant development, mainly characterized by a reduced ability to generate primary roots and a delayed chlorophyll accumulation in seedlings. Electron microscopic examination of chloroplast substructure also revealed an impaired formation of thylakoids in RNAi seedlings. Moreover, RNAi- and AtNTT1-2∷T-DNA plants showed reduced accumulation of the nuclear-encoded protein CP24 during deetiolation. Under short-day conditions reduced plastidic ATP import capacity correlates with a substantially reduced plant growth rate. This effect is absent under long-day conditions, strikingly indicating that nocturnal ATP import into chloroplasts is important. Plastidic ATP/ADP transport activity exerts significant control on lipid synthesis in developing Arabidopsis seeds. In total we made the surprising observation that plastidic ATP/ADP transport activity is not required to pass through the complete plant life cycle. However, plastidic ATP/ADP-transporter activity is required for both an undisturbed development of young tissues and a controlled cellular metabolism in mature leaves.
ATP represents the universal energy currency of all living cells. Due to both size and charge, adenylates do not cross biomembranes freely, making the involvement of highly specific transport proteins necessary. In eukaryotic cells mitochondria export ATP previously generated via oxidative phosphorylation at the matrix site in strict counter exchange to cytosolic ADP. The corresponding ADP/ATP carriers (AAC) function as dimers, comprising two identical subunits, each exhibiting six predicted transmembrane domains (Klingenberg, 1989). AAC proteins belong to the best characterized solute transporters and are the subject of numerous publications (Fiore et al., 1998).
We identified the plastidic ATP/ADP transporter as a second type of eukaryotic-adenylate carrier protein (Kampfenkel et al., 1995). Plastidic ATP/ADP transporters exist in all higher and lower plants analyzed so far (Linka et al., 2003), exhibit 11 to 12 predicted transmembrane domains (Winkler and Neuhaus, 1999), and do not show substantial structural similarities to the functional AAC homologs in mitochondria (Winkler and Neuhaus, 1999). In Arabidopsis (Arabidopsis thaliana), two plastidic ATP/ADP transporters are present, and both exhibit very similar biochemical transport properties when heterologously expressed in Escherichia coli (Möhlmann et al., 1998; Tjaden et al., 1998b).
The main function of plastidic ATP/ADP transporters is the supply of storage plastids with ATP (Schünemann et al., 1993; Kang and Rawsthorne, 1994; Neuhaus and Emes, 2000). In potato (Solanum tuberosum) tubers, the plastidic ATP/ADP transporter exerts significant control on starch accumulation (Tjaden et al., 1998a), leading to a high-flux control coefficient within this metabolic pathway (Geigenberger et al., 2001). In contrast, a recently made metabolite flux analysis on developing rapeseed (Brassica napus) embryos indicated that ATP import into corresponding plastids is not required to achieve high rates of lipid biosynthesis (Schwender et al., 2004).
All orthologs of the plastidic ATP/ADP transporter, e.g. the two isoforms from Arabidopsis, a potato ortholog, or an ortholog from the primitive red alga Galderia sulfuralia, exhibit similar transport properties in respect to substrate specificity and substrate affinity (Möhlmann et al., 1998; Tjaden et al., 1998a, 1998b; Linka et al., 2003). Therefore, the observation of similar transport properties and the contradictory information on the involvement of plastidic ATP/ADP transporters in plastidic storage product synthesis (Tjaden et al., 1998a; Schwender et al., 2004) provoke to identify reasons for the presence of plastidic nucleotide transporter (NTT) isoforms in plants and to examine the physiological impact of these ATP transporters. The analysis of the physiological implication of plastidic ATP/ADP transport activity is further encouraged since reduction of another eukaryotic ATP-transporter activity, namely the mitochondrial AAC protein, correlates with a massive, partly complete inhibition of cellular metabolism (Kolarov et al., 1990; Drgon et al., 1991) and is causative for various severe diseases (Fiore et al., 1998).
To answer the questions of expression patterns and physiological implications we started a comprehensive approach and generated in total six independent transgenic Arabidopsis plants, namely AtNTT1- and AtNTT2-promoter∷GUS lines, AtNTT1∷T-DNA-, AtNTT2∷T-DNA-, AtNTT1-2∷T-DNA-, and RNA interference (RNAi) lines. A detailed molecular analysis revealed that AtNTT2 represents a widely expressed Arabidopsis gene, whereas AtNTT1 exhibits a spatial-expression pattern. The absence of AtNTT2 or the simultaneous absence of both transporters strongly affects plant development as revealed by analysis of root formation, chloroplast maturation, and plant growth rate. Inhibition of plastidic ATP/ADP-transporter activity also exerted substantial effect on lipid content in Arabidopsis seeds. Obviously, the continuous ATP supply into developing plastids from both young seedlings and embryo tissues and into mature chloroplast at night is required for a controlled plant development.
RESULTS
Expression Analysis of AtNTT1 and AtNTT2
Arabidopsis possesses two isoforms of the plastidic ATP/ADP transporter with similar biochemical transport properties. To reveal whether the presence of two independent transporter genes correlates with an organ- or development-specific expression pattern, we analyzed both the relative mRNA accumulation by northern-blot analysis and the promoter activity in transgenic plants carrying corresponding promoter-β-glucuronidase (GUS) fusions.
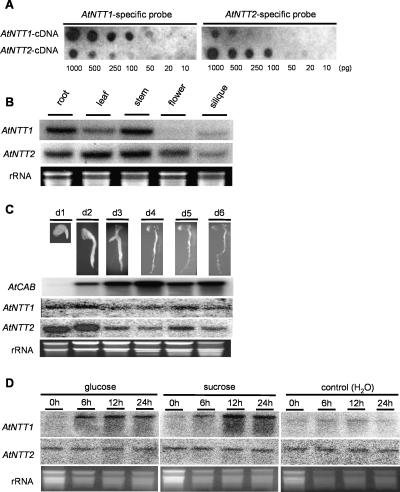
For reliable northern-blot analysis of isoform-specific mRNA accumulation it is required to use gene-specific probes. We generated probes specific for either AtNTT1- or AtNTT2 mRNA by using corresponding 3′-untranslated cDNA fragments (Fig. 1A). Although there is some minor cross hybridization, the probes used exhibited a sufficiently high specificity (Fig. 1A).
Figure 1.
Expression analysis of AtNTT-genes. Transcripts of AtNTT1 and AtNTT2 were detected by northern-blot analysis. For this, total RNA was isolated, separated by electrophoresis, and transferred to a nylon membrane. Ethidium bromide (EtBr) staining reveals equal RNA loading. A, Specificity of AtNTT1 and AtNTT2 probes. AtNTT1 and AtNTT2 cDNA was spotted onto nylon membranes (10–1,000 pg) and subsequently hybridized with an AtNTT1- or AtNTT2-specific probe, respectively. B, Tissue-specific mRNA accumulation. Total RNA (10 μg) was isolated from Arabidopsis tissues (roots, leaves, stems, flowers, and siliques) and hybridized with gene-specific probes. C, Alterations of AtCAB-, AtNTT1-, and AtNTT2-mRNA-levels during the first 6 d of development. The upper pictures represent the corresponding plant phenotypes. Total RNA was extracted from whole seedlings. Blots were hybridized with an AtCAB-, AtNTT1-, or AtNTT2-specific probe. D, Effects of sugar feeding on mRNA accumulation in source leaf discs. Source leaf discs were floated on either water (control), or 100 mm Glc or Suc. Samples were taken at the indicated time points and blots were hybridized with gene-specific probes.
AtNTT1 mRNA accumulated strongest in root and stem tissue, and less in source leaves (Fig. 1B). In flowers and siliques the level of AtNTT1 mRNA was below or close, respectively, to the detection level (Fig. 1B). In contrast, AtNTT2 mRNA accumulated to similar amounts in roots, leaves, stem, and flower tissue (Fig. 1B). Similar to AtNTT1, the AtNTT2 mRNA was much less present in siliques (Fig. 1B).
To gain first evidence on the expression pattern of both plastidic ATP/ADP-transporter genes during early germination, we monitored the relative mRNA abundance within the first 6 d of development. Within this time span Arabidopsis develops a primary root and gains photosynthetic competence as revealed by accumulation of both chlorophyll and chlorophylla/b-binding protein (CAB) mRNA (Fig. 1C). The level of AtNTT1 mRNA within this period of development remained close to the detection level without any substantial changes (Fig. 1C). This is different from the expression of AtNTT2, as latter mRNA strongly accumulated in days 1 and 2, and declined from day 3 to a level still above the AtNTT1 mRNA (Fig. 1C).
To reveal whether plastidic ATP/ADP-transporter gene expression responds on altered sugar availabilities, we floated source leaf discs in either water (control) or 100 mm Glc or Suc (Fig. 1D). The incubation of leaf discs for 24 h in water did not alter the levels of AtNTT1 or AtNTT2 mRNA (Fig. 1D). In contrast, the presence of Glc or Suc strongly increased the accumulation of AtNTT1 mRNA but did not influence AtNTT2 mRNA concentration (Fig. 1D).
