Abstract
Development of thylakoid membranes depends upon the transport of membrane vesicles from the chloroplast inner envelope and subsequent fusion of vesicles within the interior of the plastid. The Arabidopsis (Arabidopsis thaliana) Thylakoid formation1 (Thf1) gene product is shown here to control an important step required for the normal organization of these vesicles into mature thylakoid stacks and ultimately for leaf development. The Arabidopsis Thf1 gene encodes an imported chloroplast protein, as shown by in vitro import and localization of a Thf1-green fluorescent protein fusion product in transgenic plants. This gene is conserved in oxygenic photoautotrophs ranging from cyanobacteria to flowering land plants. Transcript levels for Thf1 are induced in the light and decrease under dark conditions, paralleling profiles of light-regulated nuclear genes involved in chloroplast function. Disruption of the Thf1 gene via T-DNA insertion results in plants that are severely stunted with variegated leaf patterns. Nongreen sectors of variegated leaves lacking Thf1 expression contain plastids that accumulate membrane vesicles on the interior and lack organized thylakoid structures. Green sectors of Thf1-disrupted leaves contain some chloroplasts that form organized thylakoid membranes, indicating that an inefficient compensatory mechanism supports thylakoid formation in the absence of Thf1. Genetic complementation of a Thf1 knockout line confirms the role of this gene in chloroplast and leaf development. Transgenic plants expressing the Thf1 gene in antisense orientation are stunted with altered thylakoid organization, especially in young seedlings. The data indicate that the Thf1 gene product plays a crucial role in a dynamic process of vesicle-mediated thylakoid membrane biogenesis.
The development of normal chloroplasts is a crucial step in the survival of photosynthetic eukaryotes. Maturation and activity of the chloroplast is highly dependent on its relationship with the nucleus, as the bulk of proteins that function in the chloroplast are encoded in the nucleus and are posttranslationally imported from the cytoplasm (Keegstra and Cline, 1999). Lesions in genes encoding plastid-localized proteins can have severe effects on the many metabolic processes that occur there and ultimately can prevent normal plant growth and development. Demonstrating the overriding importance of chloroplast proteins, the majority of mutations in a screen of mutagenized Arabidopsis (Arabidopsis thaliana), designed to identify genes important for seedling viability, appeared to affect chloroplast formation or function (Budziszewski et al., 2001). Environmental conditions, especially light, also play important roles in triggering chloroplast development and orchestrating gene expression required for plastid function (Anderson, 1986). In spite of the dependence of plants on the photosynthesis occurring on chloroplast thylakoid membranes, there is much to be learned regarding the mechanisms by which these internal membranes originate and how the photosynthetic machinery is assembled.
When green tissues develop in the presence of light, chloroplasts are normally formed from undifferentiated proplastids lacking internal membranes. As cells and organelles divide, new thylakoid membranes must be synthesized and organized along with the light-gathering and photosynthetic protein components (for review, see Vothknecht and Westhoff, 2001). The membrane portion of thylakoids originates in the inner envelope membrane of the chloroplast (Carde et al., 1982). Accumulation of small vesicles can readily be observed between the chloroplast envelope and thylakoid stacks in cold-treated leaves (Morré et al., 1991). It has been suggested that, in flowering land plants, both membrane and nonlipid components might be transferred to thylakoids by these vesicles originating at the inner envelope membrane (Westphal et al., 2003). In cyanobacteria, the core reaction centers for PSI and PSII are assembled at the plasma membrane and are perhaps transported to thylakoids via a membrane vesicle transit system (Zak et al., 2001). Such vesicle-transfer mechanisms are utilized in many aspects of membrane and protein transfer in biological systems, enabling cargo to move both within and to the outside of the cell (Bock et al., 2001).
Several nuclear- and plastid-encoded Arabidopsis genes have been identified that can ultimately affect thylakoid formation or chloroplast development. However, it remains to be determined whether the actions of these genes lead directly or indirectly through alteration of important metabolic processes, to plastid malformation. The apparent function of the individual proteins encoded by these genes can vary widely. For example, impairment of plastid RNA polymerases can lead to altered chloroplasts and plants with pale or variegated leaves (De Santis-Maciossek et al., 1999). On the contrary, variegated plants carrying the nuclear immutans mutation are altered in a gene encoding a plastid-localized homolog of an alternative oxidase (Aluru et al., 2001). The white or yellow sections of these variegated leaves generally contain abnormal chloroplasts.
Deletion of the Arabidopsis vesicle-inducing protein (VIPP1) shows that it is involved in the formation of vesicles leading to thylakoid biogenesis. Mutation of VIPP1 leads to pale-green plants that cannot grow as photoautotrophs and contain plastids that do not form the vesicles that are thought to be necessary for thylakoid formation (Kroll et al., 2001).
Through characterization of a T-DNA-tagged mutant line of Arabidopsis and production of transgenic lines, we have identified a nuclear-encoded gene important for the normal development of thylakoid membrane stacks and ultimately for leaf growth and development.
