Abstract
Rhizobium nodulation (Nod) factors are specific lipochito-oligosaccharide signals essential for initiating in root hairs of the host legume developmental responses that are required for controlled entry of the microsymbiont. In this article, we focus on the Nod factor signal transduction pathway leading to specific and cell autonomous gene activation in Medicago truncatula cv Jemalong in a study making use of the Nod factor-inducible MtENOD11 gene. First, we show that pharmacological antagonists that interfere with intracellular ion channel and Ca2+ pump activities are efficient blockers of Nod factor-elicited pMtENOD11-β-glucuronidase (GUS) expression in root hairs of transgenic M. truncatula. These results indicate that intracellular Ca2+ release and recycling activities, essential for Ca2+ spiking, are also required for specific gene activation. Second, pharmacological effectors that inhibit phospholipase D and phosphoinositide-dependent phospholipase C activities are also able to block pMtENOD11-GUS activation, thus underlining a central role for multiple phospholipid signaling pathways in Nod factor signal transduction. Finally, pMtENOD11-GUS was introduced into all three Nod−/Myc− dmi M. truncatula mutant backgrounds, and gene expression was evaluated in response to the mastoparan peptide agonist Mas7. We found that Mas7 elicits root hair MtENOD11 expression in dmi1 and dmi2 mutants, but not in the dmi3 mutant, suggesting that the agonist acts downstream of DMI1/DMI2 and upstream of DMI3. In light of these results and the recently discovered identities of the DMI gene products, we propose an integrated cellular model for Nod factor signaling in legume root hairs in which phospholipids play a key role in linking the Nod factor perception apparatus to downstream components such as Ca2+ spiking and ENOD gene expression.
The establishment of the symbiotic association between legumes and the soil bacteria rhizobia leading to the formation of the nitrogen-fixing root nodule requires the synthesis and mutual perception of a variety of signaling molecules. Rhizobium-secreted lipochito-oligosaccharides (LCOs), known as Nod factors, are key players in this molecular dialogue, eliciting in both epidermal and inner root tissues of the host plant developmental responses that are required for both bacterial entry and nodule organogenesis (for review, see Geurts and Bisseling, 2002; Oldroyd and Downie, 2004). Discovering how Nod factors bring about these changes provides a unique opportunity of studying signal perception and transduction in higher plants.
Until now, research on Nod factor signal transduction has essentially focused on epidermal root hairs, the initial site of Rhizobium-plant contact. At the morphological level, Nod factor solutions induce root hair deformation in growth-terminating hairs, resulting from transient tip swelling followed by renewed polar outgrowth (de Ruijter et al., 1998). In addition, it has been shown that changes in the actin cytoskeleton are associated with this Nod factor root hair response (de Ruijter et al., 1999). More recently, Esseling et al. (2003) have demonstrated that spot application of Nod factors to one side of the root hair apex induces rapid reorientation of tip growth toward the side of application. Electrophysiological studies have revealed that Nod factors trigger very rapid ion fluxes (including Ca2+, H+, Cl−, and K+) across the root hair plasma membrane (Felle et al., 1998) associated with membrane depolarization (Ehrhardt et al., 1992; Felle et al., 1995). Further direct evidence for a role for Ca2+ in Nod factor signaling has come from measurements of changes in cytoplasmic concentration, revealing both rapid increases of intracellular Ca2+ (Cárdenas et al., 1999; Felle et al., 1999; Shaw and Long, 2003) and delayed regular Ca2+ spiking (Ehrhardt et al., 1996; Wais et al., 2000; Harris et al., 2003) in response to Nod factors. A pharmacological approach based on the activation of a β-glucuronidase (GUS) reporter driven by an early nodulin gene promoter (pMtENOD12) in transgenic alfalfa (Medicago sativa) was used to study Nod factor signal transduction in root hairs, leading to a proposed pathway involving heterotrimeric G proteins mediating downstream phosphoinositide-based signaling (Pingret et al., 1998). Subsequent biochemical studies in common vetch (Vicia sativa) suggested roles for not only phospholipase C (PLC) but also phospholipase D (PLD) in transducing responses to the Nod factor symbiotic signal (den Hartog et al., 2001).
Genetic approaches have also made significant contributions to our current knowledge of Nod factor signaling mechanisms (for review, see Cook, 2004). In the case of the model legume Medicago truncatula, a number of genes have so far been identified that are essential for Rhizobium infection and nodulation and are involved in the earliest responses to Nod factor signaling in root hairs. Mutants of three of these genes (DMI1, DMI2, and DMI3) are partially defective in root hair deformation responses, and Nod factors fail to elicit ENOD gene expression (Catoira et al., 2000). dmi mutants are also resistant to endomycorrhizal fungal colonization, with root penetration blocked at the appressorium stage (Sagan et al., 1995, 1998; Calantzis et al., 2001). Since dmi1 and dmi2 mutants (but not the dmi3 mutant) are defective in Nod factor-elicited Ca2+ spiking, it has been proposed that this characteristic calcium response is an integral element of the signaling pathway and positioned downstream of DMI1 and DMI2 and upstream of DMI3 (Wais et al., 2000). In contrast to the dmi mutants, the nfp mutant (Nod−/Myc+) is defective in all known Nod factor responses (Ben Amor et al., 2003), thus placing the corresponding NFP gene upstream of the DMI genes. A probable ortholog of NFP has been cloned in Lotus japonicus (Madsen et al., 2003), suggesting that this gene encodes a putative Nod factor receptor kinase (called LYK) with a LysM-rich extracellular domain. In addition, all three M. truncatula DMI genes have recently been cloned, revealing that DMI1 is a novel transmembrane protein with unknown function (Ané et al., 2004), DMI2 is a receptor-like protein kinase (called NORK) with an extracellular domain containing Leu-rich repeats (Endre et al., 2002), and DMI3 is a putative calcium/calmodulin-dependent protein kinase (CCaMK; Lévy et al., 2004).
In this article, we extend pharmacological and genetic studies on Nod factor signaling in root hairs making use of a single-copy homozygous M. truncatula line transformed with the pMtENOD11-GUS reporter and a novel experimental protocol adapted to young seedlings. First, we show that a range of pharmacological antagonists previously shown to block Nod factor-elicited Ca2+ spiking (Engstrom et al., 2002) are also efficient inhibitors of pMtENOD11-GUS induction. Second, we provide additional evidence that multiple phospholipid signaling pathways are likely to be central to the transduction of Nod factor perception at the root hair plasma membrane into specific cellular responses such as Ca2+ spiking and gene activation. Finally, we have studied the capacity of a mastoparan peptide agonist (Mas7) to elicit pMtENOD11-GUS transcription in all three dmi mutant backgrounds, thereby allowing us to position the target for this agonist in relation to the three genes within the overall pathway. These data are discussed in the framework of our current knowledge of Nod factor signaling in root hairs and integrated into a cellular model.
RESULTS
The pMtENOD11-GUS Reporter for Studying Nod Factor Signaling in Root Hairs
MtENOD11 is a single-copy M. truncatula gene encoding a repetitive (hydroxy) Pro-rich protein believed to be a component of the plant extracellular matrix (Journet et al., 2001). To study the expression pattern of this early nodulin gene, we constructed a transcriptional promoter-GUS fusion using a 2.3-kb MtENOD11 promoter fragment. The introduction of this construct into M. truncatula cv Jemalong (A17) by means of Agrobacterium tumefaciens-mediated leaf disc transformation and the selection of a representative single-copy primary transformant line (L416) are described in “Materials and Methods.” Results presented in a previous article (Journet et al., 2001) showed that spatiotemporal expression of this early nodulin gene prior to and during Sinorhizobium meliloti root infection, as well as in the distal zone of the nodule, closely resembled that described for the related repetitive Pro-rich protein-encoding M. truncatula ENOD12 gene (Pichon et al., 1992).