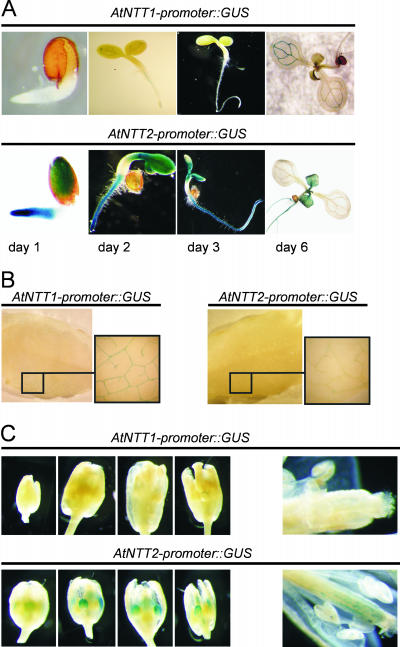
To perform a second independent approach to study regulation of gene expression, we generated transgenic plants harboring either an AtNTT1-promoter∷GUS- or AtNTT2-promoter∷GUS gene, respectively. During the first 6 d of development the AtNTT1 promoter is hardly active (Fig. 2A). At day 2 AtNTT1 promoter activity is slightly detectable in the center of the primary root (Fig. 2A), and at day 6 cells comprising the vascular structures in photosynthesizing cotyledons exhibited AtNTT1 promoter activities (Fig. 2A). In contrast to this, AtNTT2 promoter activity is very high at day 1, especially in the root tip and developing cotyledons (Fig. 2A). At day 3, highest AtNTT2 promoter activity is detectable in the root hair zone and at the basis of cotyledons. At day 6, we still observed strong AtNTT2 promoter activity in the root and in rapidly developing secondary leaves (Fig. 2A), still representing strong sinks.
Figure 2.
Histochemical localization of GUS expression under the control of either the AtNTT1 or AtNTT2 promoter in Arabidopsis seedlings and mature tissues. A, Promoter-GUS activity in developing seedlings. Seeds were sown on Murashige and Skoog agar plates and harvested after 1, 2, 3, and 6 d after imbibition and analyzed for GUS activity according to standard protocols. B, Promoter-GUS activity in source leaves. Rosette leaves were harvested from plants, grown under short-day conditions, and GUS stained. C, GUS expression in flowers at different developmental stages.
In source leaves, both promoters are barely active. AtNTT1 promoter-GUS activity is detectable only in the vascular bundles located at the edge of the leaf (Fig. 2B), whereas AtNTT2 promoter activity was hardly detectable in the leaf. This result does not necessary contradict the northern-blot analysis (Fig. 1B), as latter reflect the sum of AtNTT mRNA in total leaf tissue. In both flower tissue and developing siliques the AtNTT1 promoter activity is below the detection level (Fig. 2C). Petal crown leaves showed slight AtNTT2 promoter activity, which was, however similar to AtNTT1 promoter activity, nearly absent in developing siliques (Fig. 2C).
Generation of Arabidopsis Mutants Exhibiting Reduced or Abolished Plastidic ATP/ADP-Transporter Gene Expression
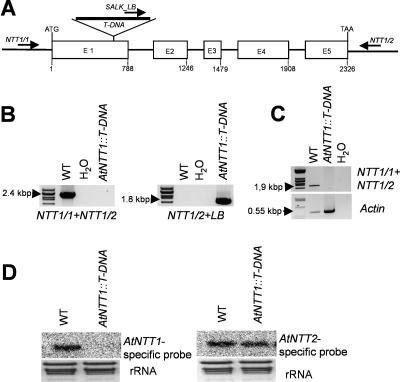
In the SALK library we identified a putative AtNTT1 knockout line exhibiting the T-DNA insertion in exon 1 (Fig. 3A). Corresponding heterozygous plants have been selfed to obtain homozygous mutants. By use of the gene-specific primers NTT1/1 and NTT1/2, we were able to amplify a PCR product of the expected size (about 2.4 kb) on wild-type DNA but not on DNA from AtNTT1∷T-DNA plants (Fig. 3B).
Figure 3.
Molecular characterization of homozygous AtNTT1 knockout mutants. A, Analysis of the AtNTT1-T-DNA-insertion line Salk_013530 (designated AtNTT1∷T-DNA). The insertion in AtNTT1∷T-DNA is localized in the first exon. The primers used for PCR analysis are marked as arrows. Primer NTT1/1 was chosen from the AtNTT1-promoter region, primer NTT1/2 from the 3′-untranslated region; SALK_LB-primer from the left border of the T-DNA. B, PCR analysis on genomic DNA of wild-type (WT) and homozygous AtNTT1∷T-DNA mutants. C, RT-PCR analysis of the expression of the AtNTT1 genes in wild-type and in AtNTT1∷T-DNA mutant plants. cDNA was isolated from rosette leaves. Actin PCR reveals correct PCR conditions. D, Northern-blot analysis of wild-type and AtNTT1∷T-DNA mutant plants. Total RNA was extracted from rosette leaves. EtBr staining revealed equal RNA loading. Blots were hybridized with AtNTT1 or AtNTT2 specific probes, respectively.
The PCR product amplified on wild-type DNA has been sequenced to confirm the correct nucleotide sequence (data not shown). Using the primers NTT1/2 and left boarder (LB) we amplified a PCR product on DNA obtained from mutant plants but not from wild-type plants (Fig. 3B). The PCR product has been sequenced to confirm the insertion site (data not shown). To check that the T-DNA insertion into the AtNTT1 gene correlates with absence of the corresponding mRNA, we performed a reverse transcription (RT)-PCR analysis (Fig. 3C). As expected, we were able to demonstrate the presence AtNTT1 mRNA in wild-type leaf tissue but not in leaves from AtNTT1∷T-DNA plants (Fig. 3C). Similarly, a northern-blot analysis demonstrated the presence of AtNTT1 mRNA in wild-type leaves but not in AtNTT1∷T-DNA leaves (Fig. 3D, left section). Remarkably, the absence of AtNTT1 mRNA (Fig. 3, C and D) is not compensated by an increase of AtNTT2 mRNA (Fig. 3D, right section).
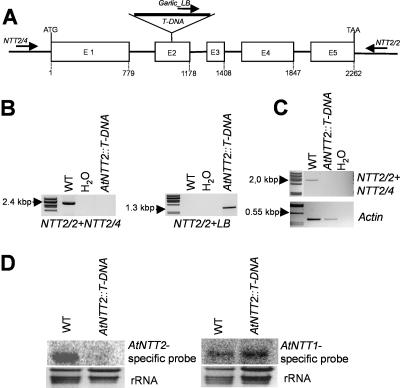
A putative AtNTT2 knockout line was available in the GARLIC library carrying the T-DNA insertion in exon 2 (Fig. 4A). Heterozygous plants have been grown and selfed to obtain a homozygous knockout line. The combination of the gene-specific primers NTT2/2 and NTT2/4 allowed amplification of a PCR product of the expected size (about 2.4 kb) on genomic wild-type DNA but not on DNA from homozygous AtNTT2∷T-DNA plants (Fig. 4B). The use of the gene-specific primer NTT2/2 and the LB primer allowed amplification of a fragment of about 1.3 kb on genomic DNA from AtNTT2:T-DNA plants but not in wild-type DNA (Fig. 4B). The PCR products have been sequenced to demonstrate that the correct DNA fragments have been amplified and to confirm the position of the T-DNA insertion (data not shown). To prove that the T-DNA insertion in the AtNTT2 gene correlates with the absence of the corresponding mRNA, we performed RT-PCR and northern-blot analysis. The gene-specific primers NTT2/2 and NTT2/4 allowed to amplify a PCR product of the expected size on cDNA prepared from wild-type leaf tissue but not on cDNA prepared from AtNTT2∷T-DNA plants (Fig. 4C). Similar to this, the northern-blot analysis revealed the absence of AtNTT2 mRNA in the homozygous knockout plants but showed the presence of this mRNA in wild-type leaf tissue (Fig. 4D, left section).
Figure 4.
Molecular characterization of homozygous AtNTT2 knockout mutants. A, Analysis of the AtNTT2-T-DNA-insertion line Garlic_288_E08.b.1a.Lb3FA (designated AtNTT2∷T-DNA). The insertion in AtNTT2∷T-DNA is localized in the second exon. The primers used for PCR analysis are marked as arrows. Primer NTT2/4 was chosen from the AtNTT2-promoter region, primer NTT2/2 from the 3′-untranslated region, and GARLIC_LB-primer from the left border of the T-DNA. B, PCR analysis on genomic DNA of wild-type (WT) and homozygous AtNTT2∷T-DNA mutants. C, RT-PCR analysis of the expression of the AtNTT2-genes in wild-type and in AtNTT2∷T-DNA mutant plants. cDNA was isolated from rosette leaves. Actin PCR revealed correct PCR conditions. D, Northern-blot analysis of wild-type and AtNTT2∷T-DNA mutant plants. Total RNA was extracted from rosette leaves. EtBr staining revealed equal RNA loading. Blots were hybridized with AtNTT1 or AtNTT2 specific probes, respectively.