RESULTS
Thf1 Is Light Regulated and Encodes a Chloroplast-Localized Protein
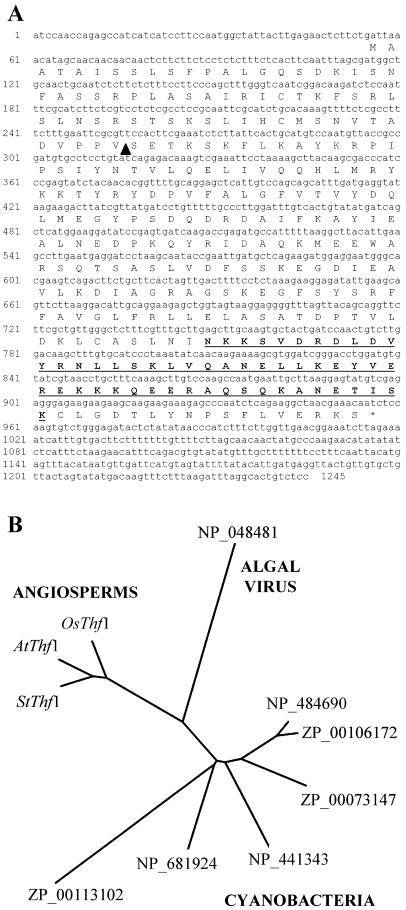
A cDNA clone for an apparent ortholog of the Arabidopsis Thylakoid formation1 (Thf1) gene was originally isolated from potato (Solanum tuberosum; accession no. AY342161) and identified as a light-regulated gene (R.W. Sullivan and K.L. Korth, unpublished data). The apparent ubiquitous nature of this gene in plants, along with the presence of a conserved strong coiled-coil motif, made it an interesting candidate for further study. A cDNA encoding Arabidopsis Thf1 was amplified using reverse-transcription (RT)-PCR based on the sequence of a full-length cDNA listed in the GenBank database (accession no. AY087394). Analysis of the Arabidopsis genome indicates that this is a single-copy gene without a closely related sequence in the remainder of the genome. The predicted amino acid sequence encoded by Thf1 suggests the presence of a chloroplast transit signal at the amino terminus and a coiled-coil domain near the carboxyl terminus in a hydrophilic protein of 33.8 kD (Fig. 1A). The predicted size of a fully processed Thf1 protein in chloroplasts is 26.8 kD. A search of the GenBank database with the predicted amino acid sequence yielded several full-length entries with a high degree of similarity to Thf1. With one exception, all of the sequences similar to Thf1 were found in photosynthetic organisms, ranging from cyanobacteria to land plants. A predicted protein with 48.9% similarity to the Thf1 sequence in the overlapping region (excluding the chloroplast transit signal) has also been identified in a large, double-stranded DNA virus (Van Etten, 2003) that infects freshwater green algae (accession no. NP_048481). Not surprisingly, measurement of similarities among the available full-length sequences shows that amino acid sequences encoded in Arabidopsis, rice, and potato are more similar to each other than to sequences of the corresponding clones from cyanobacteria or the virus (Fig. 1B). Identification of highly similar expressed sequence tags from tomato, soybean, sunflower, barley, maize, and Medicago truncatula indicates the presence of Thf1-like genes in numerous other plant species (data not shown).
Figure 1.
Sequence of the Arabidopsis Thf1 protein. A, The predicted amino acid sequence is shown along with the Thf1 cDNA as determined by DNA sequencing. The black triangle marks the putative cleavage point of the plastid-import sequence, as predicted by a ChloroP (Emanuelsson et al., 1999) score of 0.579. The underlined sequence is predicted to form a strong coiled-coil domain, as calculated by a COILSCAN (Lupas et al., 1991) probability of 0.97. B, Sequence similarity of Arabidopsis Thf1 (AtThf1) to putative full-length orthologous proteins found in other organisms. The Arabidopsis sequence (NP_565491) was compared to the predicted amino acid sequences indicated (as labeled by GenBank protein database accession numbers) from potato, AAQ19850 (StThf1); rice, AA072565 (OsThf1); the algal virus PBCV-1, NP_048481; from two different Nostoc sp., NP_484690 and ZP_00106172; Trichodesmium, ZP_00073147; Synechocystis, NP_441343; Thermosynechococcus, NP_681924; and Prochlorococcus, ZP_00113102. The tree was generated using programs in PHYLIP version 3.5c as described in “Materials and Methods.”