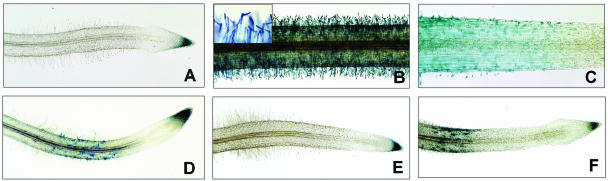
Since the pMtENOD12-GUS reporter is transcriptionally activated in the alfalfa root epidermis in response to exogenous purified Nod factors (Journet et al., 1994), similar experiments were performed using the pMtENOD11-GUS L416 line. Compared to untreated transgenic roots (Fig. 1A), 6-h treatment with 10−9 m S. meliloti Nod factors (NodSm) is sufficient to elicit strong expression of the pMtENOD11-GUS fusion in root hairs (Fig. 1B). Histochemical staining can be observed in root hairs that show marked deformations caused by Nod factor treatment (see inset). Reverse transcription-PCR experiments have shown that induction of endogenous MtENOD11 transcription can be detected within 1 to 2 h in response to NodSm (not shown), and this is illustrated using the GUS reporter in Figure 1C, where blue staining is already clearly visible in epidermal cells close to the root tip only 2 h after treatment with Nod factor. Note that GUS activity is visible in the epidermal cell layer prior to the initiation of the root hair apical outgrowth (Fig. 1C), suggesting that MtENOD11 transcription is induced early during differentiation of epidermal cells into trichoblasts.
Figure 1.
Expression of the pMtENOD11-GUS fusion in transgenic M. truncatula in response to Nod factor and mastoparan peptides. A, Untreated control L416 (pMtENOD11-GUS) root showing absence of GUS staining in the epidermis and strong constitutive staining in the root cap. B, Strong GUS staining in the L416 root epidermis following 6-h treatment with 10−9 m NodSm. Inset shows detail of typical Nod factor-elicited root hair deformation. C, Region of root hair emergence close to the root tip stained for GUS activity after only 2-h treatment with 10−9 m NodSm. Note that histochemical staining can be observed in epidermal cells prior to visible polar outgrowth. D, Reporter gene expression in L416 roots in response to 12-h treatment with 1 μm mastoparan. E, Lack of GUS expression in L416 roots after treatment with the inactive control mastoparan analog, Mas17 (1 μm, 12 h). F, pMtENOD11-GUS expression following treatment with the more active synthetic analog Mas7 (0.5 μm, 12 h).
We have previously shown that NodSm lacking the sulfate moiety is significantly less active in triggering expression of a pMtENOD12-GUS reporter in transgenic alfalfa (Journet et al., 1994). The results presented in Table I show that GUS activity in roots of L416 can be elicited at concentrations of NodSm down to 10−14 m, whereas nonsulfated Nod factors, purified from an S. meliloti nodH mutant, are only active down to 10−9 m. Thus, sulfation is also of critical importance for the transcriptional activation of MtENOD11. Since decorations at the nonreducing end of the Nod factor GlcNAc backbone are also important for biological activity, we evaluated gene-inducing activity resulting from modifications in either the O-acetyl or lipid moieties. Table I shows that factors produced by a nodF/nodL mutant, lacking the O-acetyl and with a C18:1 lipid side chain replacing C16:2, are 10- to 100-fold less active than wild-type S. meliloti Nod factors. If the lipid side chain is then shortened to C8:1 (a synthetic LCO), activity drops two to three orders of magnitude further, and if the entire lipid chain is removed [sulfated chitin tetramer; CO-IV (S, NH2)], transgene activation now requires doses in the 10−7 to 10−8 m range. On the other hand, nonsulfated chitin tetramers are inactive in eliciting pMtENOD11-GUS expression at equivalent concentrations. Thus, removing either the reducing end sulfate or the entire lipid side chain at the nonreducing end causes major reductions in Nod factor activity, and our results imply that the effects of modifications at the two ends of the sugar backbone are additive. In conclusion, pMtENOD11-GUS is a particularly convenient Nod factor-responsive reporter for studying cell autonomous signal transduction in the root epidermis. Nod factor-elicited GUS activity is highly specific with regard to both cell type (differentiating root hairs) and Nod factor structure and can be detected at concentrations of NodSm down to 10−14 m. This is at least one order of magnitude lower than that required for eliciting pMtENOD12-GUS expression (Journet et al., 1994).
Table I.
Relationship between LCO structure and activation of the pMtENOD11-GUS fusion in transgenic M. truncatula
| Elicitor
|
Elicitor Concentration (m)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 10−7 | 10−8 | 10−9 | 10−10 | 10−11 | 10−12 | 10−13 | 10−14 | |
| NodSm-IV (Ac, S, C16:2) (parental strain) | + | + | + | + | + | + | + | v.wk. |
| NodSm-IV (Ac, C16:2) (nodH mutant) | + | + | v.wk. | − | − | − | − | − |
| NodSm-IV (S, C18:1) (nodF/nodL mutant) | + | + | + | + | + | + | − | − |
| LCO-IV (S, C8:1) | + | + | + | v.wk. | − | − | − | − |
| CO-IV (S, NH2) | + | + | − | − | − | − | − | − |
| Chitin tetramer | − | − | − | − | − | − | − | − |
The homozygous single-copy pMtENOD11-GUS M. truncatula line L416 has been used for all pharmacological experiments described in this article. In addition, the protocol for the pharmacological assays was significantly modified to further improve sensitivity and reproducibility as compared with previous assays using transgenic pMtENOD12-GUS alfalfa (see “Materials and Methods”). Experiments were performed on young seedlings that had grown for 5 to 6 h in liquid growth medium prior to effector treatments. Nod factors and pharmacological effectors were then directly added to the liquid medium and incubation continued for an additional 6 h before evaluation of pMtENOD11-GUS reporter activation. In parallel, the effects of the various pharmacological agents on cell viability were evaluated by staining the treated roots with the fluorescent compound propidium iodide, which specifically labels cells that have lost membrane integrity (Engstrom et al., 2002; see “Materials and Methods”).
Ca2+ Release and Recycling Are Required for Nod Factor-Elicited MtENOD11 Expression
Since it has now been firmly established that Nod factors elicit a specific and sustained Ca2+-spiking response in host legume root hairs (see introduction), experiments were performed with pharmacological effectors interfering with either Ca2+ release from intracellular stores or the recycling of cytosolic calcium via specific organelle membrane pumps. To facilitate the comparison, we focused on effectors that had already been tested as antagonists for Nod factor induction of Ca2+ spiking (Engstrom et al., 2002). First, we evaluated two effectors, 2-aminoethyl diphenylborate (2-APB) and 8-(N,N-diethylamino) octyl-3,4,5-trimethoxybenzoate hydrochloride (TMB-8), both potentially targeting ligand-activated Ca2+ channels (Schumaker and Sze, 1987; Ma et al., 2001). Our results show that 2-APB is an efficient antagonist with respect to the induction of pMtENOD11-GUS, with a total block of transgene expression at a concentration of 20 μm (Table II). However, based on the propidium iodide cell penetration test, approximately 80% of root hairs were scored as nonviable after 6-h incubation in 20 μm 2-APB. Lower concentrations of 2-APB (5 μm) were significantly less toxic, but insufficient to block transgene activation. The second potential calcium channel blocker, TMB-8, was also found to be a potent antagonist toward Nod factor induction of pMtENOD11-GUS when applied at a concentration of 1 mm (Table II). On the other hand, a 5-fold lower concentration of TMB-8 (200 μm) failed to inhibit transgene expression. Significantly, and in contrast to 2-APB, root hair viability was unaffected following 6-h incubation with 1 mm TMB-8.
Table II.