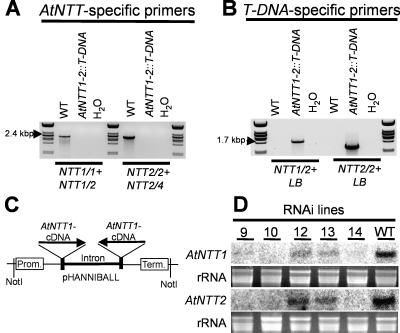
Although both genes, AtNTT1 and AtNTT2, reside on chromosome 1, we crossed homozygous AtNTT1 and AtNTT2∷T-DNA lines to receive null mutants (AtNTT1-2∷T-DNA), lacking both functional plastidic ATP/ADP-transporter genes. Due to the relatively wide distance of both genes on chromosome 1, it appeared likely to receive null mutants due to crossover during meiosis. We screened about 100 independent plants and identified 5 plants lacking intact genes from both transporters. That these plants represent homozygous null mutants has been demonstrated by PCR on genomic DNA (Fig. 5, A and B). The primer combinations NTT1/1 and NTT1/2, and NTT2/2 and NTT2/4, respectively, allowed amplification of expected PCR products on genomic DNA from wild-type but not from AtNTT1-2∷T-DNA plants (Fig. 5A). The use of the primer combinations NTT1/2 and LB, and NTT2/2 and LB, in contrast, allowed amplification of expected DNA fragments on DNA from homozygous AtNTT1-2∷T-DNA plants but not on DNA isolated from wild-type plants (Fig. 5B).
Figure 5.
Molecular characterization of the double-knockout mutant (designated AtNTT1-2∷T-DNA) and RNAi mutant. A, Amplification of AtNTT1- and AtNTT2-specific PCR products. B, Identification of the T-DNA in the AtNTT1 and AtNTT2 gene in the double-knockout mutant. C, Structure of the RNAi construct. A 418-bp fragment from AtNTT1cDNA was cloned in sense and antisense orientation into the pHANNIBALL vector. D, Northern-blot analysis of wild-type plants and different RNAi-lines. Five independent RNAi-lines were tested. Total RNA was extracted from rosette leaves. EtBr staining shows equal RNA loading. Blots were hybridized with AtNTT1 or AtNTT2 specific probes, respectively.
To prove that putative effects connected with the absence of functional AtNTT1 or AtNTT2 genes, or which are present in the double-knockout line are really due to reduced levels of corresponding gene products, we created further transgenic plant lines exhibiting strongly reduced levels of both mRNA species due to an RNAi effect. For this, we cloned a 418-bp fragment from AtNTT1 (corresponding to base positions 1,006–1,424 in the AtNTT1 cDNA; Kampfenkel et al., 1995) in sense and antisense orientation into the Hannibal vector (Fig. 5C). This cDNA fragment of AtNTT1 exhibits 92% sequence identity to the corresponding cDNA domain in AtNTT2 (see Möhlmann et al., 1998) leading to the expectation that the final RNAi construct might reduce the levels of both mRNA species simultaneously. After transformation of Arabidopsis plants, we received various independent transgenic plants with strongly reduced levels of AtNTT1 and AtNTT2 mRNA (Fig. 5D). Especially in lines 9, 10, and 14, both mRNA species were below the detection level (Fig. 5D). The absence of highly specific antisera detecting plastidic ATP/ADP transport proteins so far prevents a quantification of the final NTT protein levels in RNAi lines. However, as all RNAi lines generally exhibited physiological characteristics similar to null mutants (see below), we assume that they contained very strongly reduced transport activities, usually named knock down mutants.
Analysis of Germination and Growth Pattern of Wild-Type and Transgenic Arabidopsis Plants
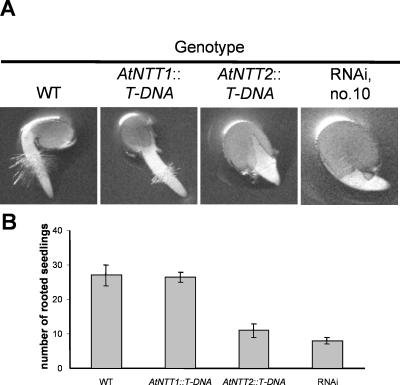
After 1 d of germination, Arabidopsis shows a primary root of about 3 to 4 mm length, exhibiting a root hair zone of about 1 mm following the root tip (Fig. 6A). Mutant plants lacking the functional transporter gene AtNTT1 showed a similar size and shape of the primary root as wild-type roots (Fig. 6A). In contrast, mutant plants lacking a functional AtNTT2 gene and RNAi line 10 exhibited less developed primary roots (Fig. 6A). RNAi lines 9 and 14 showed similarly reduced roots (data not shown). After 1 d of germination, AtNTT2∷T-DNA plants and RNAi lines showed a substantially reduced number of rooted seedlings when compared to wild-type or AtNTT2∷T-DNA plants (Fig. 6B).
Figure 6.
Root growth analysis during the first 24 h of germination. Arabidopsis seeds were sown on Murashige and Skoog agar plates. Twenty-four hours after imbibition the number of rooted seedlings was counted. A, Phenotype of wild-type and transgenic seedlings after 24 h on Murashige and Skoog agar plates. B, Number of rooted seedlings after 24 h. Per experiment, 30 seedlings were counted. Data represent the mean of two independent experiments.
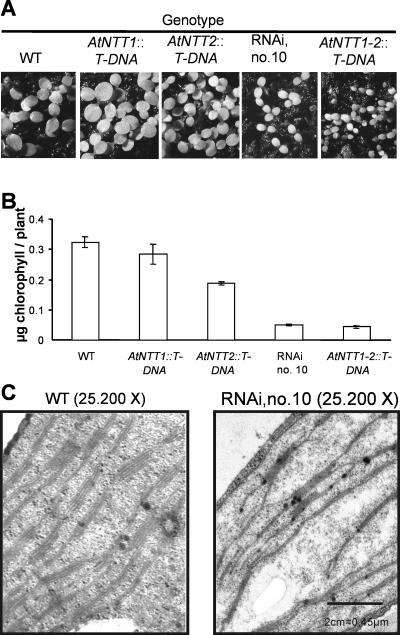
To reveal whether plastidic ATP/ADP transporters are important for development of photosynthetically competent chloroplasts we analyzed chlorophyll accumulation within the first days of development. For this we germinated wild-type and mutant seeds for 5 d in a growth chamber under short-day conditions and analyzed the resulting chlorophyll content. Wild-type and AtNTT1∷T-DNA seedlings showed similar levels of chlorophyll (Fig. 7, A and B). In contrast to this, knockout plants lacking a functional AtNTT2 gene; RNAi lines 10, 9, 14; and the null mutant showed a reduced seedling size (Fig. 7A; data not shown) and a strongly reduced average chlorophyll content (Fig. 7B; data not shown). Wild-type plants contained about 0.33 μg chlorophyll/plant, whereas chlorophyll in AtNTT2∷T-DNA plants amounted to only 0.20 μg/plant (Fig. 7B). Both plants from RNAi line 10 and null mutants contained less than one-sixth of the chlorophyll present in wild-type seedlings (Fig. 7B).
Figure 7.
Growth analysis, chlorophyll quantification, and chloroplast ultrastructure analysis from wild-type and transgenic plants. A, Growth analysis of 5-d-old wild-type and transgenic plants, grown in a climate-controlled chamber on soil under ambient conditions. B, Chlorophyll content in wild-type and transgenic seedlings. Data correspond to plants shown in A. Per measurement, 40 seedlings without root tissue were harvested. The data represent the mean of two independent experiments. sd less than 6% of the given mean. C, Chloroplast ultrastructure analyzed by transmission electron microscopy. Chloroplast substructure has been determined on 5-d-old wild-type or RNAi (line 10) seedlings, grown under short-day conditions.
To reveal whether the reduced chlorophyll level observed in some mutant lines is due to an impaired chlorophyll biosynthesis per se or might also correlate with alterations of the whole thylakoid system we examined the chloroplast ultrastructure by transmission electron microscopy. The ultrastructure of chloroplasts in 5-d-old wild-type plants exhibits a well-organized intraorganell membrane system, comprising grana and stroma thylakoids (Fig. 7C). In contrast, low-chlorophyll-containing chloroplasts from RNAi line 10 exhibited less thylakoids; especially the number of grana stacks appeared to be strongly reduced in this mutant (Fig. 7C).
