Abstract
Expression of Kaposi’s sarcoma herpesvirus vFLIP, a potent activator of NFkB signaling, promotes latency. Inhibition of NFkB signaling promotes lytic reactivation. We previously reported that lytic inducer, RTA, inhibits vFLIP induced NFkB signaling by inducing the degradation of vFLIP via the proteasome. Here we report that the cellular ubiquitin ligase, Itch, is required for RTA induced degradation of vFLIP. Expression of either Itch targeting shRNA or a dominant negative mutant of the ubiquitin ligase both increased the stability of vFLIP in the presence of RTA. Itch potently ubiquitinated vFLIP in vivo and in vitro. We provide evidence for interaction between RTA, vFLIP and Itch and we identified an RTA resistant mutant of vFLIP that is unable to interact with Itch. These observations contribute to our understanding of how RTA counteracts the activities of vFLIP.
Introduction
Kaposi’s sarcoma-associated herpesvirus (KSHV) is associated with Kaposi′s sarcoma (KS), primary effusion lymphoma and multicentric Castleman′s disease (Gaidano et al., 1999; Ganem, 2006; Said et al., 1996; Schalling et al., 1995). Following infection, the circular double stranded DNA genome of KSHV persists in a latent state, as an episome tethered to the host genome. During latency, gene expression is limited to latency associated nuclear antigen (LANA), viral cyclin (vCYC), viral FLICE inhibitory protein (vFLIP), Kaposins, and miRNAs. These latency transcripts allow for cell proliferation, evasion of apoptosis, and proangiogenic and proinfammatory signaling (Mesri et al., 2010). In KS tumor cells, the majority of cells are latently infected, with a small percentage of cells undergoing lytic replication (Zhong et al., 1996). Lytic replication is induced by activation of the ORF50 promoter, resulting in expression of the lytic transactivator RTA (Gradoville et al., 2000; Lukac et al., 1998; Sun et al., 1998). RTA induces expression of lytic genes that control viral gene expression, DNA replication, and late genes that encode viral structural proteins required for assembly of nascent virions.
vFLIP is responsible for a number of functions thought to be essential for maintaining a long term latent infection, allowing for viral genome replication and segregation, cell growth, and survival. vFLIP was previously shown to inhibit death receptor-induced apoptosis, activate NFkB and is required for lymphoid cell survival (Chaudhary et al., 1999; Field et al., 2003; Guasparri et al., 2004; Liu et al., 2002; Matta et al., 2007; Thome et al., 1997). Mice expressing vFLIP display increased levels of proinflammatory cytokines, similar to what is observed in KSHV inflammatory cytokine syndrome (Ballon et al., 2015). Inflammatory cells and cytokines are typically present in the microenvironment of KS lesions. vFLIP upregulates the miR-17-92 cluster of microRNAs, downregulates TGFβ signaling, and decreases SMAD2 protein levels (Choi et al., 2015). vFLIP inhibits apoptosis by suppressing autophagy and induces Nrf2 through a SQSTM1 dependent mechanism (Gjyshi et al., 2015; Lee et al., 2009).
vFLIP expression and NFkB signaling have inhibitory effects on lytic replication (Brown et al., 2003; Guasparri et al., 2004; Keller et al., 2000). However, vFLIP expression is reduced in infected cells due to inefficient codon usage. This low level expression is advantageous to the virus, as codon optimization resulted in inefficient lytic induction (Bellare et al., 2015). These observations suggest that vFLIP must maintain a delicate balance. High levels of vFLIP expression and NFkB activation reduce the efficiency of lytic replication, yet insufficient vFLIP expression and NFkB signaling will hamper maintenance of a latent state of infection.
RTA is the major transactivator of lytic gene expression and was shown to target repressors of lytic replication using its intrinsic ubiquitin E3 ligase activity (Gould et al., 2009; Sun et al., 1998; Yang et al., 2008; Yu et al., 2005). In addition, RTA coopts the cellular ubiquitin ligase, RAUL, to target both IRF3 and IRF7 for proteasomal degradation, thereby inhibiting type I interferon production (Yu and Hayward, 2010).
We previously reported that vFLIP-induced NFkB activation was abrogated by RTA early in lytic reactivation (Ehrlich et al., 2014). RTA induced the proteasomal degradation of vFLIP, however the ubiquitin ligase domains of RTA and RAUL were not required for this activity. Here we report that the Itch ubiquitin ligase is required for RTA-induced vFLIP degradation using both a dominant negative mutant of Itch as well as a stable shRNA induced knockdown. Using an in vitro ubiquitination assay, we unequivocally demonstrate ubiquitination of vFLIP by the Itch ubiquitin ligase. We observed increased ubiquitination of vFLIP in cells transfected with RTA, which was abrogated upon addition of a dominant negative mutant of Itch. We provide evidence for Itch interaction with both vFLIP and RTA and have identified mutants of vFLIP that are resistant to RTA induced degradation and can no longer interact with Itch. These data provide insight into the mechanism by which RTA abolishes vFLIP activity early in lytic reactivation.