Inhibitory effects of intracellular Ca2+ antagonists (ion channel and pump) on Nod factor-induced epidermal pMtENOD11-GUS expression
| Elicitor | Effector | Putative Target | PMtENOD11-GUS Expression | Root Hair Lethality |
|---|---|---|---|---|
| % | ||||
| Water | None | − | − | 0 |
| NodSm (10−9m) | None | − | + | 0 |
| NodSm (10−9m) | 2-APB (5 μm) | Store-operated calcium channels | + | 0 |
| NodSm (10−9m) | 2-APB (20 μm) | − | ∼80 | |
| NodSm (10−9m) | TMB-8 (200 μm) | IP3 receptor store channels | + | 0 |
| NodSm (10−9m) | TMB-8 (1 mm) | − | 0 | |
| NodSm (10−9m) | CPA (1 μm) | Type IIA calcium ATPase | + | 0 |
| NodSm (10−9m) | CPA (5 μm) | v. wk. | ≤5 |
To evaluate the requirement for Ca2+ recycling, we then tested the antagonist activity of cyclopiazonic acid (CPA), a putative type IIA calcium pump blocker (for review, see Sze et al., 2000). Our results show that Nod factor treatment in the presence of 5 μm CPA results in a major reduction (but not total blockage) in pMtENOD11-GUS gene activation (Table II). Furthermore, the majority of root hairs retained viability (>95%) after 6-h incubation in the presence of 5 μm CPA. In conclusion, based on the low toxicity-associated antagonist activities of TMB-8 and CPA, these pharmacological studies suggest that the activation of early nodulin gene expression in root hairs in response to Nod factors involves Ca2+ release from intracellular stores via ligand-activated calcium channels, coupled with a type IIA pump for calcium recycling. The correlation with the antagonist activities of these same effectors with respect to the induction of the Ca2+ spiking will be discussed later.
PLD and PLC Antagonists Are Inhibitors of Nod Factor-Elicited MtENOD11 Expression
Phospholipases are key enzymes in phospholipid-dependent intracellular signaling in eukaryotes since they generate potential secondary messengers such as inositol trisphosphate (IP3) and phosphatidic acid (PA) and are also frequently targets for upstream components of signaling pathways. Recently, den Hartog et al. (2001, 2003) have provided direct biochemical evidence for the activation of both PLD and phosphoinositide-dependent phospholipase C (PI-PLC) in response to Nod factors in vetch roots and alfalfa cell suspensions by studying the accumulation of the potential lipid secondary messengers PA and diacylglycerol pyrophosphate (DGPP). In the case of PLD, n-butanol can be used as an antagonist for PA production by PLD by competing for the phosphatidyl group (Munnik et al., 1995). We therefore evaluated the capacity of this primary alcohol to inhibit Nod factor-induced MtENOD11 expression. Our results show that 68 mm (0.5%) n-butanol totally blocks pMtENOD11-GUS activation in root hairs without affecting root hair cell viability (Table III). As an important control, we also tested the pharmacological activity of tert-butanol, since tertiary butanols are unable to compete for the phosphatidyl group (Munnik et al., 1995). Our experiments confirm that 68 mm (0.5%) tert-butanol is unable to inhibit Nod factor-elicited gene activation (Table III). To our knowledge, this is the first demonstration that a PLD antagonist can inhibit Nod factor-elicited gene expression in root hairs.
Table III.
Inhibitory effects of PLD and PLC antagonists on pMtENOD11-GUS induction
| Elicitor | Effector | Putative Target | pMtENOD11-GUS Expression | Root Hair Lethality |
|---|---|---|---|---|
| % | ||||
| NodSm (10−9m) | None | − | + | 0 |
| NodSm (10−9m) | n-Butanol (68 mm) | PLD | − | 0 |
| NodSm (10−9m) | tert-Butanol (68 mm) | Control | + | 0 |
| NodSm (10−9m) | Neomycin (100 μm) | PI-PLC | − | 0 |
| NodSm (10−9m) | U73122 (5 μm) | PI-PLC | − | ∼80 |
In a previous article, we showed that two classic antagonists of PI-PLC in eukaryotes, the aminoglycoside neomycin and the aminosteroid U73122, are both inhibitors of Nod factor-elicited pMtENOD12-GUS expression in transgenic alfalfa (Pingret et al., 1998). To confirm that PI-PLC activity is also required for MtENOD11 gene activation, we evaluated the antagonist activities of identical concentrations of these two effectors with respect to pMtENOD11-GUS reporter induction by Nod factors. Table III shows that the inclusion of either 100 μm neomycin or 5 μm U73122 results in a total block of gene activation. However, it is important to underline that while neomycin treatment does not affect root hair viability, 80% of root hairs became permeable to propidium iodide after 6-h treatment with 5 μm U73122. Thus, as for 2-APB, the significant toxicity of the aminosteroid means that results obtained using this pharmacological agent should be treated with considerable precaution. In conclusion, our findings are consistent with the biochemical data of den Hartog et al. (2001) and imply that both PLD and PI-PLC activities are essential components of the Nod factor signal transduction pathway leading to MtENOD11 gene activation in root hairs.
Mastoparan Peptide Agonists Target the Nod Factor Signaling Pathway between DMI1/2 and DMI3
Previously, we demonstrated that it was possible to activate pMtENOD12-GUS transcription in epidermal tissues of alfalfa by treatment with the amphiphilic peptide mastoparan, a well-characterized heterotrimeric G-protein activator in animal cells (Pingret et al., 1998). To study the activity of this potential Nod factor agonist in more detail, we first asked whether mastoparan is also able to specifically induce pMtENOD11-GUS reporter expression in epidermal tissues of transgenic L416 M. truncatula plants. Figure 1D shows that 12-h incubation with 1 μm mastoparan activates the GUS reporter gene in epidermal tissues localized in the region of root hair emergence and early development. Furthermore, in line with our previous findings for MtENOD12 induction, (1) treatment with the control inactive synthetic peptide Mas17 (0.25–1 μm) failed to induce GUS activity in transgenic L416 (Fig. 1E); (2) the more active synthetic peptide Mas7 elicited GUS expression at lower concentrations (0.25–0.5 μm) than for mastoparan (Fig. 1F); and (3) the intensity of mastoparan-induced GUS staining is significantly lower by comparison with that elicited by Nod factors (compare Fig. 1, B and D).
In attempting to induce Ca2+ spiking in pea and Medicago root hairs with mastoparan, both Walker et al. (2000) and Engstrom et al. (2002) have reported toxicity problems when applying peptide concentrations of 1 μm and above. In our own experimental system, the propidium iodide test revealed that both mastoparan (1 μm) and Mas7 (0.5 μm) significantly compromise root hair integrity, with approximately 50% of the hairs staining fluorescent after 6 h of peptide treatment (Table IV). We believe that this loss of root hair viability is an indirect consequence of lengthy global applications of mastoparan peptides to the entire root, since spot applications (Esseling et al., 2003) of mastoparan to individual root hairs of the L416 line elicit pMtENOD11-GUS expression in 100% of hairs and, furthermore, do not inhibit root hair growth (J.J. Esseling and A.M.C Emons, personal communication). Indeed, this may explain in part the relatively low numbers of root hairs that express pMtENOD11/12-GUS in response to global mastoparan peptide treatments. Thus, despite the toxicity problems associated with such global treatments, we consider that the tissue-specific nature of the response, the fact that at least two Nod factor-responsive genes are activated by mastoparan agonists, and the relative activities of the different mastoparan peptides, together argue that this response is relevant to Nod factor signaling.
Table IV.
Comparison of the elicitor activities of Nod factor and Mas7 in wild-type and dmi mutant backgrounds
| Elicitor | Genetic Backgound | pMtENOD11-GUS Expression | Root Hair Lethality |
|---|---|---|---|
| % | |||
| NodSm (10−9m) | WT | + | 0 |
| Mas7 (0.5 μm) | WT | +a | ∼50 |
| NodSm (10−9m) | dmi1-1 | − | 0 |
| Mas7 (0.5 μm) | dmi1-1 | +a | ∼50 |
| NodSm (10−9m) | dmi2-1 | − | 0 |
| Mas7 (0.5 μm) | dmi2-1 | +a | ∼50 |
| NodSm (10−9m) | dmi3-1 | − | 0 |
| Mas7 (0.5 μm) | dmi3-1 | − | ∼50 |
| Mas7 (0.5 μm) + TMB-8 (1.0 mm) | WT | − | ∼50 |
| dmi1-1 | − | ND | |
| dmi2-1 | − | ND | |
| Mas7 (0.5 μm) + n-butanol (68 mm) | WT | − | ∼50 |
| dmi1-1 | − | ND | |
| dmi2-1 | − | ND | |
| Mas7 (0.5 μm) + neomycin (100 μm) | WT | − | ∼50 |
| dmi1-1 | − | ND | |
| dmi2-1 | − | ND |
Typical response to mastoparan (see text and Fig. 1, D and F). ND, Not determined; WT, wild type.