To complete our picture on the effects of altered NTT gene expression on chloroplast development, we additionally analyzed the accumulation of nuclear-encoded chloroplast protein during initiation of deetiolation. For this, we germinated wild-type, RNAi, and null mutant seedlings for 6 d in the dark and illuminated etiolated seedlings for 8 or 24 h (at 100 μmol quanta m2 s−1). Subsequently, the change in the chlorophyll content was monitored, and the accumulation of chlorophyll-binding protein CP24, as indicator for altered plastidic protein import/maturation capacity, was examined by western-blot analysis.
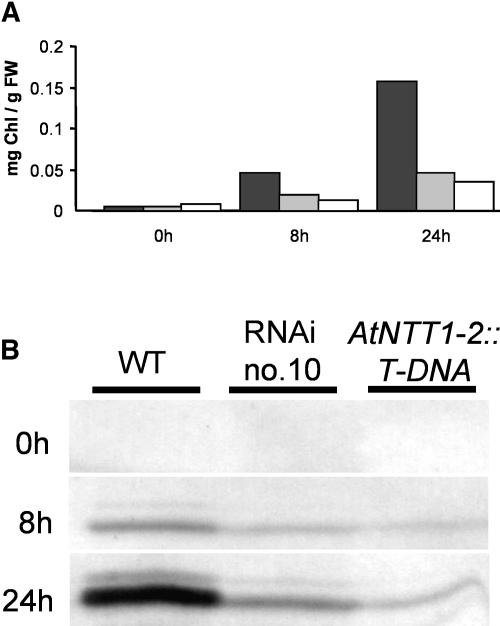
After 6 d of dark incubation, the chlorophyll levels in wild-type and all mutant seedlings were similarly low and amounted to less than 0.01 mg/plant (Fig. 8A). After 8 and 24 h of illumination the chlorophyll level in wild-type leaves increased already to about 0.045 mg/g fresh weight (FW) and 0.160 mg/g FW, respectively (Fig. 8A). In contrast, RNAi lines 10, 9, 14, and the null mutant showed much less chlorophyll accumulation, amounting to only about 0.050 mg/g FW after 24 h of illumination (Fig. 8A; data no shown). This reveals that also during the sudden induction of deetiolation, mutant plants showed a reduced capacity for chlorophyll synthesis.
Figure 8.
Chlorophyll content and accumulation of nuclear-encoded CP24 protein in wild-type and mutant plants. Wild-type and transgenic seedlings were germinated for 6 d in the dark, and etiolated seedlings were subsequently illuminated for up to 24 h (at 100 μmol quanta m2 s−1). A, Change in the chlorophyll content during illumination. For each measurement, 0.1-g seedlings without root tissue were harvested (wild type, black bar; RNAi, gray bar; AtNTT1-2∷T-DNA, white bar). B, Accumulation of chlorophyll-binding protein CP24 during illumination. Western-blot analysis was carried out using a polyclonal antiserum raised against the purified CP24 protein.
In none of the Arabidopsis lines analyzed was CP24 detectable after 6 d of dark germination (Fig. 8B). However, appreciable levels of CP24 were present in wild-type seedlings after already 8 h of light induction (Fig. 8B). CP24 levels in corresponding RNAi tissue (lines 10, 9, and 14) and null mutants were significantly lower than in wild-type tissue (Fig. 8B, and data not shown). After 24 h of light incubation, CP24 levels were high in wild-type tissue and still significantly lower in RNAi- (lines 10, 9, and 14) and null mutants (Fig. 8B; data not shown).
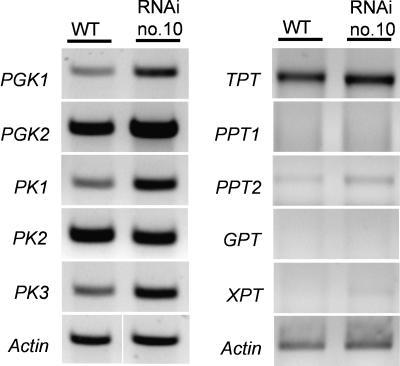
Besides the direct import of ATP via a plastidic ATP/ADP transporter, plastids may regenerate endogenous ATP via glycolytic enzyme activities. To raise evidence on an up-regulation of plastidic glycolytic activity during deetiolation in mutants, we quantified the level of mRNAs encoding enzymes and transporters involved. For this we germinated wild-type or RNAi plants for 6 d in the dark and illuminated subsequently etiolated seedlings for 8 h before cDNA was prepared. Gene-specific primers were chosen to amplify about 500-bp fragments coding for either plastidic phosphoglycerate kinases 1 and 2; pyruvate kinases 1, 2, and 3; or plastidic triose P/P, Glc 6-P/P, phosphoenolpyruvate/P transporters 1 and 2, and xyluose 5-P/P transporter. We observed increased mRNA levels of plastidic PGK1, PK1, and PK3 in RNAi plants compared to wild-type plants (Fig. 9), whereas the mRNA levels of the PGK2, PK2, and all plastidic phosphate transporters have not been changed substantially in mutant tissues (Fig. 9).
Figure 9.
RT-PCR expression analysis of genes coding for various plastidic glycolytic enzymes and different plastidic phosphate transporters. Wild-type and RNAi seedlings were germinated for 6 d in the dark and subsequently illuminated for 8 h. mRNA was extracted from seedlings without root tissue and converted to cDNA by RT. Gene-specific primers were chosen to amplify about 500-bp fragments coding for plastidic phosphoglycerate kinases 1 and 2 (AtPGK1, AtPGK2); pyruvate kinase 1, 2, and 3 (AtPK1, AtPK2, AtPK3); plastidic triose phosphate- (AtTPT); Glc-6-phosphate- (AtGPT); xylulose-5-phosphate/phosphate translocator (AtXPT); or phosphoenolpyruvate/phosphate transporters 1 and 2 (AtPPT1, AtPPT2). AtActin is given as control.
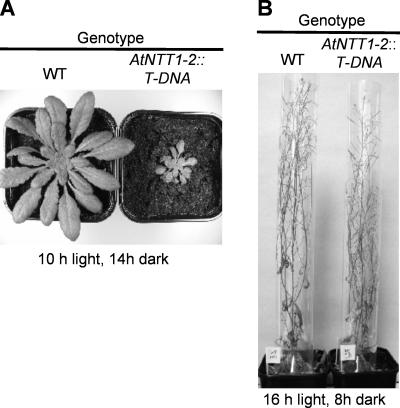
Wild-type and AtNTT1∷T-DNA plants grown for 50 d under short-day conditions exhibited an average rosette size of about 12 cm (Fig. 10A; data not shown). AtNTT2∷T-DNA plants, RNAi, or null mutants showed, however, a strongly reduced average size of the leaf rosette, approaching only 6 and 3 cm on average (Fig. 10A; data not shown). Interestingly, under long-day conditions (16 h light/d), the growth difference between wild-type plants and null mutants is nearly abolished (Fig. 10B).
Figure 10.
Growth analysis of wild-type and null mutants. Plants were grown on soil for 50 d in a climate-controlled growth chamber at 22°C and 100 μmol quanta m2 s−1. A, Growth pattern under short-day conditions (10 h light). B, Growth pattern under long-day conditions (16 h light).
The observation that RNAi and null mutants exhibited severely impaired growth tempted us to study physiological and morphological changes in these mutants in more detail. As the impaired growth is due to processes connected to a reduced plastidic ATP supply under conditions of long-night phases we first focused on changes in starch levels at the end of the day and night phase. However, we did not observe specific changes in transitory starch metabolism in AtNTT1-; AtNTT2∷T-DNA; RNAi lines 10, 9, and 14; or in null mutants when compared to wild-type leaves (plants were grown under short-day conditions). All plant lines exhibited starch contents equivalent to about 30 μmol C6/mg chlorophyll at the end of the day and about 7.5 μmol C6/mg chlorophyll at the end of the night.
Seed Quality Produced by Arabidopsis Plants with Reduced Plastidic ATP/ADP-Transporter Activity
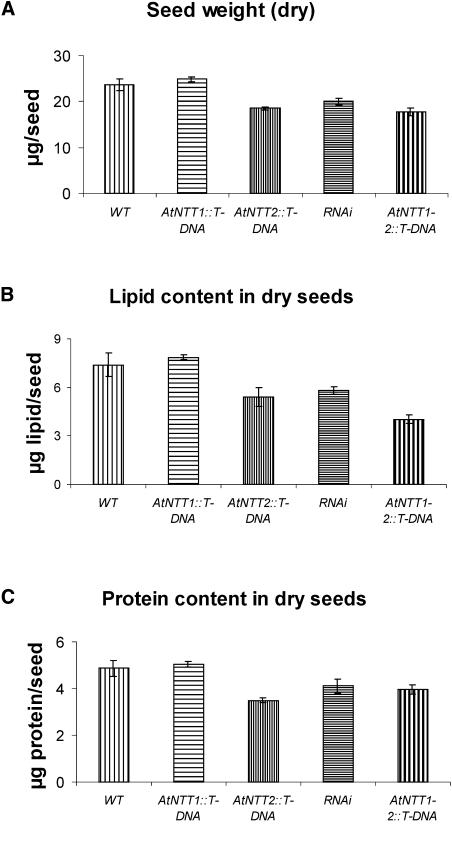
To analyze the effect of reduced plastidic ATP/ADP-transporter activity on seed quality, we grew Arabidopsis wild-type and mutant plants under long-day conditions, a light period required for induction of flowering. Fully developed seeds from wild type, AtNTT1- and AtNTT2∷T-DNA-, RNAi, and AtNTT1-2∷T-DNA mutants were collected from opened siliques, and the seed weight, the lipid content, and the protein levels were quantified (Fig. 11).