Results
The Itch ubiquitin ligase is required for degradation of vFLIP by RTA
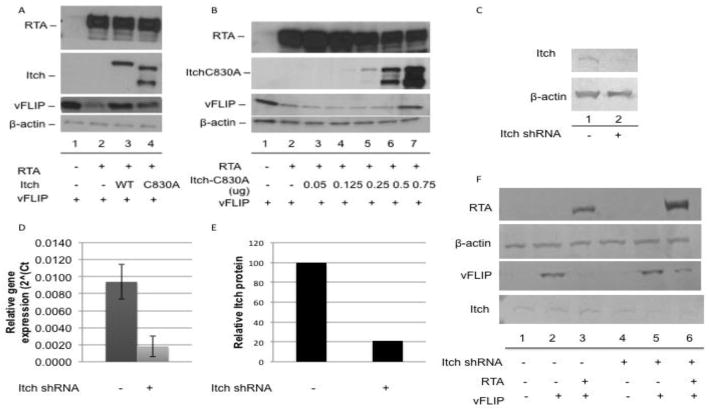
Itch is a cellular ubiquitin E3-ligase that was reported to target the cellular FLICE inhibitory protein (cFLIP), among other proteins, for proteasomal degradation (Chang et al., 2006). Due to the homology between cFLIP and vFLIP, we hypothesized that Itch might target vFLIP. To determine whether the ubiquitin E3 ligase activity of Itch was required for RTA induced vFLIP degradation we utilized a dominant negative mutant, Itch-C830A (Angers et al., 2004). In the presence of RTA and wild-type Itch, we observed degradation of vFLIP, however in the presence of Itch-C830A, vFLIP exhibited increased stability (Fig. 1a). vFLIP stability in the presence of Itch-C830A was dose dependent; as vFLIP stabilization increases with increasing amounts of Itch-C830A (Fig. 1b).
Figure 1.
Itch is required for RTA induced degradation of vFLIP. A. vFLIP is stabilized in the presence of both wild type and a dominant negative mutant (C830A) of Itch. B. vFLIP is stabilized by Itch C830A in a dose dependent manner. (A and B) 293T cells were transfected with myc-vFLIP, RTA, and Flag tagged wild-type or mutant Itch where indicated. 48hrs post transfection, cells were harvested and lysates were analyzed by SDS-PAGE followed by immunoblot using antibodies against myc, RTA, and Flag. Beta actin was used as a loading control. C–E. Cells expressing Itch targeting shRNA display reduced levels of Itch protein and mRNA. Cells were transduced with lentivirus containing shRNA targeting Itch where indicated. Cells were selected with 0.9ug/ml puromycin and harvested for analysis of (C) Itch protein or (D) mRNA. (E) Itch protein levels were quantified using ImageJ. F. vFLIP exhibited increased stability in the presence of RTA in cells with reduced Itch. Control and Itch shRNA expressing 293T cells were transfected with myc-vFLIP and RTA where indicated. 48hrs post transfection, cells were harvested and lysates were analyzed by SDS-PAGE followed by immunoblot using antibodies against myc, RTA, and Itch. Beta actin was used as a loading control.
Interestingly, vFLIP was also stabilized in the presence of wild-type Itch and RTA, suggesting that Itch may be acting to degrade vFLIP as part of a complex (Fig. 1a, lane 3). An increase in one component of a complex would affect the stoichiometry, resulting in a dominant negative phenotype. Itch has been shown target multiple substrates, a number of which are involved in NFkB signaling. Itch has been shown to function both as a monomer and as part of a multiprotein complex (Melino et al., 2008). To further examine whether Itch is required for vFLIP degradation, we constructed 293T cells stably expressing Itch-targeting shRNA. Itch knockdown cells exhibited a five-fold reduction in Itch mRNA and protein compared to control cells (Fig. 1c–e). While transfection of control cells with vFLIP and RTA resulted in near complete degradation of vFLIP, Itch knockdown resulted in increased vFLIP stability in the presence of RTA (Fig. 1f, compare lanes 3 and 6).
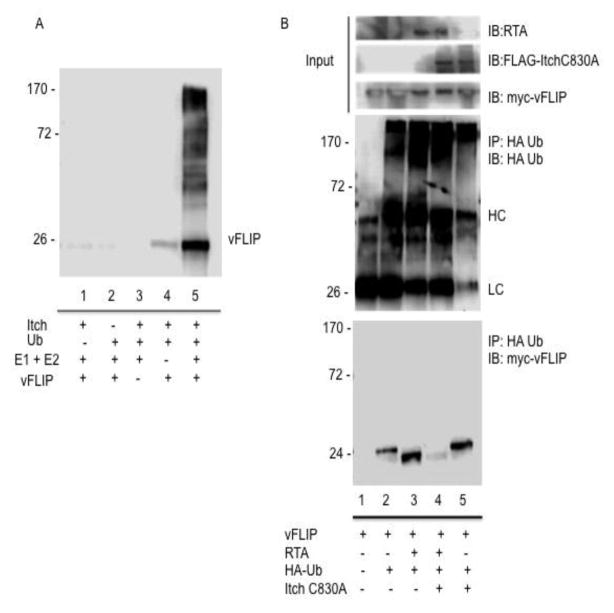
To further examine whether the Itch ubiquitin ligase is required for ubiquitination of vFLIP, we performed an in vitro ubiquitination assay using purified Itch and vFLIP. While reactions lacking ubiquitin, E1 and E2 enzymes, Itch, or vFLIP displayed no vFLIP:ubiquitin conjugates (Fig. 2a, lanes 1–4), reactions containing all reaction components exhibited robust vFLIP ubiquitination (Fig. 2a, lane 5).
Figure 2.