As mentioned earlier, a number of M. truncatula nodulation mutants defective in the Nod factor signal transduction pathway leading to MtENOD11 expression have been identified and partially characterized. Based on several epidermal Nod factor-dependent responses, including rapid calcium influx, Ca2+ spiking, and root hair deformation, the corresponding genes have been placed in the following order: NFP to DMI1, DMI2 to DMI3. Ben Amor et al. (2003) have recently shown that addition of mastoparan to a transgenic pMtENOD11-GUS line in an nfp mutant background elicits similar GUS expression as compared to peptide treatment of the L416 line. There are two possible interpretations for this result. Either mastoparan activates MtENOD11 expression via a signaling pathway independent of the Nod factor pathway, or the peptide agonist acts on a component of the Nod factor pathway that is downstream of NFP. To attempt to distinguish between these two alternatives, we have examined the capacity of mastoparan peptides to induce MtENOD11 expression in dmi1, dmi2, and dmi3 backgrounds by introducing the transgene into mutant alleles of all three genes (see “Materials and Methods”).
First, control experiments with Nod factor treatment (10−9 m) confirmed that pMtENOD11-GUS induction is totally blocked in all three dmi mutant transgenic lines (Table IV). Second, to evaluate the capacity of mastoparan peptides to elicit transgene expression in these mutants, we chose the synthetic peptide Mas7 because of its superior agonist activity. Table IV shows that, as for the nfp mutant, 0.5 μm Mas7 is able to elicit pMtENOD11-GUS expression in both dmi1 and dmi2 mutant backgrounds. The result for the dmi2 mutant TR25 (dmi2-1) was previously reported in the proceedings of the International Society for Molecular Plant-Microbe Interactions (ISMPMI) meeting in Amsterdam (Vernoud et al., 1999b). In contrast, however, no activation of the pMtENOD11-GUS transgene was observed in the dmi3 background (Table IV). This latter result is particularly important because it provides genetic evidence that at least part of the mastoparan-activated signaling pathway leading to pMtENOD11-GUS expression is shared with the Nod factor pathway. In this context, these findings also imply that mastoparan acts downstream of NFP/DMI1/DMI2 and upstream of DMI3. Note that loss of root hair viability resulting from Mas7 treatment is not modified in any of the mutant backgrounds.
Finally, combined pharmacological experiments were performed to examine the effect of antagonists that interfere with either Ca2+ release/recycling or phospholipase activities on mastoparan-elicited gene expression in wild-type transgenic M. truncatula. TMB-8, n-butanol, and neomycin were selected for these experiments because these pharmacological agents are not associated with loss of root hair viability. Our results clearly show that 1 mm TMB-8, 68 mm n-butanol, and 100 μm neomycin are also efficient antagonists with respect to pMtENOD11-GUS activation by Mas7 (Table IV). Assuming that the targets for these antagonists are the same for both Nod factor and mastoparan-activated pathways, these findings reinforce the hypothesis that the two pathways are common downstream of the site of mastoparan action. These combined agonist/antagonist experiments have also been performed for both dmi1 and dmi2 mutants, with identical results as for the wild-type L416 line (Table IV). Taken together, these results argue that, in addition to the signal transduction components involving Ca2+ release from intracellular stores, both PLD and PI-PLC activities can be positioned downstream of DMI1/DMI2.
DISCUSSION
Is Ca2+ Spiking Part of the Signal Transduction Pathway Leading to MtENOD11 Expression in Root Hairs?
This article describes experiments aimed at furthering our understanding of the mechanism of Rhizobium Nod factor signal transduction in root hairs, the initial cellular target on the legume host root for these symbiotic LCO elicitor molecules. Pharmacological tests to identify potential components of the signaling pathway required for activating specific gene expression were performed using a novel protocol adapted to young seedlings (see “Materials and Methods”) in conjunction with a single-copy homozygous transgenic M. truncatula line (L416) expressing the Nod factor-inducible root epidermis-specific pMtENOD11-GUS reporter gene. This experimental system is superior in terms of sensitivity and reproducibility as compared to that previously described for transgenic alfalfa expressing the pMtENOD12-GUS reporter, for which homozygous lines cannot be obtained (Pingret et al., 1998).
Initially, we evaluated the antagonist activities (inhibition of Nod factor-elicited pMtENOD11-GUS expression) of several pharmacological effectors known to interfere either with release of calcium ions from intracellular stores or the recycling of Ca2+ from the cytosol to the store. Our results show that two potential ligand-activated calcium channel antagonists, 2-APB and TMB-8, are able to totally block Nod factor gene activation in root hairs at concentrations of 20 μm and 1 mm, respectively (Table II). Such calcium channels are essential for the initiation and maintenance of cytosolic calcium oscillations. Furthermore, Engstrom et al. (2002) have reported that 2-APB (50 μm) is an efficient antagonist of Nod factor-elicited calcium oscillations in M. truncatula root hairs. However, these authors also observed that 50 μm 2-APB severely compromised root hair viability even after relatively short exposures (30 min) and our own results confirm the toxicity of this effector even at the lower concentration of 20 μm (Table II). On the other hand, as judged by the propidium iodide test, the benzoic acid derivative TMB-8 can be considered a superior intracellular calcium channel antagonist since 1 mm TMB-8 lacks toxic side effects on root hairs even after 6-h incubation (Table II). Unfortunately, evaluation of the antagonist activity of TMB-8 with respect to Ca2+ spiking was only performed at 200 μm (Engstrom et al., 2002), and our results show that this concentration is insufficient to inhibit Nod factor-elicited transgene induction (Table II). Since there is evidence that TMB-8 targets intracellular IP3-responsive channels in plant cells (Schumaker and Sze, 1987), our data consolidate the hypothesis that IP3 generated by PI-PLC activity is a key secondary messenger in the Nod factor signaling pathway (Pingret et al., 1998; den Hartog et al., 2001, 2003). On the other hand, it should be noted that IP3-gated calcium channels have not yet been identified in the Arabidopsis (Arabidopsis thaliana) genome and, as suggested by Meijer and Munnik (2003), it is conceivable that the IP3 generated may in fact be phosphorylated to inositol hexakisphosphate (IP6), which then acts as the true secondary messenger releasing Ca2+ from internal stores.
CPA is a highly specific inhibitor of the sarcoplasmic/endoplasmic reticulum (ER) calcium ATPase class of calcium recycling ATPase pumps located either in the sarcoplasmic or ER membrane in animal cells, for which homologs (called type IIA calcium pumps) have been identified in plants (for review, see Sze et al., 2000). Engstrom et al. (2002) have shown that 5 μm CPA blocks Nod factor-elicited calcium spiking, and here we report that the identical concentration of CPA is an efficient inhibitor of MtENOD11 induction (Table II). In both experimental systems, CPA-induced cell lethality is below the 5% level. The fact that CPA is believed to target the plant ER membrane (Liang and Sze, 1998) is in line with the observation that the regular oscillations in cytosolic calcium are initiated at the periphery of the root hair nucleus (Ehrhardt et al., 1996; Shaw and Long, 2003). Taken together, the pharmacological data presented in this paper, making use of intracellular calcium channel and pump antagonists, are consistent with the genetic model positioning the calcium spiking signaling element upstream of ENOD gene activation in the Nod factor signaling pathway (Wais et al., 2000).
Finally, a striking correlation between Nod factor activation of ENOD gene expression and Ca2+ spiking emerges when comparing the relative activities of modified NodSm LCOs. The results presented in this article show that the replacement of C16:2 by C18:1 (nodF/nodL mutant), removal of the critical sulfate decoration (nodH mutant), or total removal of the fatty acid chain (sulfated chitotetraose) drops pMtENOD11-GUS induction activity by 10- to 100-fold, 105-fold, and 105- to 106-fold, respectively (Table I). The fact that almost identical results were reported by Oldroyd et al. (2001) in relation to the capacity of the same molecules to trigger Ca2+ spiking provides an additional argument that the Nod factor perception machinery is identical for the two responses.
A Central Role for Phospholipases and Phospholipid Second Messengers in Nod Factor Signaling?