Figure 11.
Seed quality analysis of wild-type and transgenic plants. Plants were grown for 35 d on soil under short-day conditions. Subsequently the light phase was prolonged to induce flowering (16 h light) until the life cycle was completed. Seeds from wild-type, AtNTT1∷T-DNA, AtNTT2∷T-DNA, AtNTT1-2∷T-DNA, and RNAi were collected, and seed weight (A), lipid content (B), and protein content (C) in dry seeds were quantified. Data represent the mean of three independent experiments.
Wild-type and AtNTT1∷T-DNA seeds exhibited an average weight of 23 μg/seed (Fig. 11A). AtNTT2∷T-DNA, RNAi, and AtNTT1-2∷T-DNA seeds exhibited reduced average weights leading to 19, 20, and 18.5 μg/seed, respectively (Fig. 11A). Lipids represent the main storage product in Arabidopsis seeds. Both wild-type and AtNTT1∷T-DNA seeds accumulated similar levels of storage lipids amounting to about 7.2 to 7.5 μg lipid/seed (Fig. 11B). In contrast, AtNTT2∷T-DNA and RNAi seeds exhibited only 5.8 and 6.0 μg lipid/seed, respectively, and seeds from AtNTT1-2∷T-DNA plants still showed only 4.5 μg lipid/seed (Fig. 11B). The protein in wild-type and AtNTT1∷T-DNA seeds has been estimated to be about 4.8 μg/seed (Fig. 11C). AtNTT2∷T-DNA, RNAi, and null mutants showed reduced protein levels approaching 3.3, 3.9, and 3.8 μg/seed, respectively (Fig. 11C).
DISCUSSION
Regulation of NTT Isoform Expression
ATP represents a uniquely important cellular-energy source and is required in most cell compartments to energize a wide number of anabolic and catabolic reactions. Up to now three structurally unrelated types of intracellular ATP transporters have been identified, namely the mitochondrial AAC proteins, the plastidic ATP/ADP-transporters NTT, and the peroxisomal ATP/AMP carrier found in yeast (Saccharomyces cerevisiae; Fiore et al., 1998; Winkler and Neuhaus, 1999; Palmieri et al., 2002).
The observation that the Arabidopsis genome encodes two isoforms of plastidic ATP/ADP transporters with very similar biochemical properties (Möhlmann et al., 1998) is not surprising given that even in the unicellular bakers' yeast three AAC isoforms exist (Kalorov et al., 1990). It is assumed that AAC isoforms in all eukaryotic cells occur to allow an optimal adaptation of mitochondrial ATP transport activity on changing environmental and developmental conditions (Kalorov et al., 1990; Fiore et al., 1998). Obviously, the same holds true for the two plastidic ATP/ADP-transporter isoforms, as they also exhibit a spatially and developmentally regulated expression pattern (Figs. 1, B and D, and 2, A–C).
AtNTT1 and AtNTT2 mRNA accumulate in photosynthesizing leaf and stem cells (Fig. 1B). We assume that the nocturnal ATP import into the chloroplasts is the main reason for expression of both plastidic ATP/ADP-transporter genes in photosynthesizing cells. This assumption is strengthened for the following reasons: (1) Heldt (1969) demonstrated already 35 years ago that the biochemical properties of the chloroplastic ATP/ADP transporter prevents ATP export into the cytosol but allows ATP import during the night phase; and (2) the growth differences of null mutants grown under either short- or long-day conditions strikingly demonstrate that nocturnal ATP uptake into the chloroplasts is required for proper plant development (Fig. 10, A and B). This ATP is, however, in Arabidopsis not required for degradation of transitory starch (see “Results”; Nittyla et al., 2004) but for other, still unidentified processes.
The strong accumulation of AtNTT1 mRNA in leaf discs incubated on high Glc or Suc concentrations (Fig. 1D) indicates that this gene belongs to a large group of sugar up-regulated genes (Koch, 1996) required to reprogram chloroplasts into starch-accumulating ATP-importing storage plastids. In this respect AtNTT1 is, beside the plastidic Glc6P/Pi transporter (Quick et al., 1995), a second plastid envelope transporter protein up-regulated upon sugar application. The specific need for ATP supply into heterotrophic plastids is further indicated by the demonstration of AtNTT2 promoter activity in rapidly developing root tips and cotyledons and in petals (Fig. 2, A and C). This is indicative of a high ATP import into corresponding plastids, which is substantiated by the demonstration of ATP uptake and the presence of several ATP-dependent anabolic reactions in isolated chromoplasts, root plastids, and premature chloroplasts (Robinson and Wiskich, 1977; Kleinig and Liedvogel, 1980; Kleppinger-Sparace et al., 1992).
Why Does Reduced ATP Supply into Developing Plastids Impair Leaf and Root Development?
Both the northern-blot analysis and the promoter-GUS analysis indicate that especially AtNTT2 expression is high in root tips and cotyledons of developing seedlings (Fig. 1C). This observation tempted us to study the effect of altered plastidic ATP/ADP-transporter activity on both root formation and establishment of photosynthetic competence (Figs. 6, A and B, 7, A–C, and 8). The deletion of a functional AtNTT1 gene in Arabidopsis (Fig. 3, A–C) does not result in an impaired root formation of young seedlings (Fig. 6, A and B), nor did it appear that chlorophyll accumulation or seedling development was affected (Fig. 7, A and B). This observation nicely correlates with the relatively low expression of AtNTT1 in corresponding tissues (Figs. 1C and 2A). In strong contrast, the absence of a functional AtNTT2 gene (Fig. 4, A–C) or the reduction of both mRNA species (AtNTT1 and AtNTT2) in RNAi mutants (Fig. 5D) led to a strongly decreased formation of primary roots in young seedlings (Fig. 6, A and B) and a retarded chlorophyll accumulation (Fig. 7B) corresponding to a reduced growth rate (Figs. 7A and 10A).
The impaired root development is most likely due to an inhibited rate of fatty-acid synthesis. In plants this process takes place exclusively in plastids, and in case of root plastids the process has been characterized to be strictly dependent upon ATP import rather than on internal ATP regeneration via glycolytic reactions (Kleppinger-Sparace et al., 1992). The localization of AtNTT2 expression in the root tip, representing the meristematic zone of cell division and elongation, is therefore fully consistent with a high demand for fatty-acid synthesis in this tissue. In addition, heterotrophic plastids are known as a cellular site for energy-consuming amino acid biosynthesis (Neuhaus and Emes, 2000). Therefore, reduced rates of amino acid synthesis in mutant lines might also contribute to an impaired root development.
In the case of developing leaf tissue several processes are negatively affected by reduced plastidic ATP/ADP-transporter activity. First, the accumulation of chlorophyll is delayed in AtNTT2∷T-DNA-, AtNTT1-2∷T-DNA, and RNAi seedlings (Figs. 7A and 8A). Second, the generation of functional thylakoid structures is impaired in plants with strongly reduced plastidic ATP import capabilities (Fig. 7C); and third, the accumulation of nuclear-encoded proteins in developing-mutant chloroplasts is reduced (Fig. 8B). Both chlorophyll synthesis and protein import are dependent upon the presence of ATP at the stromal site (Soll and Tien, 1998; Buchanan et al., 2000), and the inhibition of both processes in mutant plastids strikingly show that alternative routes for ATP regeneration do not compensate for insufficient import capacity. Interestingly, we did not observe an accumulation of CP24 preprotein in mutant tissues exhibiting impaired accumulation of the mature CP24 protein (Fig. 8B). This observation might indicate that a so-far unknown signaling exists between the developing chloroplast and the nucleus regulating the expression of genes encoding for chloroplastic proteins. It should be mentioned here that we were not able to compare altered NTT mRNA accumulation in transgenic mutant lines with alterations of corresponding transport protein levels. For such analysis we will attempt to generate isoform-specific antisera in the near future.
Reduced ATP Supply into Developing Seed Plastids Limits Lipid Accumulation
We showed in the past that starch accumulation in potato tubers is strongly affected by altering the plastidic ATP/ADP-transporter activity (Tjaden et al., 1998a) leading to a high metabolic-flux control coefficient (Geigenberger et al., 2001). Therefore, we analyzed whether reduced plastidic ATP import capacity governs the end-product accumulation in Arabidopsis embryos to a similar extent as observed in potato. This analysis was further encouraged, since experiments on isolated rapeseed seed-embryo plastids showed that the highest rates of fatty-acid synthesis depend upon the supply with exogenous ATP (Eastmond and Rawsthorne, 1998; Rawsthorne, 2002), whereas a recently developed mathematical carbon-flux model indicated that net ATP import is not required for maximal fatty-acid synthesis in rapeseed embryos (Schwender et al., 2004).