Itch ubiquitinates vFLIP. A. Itch ubiquitinates vFLIP in vitro. In 30ul reactions, the following reagents were added where indicated: Itch (0.5ug), Ub aldehyde (5 ug), fraction B (E1+E2) 12ul, 10X energy solution (3ul), vFLIP (5.5ug). Reactions were incubated at 37°C for 1.5h and stopped by addition of 2x Laemmli loading buffer. Reactions were analyzed by SDS PAGE followed by detection of vFLIP:ubiquitin conjugates with antibody against V5-vFLIP. B. Itch is required for RTA induced ubiquitination of vFLIP. 293T cells were transfected with HA tagged ubiquitin, myc tagged vFLIP, RTA, and Flag tagged Itch C830A where indicated. Cells were treated with MG132 4h prior to harvesting. Cell lysates were harvested for immunoprecipitation with anti HA. Immunoprecipitates were washed with RIPA lysis buffer containing 500mM NaCl and analyzed via immunoblot against HA, myc, FLAG, and RTA. (HC = heavy chain, LC = light chain)
To determine whether Itch is required for RTA induced ubiquitination of vFLIP, we transfected 293T cells with HA tagged ubiquitin and vFLIP in the presence of RTA and/or Itch C830A and immunoprecipitated HA tagged ubiquitin conjugates of vFLIP (Fig. 2b). Interestingly, we detected robust ubiquitination of vFLIP in the absence of RTA (Fig. 2b, lane 2). These ubiquitin conjugates were stable (data not shown) and migrated at a slower rate than the RTA induced ubiquitin conjugates (compare lanes 2 and 3). Upon addition of Itch C830A, in the presence of RTA, we observed a near complete loss of ubiquitinated vFLIP, however the dominant negative mutant of Itch had no effect on the RTA independent vFLIP ubiquitination (compare lanes 4 and 5). Taken together, this data strongly suggests that the Itch ubiquitin ligase is required for RTA induced ubiquitination and degradation of vFLIP. This data also suggests that an alternative ubiquitin ligase is responsible for RTA independent ubiquitination of vFLIP and that these ubiquitin conjugates do not target vFLIP for degradation via the proteasome.
RTA and vFLIP interact with endogenous Itch
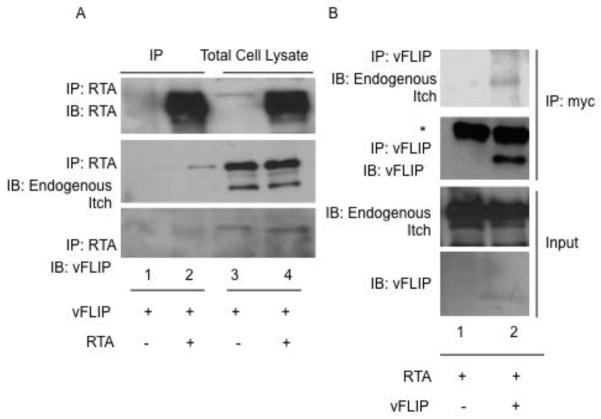
We next assessed the composition of the Itch containing ubiquitin ligase complex. 293T cells were transfected with vFLIP, RTA and/or empty vector and treated with proteasome inhibitor (MG132) 16 hours prior to harvesting. Through a series of co-immunoprecipitations, we were able to detect interaction between RTA, vFLIP and endogenous Itch (Fig. 3a and b). Immunoprecipitation of RTA and vFLIP, respectively, resulted in detection of endogenous Itch in RTA and vFLIP transfected cells compared to empty vector-transfected controls (Fig. 3a and b). While all samples were treated with MG132 16h prior to harvesting, it was difficult to detect high levels of vFLIP in RTA immunoprecipitates, possibly due to incomplete proteasome inhibition.
Figure 3.
vFLIP and RTA interact with endogenous Itch. A. RTA interacts with endogenous Itch and vFLIP. 293T cells were transfected with vFLIP and RTA. 24h post-transfection, cells were treated with 1uM MG132. 48h post transfection, cells were harvested and processed for immunoprecipitation. Precleared lysates were incubated with antibodies against RTA followed by addition of protein A/G agarose and washed with RIPA buffer. Immunoprecipitates were analyzed by SDS-PAGE followed by immunoblot using antibodies against myc, RTA, and Itch. B. vFLIP interacts with endogenous Itch. 293T cells were transfected with vFLIP and RTA. 24h post-transfection, cells were treated with 1uM MG132. 48h post transfection, cells were harvested and processed for immunoprecipitation. Precleared lysates were incubated with antibodies against myc-vFLIP followed by addition of protein A/G agarose and washed with RIPA buffer. Immunoprecipitates were analyzed by SDS-PAGE followed by immunoblot using antibodies against myc and Itch. *Asterisk denotes light chain (LC) band.
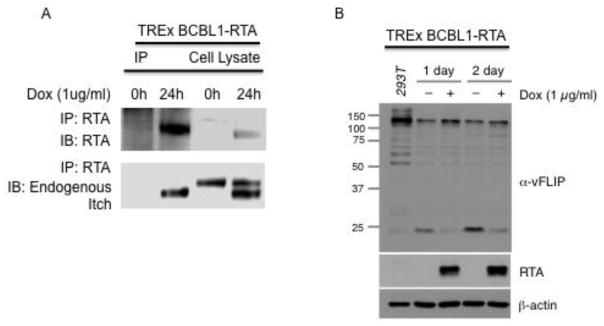
To further evaluate the interaction between Itch and RTA, we utilized the TREx BCBL1-RTA cells. Following treatment with doxycycline (1ug/ml) for 0 or 24 hours, RTA was immunoprecipitated from lysates. We observed Itch and RTA interaction at the 24-hour time point (Fig. 4a). Interestingly, in the TREx BCBL1-RTA cells we detected two forms of Itch in the cell lysate. Two bands were detected in the following 24h treatment with doxycycline, compared to one faster migrating band in untreated t=0 cells. This high molecular weight band likely represents a modified form of Itch. Only lower molecular weight Itch was detected in RTA containing immunoprecipitates.