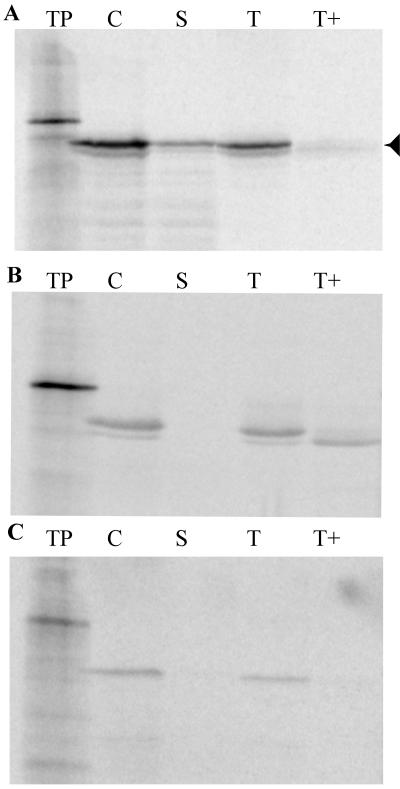
The presence of a putative transit signal on Thf1 led us to examine intact chloroplasts for the ability to import the protein. Radiolabeled Thf1 was efficiently imported into chloroplasts as judged by protease protection of the chloroplast-localized protein. Furthermore, the labeled Thf1 was processed to the predicted size (26.8 kD) after cleavage of the transit peptide (Fig. 2A). Fractionation of protease-treated chloroplasts containing imported Thf1 showed that the protein is localized to both soluble stroma and membranes. Addition of the protease thermolysin to membrane fractions results in the degradation of the bulk of Thf1, indicating that it is most likely bound at the periphery and not integrated into the lipid bilayer. This result is consistent with the absence of a predicted transmembrane region in the Thf1 sequence. However, a weak signal for Thf1 remains in the thermolysin-treated thylakoid fraction (Fig. 2A), suggesting that Thf1 is partially protected from protease treatment. Localization of chloroplast proteins cab80 (light-harvesting complex protein [LHCP]; Cashmore, 1984) and p36 (Schnell et al., 1990) was used as a measure of the effectiveness of fractionation. The LHCP protein is known to integrate into thylakoids and is only partially accessible to added protease (Fig. 2B). The p36 phosphate translocator protein is localized to the chloroplast inner envelope membrane, and the localization patterns for both imported LHCP (Fig. 2B) and p36 (Fig. 2C) confirm that there was no contamination of soluble fractions with either thylakoid or envelope membrane fragments. Both of the control proteins migrated on the gel at the predicted positions; the size of the processed LHCP protein is 27 kD and of the processed p36 protein is 36 kD.
Figure 2.
Thf1 encodes a light-regulated plastid-localized protein. The figure demonstrates import of 35S-radiolabeled protein into isolated chloroplasts and association of Thf1 with both stromal and thylakoid fractions. Labeled proteins were produced via in vitro transcription/translation and products were separated on SDS-polyacrylamide gels. The largest proteins represent the precursor form, whereas the next smaller band represents the mature, processed form of each protein (Thf1, 26.8 kD; LHCP, 27 kD; p36, 36 kD). In addition to labeling Thf1 (A), 35S-labeled LHCP, a protease-accessible thylakoid protein where a degraded form of the protein is found in the thermolysin-treated thylakoid fraction (B), and p36, a protein found in the inner envelope membrane (C), were prepared. For each labeled protein, further fractionation and/or treatment were performed and products were separated; they represent translation product (TP), chloroplasts after import (C), stroma fraction (S), thylakoids (T), and thylakoids treated with thermolysin protease (T+).
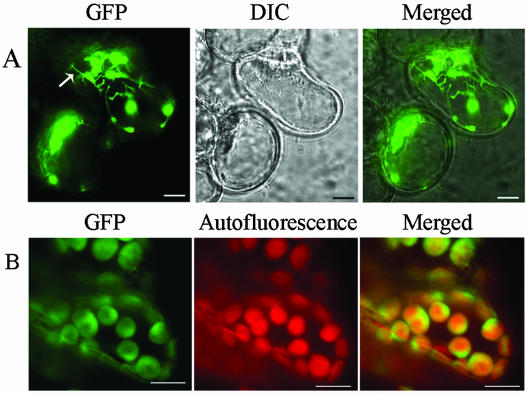
Fusion of the Thf1 protein at the carboxyl terminus with green fluorescent protein (GFP) verified chloroplast localization of the protein in planta (Fig. 3). To confirm the subcellular localization of Thf1 in plastids or chloroplasts, we made the construct 35S:Thf1-GFP and transformed it into suspension cells and plants. Thf1-GFP was localized in plastids and stromules, which seem to connect plastids together (Fig. 3A). Some stromules extended to the plasma membrane, indicating that communication might take place between the plasma membrane and plastids. In mesophyll cells, Thf1-GFP was distributed throughout the chloroplasts but predominantly in uneven patterns around the periphery, perhaps associated with the chloroplast envelope (Fig. 3B). This result is consistent with the above in vitro assay. These data showed that Thf1 was localized not only in plastids and chloroplasts but also in stromules.
Figure 3.
Thf1-GFP fusion demonstrates chloroplast localization in planta. Thf1-GFP is localized in plastids/chloroplasts and stromules in plant cells. A, Thf1-GFP localized in plastids and stromules in suspension cells. Arrow indicates stromules. B, Thf1-GFP localized in chloroplasts of mesophyll cells. Ten-day-old seedlings grown in constitutive light were used. Scale bar = 20 μm. DIC, Differential-interference-contrast.