As given in Figure 11, AtNTT1∷T-DNA did not show altered seed weight, lipid, and protein content when compared to wild-type seeds, whereas AtNTT2∷T-DNA seeds showed reduced weight, which correlates with reduced levels of lipids and storage protein (Fig. 11, A–C). Strongest reduction of the lipid content showed seeds generated from double-knockout mutants as these seeds contained only about 50% of the lipid content present in wild-type seeds (Fig. 11B). This result is surprising, because the expression level of NTT1 and NTT2 mRNA in developing siliques and seeds is remarkable low (Figs. 1B and 2C). Obviously, even low mRNA levels allow the maintenance of sufficient plastidic ATP import capacity.
It is important to note that the reduced seed oil phenotype is evident under long-day conditions, where the effects of gene knockout on whole-plant physiology, and hence maternal carbon supply to the embryo, were absent. These effects on storage product content are therefore likely to be specific to alterations to NNT gene expression in the seed. From this result we conclude that, similar to rapeseed and cauliflower (Brassica oleracea) inflorescence plastids (Möhlmann et al., 1994; Eastmond and Rawsthorne, 1998), Arabidopsis embryo plastids need to import cytosolic ATP to achieve the highest rates of lipid synthesis. This conclusion is fully consistent with recent observations that the overall energy status of developing rapeseed seeds correlate with lipid synthesis (Vigeolas et al., 2003).
However, Arabidopsis embryo plastids obviously possess, in addition to ATP/ADP-transporter proteins, endogenous sources for ATP regeneration because the absence of both transporter activities does not correlate with a total loss of storage product (Fig. 11B). In general two other metabolic pathways might allow stromal regeneration of ATP: First, chlorophyll-containing embryo plastids might regenerate ATP by photo-phosphorylation. Secondly, stromal-located glycolytic sequences might regenerate ATP at the enzymic steps catalyzed by phosphoglycerate kinase (PGK), or pyruvate kinase (PK). We would like to exclude the first possibility, since in case of rapeseed the light transmission into the developing seed tissue is supposed to be too low (Eastmond et al., 1996). Moreover, rapeseed mutants showing strongly reduced chlorophyll levels in developing embryos did not contain less lipids than wild-type plants (Tsang et al., 2003).
These independent observations point to stromal glycolysis as the alternative source for endogenous ATP resynthesis. This assumption is substantiated by the demonstration that Glc 6-phosphate is a very suitable carbon precursor for fatty-acid synthesis in rapeseed plastids (Kang and Rawsthorne, 1996). In addition, the observation that developing leaf plastids from RNAi plants exhibited increased accumulation of plastidial PGK- and PK mRNAs during deetiolation (Fig. 9) might indicate that heterotrophic Arabidopsis plastids use endogenous glycolysis for ATP resynthesis. The latter observation is moreover in full agreement with the demonstration of a DHAP-driven ATP production in etioplasts from dark-grown barley leaves (Batz et al., 1992). Obviously, some types of heterotrophic plastids use endogenous glycolysis for stromal ATP production, which contribute to energize anabolic reactions and compensate partly the lack of plastidic ATP import capacity in mutant plants (Figs. 6, 7, 8, 10, and 11). Moreover, the observation that mRNA coding for plastidic glycolytic enzymes specifically accumulated in deetiolating plastids from RNAi plants (Fig. 9) might indicate that the stromal ATP (energy) status is sensed and governs expression of genes allowing regeneration of ATP by alternative sources.
CONCLUSION
Arabidopsis contains two isoforms of plastidic ATP/ADP transporter to allow an optimal spatially and developmentally regulated adaptation of gene expression. Surprisingly, Arabidopsis does not need plastidic ATP/ADP-transporter activity to pass through the complete developmental cycle. However, plastidic ATP/ADP-transporter activity is required for a controlled development of young tissues, especially shown for roots and cotyledons, and is required in mature chloroplasts at night. The absence of plastidic ATP import in developing embryo tissue correlates with a reduction of lipid accumulation, which however still occurs at appreciable levels. This observation points to an ATP regeneration by stromal-located glycolytic enzymes, which seems to participate on ATP provision.
MATERIALS AND METHODS
AtNTT1 (AtNTT1∷T-DNA) and AtNTT2 (AtNTT2∷T-DNA) Knockout Mutant Plants
The heterozygous AtNTT1∷T-DNA mutant plant (Salk_013530) was provided by the SALK library. In that mutant the T-DNA is located in the first exon of AtNTT1 (locus At1g80300) on bp position 777. The heterozygous AtNTT2∷T-DNA mutant (GARLIC_ 288_E08.b.1a.Lb3Fa) was provided by the Torrey Mesa Research Institute (San Diego). In that mutant the T-DNA is located in the second exon of AtNTT2 (locus At1g15500) on bp position 1,015.
To confirm that we generated homozygous mutants after backcrossing, we used gene- and T-DNA-specific primers. For PCR on genomic DNA the following primers were used: NTT1/1(5′-TTTCTTCTGTGTATCTGCGGGAGAGAGTG-3′); NTT1/2 (5′-CTTTCTTTCCCCCCCAACAAAACCAAATA-3′); SALK_LB (5′-ACTCAACCCTATCTCGGGCTATTC-3′); NTT2/4 (5′-TCTCTTCTCCTCTCTACCCAGAGC-3′); NTT2/2 (5′-CCAAATCCCAAAACCCTTTTATTCATC-3′); and GARLIC_LB (5′-TAGCATCTGAATTTCATAACCAATCCGATACAC-3′).
Generation of Double-Knockout Plants (AtNTT1-2∷T-DNA)
To generate double-knockout mutants (designated AtNTT1-2∷T-DNA) lacking both functional plastidic ATP/ADP-transporter genes, homozygous AtNTT1∷T-DNA and homozygous AtNTT2∷T-DNA mutant plants were crossed. Although both genes reside on chromosome 1, we were able to identify double-knockout mutants by use of the primers given above.
Generation of RNAi Mutants
Transgenic RNAi plants were generated to achieve strongly reduced mRNA-levels of both AtNTT1 and AtNTT2. For Arabidopsis (Arabidopsis thaliana) transformation the pART27 vector (Gleave, 1992) was used. For this we cloned a 418-bp fragment from AtNTT1 (corresponding to bp positions 1,006–1,424) in sense and antisense orientation into the pHANNIBALL vector (Wesley et al., 2001). This cDNA sequence of AtNTT1 exhibits 92% identity to the corresponding cDNA sequence of AtNTT2. For the sense construct, the following primers were used: RNAi-sense-XhoI-fp, 5′-GATATGTTCCTCGAGCAACCCGTA-3′; and RNAi-sense-EcoRI-rp, 5′-GGAATTCCAAGTAGCGGTGTCATACC-3′. For the antisense construct, the following primers were used: RNAi-antisense-XbaI-fp, 5′-GATATGTTCCTCTAGAAACCCGTA-3′; and RNAi-antisense-BamHI-rp, 5′-CGGGATCCCGAAGTAGCGGTGTCATACC-3′. The sense construct was restricted with XhoI and EcoRI and the antisense construct with XbaI and BamHI. Restriction sites added by the primers ensured the correct orientation of the resulting sense and antisense constructs. The resulting pHANNIBALL constructs were subcloned as NotI fragments into pART27, and the final plasmid was subsequently transformed into Agrobacterium. Transformation of Arabidopsis was conducted according to the floral-dip method.
Generation of AtNTT1∷Promoter-GUS and AtNTT2∷Promoter-GUS Plants
For the generation of promoter-GUS constructs the binary vector pGPTV (Becker et al., 1992) containing the β-glucuronidase-(uidA-) gene from Escherichia coli was used. For the generation of promoter-GUS fusion constructs a promoter region of about 1.4 kb of either the AtNTT1 or AtNTT2 gene was cloned upstream of the GUS gene. The promoter region of the AtNTT1 or AtNTT2 gene (including 21 bp of the coding region) was amplified by PCR from genomic DNA. Both promoters were sequenced to check that the correct products were amplified. For amplification of the AtNTT1 promoter the following primers were used: JR1-sense, 5′-TGGACCTACATATGGGTTCGATTCGACTCC-3′; and JR2ant, 5′-AAGAGAGAAGCCCCCGGGTTTGAATCACAGC-3′. For amplification of the AtNTT2 promoter the following primers were used: JR3-sense, 5′-GGAAGAATCTGAAGTTTTGGAACCC-3′; and JR4-anti, 5′-GAGAGAATTCCCCGGGTTTGAATCAG-3′. After blunt-end ligation of the PCR products in T7 orientation into the SmaI-restricted pBSK vector, both promoters were restricted with HindIII and SmaI and subsequently inserted in frame with the GUS gene. The resulting constructs were used for Agrobacterium transformation. Transformation of Arabidopsis was as given above.