Figure 4.
RTA interacts with Itch and induces the degradation of vFLIP in naturally infected TREx BCBL1-RTA cells. A. RTA interacts with Itch. TREx BCBL1-RTA cells were treated with doxycycline (1ug/ml) for 0 or 24h to induce RTA expression. Cells were harvested and processed for immunoprecipitation with antibodies against RTA. Immunoprecipitates were analyzed by SDS PAGE followed by immunoblot against RTA and endogenous Itch. B. RTA induces the degradation of vFLIP in naturally infected cells. TREx BCBL1-RTA cells were treated with doxycycline (1ug/ml) for 0, 24 or 48h to induce RTA expression. Cells were harvested and analyzed by SDS PAGE followed by immunoblot against vFLIP, RTA and endogenous beta actin.
We observed reduced levels of vFLIP in the TREx BCBL1-RTA cell lysates at 24 and 48 hours post doxycycline induction of RTA (Fig. 4b).
Taken together, this data suggests that RTA interacts with Itch in both 293T and TREx BCBL1-RTA cells.
An RTA resistant mutant of vFLIP is unable to interact with Itch
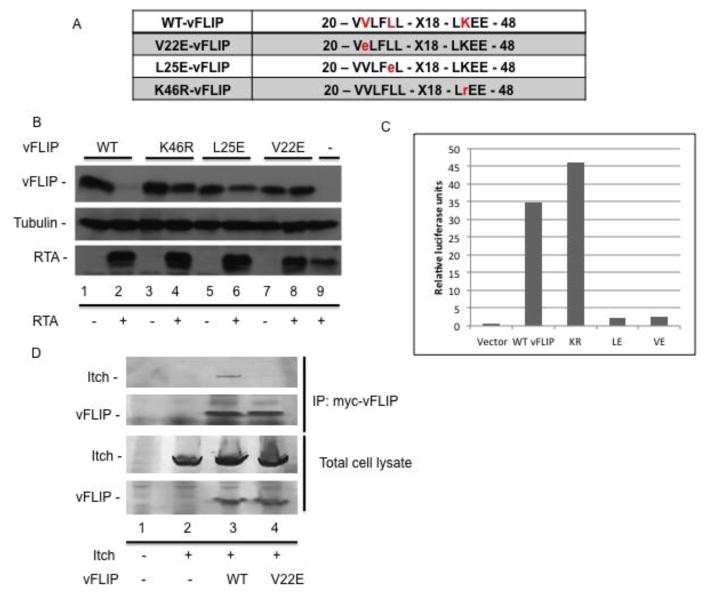
We reasoned that if RTA is inducing degradation of vFLIP through Itch, we should be able to identify a vFLIP mutant that is resistant to degradation by RTA and this mutant should be unable to interact with Itch. We generated a series of point mutants in vFLIP and screened them for sensitivity to RTA. We identified three mutants of vFLIP that were resistant to RTA - V22E, L25E, and K46R (Fig. 5a). Transfection of 293T cells with wild-type (WT) vFLIP and RTA resulted in degradation of vFLIP, however mutation of amino acids 22, 25, or 46, resulted in complete stability, suggesting that these residues are important for interaction with the ubiquitin proteasome machinery (Fig. 5b). Both vFLIP V22E and L25E were unable to activate NFkB via luciferase reporter assay, while K46R was unaffected (Fig. 5c). Following further characterization of these vFLIP point mutants, we chose to examine whether the interaction between vFLIP and Itch was affected by mutation of V22 to E. 293T cells were transfected with wild-type, mutant vFLIP V22E, or empty vector control plasmid and FLAG tagged Itch. We immunoprecipitated myc tagged wild type or mutant vFLIP and were only able to detect Itch bound to wild type vFLIP (Fig. 5d). Taken together this data suggests that a motif spanning V22 to L25 is important for both vFLIP interaction with Itch and degradation induced by RTA.
Figure 5.
Valine 22 is required for vFLIP interaction with Itch and sensitivity to degradation in the presence of RTA. A. Illustration of RTA resistant mutants of vFLIP. B. Mutations of amino acids 22, 25, and 46 in vFLIP confer resistance to RTA induced degradation. 293T cells were transfected with RTA and wild type or mutant vFLIP where indicated. 48h post transfection, cells were harvested and lysates were analyzed via immunoblot against myc-vFLIP, RTA, and tubulin. C. vFLIP V22E and L25E are unable to activate NFκB. NFκB activation was quantified using the Dual Luciferase reporter assay system. 293T cells were transfected with wild type or mutant vFLIP plus the two reporter plasmids pNFκB-Luc and pGL4.70 Renilla luciferase. Luciferase was measured and data were taken as a ratio of firefly/renilla luciferase. D. Valine 22 in vFLIP is required for vFLIP interaction with RTA. 293T cells were transfected with Flag tagged Itch and wild type or mutant vFLIP where indicated. 48h post transfection, cells were harvested and processed for immunoprecipitation. Precleared lysates were incubated with antibodies against myc-vFLIP followed by addition of protein A/G agarose and washed with RIPA buffer. Immunoprecipitates were analyzed by SDS-PAGE followed by immunoblot using antibodies against myc-vFLIP and Flag-Itch.