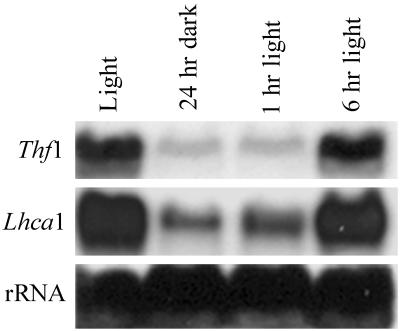
Transcripts for Thf1 accumulate in leaves of plants grown under normal long daylight (16 h)/dark (8 h) conditions. Leaves collected 4 h into the light cycle contain abundant transcripts, whereas transcript levels are greatly reduced in leaves from plants placed in complete darkness for 24 h (Fig. 4). Transcript levels return to normal within 6 h after plants are returned to normal light levels. Alterations in Thf1 transcript levels closely parallel those of a characterized light-regulated nuclear gene encoding a chloroplast protein, Lhca1 (Kim et al., 1999), in response to light conditions (Fig. 4).
Figure 4.
RNA blot demonstrating the effects of light on Thf1 transcript accumulation. Identical RNA blots were hybridized with labeled probes for Thf1 or the LhcA1 transcript. Total RNA was collected from lane 1, normal light/dark-grown plants 4 h into the light cycle; lane 2, plants in darkness for 24 h; lane 3, dark-treated plants returned to the light for 1 h; and lane 4, dark-treated plants returned to the light for 6 h. The 18S rRNA probe serves as a loading control.
A T-DNA-Tagged Thf1 Mutant Is Stunted with Variegated Leaves
An Arabidopsis line with a potential T-DNA insertion in the Thf1 gene (SALK_094925) was acquired after a search of the Salk Institute Genomic Analysis Laboratory Arabidopsis gene-mapping database (http://signal.salk.edu/cgi-bin/tdnaexpress). Seedlings were screened via PCR for the presence of a T-DNA insertion in chromosome 2 within the gene designated At2g20890. Amplification of a specific band in several individual plants confirmed incorporation of the T-DNA in the Thf1 gene (data not shown). We found no insertions in another plant line (SALK_094926) listed as having a T-DNA insertion at the same location. Sequencing of the PCR products showed that the insertion was at nucleotide position 299 in the Thf1 cDNA, near the 3′ end of exon 1 (Fig. 5A). With the exception of one plant, individuals that grew to maturity from the SALK_094925 seed stock were indistinguishable from wild-type Arabidopsis ecotype Columbia plants grown alongside. Among the plants of normal appearance, there was a mixture of PCR-positive and -negative individuals, indicative of segregation of the T-DNA tag within the population. One plant, designated 25-7, was severely stunted with leaves containing green and yellow/white sectors (Fig. 5 B and C). Young seedlings of this line, less than 10 d old, were yellow, but, as the plants aged, they eventually began to develop interspersed green patches. Although the timing was delayed, line 25-7 flowered and formed viable seed. When reciprocal crosses were made between line 25-7 and wild-type Columbia plants, the resulting F1 progeny all grew normally and were indistinguishable from the wild-type parent (Fig. 5D). Segregation patterns in subsequent generations showed that the mutation leading to the abnormal leaf phenotype behaved as a single, recessive gene.
Figure 5.
A T-DNA insertion in the Thf1 gene leads to altered plant morphology. A, Genomic organization of the Arabidopsis Thf1 gene, At2g20890, with the position of the T-DNA insertion in line 25-7 indicated with a black triangle. Boxes indicate exons confirmed by comparison of cDNA and genomic sequences and the scale indicates nucleotide distances. B, The stunted phenotype of T-DNA-tagged line 25-7 (left) as shown in comparison to a wild-type Columbia plant (right) of the same age grown under the same conditions. C, The variegated coloration pattern in leaves of line 25-7 is readily observed. D, Plants in the F1 generation of a cross between the homozygous-tagged line 25-7 and wild-type Columbia plants all grow normally and are phenotypically indistinguishable from wild-type control plants grown alongside. E, Gel-blot analysis of total RNA, indicating transcript accumulation of Thf1 and another light-regulated gene, LhcA1, in Arabidopsis ecotype Columbia (wild type) and individual lines derived from SALK_094925 seed stock. The only homozygous T-DNA-tagged mutant line, 25-7, does not accumulate Thf1 transcripts. The 18S rRNA probe serves as a loading control.
Genomic DNA-blot analysis confirmed that line 25-7 contained a unique restriction enzyme pattern when hybridized with a radiolabeled Thf1 cDNA and compared to the other lines (data not shown). The DNA restriction profiles were consistent with line 25-7 being homozygous for the T-DNA insertion, whereas other PCR-positive lines appeared to be heterozygous and PCR-negative lines contained no T-DNA insertion. Segregation patterns of the yellowed/stunted phenotype in seedlings derived by selfing PCR-positive lines also indicate that 25-7 is homozygous for the T-DNA insertion and that the other original lines are heterozygous.
Even upon prolonged exposure of leaf RNA blots, transcripts hybridizing to Thf1 appear to be completely absent in line 25-7, whereas they are found at normal levels in the other Arabidopsis lines tested (Fig. 5E). The profiles of transcripts for other light-inducible nuclear-encoded genes encoding chloroplast proteins such as LhcA1 are not altered in 25-7, indicating that the deficiency of Thf1 mRNA is not due to a lesion affecting transcript accumulation for this class of genes. Leaves containing a mixture of green and yellow sectors were used for RNA extractions; the results therefore show that, even in normal-appearing green sectors of line 25-7, there is no Thf1 expression.