Generation of AtNTT1- and AtNTT2-Specific Probes for Northern-Blot Analysis
Gene-specific probes, each corresponding to the respective 3′untranslated region, were generated by PCR on cDNA. AtNTT1-specific probes were amplified with the following primers: At1oligo3sense, 5′-GGAGAAATCTGCTCC-3′; and At1oligo1anti, 5′-ACTTCAACGATACACACAAAGG-3′. AtNTT2-specific probes were amplified with the following primers: At2oligo3sense, 5′-ACTGGCATTTAGACG-3′; and At2oligo1anti, 5′-CTAGTTTGGTATTGG-3′. The PCR products were subsequently cloned into the pGEMTeasy vector. For northern-blot analysis the cloned fragments were excised, separated by gel electophoresis, gel purified, and radioactively labeled with [α32P]-dCTP by random priming, using the Ready To Go kit (Amersham-Pharmacia, Freiburg, Germany).
Gene Expression Studies
Poly(A+) mRNA was isolated from rosette leaves or whole seedlings (0.1 g each) by use of Dynabeads (Dynal AS, Oslo) and converted to cDNA by reverse transcription (SuperscriptII, Invitrogen, Carlsbad, CA). For semiquantitative RT-PCR reactions were carried out with 1 μL template and 1 unit Taq-Polymerase in a total volume of 50 μL. PCR conditions were 3 min at 95°C, followed by 25 cycles of 30 s at 96°C, 30 s at 50°C, and 60 s at 72°C. Primers used for amplification are listed. Gene-specific primers were used to amplify PCR products of 450 to 550 bp. The following primers were used: pyruvate kinase I-fp, (At1g32440) 5′-GTGCGACTCTTCCATCCATT-3′; pyruvate kinase I-rp, (At1g32440) 5′-GGTTCTCAACGCCACAGTAT-3′; pyruvate kinase II-fp, (At3g22960) 5′-CAAGGCGCTCACGGTTCTAA-3′; pyruvate kinase-II-rp, (At3g22960) 5′-CATGTCCGAGACTGCGATCA-3′; pyruvate kinase III-fp, (At3g52920) 5′-CTCCTGAAGATGTGCCTAAC-3′; pyruvate kinase III-rp, (At3g52920) 5′-CCAGTCCTTCTCAGTGATTG-3′; phosphoglycerate kinase I-fp, (At1g56190) 5′-ACGATGGCGAAGAAGAGTGT-3′; phosphoglycerate kinase I-rp, (At1g56190) 5′-ACCACCAAGCAGAAGGATGT-3′; phosphoglycerate kinase II-fp, (At3g12780) 5′-CTACCGAAGGAGTCACTAAG-3′; phosphoglycerate kinase II-rp, (At3g12780) 5′-CTGAGTCTCCTCCTCCTATT-3′; phosphoenolpyruvate translocator I-fp, (At5g33320) 5′-CATCTTGCCGCTTGCTGTTGT-3′; phosphoenolpyruvate translocator I-rp, (At5g33320) 5′-TGGAAGCAGAGTGCAGCGATAA-3′; phosphoenolpyruvate translocator II-fp, (At3g01550) 5′-TCAGCGTTGAGCAGAAGAAG-3′; phosphoenolpyruvate translocator II-r, (At3g-01550) 5′-TCACCGAGCAAGAGAACAGA-3′; triose-phosphate translocator-fp, (At5g46110) 5′-CTGAAGGTGGAGATACCGCTG-3′; triose-phosphate translocator-rp, (At5g46110) 5′-GAGTGCGATGATGGAGATGTA-3′; Glc 6-phosphate translocator-fp, (At5g54800) 5′-TTCCATCGACGGAGCTTCCA-3′; Glc-6-phosphate translocator-rp, (At5g54800) 5′-ACGCAGGTTCACCACTCTTG-3′; xylulose-5-phosphate translocator-fp, (At5g17630) 5′-CCGTTGGCTCATCGGATTCAA-3′; xylulose-5-phosphate translocator-rp, (At5g17630) 5′-GCTCTGTAAGCTACGTTTAGA-3′; Actin-fp, 5′-TGTACGCCAGTGGTCGTACAACC-3′; and Actin-rp, 5′-GGAGCAAGAATGGAACCACCG-3′.
Cultivation of Plant Material
Wild-type and transgenic Arabidopsis plants (ecotype Columbia) were grown in a climate-controlled chamber on soil at 22°C and 100 μmol quanta m2 s−1. Prior to germination, seeds were incubated for 2 d in the dark at 4°C for imbibition (Weigel and Glazebrook, 2002). For short-day growing conditions, the light was given for 10 h/d; for long-day conditions light was present for 16 h/d. For root growth and seedling analysis, surface-sterilized seeds were sown on half-concentrated Murashige and Skoog (1962) plates, containing 0.8% Agar, 0.05% MES (adjusted to pH 5.7 with KOH), and 1% Suc. Prior to germination, plates were incubated at 4°C for 2 d in the dark and subsequently transferred to the growth chamber, and growth was continued for 24 h under long-day conditions.
Extraction of Total RNA and RNA Gel-Blot Hybridization
Total RNA was isolated from frozen tissue samples (liquid nitrogen) by using the Purescript extraction kit (Gentra Systems, North Minneapolis, MN), according to the manufacturer's instructions. For RNA gel-blot hybridization analysis, standard methods (Sambrook et al., 1989) were used. Blots were visualized by a Phospho-Imager (Packard, Frankfurt).
Histochemical Localization of GUS
Whole seedlings or tissue from transgenic plants were collected in glass scintillation vials, filled with ice-cold 90% acetone, and incubated for 20 min at room temperature. Subsequently, the samples were stained according to standard protocols (Weigel and Glazebrook, 2002).
Immunological Analysis
Accumulation of chlorophyll-binding protein CP24 during illumination was examined by western-blot analysis. Antibodies were kindly provided by Prof. R.B. Klösgen (Pflanzenphysiologie, Martin-Luther Universität Halle, Germany). Plant tissue (0.5 g) frozen in liquid nitrogen was homogenized in 250 μL buffer A (50 mm HEPES, 5 mm MgCl2, pH 7.5, 2% SDS, 1% Triton X-100, 15% glycerol, 1 mm EDTA, phenylmethylsulfonyl fluoride [PMSF] 1/100 [v/v]) at room temperature. SDS-PAGE, northern transfer, and immunodetection were conducted according to standard protocols.
Transmission Electron Microscopy
For chloroplast ultrastructure analysis from wild-type and RNAi mutants cotyledons from 5-d-old seedlings, grown under short-day conditions, were used. The seedlings were fixed with solution 1 (3% [v/v] glutaraldehyd, 30 mm PIPES, pH 7.0) for 1 h and subsequently washed two times for 10 min in cacodylat buffer, pH 7.0 (50 mm sodium cacodylat, 6.4 mm HCl). The samples were post fixed in solution 2 (1% [w/v] osmium tetroxide, 50 mm sodium cacodylat, 6.4 mm HCl, pH 7.0) for 1 h and washed as described above. Subsequently, samples were incubated for 1 h in 0.5% uranylacetat, followed by a serial dehydration with 30%, 50%, 70%, 90%, and 100% (v/v) of acetone in water. The specimens were infiltrated with a series of 25%, 50%, 75%, and 100% Spurr (Ted Pella, Redding, CA) in acetone. After embedding in Spurr the blocks were sectioned and stained with 2% uranyl acetate and lead citrate before viewing in a transmission electron microscope (Zeiss, Oberkochen, Germany).
Seed Analysis
For lipid quantification, 0.1 g completely mature and air-dried seeds were homogenized in a mortar in liquid nitrogen. Subsequently, 1.5 mL isopropanol was added and further homogenized. The suspension was transferred into a 1.5-mL reaction tube and incubated for 12 h at 4°C on a laboratory shaker at 100 rpm. Subsequently, samples were centrifuged at 12,000g for 10 min and the supernatant was transferred into preweighted 1.5-mL reaction tube. Tubes were incubated at 60°C for 8 h to evaporate the isopropanol. Subsequently, total lipid was quantified gravitometrically.
For seed protein quantification 0.1-g seeds were homogenized in a mortar at room temperature. Subsequently, 1,000 μL buffer medium 1 (50 mm HEPES, 5 mm MgCl2, pH 7.5, 1% Triton X-100, 15% glycerol, 2% SDS, 1 mm EDTA, PMSF, 1/100 [v/v]) was added and further homogenized. The suspension was transferred into 1.5-mL reaction tubes, and samples were centrifuged at 12,000g at room temperature for 10 min. The supernatant was transferred into new 1.5-mL reaction tubes, and proteins were quantified with bicinchoninic acid reagent (Pierce Chemical, Rockford, IL) according to manufacturer's instructions.