Experimental procedures
Cell Line Maintenance and Transfection
HEK 293T cells were obtained from ATCC and cultured in DMEM media supplemented with 10% FBS and were grown at 5 % CO2 at 37 °C. Cells were transfected at 60–70% confluence using 1mg/ml polyethyleneimine (PEI) linear, MW~25,000 (Polysciences, Inc. Cat#23966) at a ratio of 1ug plasmid DNA:3ul PEI. For co-immunoprecipitations (co-IPs), cells were transfected using the Calcium Phosphate or Mirus LT1. TREx BCBL1-RTA cells were (a generous gift from Jae U. Jung) were maintained in RPMI supplemented with 15% FBS. Doxycycline (1ug/ml) was added for 24 or 48h to induce RTA expression (Nakamura et al., 2003).
Reagent, Plasmids, and Antibodies
MG132 (Boston Biochem) was used for proteasome inhibition. The following plasmids were generously provided by Annie Angers Flag-Itch and Flag-Itch-C830A (Angers et al., 2004). myc-vFLIP (pYNC989) and RTA (pSEW-R01) were provided by Gary Hayward and were described previously (Ehrlich et al., 2014; Wang et al., 2003; Yu et al., 2005). Recombinant V5-His-vFLIP: vFLIP was cloned into pET-DEST42 using Gateway cloning technology (Invitrogen). HA tagged ubiquitin was provided by Joanna Shisler.
The following antibodies were used: anti-RTA (G. Hayward), anti-cmyc (Millipore), anti-Itch (BD Biosciences), anti-β-actin (ThermoScientific), anti-tubulin (ThermoScientific), anti-Flag (Sigma-Aldrich) Secondary antibodies were either HRP or AP labled and obtained from Jackson ImmunoResearch.
Immunoblot Analysis
Cells were transfected with the indicated constructs and harvested 48h post-transfection either by direct addition of 2X Laemmli Buffer or addition of RIPA lysis buffer followed by sonication. Lysates were run on a 10% SDS-PAGE or Any kD Mini-PROTEAN TGX Precast Gel (Biorad) and Tris-glycine running buffer. Proteins were transferred to a PVDF membrane using a semi-dry transfer system at 25V for 25min. Membranes were blocked in 5% non-fat dry milk in PBS for 1 hr followed by incubation with primary antibody overnight at 4°C and secondary antibody for 1 hr at room temperature. Proteins were visualized by addition of ECL substrate and detection of chemiluminescent signal on x-ray film or via Licor C-DiGit or BCIP/NBT and direct visualization.
Immunoprecipitation
Cells were transfected with the indicated constructs and harvested 48h post-transfection. RIPA lysis buffer containing NEM and protease inhibitor was added to cell pellets. Cell lysates were sonicated, followed by centrifugation to clarify lysates. 10% of cell lysate was saved for input analysis. Samples were precleared with protein A/G PLUS-agarose (Santa Cruz) for 30 min at 4°C. Lysates were incubated with 1ug antibody overnight at 4°C. Proteins were immunoprecipitated by addition of 25μl protein A/G-agarose 1 hr at 4°C. Immunoprecipitates were washed 4x with RIPA lysis buffer. 50μl of 2X Laemmli Buffer was added and samples were incubated at 100°C for 5 min. Immunoprecipitates and reserved input lysates were analyzed via immunoblot.
Stable cell lines
293T cells stably expressing shRNA were produced using Mission shRNA lentivirus particles according to manufacturers instructions (Sigma). 24h following transduction, media was changed and replaced with puromycin (0.9ug/ml). Cells were selected for two weeks and maintained in DMEM + 10% FBS +0.9ug/ml puromycin. Knockdown was confirmed via western blot and qPCR.
RNA extraction, reverse transcription, and qPCR
RNA was extracted using the Promega SV Total RNA Extraction Kit. DNase-treated RNA was reverse transcribed using the RevertAid First Strand cDNA Synthesis Kit (Fermentas) according to manufacturer’s instructions. The resulting cDNA was used for qPCR performed on Biorad CFX Connect using iTaq Universal SYBR green supermix (Biorad) as per manufacturer's specifications. Itch, GAPDH, and HSPCB were amplified using the following primers: Itch F, ACCGGCTGCCATCTTAGTCT; Itch R, GGAAAACCTGAAGTTCTCACAGT; GAPDH F AATCCCATCACCATCTTCCAG; GAPDH R AAATGAGCCCCAGCCTTC; HSPCB F TCTGGGTATCGGAAAGCAAGCC; HSPCB R GTGCACTTCCTCAGGCATCTTG. Relative gene expression was calculated using the ΔCT method (2^(CT HSPCB-CT ITCH)) using the HSPCB housekeeping gene for normalization. Error bars represent the standard error.
In vitro ubiquitination assay
V5-His tagged vFLIP was expressed in E. coli (BL21) and purified using Ni-NTA Resin (ThermoFisher). Purified Itch, ubiquitin aldehyde, Fraction B (source of E1 and E2) and 10x energy regeneration solution was purchased from Boston Biochem. The following reagents were added to 30ul reactions where indicated: Itch (0.5ug), Ub aldehyde (5 ug), fraction B (E1+E2) 12ul, 10X energy solution (3ul), vFLIP (5.5ug). Reactions were incubated at 37°C for 1.5h followed by addition of 2x Laemmli loading buffer. Reactions were analyzed by SDS PAGE followed by immunoblot with the indicated antibodies.
Luciferase reporter assay
NFκB activation was quantified using the Dual Luciferase reporter assay system (Promega). Briefly cells were transfected with indicated plasmids plus the two reporter plasmids pNFκB-Luc (Stratagene) and pGL4.70[hRluc]. Cell lysates were prepared according to the manufacturer’s protocol. Luciferase was measured on a GloMax-Multi Microplate Multimode Reader (Promega). Data were taken as a ratio of firefly/renilla luciferase.