Thf1 Is Involved in Thylakoid Membrane Stacking
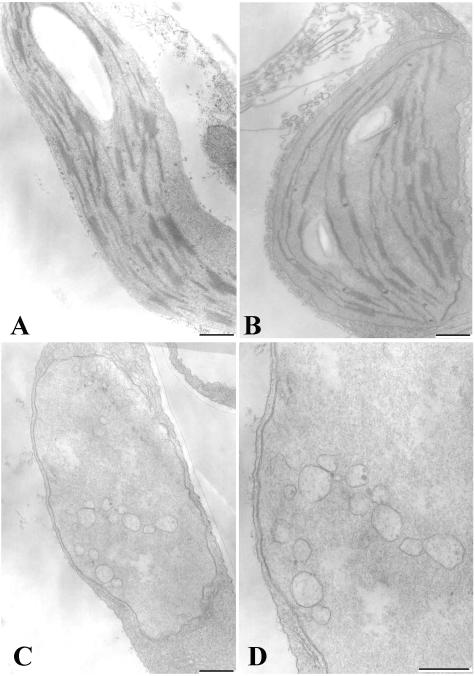
Ultrastructural examination via transmission electron microscopy (TEM) shows that some chloroplasts from green sectors of leaves in line 25-7 appear to have normal thylakoid development in terms of abundance and structure. However, many plastids in the same tissue had reduced amounts of thylakoid stacking, and distances between grana stacks were greater than in wild-type chloroplasts (Fig. 6 A and B, ). Although a few chloroplasts of relatively normal appearance were found, most plastids in yellow/white sectors of line 25-7 lacked thylakoid membranes, grana, or starch granules. However, the envelope membrane structure, size, and general shape of these plastids appear to be normal (Fig. 6C). These plastids accumulated numerous membrane vesicles of varying sizes in the interior soluble region (Fig. 6D), but the vesicles do not coalesce to form thylakoids.
Figure 6.
Chloroplasts in Thf1-tagged plants lack normal thylakoid formation. Chloroplast structure and thylakoid organization as observed via transmission electron microscopy. A, Normal chloroplasts seen in wild-type Columbia leaves. B, Chloroplasts from a green sector of the Thf1-tagged line 25-7 form thylakoid membranes, although the grana stacks are generally shorter. C and D, Plastids from a yellow sector of line 25-7 lack organized thylakoids and accumulate membrane vesicles of varying sizes, although the envelope membranes appear to maintain a normal structure. Bar = 500 nm.
Transgenic Lines Confirm the Role of Thf1 in Chloroplast and Leaf Development
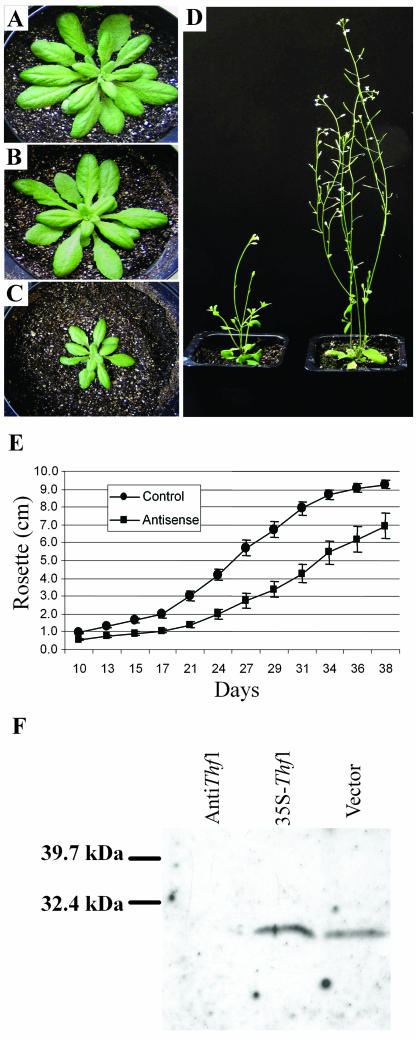
The Thf1 cDNA was integrated into the Arabidopsis genome in both sense and antisense orientations under the control of a cauliflower mosaic virus 35S promoter using Agrobacterium-mediated transformation. This allowed us to test whether changing levels of Thf1 expression leads to altered leaf and/or chloroplast development. Progeny of selfed, transformed lines expressing the antisense Thf1 grew more slowly than offspring of plants transformed with either the sense construct or the binary vector alone (Fig. 7, A–D). Although they were initially smaller, Thf1 antisense lines in short daylight (12 h) ultimately grew to nearly equal size of the sense lines or vector-transformed controls. We observed no differences in plant growth or development when comparing the progeny of independent selfed lines transformed with either the sense construct or the vector alone. When antisense plants were grown under long daylight conditions (16 h), their leaves had a more wrinkled appearance, and although they flowered, they were smaller than control lines and formed less siliques (Fig. 7D). The overall size of Thf1 antisense lines was reduced compared to vector-transformed control lines, as indicated by smaller rosettes over the life of the plant (Fig. 7E). Unlike the homozygous knockout mutant line 25-7, the antisense lines did not always display yellow-sectored leaves, and when variegation appeared, the phenotype was not as pronounced as in line 25-7. The level of Thf1 protein is severely reduced in antisense chloroplasts, as indicated by immunoblot analysis (Fig. 7F). The time to bolting was also delayed in the antisense lines. The vector-transformed control lines all bolted within 41 d after planting, whereas not all antisense lines bolted until 52 d. The plants containing the antisense construct eventually developed normal flowers and viable seeds.