Chlorophyll Quantification
Chlorophyll quantification was carried out according to a standard protocol (Arnon, 1949).
Acknowledgments
We thank Prof. R.B. Klösgen and Dr. M. Gutensohn (Martin-Luther Universität Halle, Germany) for kindly supplying CP24 antiserum. We are grateful to Dr. H. Fuge (Zellbiologie, Universität Kaiserslautern) for his support during electron microscopy.
This work was supported by the Deutsche Forschungsgemeinschaft, Schwerpunktprogramm 1108 (Pflanzenmembrantransport).
This paper is dedicated to a nestor of plant physiology and great biologist, Prof. Dr. Dr. hc Erwin Latzko (Kranzberg, Germany), on the occasion of his 80th birthday.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.049502.
References
- Arnon DI (1949) Copper enzymes in isolated chloroplasts: polyphenol oxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batz O, Scheibe R, Neuhaus HE (1992) Transport processes and corresponding changes in metabolite levels in relation to starch synthesis in barley (Hordeum vulgare L.) etioplasts. Plant Physiol 100: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20: 1195–1197 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Gruissem W, Jones RL (2000) Biochemistry & Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD
- Drgon T, Sabova L, Nelson N, Kolarov J (1991) ADP/ATP translocator is essential only for anaerobic growth of yeast Saccharomyces cerevisiae. FEBS Lett 289: 159–162 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Kolacna L, Rawsthorne S (1996) Photosynthesis by developing embryos of oilseed rape (Brassica napus L.). J Exp Bot 47: 1763–1769 [Google Scholar]
- Eastmond PJ, Rawsthorne S (1998) Comparison of the metabolic properties of plastids isolated from developing leaves and embryos of Brassica napus L. J Exp Bot 49: 1105–1111 [Google Scholar]
- Fiore C, Trézéguet V, Le Saux A, Roux P, Dianoux AC, Noel F, Lauquin GJM, Brandolin G, Vignais PV (1998) The mitochondrial ADP/ATP carrier: structural, physiological and pathological aspects. Biochimie 80: 137–150 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Stamme C, Tjaden J, Schulze A, Quick WP, Betsche T, Kersting HJ, Neuhaus HE (2001) Tuber physiology and properties of starch from tubers of transgenic potato plants with altered plastidic adenylate transporter activity. Plant Physiol 125: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1210 [DOI] [PubMed] [Google Scholar]
- Heldt HW (1969) Adenine nucleotide translocation in spinach chloroplasts. FEBS Lett 5: 11–14 [DOI] [PubMed] [Google Scholar]
- Kalorov J, Kalorova N, Nelson N (1990) A third ADP/ATP translocator gene in yeast. J Biol Chem 265: 12711–12716 [PubMed] [Google Scholar]
- Kampfenkel K, Möhlmann T, Batz O, van Montagu M, Inzé D, Neuhaus HE (1995) Molecular characterization of an Arabidopsis thaliana cDNA encoding a novel putative adenylate translocator of higher plants. FEBS Lett 374: 351–355 [DOI] [PubMed] [Google Scholar]
- Kang F, Rawsthorne S (1994) Starch and fatty acid synthesis in plastids from developing embryos of oilseed rape (Brassica napus L.). Plant J 6: 795–805 [Google Scholar]
- Kang F, Rawsthorne S (1996) Metabolism of glucose-6-phosphate and utilization of multiple metabolites for fatty acid synthesis by plastids from developing oilseed rape embryos. Planta 199: 321–327 [Google Scholar]
- Kleinig H, Liedvogel B (1980) Fatty acid synthesis by isolated chromoplasts from the daffodil: energy source and distribution patterns of the acids. Planta 150: 166–169 [DOI] [PubMed] [Google Scholar]
- Kleppinger-Sparace KF, Stahl RJ, Sparace SA (1992) Energy requirements for fatty acid synthesis and glycerolipid biosynthesis from acetate by isolated pea root plastids. Plant Physiol 98: 723–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg M (1989) Molecular aspects of adenine nucleotide carrier from mitochondria. Arch Biochem Biophys 270: 1–14 [DOI] [PubMed] [Google Scholar]
- Koch KE (1996) Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 509–540 [DOI] [PubMed] [Google Scholar]
- Kolarov J, Kolarova N, Nelson N (1990) A third ADP/ATP translocator gene in yeast. J Biol Chem 265: 12711–12716 [PubMed] [Google Scholar]
- Linka N, Hurka H, Lang BF, Burger G, Winkler HH, Stamme C, Urbany C, Seil I, Kusch J, Neuhaus HE (2003) Phylogenetic relationship of non-mitochondrial nucleotide transport proteins in bacteria and eukaryotes. Gene 306: 27–35 [DOI] [PubMed] [Google Scholar]
- Möhlmann T, Scheibe R, Neuhaus HE (1994) Interaction between fatty-acid and starch synthesis in isolated amyloplasts from cauliflower floral buds. Planta 194: 492–497 [Google Scholar]
- Möhlmann T, Tjaden J, Schwöppe C, Winkler HH, Kampfenkel K, Neuhaus HE (1998) Occurrence of two plastidic ATP/ADP transporters in Arabidopsis thaliana L.: molecular characterisation and comparative structural analysis of similar ATP/ADP translocators from plastids and Rickettsia prowazekii. Eur J Biochem 252: 353–359 [DOI] [PubMed] [Google Scholar]
- Murashige F, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco tissue cultures. Plant Physiol 15: 473–496 [Google Scholar]
- Neuhaus HE, Emes MJ (2000) Nonphotosynthetic metabolism in plastids. Annu Rev Plant Physiol Plant Mol Biol 51: 111–140 [DOI] [PubMed] [Google Scholar]
- Nittyla T, Messerli G, Trevisan M, Chen J, Smith A, Zeeman SC (2004) A previously unknown maltose transporter essential for starch degradation in leaves. Science 203: 87–89 [DOI] [PubMed] [Google Scholar]
- Palmieri L, Rottensteiner H, Girzalsky W, Scarcia P, Palmieri F, Erdmann R (2002) Identification and functional reconstitution of the yeast peroxisomal adenine nucleotide transporter. EMBO J 20: 5049–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick WP, Scheibe R, Neuhaus HE (1995) Induction of hexose-phosphate translocator activity in spinach chloroplasts. Plant Physiol 109: 113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawsthorne S (2002) Carbon flux and fatty acid synthesis in plants. Prog Lipid Res 41: 182–196 [DOI] [PubMed] [Google Scholar]
- Robinson SP, Wiskich JT (1977) Uptake of ATP analogs by isolated pea chloroplasts and their effect on CO2 fixation and electron transport. Biochim Biophys Acta 461: 131–140 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schünemann D, Borchert S, Flügge UI, Heldt HW (1993) ATP/ADP translocator from pea root plastids. Comparison with translocators from spinach chloroplasts and pea leaf mitochondria. Plant Physiol 103: 131–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender J, Ohlrogge JB, Shachar-Hill Y (2004) A flux model of glycolysis and the oxidative pentosephosphate pathway in developing Brassica napus embryos. J Biol Chem 278: 29442–29453 [DOI] [PubMed] [Google Scholar]
- Soll J, Tien R (1998) Protein translocation into and across the chloroplastic envelope membranes. Plant Mol Biol 39: 191–207 [PubMed] [Google Scholar]
- Tjaden J, Möhlmann T, Kampfenkel K, Henrichs G, Neuhaus HE (1998. a) Altered plastidic ATP/ADP-transporter activity influences potato (Solanum tuberosum) tuber morphology, yield and composition of tuber starch. Plant J 16: 531–540 [Google Scholar]
- Tjaden J, Schwöppe C, Möhlmann T, Neuhaus HE (1998. b) Expression of the plastidic ATP/ADP transporter gene in Escherichia coli leads to a functional adenine nucleotide transport system in the bacterial cytoplasmic membrane. J Biol Chem 273: 9630–9636 [DOI] [PubMed] [Google Scholar]
- Tsang EWT, Yang J, Chang Q, Nowak G, Kolenosky A, McGregor DI, Keller WA (2003) Chlorophyll reduction in the seed of Brassica napus with a glutamate 1-semialdehyde aminotransferase antisense gene. Plant Mol Biol 51: 191–201 [DOI] [PubMed] [Google Scholar]
- Vigeolas H, van Dongen JT, Waldeck P, Hühn D, Geigenberger P (2003) Lipid storage metabolism is limited by the prevailing low oxygen concentrations within developing seeds of Brassica napus L. Plant Physiol 133: 2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Wesley SV, Helliwell CA, Smith SA, Wang NB, Rouse DT, Liu Q, Gooding PS, Sing SP, Abbott P, Stoutjesdijk PA, et al (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Winkler HH, Neuhaus HE (1999) Non-mitochondrial ATP transport. Trends Biochem Sci 24: 64–68 [DOI] [PubMed] [Google Scholar]