Pharmacological experiments performed in our laboratory several years ago provided the first evidence that PI-PLC activity is required for the activation of root hair-specific gene expression in response to the Nod factor signal (Pingret et al., 1998). More direct biochemical evidence for phospholipid signaling was subsequently provided by den Hartog et al. (2001), who were able to show that Nod factors stimulate both PI-PLC and PLD activity in vetch roots, as well as the accumulation of the potential lipid second messengers PA and DGPP. In this article, we have made use of the PLD antagonist n-butanol (68 mm) and the PI-PLC antagonists neomycin (100 μm) and U73122 (5 μm) to provide evidence that both phospholipases are required for Nod factor activation of the MtENOD11 gene in M. truncatula root hairs (Table III). Although Engstrom et al. (2002) have shown that U73122 is an antagonist of Nod factor-induced Ca2+ oscillations, no data are available concerning the relationship between PLD activity and Ca2+ spiking.
A central role for both PLD and PI-PLC in phospholipid-based signaling pathways in plants is now clearly emerging from current research in a number of model systems (for review, see Meijer and Munnik, 2003). The sequencing of the Arabidopsis genome has revealed a total of 12 PLD genes (subdivided into five groups based on amino acid sequence and biochemical properties) and at least 7 PI-PLC genes (Mueller-Roeber and Pical, 2002; Qin and Wang, 2002). Both enzymes are capable of generating PA and DGPP (via subsequent phosphorylation). In the case of PI-PLC, cleavage of PtdInsP2 initially generates the soluble messenger IP3 and diacylglycerol, which can then be converted to PA after phosphorylation by diacylglycerol kinase. It has been shown that PA is capable of regulating a number of cellular functions in plants, although the precise molecular targets in most cases remain to be identified (for review, see Munnik, 2001). By analogy with animal cells, it is proposed that PA can either directly modify target enzyme activities, such as kinases and phosphatases, or function to assemble membrane-located signaling complexes. Recently, Zhang et al. (2004) have demonstrated that a direct interaction between PA and the ABI1 phosphatase 2C is implicated in abscisic acid signaling. In addition to generating potential lipid second messengers, PLD itself can play an important role in intracellular signaling, on the one hand because enzyme activity can be stimulated by a wide variety of factors including Ca2+, polyphosphoinositides such as PtdInsP2, G proteins (see below), membrane lipids, and phosphorylation (e.g. Novotná et al., 2003), and, on the other hand because certain PLDs can translocate to the membrane, modify vesicle trafficking, and even directly interact with the microtubular cytoskeleton (e.g. Dhonukshe et al., 2003). Possible roles of PI-PLC/PLD lipases in Nod factor signaling, as well as the second messengers generated by their activity, are proposed in the integrated model discussed below.
Does the Nod Factor Signaling Pathway Involve Activation of a Heterotrimeric G Protein?
The proposition that a plant heterotrimeric G protein is required for transduction of the Nod factor signal in legume root hairs emerged principally from experiments making use of mastoparan, a dodecapeptide isolated from wasp venom and known to activate G proteins in animal cells in the absence of normal ligand activation of the appropriate heptahelical G-protein-coupled receptor (GPCR; Higashijima et al., 1990; Ross and Higashijima, 1994). It was initially demonstrated that mastoparan (and its more active synthetic variant Mas7) is able to mimic Nod factor activation of MtENOD12 in alfalfa root hairs (Pingret et al., 1998) and subsequently that these peptide agonists were also capable of eliciting both PI-PLC and PLD activity in vetch roots, with a similar accumulation of the phospholipids PA and DGPP as for Nod factors (den Hartog et al., 2001). In this article, we show first that 0.5 to 1.0 μm mastoparan and Mas7 (but not the less active peptide control Mas17) are also able to activate the Nod factor-responsive pMtENOD11-GUS reporter in root hairs of transgenic M. truncatula (Fig. 1, D–F). In addition, we have tested mastoparan activity in M. truncatula mutant lines, revealing that whereas agonist-induced reporter gene expression is unaffected in dmi1 and dmi2 mutants, pMtENOD11-GUS induction is totally blocked in a dmi3 mutant (Table IV). This last result is particularly important because it implies that the signaling pathways activated in root hairs by both Nod factors and mastoparan leading to ENOD gene activation share common elements. Furthermore, the fact that the DMI3 gene acts downstream of NFP, DMI1, and DMI2 is entirely consistent with the mastoparan/Mas7 agonist results obtained here for dmi1 and dmi2 mutants and the nfp mutant by Ben Amor et al. (2003), and indicates that mastoparan acts at a step in the pathway between DMI1/DMI2 and DMI3. The combined agonist/antagonist experiments performed with Mas7 and the inhibitors of PLD (n-butanol), PLC (neomycin), and intracellular calcium-channel activity (TMB-8; Table IV) are also consistent with this positioning of mastoparan action. However, are heterotrimeric G proteins the target for mastoparan peptides in the legume root hair?
The complete sequencing of the Arabidopsis genome has so far revealed only a single potential canonical heterotrimeric G protein (based on the unique G-protein α-subunit GPA1) and only a very few candidate GPCRs compared to the large numbers present in animal cells. Significantly, Pandey and Assman (2004) have recently demonstrated that a direct association between the GPA1 α-subunit and a candidate Arabidopsis GPCR known as GCR1 participates in the regulation of abscisic acid signaling. While these data clearly argue in favor of a role for GPCR-coupled heterotrimeric G proteins in plant signaling, genetic results also suggest that GCR1 can act independently of G proteins in certain signaling responses related to seed germination (Chen et al., 2004). To add to the complexity, Zhao and Wang (2004) have recently reported that GPA1 can also bind directly to PLDα1 via a structural motif analogous to the intracellular motif in animal GPCRs involved in activating the appropriate G protein. Furthermore, the authors demonstrate that this interaction reciprocally modulates the activities of the two proteins. Since mastoparan is believed to act as a mammalian G-protein agonist by mimicking the activated form of a cytoplasmic GPCR loop (Higashijima et al., 1988), it is conceivable that these peptide agonists target such receptor-independent interactions. Finally, mastoparan peptides may also target signaling components in plant cells that are totally independent of G proteins, as suggested by the recent studies by Miles et al. (2004), who have shown that mastoparan is able to activate an Arabidopsis mitogen-activated protein kinase in lines disrupted for both G-protein α- and β-subunits. It should be noted, however, that relatively high concentrations of mastoparan (5 μm) were used in these experiments. Although further studies are clearly required to determine how mastoparan functions in plants, our combined pharmacological data strongly suggest a membrane-associated target for the peptide since both phospholipase antagonists block mastoparan-elicited gene expression.
An Integrated Cellular Model for Nod Factor Signaling in Root Hairs
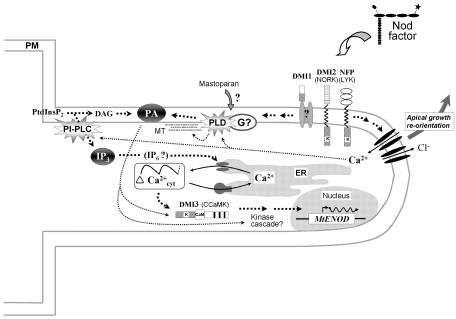
In Figure 2 we propose a schematic cellular model for Nod factor signal transduction required for the earliest known responses in root hairs, including rapid ion fluxes across the plasma membrane, the initiation and maintenance of intracellular calcium oscillations, modifications in root hair morphology, and induction of specific gene expression. The salient features of this model are the following. First, the two receptor-like kinases LYK and NORK, encoded by the NFP and DMI2 genes, respectively, are presumed to be involved in Nod factor perception and indeed may be components of a single receptor complex. In this context, the potential sugar-binding LysM extracellular domains of LYK are the likely recognition site for the LCO ligand (Cullimore and Dénarié, 2003). By analogy with equivalent Lotus mutants (Radutoiu et al., 2003), the Nod factor receptor complex may in fact comprise two members of the LYK family. The function of the DMI1 protein is currently unclear, although the presence of transmembrane domains and a limited homology to ligand-gated cation channels suggests a possible role in transmembrane ion transport (Ané et al., 2004).
Figure 2.