Figure 7.
Normal leaf growth and development is dependent on Thf1 expression. Transgenic Arabidopsis transformed with the binary vector alone (A), an overexpression sense construct of Thf1 (B), and a Thf1 antisense construct (C). A to C, Plants grown under short daylight (12 h) conditions. D, Thf1 antisense plants (left) grown under longer daylight (16 h) conditions are smaller and form less siliques than vector-transformed control plants (right); this is also demonstrated by a smaller diameter of the rosette when measured over the life of the plant (E); error bars indicate sd, n = 10. Protein immunoblots probed with Thf1-antiserum (F) indicate that plants transformed with the antisense construct contain a reduced amount of Thf1 protein, as compared to those with the sense construct (35S-Thf1) or the binary vector.
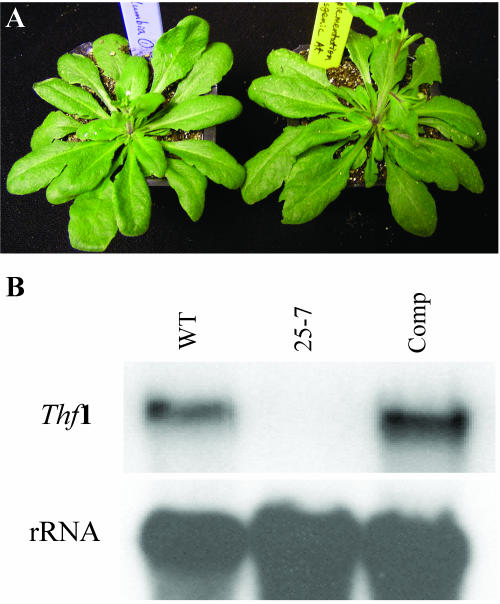
When the knockout line 25-7 was transformed with the Thf1 sense construct, the resulting lines grew normally (Fig. 8A) and transgenic lines accumulated high levels of Thf1 transcripts (Fig. 8B). This complementation confirms that disruption of the Thf1 gene leads to the severe phenotype observed in line 25-7.
Figure 8.
Expression of a Thf1 cDNA can complement the mutation in line 25-7. A, Progeny of the Thf1-tagged line 25-7 transformed with the sense construct (right) show complementation of the mutant phenotype and are indistinguishable from wild-type Columbia ecotype plants (left). B, RNA-blot analysis confirms the accumulation of transcripts for the Thf1 gene expressed in the transgenic, complemented line (Comp) at levels at least as great as in the wild-type control (WT). No transcripts accumulate in line 25-7.
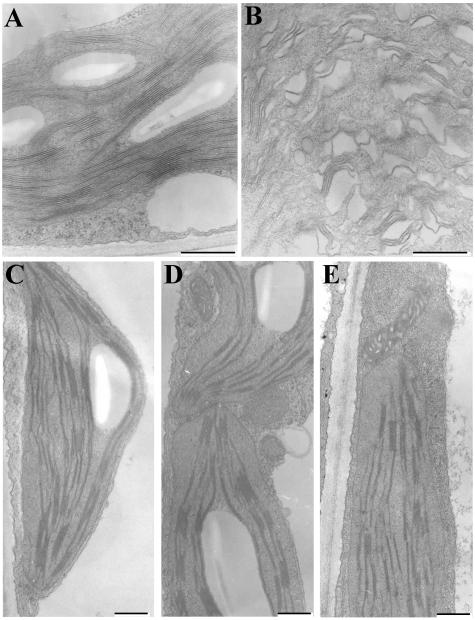
Analysis of chloroplasts via TEM of transgenic lines suggests that thylakoid organization is severely affected in young plants transformed with the Thf1 antisense construct. Although loosely organized thylakoid membranes seem to form in 10-d-old antisense-construct seedlings, the chloroplasts lack normal grana stacks when compared to chloroplasts from control lines of the same age (Fig. 9 A and B). Abundant membranes and plastoglobules accumulate in these chloroplasts, but the membrane matrix is severely disrupted, giving the interior a sponge-like appearance. As the plants age, chloroplasts in antisense lines take on a more normal appearance (Fig. 9, C–E). However, as in chloroplasts from the green sectors of the 25-7 knockout line, in antisense lines the grana stacks appear shorter and there is less starch granule accumulation. As in line 25-7 tissues, the structure and size of other cell components and organelles such as mitochondria appear normal in the sense and antisense lines.
Figure 9.
Thf1 antisense lines contain abnormal chloroplasts early in leaf development. In 10-d-old seedlings, vector-transformed plants (A) have normal thylakoid and grana organization, whereas Thf1 antisense chloroplasts (B) contain loosely stacked thylakoid membranes. When compared to chloroplasts from vector-transformed plants (C) and Thf1 sense transformed plants (D), chloroplasts from Thf1 antisense plants (E) take on a more normal appearance after 25 d in short daylight (12 h) conditions. Bar = 500 nm.