In vivo ubiquitin pull down
Cells were transfected with HA tagged ubiquitin as described above. Cells were treated with 10uM MG132 4h prior to harvest. Lysates were prepared using RIPA lysis buffer containing protease inhibitor, MG132, and NEM. Cell lysates were sonicated, followed by centrifugation to clarify lysates. 10% of cell lysate was saved for input analysis. Samples were precleared with protein A/G PLUS-agarose (Santa Cruz) for 30 min at 4°C. Lysates were incubated with 1ug HA antibody for 2 hours at 4°C. Proteins were immunoprecipitated by addition of 25μl protein A/G-agarose 1 hr at 4°C. Immunoprecipitates were washed 4x with RIPA lysis buffer containing 500mM NaCl. 50μl of 2X Laemmli Buffer was added and samples were incubated at 100°C for 5 min. Immunoprecipitates and reserved input lysates were analyzed via immunoblot.
Discussion
We previously reported that RTA inhibits NFkB signaling early in lytic reactivation by targeting vFLIP for degradation via the proteasome, however the exact mechanism responsible for RTA induced vFLIP degradation remained unclear (Ehrlich et al., 2014). Here we provide evidence suggesting that the cellular ubiquitin ligase, Itch, is required for RTA induced degradation of vFLIP. We demonstrate vFLIP and RTA interaction with Itch and we provide evidence of increased vFLIP stability in the presence of both a dominant negative mutant of Itch and Itch targeting shRNA. We demonstrate ubiquitination of vFLIP by purified Itch in vitro and provide evidence for RTA interaction with Itch in TREx BCBL1-RTA cells. We have also identified three vFLIP mutants that are resistant to degradation in the presence of RTA. We were able to detect interaction between wild-type vFLIP and Itch even in the absence of RTA. This interaction is dependent on V22 in vFLIP, as mutation of this amino acid abolished interaction. Based on these observations, we propose a model where vFLIP interacts with Itch in latency. Following lytic reactivation, RTA is recruited to the vFLIP/Itch complex, resulting in the degradation of vFLIP.
Interestingly, we observed increased vFLIP stability in the presence of wild-type Itch. This was surprising as one would expect overexpression of a ubiquitin ligase to result in increased degradation of the substrate. We have observed a similar result in previous work with Cullin 5, a member of the Cullin-RING family of ubiquitin ligases (CRL) (personal communication with Xiao-Fang Yu). CRLs are modular ubiquitin ligases that contain multiple proteins. Overexpression of one member of the complex resulted in stabilization of the substrate due to the disruption of the stoichiometry of the ubiquitin ligase components, producing a dominant negative phenotype. The stabilizing effect of wild-type Itch suggests that this ubiquitin ligase is essential for RTA induced degradation of vFLIP and is likely working as part of a multi-protein complex, however the exact nature and regulation of this complex remains unclear.
In light of the observation that wild-type Itch was causing a stabilizing effect on vFLIP, we constructed 293T cells stably expressing shRNA targeting Itch to further examine the requirement for Itch. We observed a five-fold decrease in Itch mRNA and protein in cells expressing Itch shRNA compared to control cells and a corresponding increase in the stability of vFLIP.
To further examine the role of Itch in vFLIP degradation, we performed an in vitro ubiquitination assay, where we observed robust ubiquitination of vFLIP by purified Itch. When we carried out a ubiquitin pull down assay using transfected 293T cells, we got an unexpected yet very interesting result. In initial experiments, we immunoprecipitated vFLIP in the presence or absence of Itch C830A and RTA and we repeatedly observed strong ubiquitination of vFLIP in the absence of RTA, however when we transfected RTA with vFLIP, we observed rapid degradation of vFLIP (data not shown). This RTA independent ubiquitinated vFLIP was stable and only upon addition of RTA did we observe degradation. To further understand this result, we immunoprecipitated HA tagged ubiquitin from cells transfected with vFLIP, RTA, and/or Itch C830A. Again, in the absence of RTA we detected ubiquitinated vFLIP. Interestingly, Itch C830A expression, had no effect on these RTA independent vFLIP:ubiquitin conjugates, suggesting that vFLIP is ubiquitinated by another ubiquitin ligase during latency. Addition of RTA resulted in increased ubiquitnation of vFLIP and these vFLIP:ubiquitin conjugates migrated at a faster rate than the RTA independent vFLIP:ubiquitin conjugates. Addition of the Itch dominant negative mutant C830A abolished the RTA dependent vFLIP:ubiquitin conjugates. This data along with the dominant negative mutant data strongly suggest that Itch is required for RTA induced vFLIP ubiquitination and degradation. Our data implies that RTA independent ubiquitination of vFLIP occurs and that this ubiquitination occurs independent of Itch and is likely not K48 linked. Future studies are needed to address the role of this RTA and Itch independent ubiquitination of vFLIP.
One question remains; what is the mechanistic role of RTA in vFLIP degradation? Our data clearly demonstrates reduced vFLIP protein only in the presence of RTA in both naturally infected TREx BCBL1-RTA cells and 293T cells overexpressing vFLIP and RTA. This decrease in vFLIP is dependent on co-expression of RTA and Itch. However, we observed a strong interaction between Itch and vFLIP in the absence of RTA, suggesting that vFLIP may interact with Itch in latency and the expression of RTA during lytic reactivation may activate Itch dependent ubiquitination of vFLIP. Itch is a member of the homologous to E6-AP carboxy terminus (HECT) family of ubiquitin ligases. Deletion of the gene in mice results in an Itchy phenotype due to defects in the immune system and dysregulation of the inflammatory response. Itch has been reported to target substrates involved in the regulation of multiple pathways involved in immune regulation such as c-Jun and Jun-B, Notch, Deltex, p63, p73, cFLIP, SMAD2 and A20 (Melino et al., 2008). The microenvironment typical to KS lesions is rich in inflammatory cells and cytokines making in a role for dysregulated Itch appealing. Further studies evaluating Itch activity in naturally infected cells are needed to determine whether this ubiquitin ligase plays a role in formation of the well documented inflammatory microenvironment in KS.