Hypothetical cellular model for Rhizobium Nod factor signal transduction in the host plant root hair. In this model, we propose that phospholipid signaling provides the essential link between perception of exogenous Nod factors via a putative plasma membrane complex including LYK and NORK receptor-like kinases and well-characterized intracellular responses such as calcium spiking and specific gene (e.g. MtENOD11) transcription. Activation of PLD and PI-PLC by so far unknown mechanisms generates secondary messengers such as PA and IP3 from membrane lipids, which in turn either activate intracellular targets such as kinases/phosphatases or open store Ca2+ channels (see text for detailed description). Note that Ca2+ release from intracellular stores may involve more than one type of channel activity (Pingret et al., 1998). The precise target of the mastoparan agonist is currently unclear, and may involve either a heterotrimeric G protein (G) or an alternative membrane-associated protein such as PLD. The rapid opening of plasma membrane ion channels in response to Nod factor perception is believed to lie on a separate pathway leading to reorientation of root hair tip growth. With the exception of ion fluxes, all proposed hypothetical pathways (including those that may provide signaling cross-talk) are indicated by broken arrows. PM, Plasma membrane; MT, microtubular cytoskeleton.
Evidence presented in this and other articles argues strongly that PLD and PLC, as well as the potential second messengers PA and IP3 (or IP6), provide the crucial link between signal perception at the plasma membrane and intracellular responses such as calcium oscillations and gene expression. In particular, we have placed PLD at a central location in this proposed pathway, based both on the numerous factors that can modulate its activity and the many potential downstream targets for PLD and/or PA, as discussed above. Note that the PA generated via PLD hydrolysis of structural lipids is different from that derived from PtdInsP2 via PI-PLC hydrolysis and subsequent phosphorylation by diacylglycerol kinase, and therefore may have distinct downstream targets (Munnik, 2001). These targets could include kinases and/or phosphatases (e.g. Anthony et al., 2004; Zhang et al., 2004) and possibly even the CCaMK encoded by the DMI3 gene (Lévy et al., 2004; Mitra et al., 2004). It has been proposed by these authors that this particular CCaMK could provide the calcium sensor required to translate Nod factor-elicited calcium oscillations into a specific downstream signaling cascade.
Finally, the rapid plasma membrane ion fluxes and root hair growth reorientation elicited by Nod factors probably lie on a separate pathway downstream of NFP since (1) dmi1 and dmi2 mutants are not defective in the rapid calcium influx (Shaw and Long, 2003), and (2) dmi mutants are not affected in their capacity for reorientating root hair apical growth following spot application of Nod factor (Esseling et al., 2004). In addition, it is conceivable that the rapid Ca2+ influx could play a role in modulating phospholipase activities, since Ca2+-dependent isoforms are known for both PI-PLC (Mueller-Roeber and Pical, 2002) and PLD (Qin and Wang, 2002). This would be consistent with the rapidity of phospholipase induction in response to Nod factors (den Hartog et al., 2003) and the fact that antagonists of plasma membrane calcium influxes also block ENOD gene activation (Pingret et al., 1998). Nevertheless, it is clear that much still needs to be uncovered in order to fully understand the cellular mechanism of Nod factor signaling in root hairs and, in particular, to determine precisely how Nod factor perception is linked to phospholipase activation and how the activation of cytosolic kinases, such as CCaMK, can lead to specific gene expression. In addition to providing a basis for future research, this model suggests that cross-talk can occur between different segments of the pathway, thus providing the means to modulate and coordinate the various cellular responses required for preparing this specialized cell for subsequent infection by Rhizobium.
MATERIALS AND METHODS
Plant Materials
The Medicago truncatula cv Jemalong single-seed descendent line A17 was used for genetic transformation. The Nod−/Myc− M. truncatula mutants TR25 (dmi2-1; Sagan et al., 1995) and TRV25 (dmi3-1; Sagan et al., 1998) were kindly provided by G. Duc (INRA, Dijon, France), and the mutant C71 (dmi1-1; Catoira et al., 2000) by D. Cook (University of California, Davis, CA).
Construction of a MtENOD11 Promoter-GUS Reporter Gene Fusion
A precise transcriptional gene fusion between the promoter of the MtENOD11 gene and the Escherichia coli GUS gene was constructed using a strategy similar to that described in Pichon et al. (1992). A genomic clone containing the entire MtENOD11 gene (Journet et al., 2001) served as a template for PCR using a forward primer (5′-GTTTACTTGCATTACCCCCGC-3′) and a reverse primer (5′-GGAAGCCATGG(NcoI)TAGGTAGTGATTTT-3′) to amplify a 2.35-kb promoter fragment (single underlining indicates substitutions required to create an NcoI restriction site at the 3′ end of the fragment, and double underlining indicates the MtENOD11 translation initiation codon). After complete digestion with HindIII (a single site is located 50 nucleotides downstream of the 5′ primer) and partial digestion with NcoI (a second site is present 300 nucleotides upstream of the 3′ primer), the promoter fragment (2.3 kb) was cloned into the pBSGUS vector (Vernoud et al., 1999a) to generate a precise transcriptional fusion. The chimeric pMtENOD11-GUS reporter construction was then excised from the Bluescript vector following HindIII/SacI digestion and cloned between the corresponding unique sites of the binary plant vector pLP100 (Szabados et al., 1995).
Introduction of pMtENOD11-GUS into M. truncatula and Genetic Crosses with Nod− Mutants
The pMtENOD11-GUS reporter gene construction was introduced into M. truncatula Jemalong A17 using an Agrobacterium tumefaciens (strain LBA4404) leaf disc transformation protocol and subsequent regeneration via somatic embryogenesis (Chabaud et al., 1996). The 15 independent primary transformants thus obtained were screened for expression of the transgene in both symbiotic and nonsymbiotic contexts (Journet et al., 2001). Several high-expressing lines were then analyzed by Southern blotting to evaluate transgene copy number. This led to the selection of the single-copy line L416, which was then self-pollinated to obtain S1 plants homozygous for the pMtENOD11-GUS construction. Expression of the transgene was found to be completely stable in S2, S3, and S4 descendants. Manual genetic crosses between L416-S2 and Nod− mutants were performed following the procedure described in Thoquet et al. (2002), using the transgenic parent as pollen donor. Note that the expression of pMtENOD11-GUS in nonsymbiotic tissues provided a convenient screen for the presence of the transgene in Nod− F2 descendants, and the segregation of GUS expression in F3 maturing embryos allowed rapid selection for Nod− F2 plants homozygous for the transgene. Homogeneous transgenic mutant F3 and F4 progenies were then obtained from such plants and used in this study.
Nod Factors and Pharmacological Effectors
Sinorhizobium meliloti Nod factors purified from wild-type and mutant strains [NodSm-IV (Ac, S, C16:2), NodSm-IV (Ac, C16:2), and NodSm-IV (S, C18:1)] were kindly provided by F. Maillet (J. Dénarié's group, LIPM, Castanet-Tolosan, France). Synthetic LCO-IV (S, C8:1) and CO-IV (S, NH2) were kindly provided by Jean-Marie Beau (Université Paris-Sud, Orsay, France; Demont-Caulet et al., 1999). Chitin tetramers (N,N′,N″,N‴-tetraacetylchitotetraose) were purchased from Sigma (St. Louis). Lyophilized LCOs were dissolved at 10−3 m in 50% ethanol and then serially diluted in water. The peptides mastoparan, Mas7, and Mas17 (RBI; Sigma, Natick, MA) were dissolved at 1 mm in water and stored at −20°C. The three putative intracellular calcium antagonists, the benzoic acid derivative TMB-8 (RBI; Sigma), the borate derivative 2-APB (Calbiochem, San Diego), and the indole tetrameric acid CPA (Sigma), were prepared as 50 mm water, 100 mm DMSO, and 50 mm DMSO stock solutions, respectively. The PI-PLC inhibitor U73122 (Calbiochem) was freshly prepared as a 4-mm stock solution in DMSO. Neomycin sulfate (Sigma) was prepared as a 10-mm aqueous stock solution. n-Butanol, the specific PLD antagonist, and tert-butanol, its tertiary isomer, were obtained from Fluka (Bellevue, WA) and diluted in water before use. The concentrations of DMSO never exceeded 0.125% in the plant growth medium during pharmacological treatments and we confirmed that these doses had no effect on plant growth, root hair integrity, or the induction of pMtENOD11-GUS transgene expression in the presence or absence of Nod factors (not shown).