DISCUSSION
Through analysis of T-DNA-tagged Arabidopsis mutants and transgenic plants, we have determined a role in chloroplast and leaf development for the previously undefined Arabidopsis Thf1 gene. A conserved form of Thf1 appears to be present in all plants and cyanobacteria species that were examined via gene database searches, suggesting that it has a vital function in photoautotrophs. A highly similar form of the sequence is also present in a large double-stranded DNA virus that infects freshwater green algae. It is possible that this viral form of the gene was transferred from a photosynthetic organism to the viral genome. Although the gene is well conserved among cyanobacteria, examination of cyanobacterial genomes (http://www.kazusa.or.jp/cyano/cyano.html) does not indicate that it is part of an operon of known function. There are abundant examples of genes of cyanobacterial origin being ultimately encoded in the nuclear genome of plants (Martin et al., 1998), and it is likely that transfer of the Thf1 gene occurred from the chloroplast genome to the nuclear genome.
There is mounting evidence that a dynamic system of vesicle transport is involved in the biogenesis of thylakoids in mature chloroplasts (Westphal et al., 2001; Andersson and Sandelius, 2004), but the protein components acting in this system remain to be identified. A protein involved in the organization of plastid membranes in pepper (Capsicum annuum) fruit plastids has been identified (Hugueney et al., 1995), and it is similar to the VAR2-encoded protein (Chen et al., 2000).
The characteristics of phenotype in Thf1 knockout lines and Thf1 antisense lines in several ways parallel those of other known mutants of chloroplast development. The variegated leaf phenotype of Thf1 knockout lines can be observed in several characterized Arabidopsis mutants. For example, lesions in several chloroplast RNA polymerase genes lead to variegated leaves and the accumulation of vesicles in plastids of young leaf tissue (De Santis-Maciossek et al., 1999). Within a variegated leaf, there must be a mechanism for triggering the development of normal chloroplasts, and this has to occur even in the absence of a seemingly critical gene like Thf1. The aforementioned immutans mutation in Arabidopsis is caused by a defect in a chloroplastic homolog of an alternative oxidase enzyme and leads to plants with variegated leaves. It is proposed that green sectors arise in immutans leaves because of the action of a redundant electron transfer system (Wu et al., 1999). Mutations of the VAR2 gene of Arabidopsis also lead to variegation; in this case, the lesion lies in a gene encoding a proposed metalloprotease associated with thylakoids, and again, the ability to form green sectors is proposed to be derived from a redundant activity that can compensate for the loss of the specific VAR2 allele (Chen et al., 2000). In the case of immutans and VAR2 mutants, potentially redundant activities/enzymes within the cell can be identified. In the case of Thf1, however, sequence comparisons with the remainder of the Arabidopsis genome do not readily identify any genes that are obviously similar. Still, the most likely explanation for green sectors forming in Thf1 knockout lines is the action of a compensating mechanism for thylakoid formation, but it is not clear where this activity originates. Both RNA blots and protein immunoblots indicate that no Thf1 is expressed in any tissue of the knockout line 25-7 and yet green sectors form on leaves.
There are examples to suggest that variegation and vesicle formation characteristics are not necessarily a general response to interference with chloroplast function. A severe decrease of the light-harvesting complex of PSII, brought about via antisense-mediated decrease in the light-harvesting complex of PSII subunits, did not result in altered chloroplast formation, and thylakoid stacks readily formed in such plants (Andersson et al., 2003). However, because mutations in several genes can lead independently to the accumulation of vesicles in plastids, it leaves open the possibility that Thf1 does not directly participate in vesicle fusion to form mature thylakoids, but rather that this is an indirect effect stemming from interference with some step of chloroplast biogenesis. It is interesting to note that, in a survey of vesicle-transport systems, vesicle-mediated formation of thylakoids was observed in embryophytes but not among other photosynthetic eukaryotes or the cyanaobacteria that were tested (Westphal et al., 2003). Also noteworthy is that a protein with high sequence similarity to Thf1 is encoded in the species Gloeobacter violaceous PCC 7421, a cyanobacterium that lacks distinct thylakoid membranes. It could be that the ancestral form of Thf1 is not necessary for thylakoid formation per se, but that it is important for assembly of membrane substrate for photosystems. The observation might also be explained by a modification of function for Thf1 since its transfer to flowering plants.