Itch has been reported to work alone, binding to substrates through its WW domains, and in concert with other ubiquitin ligases and adaptor proteins. The WW domain in Itch interacts with a PY consensus sequence in the substrate or adaptor protein. Itch has also been shown to interact with substrates and adaptor proteins through a pSP or pTP motif as well as through other noncannonical motifs. Itch is reported to exist in either an active or inactive conformation that is dependent on whether the ubiquitin ligase is interacting with a PY motif containing protein. While vFLIP has no apparent PY motif, RTA has a number of proline rich regions, none of which fit the canonical PPxY, but one of which sits with in the region of RTA that we previously reported was required for degradation of vFLIP (amino acids 11–149) (Ehrlich et al., 2014). These observations support our model where vFLIP interacts with Itch either directly or through an adaptor protein and binding of RTA activates Itch to induce the degradation of vFLIP.
Interestingly, we observed two forms of Itch in the TREx BCBL1-RTA cells, a higher molecular weight form in uninduced cells and the appearance of a lower molecular weight species at 24h post induction. RTA, only interacts with the lower molecular weight species. Further examination of these different forms of Itch will provide insight into the role of RTA in vFLIP degradation.
Future studies examining effect of vFLIP and RTA on the activity of Itch and the stability of Itch substrates will increase our understanding of how cellular processes are influenced by both latent and lytic phases of KSHV replication. This knowledge will aid in the development of novel strategies for treating KSHV associated malignancies.
Research highlights.
The cellular ubiquitin ligase Itch is required for RTA induced vFLIP degradation
Itch interacts with RTA in both 293T cells and naturally infected TREx BCBL1-RTA cells
Itch ubiquitinates vFLIP in vitro and in vivo
Mutant vFLIP that cannot interact with Itch is stable in the presence of RTA
Acknowledgments
The authors would like to acknowledge Gary Hayward for generously providing RTA and vFLIP expression vectors and RTA antibody, Edward Harhaj for reagents and helpful discussion, Jae U. Jung for the TREx BCBL1-RTA cells, Joanna Shisler for the HA tagged ubiquitin expression vector and Annie Angers for the Itch and Itch C830A expression vectors. This work was funded by NIH grant 1R15GM118011 as well as Fisher Endowed Chair funds from Towson University.
Footnotes
Conflicts of interest
The authors indicate no potential conflicts of interest.
Author contributions
J.C., K.H., A.R., Y.F., Y.B.C.: collection and/or assembly of data, data analysis and interpretation; T.A. and A.M.: data analysis and interpretation; E.E.: conception and design, collection and/or assembly of data, data analysis and interpretation, and manuscript writing;
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angers A, Ramjaun AR, McPherson PS. The HECT domain ligase itch ubiquitinates endophilin and localizes to the trans-Golgi network and endosomal system. J Biol Chem. 2004;279:11471–11479. doi: 10.1074/jbc.M309934200. [DOI] [PubMed] [Google Scholar]
- Ballon G, Akar G, Cesarman E. Systemic expression of Kaposi sarcoma herpesvirus (KSHV) Vflip in endothelial cells leads to a profound proinflammatory phenotype and myeloid lineage remodeling in vivo. PLoS Pathog. 2015;11:e1004581. doi: 10.1371/journal.ppat.1004581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellare P, Dufresne A, Ganem D. Inefficient Codon Usage Impairs mRNA Accumulation: the Case of the v-FLIP Gene of Kaposi's Sarcoma-Associated Herpesvirus. J Virol. 2015;89:7097–7107. doi: 10.1128/JVI.03390-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown HJ, Song MJ, Deng H, Wu TT, Cheng G, Sun R. NF-kappaB inhibits gammaherpesvirus lytic replication. J Virol. 2003;77:8532–8540. doi: 10.1128/JVI.77.15.8532-8540.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Kamata H, Solinas G, Luo JL, Maeda S, Venuprasad K, Liu YC, Karin M. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Chaudhary PM, Jasmin A, Eby MT, Hood L. Modulation of the NF-kappa B pathway by virally encoded death effector domains-containing proteins. Oncogene. 1999;18:5738–5746. doi: 10.1038/sj.onc.1202976. [DOI] [PubMed] [Google Scholar]
- Choi HS, Jain V, Krueger B, Marshall V, Kim CH, Shisler JL, Whitby D, Renne R. Kaposi's Sarcoma-Associated Herpesvirus (KSHV) Induces the Oncogenic miR-17-92 Cluster and Down-Regulates TGF-beta Signaling. PLoS Pathog. 2015;11:e1005255. doi: 10.1371/journal.ppat.1005255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich ES, Chmura JC, Smith JC, Kalu NN, Hayward GS. KSHV RTA abolishes NFkappaB responsive gene expression during lytic reactivation by targeting vFLIP for degradation via the proteasome. PLoS One. 2014;9:e91359. doi: 10.1371/journal.pone.0091359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field N, Low W, Daniels M, Howell S, Daviet L, Boshoff C, Collins M. KSHV vFLIP binds to IKK-gamma to activate IKK. J Cell Sci. 2003;116:3721–3728. doi: 10.1242/jcs.00691. [DOI] [PubMed] [Google Scholar]