Pharmacological Study of pMtENOD11-GUS Induction in Transgenic M. truncatula Seedlings
Following germination, transgenic seedlings were grown for approximately 16 h at 25°C on a moist paper support in plastic pouches (Mega International, Minneapolis) imbibed in Fåhraeus medium. These seedlings (with 3- to 4-cm-long roots) were then transferred to a 30-mL flat-bottomed polystyrene tube (four seedlings per tube) containing 15 mL of Fåhraeus medium at 25°C. The seedlings, adhering to the inner surface of the tube, were positioned vertically with the lower 1 to 2 cm of the root immersed in the liquid medium. Prior to Nod factor/pharmacological treatment, seedlings were allowed to grow for 5 to 6 h in order to recover from the stress caused by their transfer. Agonist experiments (Nod factor or mastoparan peptides alone) were then initiated by the direct addition of 50-fold concentrated solutions to the growth medium with very gentle mixing, followed by 6-h incubation at 25°C. In the case of antagonist experiments, seedlings were pretreated with the antagonist for 15 min prior to the addition of Nod factor. For each elicitor/effector treatment, a minimum of 10 to 20 plants were examined and all experiments were repeated at least twice. It should be emphasized that the use of homozygous single-copy transgenic lines of M. truncatula and the new seedling/liquid medium assay system described here resulted in very significant improvement of the homogeneity of epidermal gene reporter induction, as compared to the previously described assays performed on transgenic alfalfa plantlets (Pingret et al., 1998). Note that the Nod factor/mastoparan experiments presented in Figure 1 were performed on 7- to 10-d-old transgenic plantlets grown either in pouches or in aeroponic culture. Following incubation, roots were stained for GUS activity for approximately 16 h according to the technique described in Journet et al. (1994). To simplify the presentation of antagonist activities and responses induced by modified LCOs (Table I), histochemical GUS staining is indicated as positive (+), very weak (v. wk.), or negative (−).
Evaluation of Root Hair Lethality
Root hair integrity was examined following the incubation period with each pharmacological effector/elicitor and prior to the histochemical GUS staining. Roots were stained directly in the liquid medium with 10 μg/mL propidium iodide (Sigma) for 5 min in the dark. The segment of the root with growing root hairs at the time of elicitor/effector addition was observed under fluorescent light (emission filter 520 nm LP) with a binocular microscope (Leica MZFLIII, Wetzlar, Germany). The number of root hairs with a fluorescent-labeled nucleus was counted within a defined area on the root surface, and this figure was divided by the total number of root hairs to determine the percentage of root hair lethality. For each elicitor/effector treatment, a total of 10 to 20 plants were analyzed and all experiments were performed at least twice.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial purposes, subject to the requisite permission from any third-party owners of all or parts of the material.
Acknowledgments
We thank Gérard Duc (INRA, Dijon, France) and Doug Cook (University of California, Davis, CA) for the various dmi mutants used in this study, and both Fabienne Maillet (LIPM, Castanet-Tolosan, France) and Jean-Marie Beau (Université Paris-Sud, Orsay, France) for kindly supplying purified and synthetic LCOs, respectively. We are also very grateful to Fernanda de Carvalho-Niebel and Aurélien Boisson-Dernier for helpful criticisms of the manuscript.
This work was supported by the French Ministère de la Recherche (grants to D.C. and J.-L.P.), which also provided financial support in the framework of an ACI project (Developmental Biology and Integrative Physiology).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.051110.
References
- Ané JM, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GE, Ayax C, Lévy J, Debellé F, Baek JM, Kalo P, et al (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367 [DOI] [PubMed] [Google Scholar]
- Anthony RG, Henriques R, Helfer A, Meszaros T, Rios G, Testerink C, Munnik T, Deak M, Koncz C, Bögre L (2004) A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J 23: 572–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Amor B, Shaw SL, Oldroyd GE, Maillet F, Penmetsa RV, Cook D, Long SR, Dénarié J, Gough C (2003) The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J 34: 495–506 [DOI] [PubMed] [Google Scholar]
- Calantzis C, Morandi D, Arnould C, Gianinazzi PV (2001) Cellular interactions between G-mosseae and a Myc(-) dmi2 mutant in Medicago truncatula. Symbiosis 30: 97–108 [Google Scholar]
- Cárdenas L, Feijó JA, Kunkel JG, Sánchez F, Holdaway-Clarke T, Hepler PK, Quinto C (1999) Rhizobium Nod factors induce increases in intracellular free calcium and extracellular calcium influxes in bean root hairs. Plant J 19: 347–352 [DOI] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Dénarié J (2000) Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell 12: 1647–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabaud M, Larsonneau C, Marmouget C, Huguet T (1996) Transformation of barrel medic (Medicago truncatula Gaertn.) by Agrobacterium tumefaciens and regeneration via somatic embryogenesis of transgenic plants with the MtENOD12 nodulin promoter fused to the GUS reporter gene. Plant Cell Rep 15: 305–310 [DOI] [PubMed] [Google Scholar]
- Chen J-G, Pandey S, Huang J, Alonso JM, Ecker JR, Assmann SM, Jones AM (2004) GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination. Plant Physiol 135: 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DR (2004) Unravelling the mystery of Nod factor signaling by a genomic approach in Medicago truncatula. Proc Natl Acad Sci USA 101: 4339–4340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullimore J, Dénarié J (2003) How legumes select their sweet talking symbionts. Science 302: 575–578 [DOI] [PubMed] [Google Scholar]
- Demont-Caulet N, Maillet F, Tailler D, Jacquinet JC, Promé JC, Nicolaou KC, Truchet G, Beau JM, Dénarié J (1999) Nodule-inducing activity of synthetic Sinorhizobium meliloti nodulation factors and related lipochito-oligosaccharides on alfalfa. Importance of the acyl chain structure. Plant Physiol 120: 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hartog M, Musgrave A, Munnik T (2001) Nod factor-induced phosphatidic acid and diacylglycerol pyrophosphate formation: a role for phospholipase C and D in root hair deformation. Plant J 25: 55–65 [DOI] [PubMed] [Google Scholar]
- den Hartog M, Verhoef N, Munnik T (2003) Nod factors and elicitors activate different phospholipid signalling pathways in suspension-cultured alfalfa cells. Plant Physiol 132: 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ruijter NCA, Bisseling T, Emons AMC (1999) Rhizobium Nod factors induce an increase in sub-apical fine bundles of actin filaments in Vicia sativa root hairs within minutes. Mol Plant Microbe Interact 12: 829–832 [Google Scholar]
- de Ruijter NCA, Rook MB, Bisseling T, Emons AMC (1998) Lipochito-oligosaccharides re-initiate root hair tip growth in Vicia sativa with high calcium and spectrin-like antigen at the tip. Plant J 13: 341–350 [Google Scholar]
- Dhonukshe P, Laxalt AM, Goedhart J, Gadella TW, Munnik T (2003) Phospholipase D activation correlates with microtubule reorganization in living plant cells. Plant Cell 15: 2666–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt DW, Atkinson EM, Long SR (1992) Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256: 998–1000 [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Wais R, Long SR (1996) Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85: 673–681 [DOI] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kaló P, Kiss GB (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Engstrom EM, Ehrhardt DW, Mitra RM, Long SR (2002) Pharmacological analysis of Nod factor-induced calcium spiking in Medicago truncatula. Evidence for the requirement of type IIA calcium pumps and phosphoinositide signaling. Plant Physiol 128: 1390–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esseling JJ, Lhuissier FG, Emons AM (2003) Nod factor-induced root hair curling: continuous polar growth towards the point of Nod factor application. Plant Physiol 132: 1982–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esseling JJ, Lhuissier FG, Emons AM (2004) A nonsymbiotic root hair tip growth phenotype in NORK-mutated legumes: implications for nodulation factor-induced signaling and formation of a multifaceted root hair pocket for bacteria. Plant Cell 16: 933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felle HH, Kondorosi E, Kondorosi A, Schultze M (1995) Nod signal-induced plasma membrane potential changes in alfalfa root hairs are differentially sensitive to structural modifications of the lipochitooligosaccharide. Plant J 7: 939–947 [Google Scholar]