- Gaidano G, Castanos-Velez E, Biberfeld P. Lymphoid disorders associated with HHV-8/KSHV infection: facts and contentions. Med Oncol. 1999;16:8–12. doi: 10.1007/BF02787352. [DOI] [PubMed] [Google Scholar]
- Ganem D. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu Rev Pathol. 2006;1:273–296. doi: 10.1146/annurev.pathol.1.110304.100133. [DOI] [PubMed] [Google Scholar]
- Gjyshi O, Flaherty S, Veettil MV, Johnson KE, Chandran B, Bottero V. Kaposi's sarcoma-associated herpesvirus induces Nrf2 activation in latently infected endothelial cells through SQSTM1 phosphorylation and interaction with polyubiquitinated Keap1. J Virol. 2015;89:2268–2286. doi: 10.1128/JVI.02742-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould F, Harrison SM, Hewitt EW, Whitehouse A. Kaposi's sarcoma-associated herpesvirus RTA promotes degradation of the Hey1 repressor protein through the ubiquitin proteasome pathway. J Virol. 2009;83:6727–6738. doi: 10.1128/JVI.00351-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradoville L, Gerlach J, Grogan E, Shedd D, Nikiforow S, Metroka C, Miller G. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J Virol. 2000;74:6207–6212. doi: 10.1128/jvi.74.13.6207-6212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasparri I, Keller SA, Cesarman E. KSHV vFLIP is essential for the survival of infected lymphoma cells. J Exp Med. 2004;199:993–1003. doi: 10.1084/jem.20031467. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Keller SA, Schattner EJ, Cesarman E. Inhibition of NF-kappaB induces apoptosis of KSHV-infected primary effusion lymphoma cells. Blood. 2000;96:2537–2542. [PubMed] [Google Scholar]
- Lee JS, Li Q, Lee JY, Lee SH, Jeong JH, Lee HR, Chang H, Zhou FC, Gao SJ, Liang C, Jung JU. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol. 2009;11:1355–1362. doi: 10.1038/ncb1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Eby MT, Rathore N, Sinha SK, Kumar A, Chaudhary PM. The human herpes virus 8-encoded viral FLICE inhibitory protein physically associates with and persistently activates the Ikappa B kinase complex. J Biol Chem. 2002;277:13745–13751. doi: 10.1074/jbc.M110480200. [DOI] [PubMed] [Google Scholar]
- Lukac DM, Renne R, Kirshner JR, Ganem D. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- Matta H, Mazzacurati L, Schamus S, Yang T, Sun Q, Chaudhary PM. Kaposi's sarcoma-associated herpesvirus (KSHV) oncoprotein K13 bypasses TRAFs and directly interacts with the IkappaB kinase complex to selectively activate NF-kappaB without JNK activation. J Biol Chem. 2007;282:24858–24865. doi: 10.1074/jbc.M700118200. [DOI] [PubMed] [Google Scholar]
- Melino G, Gallagher E, Aqeilan RI, Knight R, Peschiaroli A, Rossi M, Scialpi F, Malatesta M, Zocchi L, Browne G, Ciechanover A, Bernassola F. Itch: a HECT-type E3 ligase regulating immunity, skin and cancer. Cell Death Differ. 2008;15:1103–1112. doi: 10.1038/cdd.2008.60. [DOI] [PubMed] [Google Scholar]
- Mesri EA, Cesarman E, Boshoff C. Kaposi's sarcoma and its associated herpesvirus. Nat Rev Cancer. 2010;10:707–719. doi: 10.1038/nrc2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Lu M, Gwack Y, Souvlis J, Zeichner SL, Jung JU. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J Virol. 2003;77:4205–4220. doi: 10.1128/JVI.77.7.4205-4220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said W, Chien K, Takeuchi S, Tasaka T, Asou H, Cho SK, de Vos S, Cesarman E, Knowles DM, Koeffler HP. Kaposi's sarcoma-associated herpesvirus (KSHV or HHV8) in primary effusion lymphoma: ultrastructural demonstration of herpesvirus in lymphoma cells. Blood. 1996;87:4937–4943. [PubMed] [Google Scholar]
- Schalling M, Ekman M, Kaaya EE, Linde A, Biberfeld P. A role for a new herpes virus (KSHV) in different forms of Kaposi's sarcoma. Nat Med. 1995;1:707–708. doi: 10.1038/nm0795-707. [DOI] [PubMed] [Google Scholar]
- Sun R, Lin SF, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci U S A. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer JL, Schroter M, Scaffidi C, Krammer PH, Peter ME, Tschopp J. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- Wang SE, Wu FY, Fujimuro M, Zong J, Hayward SD, Hayward GS. Role of CCAAT/enhancer-binding protein alpha (C/EBPalpha) in activation of the Kaposi's sarcoma-associated herpesvirus (KSHV) lytic-cycle replication-associated protein (RAP) promoter in cooperation with the KSHV replication and transcription activator (RTA) and RAP. J Virol. 2003;77:600–623. doi: 10.1128/JVI.77.1.600-623.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yan Z, Wood C. Kaposi's sarcoma-associated herpesvirus transactivator RTA promotes degradation of the repressors to regulate viral lytic replication. J Virol. 2008;82:3590–3603. doi: 10.1128/JVI.02229-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Hayward GS. The ubiquitin E3 ligase RAUL negatively regulates type i interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity. 2010;33:863–877. doi: 10.1016/j.immuni.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Wang SE, Hayward GS. The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity. 2005;22:59–70. doi: 10.1016/j.immuni.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci U S A. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]