- Felle HH, Kondorosi E, Kondorosi A, Schultze M (1998) The role of ion fluxes in Nod factor signalling in Medicago sativa. Plant J 13: 455–463 [Google Scholar]
- Felle HH, Kondorosi E, Kondorosi A, Schultze M (1999) Elevation of the cytosolic free [Ca2+] is indispensable for the transduction of the Nod factor signal in alfalfa. Plant Physiol 121: 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts R, Bisseling T (2002) Rhizobium Nod factor perception and signalling. Plant Cell 14(Suppl): S239–S249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JM, Wais R, Long SR (2003) Rhizobium-induced calcium spiking in Lotus japonicus. Mol Plant Microbe Interact 16: 335–341 [DOI] [PubMed] [Google Scholar]
- Higashijima T, Burnier J, Ross EM (1990) Regulation of Gi and Go by mastoparan, related amphiphilic peptides, and hydrophobic amines. Mechanism and structural determinants of activity. J Biol Chem 265: 14176–14186 [PubMed] [Google Scholar]
- Higashijima T, Uzu S, Nakajima T, Ross EM (1988) Mastoparan, a peptide toxin from wasp venom, mimics receptors by activating GTP-binding regulatory proteins (G proteins). J Biol Chem 263: 6491–6494 [PubMed] [Google Scholar]
- Journet EP, El-Gachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Arnould C, Morandi D, Barker DG, Gianinazzi-Pearson V (2001) Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol Plant Microbe Interact 14: 737–748 [DOI] [PubMed] [Google Scholar]
- Journet EP, Pichon M, Dedieu A, de Billy F, Truchet G, Barker DG (1994) Rhizobium meliloti Nod factors elicit cell-specific transcription of the ENOD12 gene in transgenic alfalfa. Plant J 6: 241–249 [DOI] [PubMed] [Google Scholar]
- Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ané JM, Lauber E, Bisseling T, et al (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364 [DOI] [PubMed] [Google Scholar]
- Liang F, Sze H (1998) A high-affinity Ca2+ pump, ECA1, from the endoplasmic reticulum is inhibited by cyclopiazonic acid but not by thapsigargin. Plant Physiol 118: 817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma HT, Venkatachalam K, Li HS, Montell C, Kurosaki T, Patterson RL, Gill DL (2001) Assessment of the role of the inositol 1,4,5-trisphosphate receptor in the activation of transient receptor potential channels and store-operated Ca2+ entry channels. J Biol Chem 276: 18888–18896 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Meijer HJ, Munnik T (2003) Phospholipid-based signaling in plants. Annu Rev Plant Biol 54: 265–306 [DOI] [PubMed] [Google Scholar]
- Miles GP, Samuel MA, Jones AM, Ellis BE (2004) Mastoparan rapidly activates plant MAP kinase signaling independent of heterotrimeric G proteins. Plant Physiol 134: 1332–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra RM, Gleason CA, Edwards A, Hadfield J, Downie JA, Oldroyd GE, Long SR (2004) A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc Natl Acad Sci USA 101: 4701–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Roeber B, Pical C (2002) Inositol phospholipid metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol 130: 22–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T (2001) Phosphatidic acid: an emerging plant lipid second messenger. Trends Plant Sci 6: 227–233 [DOI] [PubMed] [Google Scholar]
- Munnik T, Arisz SA, de Vrije T, Musgrave A (1995) G protein activation stimulates phospholipase D signaling in plants. Plant Cell 7: 2197–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotná Z, Linek J, Hynek R, Martinec J, Potocký M, Valentová O (2003) Plant PIP2-dependent phospholipase D activity is regulated by phosphorylation. FEBS Lett 554: 50–54 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA (2004) Calcium, kinases and nodulation signalling in legumes. Nat Rev Mol Cell Biol 5: 566–576 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Mitra RM, Wais RJ, Long SR (2001) Evidence for structurally specific negative feedback in the Nod factor signal transduction pathway. Plant J 28: 191–199 [DOI] [PubMed] [Google Scholar]
- Pandey S, Assmann SM (2004) The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein α subunit GPA1 and regulates abscisic acid. Plant Cell 16: 1616–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon M, Journet EP, Dedieu A, de Billy F, Truchet G, Barker DG (1992) Rhizobium meliloti elicits transient expression of the early nodulin gene ENOD12 in the differentiating root epidermis of transgenic alfalfa. Plant Cell 4: 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingret JL, Journet EP, Barker DG (1998) Rhizobium Nod factor signaling: evidence for a G protein-mediated transduction mechanism. Plant Cell 10: 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C, Wang X (2002) The Arabidopsis phospholipase D family. Characterization of a calcium-independent and phosphatidylcholine-selective PLDζ 1 with distinct regulatory domains. Plant Physiol 128: 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Gronlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Ross EM, Higashijima T (1994) Regulation of G-protein activation by mastoparans and other cationic peptides. Methods Enzymol 237: 26–37 [DOI] [PubMed] [Google Scholar]
- Sagan M, de Larambergue H, Morandi D (1998) Genetic analysis of symbiosis mutants in Medicago truncatula. In C Elmerich, A Kondorosi, WE Newton, eds, Biological Nitrogen Fixation for the 21st Century, Vol 31. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 317–318
- Sagan M, Morandi D, Tarenghi E, Duc G (1995) Selection of nodulation and mycorrhizal mutants in the model plant Medicago truncatula (Gaertn.) after γ-ray mutagenesis. Plant Sci 111: 63–71 [Google Scholar]
- Schumaker KS, Sze H (1987) Inositol 1,4,5-trisphosphate releases Ca2+ from vacuolar membrane vesicles of oat roots. J Biol Chem 262: 3944–3946 [PubMed] [Google Scholar]
- Shaw SL, Long SR (2003) Nod factor elicits two separable calcium responses in Medicago truncatula root hair cells. Plant Physiol 131: 976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabados L, Charrier B, Kondorosi A, de Bruijn FJ, Ratet P (1995) New plant promoter and enhancer testing vectors. Mol Breed 1: 419–423 [Google Scholar]
- Sze H, Liang F, Hwang I, Curran AC, Harper JF (2000) Diversity and regulation of plant Ca2+ pumps: insights from expression in yeast. Annu Rev Plant Physiol Plant Mol Biol 51: 433–462 [DOI] [PubMed] [Google Scholar]
- Thoquet P, Ghérardi M, Journet EP, Kereszt A, Ané JM, Prosperi JM, Huguet T (2002) The molecular genetic linkage map of the model legume M. truncatula: an essential tool for comparative legume genomics and the isolation of agronomically important genes. BMC Plant Biol 2: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoud V, Journet E-P, Barker DG (1999. a) MtENOD20, a Nod factor-inducible molecular marker for root cortical cell activation. Mol Plant Microbe Interact 12: 604–614 [Google Scholar]
- Vernoud V, Pingret J-L, Chabaud M, Dedieu A, de Carvalho Niebel F, Journet EP, Barker DG (1999. b) Nod factor signal transduction in the Medicago truncatula Nod−/Myc− mutants TR25/26. In PJGM De Wit, T Bisseling, WJ Stiekema, eds, Proceedings of the Ninth International Congress on Molecular Plant-Microbe Interactions. ISMPMI, St. Paul, pp 114–119
- Wais RJ, Galera C, Oldroyd G, Catoira R, Penmetsa RV, Cook D, Gough C, Dénarié J, Long SR (2000) Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc Natl Acad Sci USA 97: 13407–13412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SA, Viprey V, Downie JA (2000) Dissection of nodulation signaling using pea mutants defective for calcium spiking induced by Nod factors and chitin oligomers. Proc Natl Acad Sci USA 97: 13413–13418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Qin C, Zhao J, Wang X (2004) Phospholipase Dα1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc Natl Acad Sci USA 101: 9508–9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Wang X (2004) Arabidopsis phospholipase Dα1 interacts with the heterotrimeric G-protein α-subunit through a motif analogous to the DRY motif in G-protein-coupled receptors. J Biol Chem 279: 1794–1800 [DOI] [PubMed] [Google Scholar]