In the case of Thf1 antisense lines, it is likely that low amounts of Thf1 protein are produced. This explains the observed differences in phenotype between the severely stunted, variegated knockout line 25-7 (Fig. 5) and the antisense lines (Fig. 7). As we have also observed of the Thf1 antisense lines, there are several reports of chloroplast-linked mutations that are most pronounced in the early stages of plant development but that are eventually overcome as plants mature. In low-light conditions, immutans leaves are not variegated, but the slow-growing mutants do eventually reach the same size as wild-type plants (Aluru et al., 2001). Mutation of the Toc33 gene involved in plastid protein import results in plants that are pale in the early stages of growth but appear more normal as they age (Jarvis et al., 1998). Likewise, as in Thf1 antisense lines, chloroplasts in Toc33-deficient plants had smaller and fewer grana stacks than wild-type plants, and the chloroplast structural effects of the mutation were more evident in 5-d-old plants than in older plants (Jarvis et al., 1998). Similar observations were made in a study in which a 54-kD subunit of a chloroplast signal recognition particle (cpSRP54) was inactivated, resulting in plants that had pale cotyledons and yellowed first true leaves but took on a normal appearance as they aged (Pilgrim et al., 1998). In addition, cpSRP54 mutants had chloroplasts with fewer and irregular thylakoid membranes as compared to wild type at early stages of plant growth, but chloroplasts took on a normal appearance as plants aged (Pilgrim et al., 1998). These data again suggest that plants are able to compensate for the reduction of gene products that are otherwise important for normal chloroplast development.
It is difficult to predict the function of Thf1 based solely on the derived sequence, although the prediction of a strong coiled-coil motif might provide some clues as to a role for the protein. Coiled coils are most often associated with peptide oligomerization and can be found in proteins with a range of functions (Burkhard et al., 2001). Interestingly, a number of important steps in intercellular vesicle fusion are dependent on proteins with coiled-coil domains (Skehel and Wiley, 1998). Included in this class are so-called SNAP receptors, or SNARE proteins, that form a complex between vesicles and target membranes through interactions of coiled-coil structures on several proteins (Rizo and Südhof, 2002). It is noteworthy that the yeast (Saccharomyces cerevisiae)-encoded protein with greatest similarity (22%) to Arabidopsis Thf1 is Sec17, or α-SNAP (accession no. NP_594169). This protein interacts with SNARE proteins in a dynamic complex known to control membrane vesicle fusion in yeast vacuoles (Ungermann et al., 1998). Deletion of a protein with similar functions in the chloroplast might lead to the configuration observed in Thf1 knockout plastids.
It has been suggested that SNARE proteins play a critical role in virtually all examples of eukaryotic vesicle fusion that are known (Rizo and Südhof, 2002). Likewise, coiled-coil-containing dynamin proteins have been proposed to be important for vesicle scission and endocytosis mechanisms (Praefcke and McMahon, 2004). There is precedence for the role of a protein with coiled-coil motifs in thylakoid formation in plants. A dynamin-like protein in Arabidopsis (ADL1) has been identified as critical in thylakoid biogenesis, and ADL1 mutants are stunted, yellowed, and contain chloroplasts with reduced levels of thylakoid membranes and no grana stacks (Park et al., 1998). It was suggested that vesicle formation in the ADL1 mutants is blocked due to the reduction in dynamin-like activity (Park et al., 1998). This ADL1 mutant shares many characteristics with the Thf1-altered plants, and thus it is tempting to speculate that Thf1 might function in the same or a related pathway as ADL1. Plastid development and thylakoid biogenesis are highly integrated processes. Therefore, it is difficult at this juncture to pinpoint the precise step at which the Thf1 gene product acts. However, the seemingly normal size, shape, and integrity of envelope membranes in plastids that lack thylakoids in line 25-7 indicate that the gene product possibly functions in a step late in thylakoid formation and is not required for all the steps of plastid formation. Alteration of several genes with seemingly varied function can lead to variegated leaves and sometimes to plastids where membrane vesicles accumulate. Deletion of the VIPP1 gene in Arabidopsis results in plastids that do not form interior vesicles (Kroll et al., 2001), and so if they are acting within the same pathway, Thf1 functions downstream from the action of VIPP1. Like the VIPP1 protein, Thf1 is found at more than one location in fractionated chloroplasts. Even though both proteins are predicted to be hydrophilic in nature, VIPP1 is found to be associated with both the inner envelope and thylakoid membranes (Kroll et al., 2001), whereas Thf1 is found in stromal and thylakoid fractions of isolated chloroplasts (Fig. 2A) and in interconnecting stromules (Fig. 3).These observations could be explained if Thf1 is a component of a mobile complex, associated with membranes at the point of vesicle origin in the envelope and at the point of formation of thylakoids.
Comparisons of the Arabidopsis Thf1 sequence with genomes of other organisms offer us a seeming paradox. Cyanobacteria clearly contain sequences that are similar to Arabidopsis Thf1, yet these species might lack a vesicle transport system (Westphal et al., 2003). On the other hand, the structural motifs and sequence similarity with animal and yeast proteins suggest a possible role for the protein in a vesicle transport system, a hypothesis that is supported by the accumulation of membrane vesicles in the Thf1 knockout plants. The vesicles thought to give rise to thylakoid membranes are seldom observed in mature chloroplasts of plants growing under standard conditions, and one generally needs to expose plants to cold temperatures to capture images of such structures. The seemingly transient and dynamic nature of vesicle formation and fusion in this system, both in terms of chloroplast biogenesis and leaf development, make it inherently difficult to follow. Mutations that block the organization of thylakoids and lead to accumulation of such vesicles might be particularly useful in learning more about how these complex structures are formed. The similarities of Arabidopsis Thf1 with proteins involved in vesicle transport in other systems, along with the dramatic alteration of plastid appearance in Thf1 mutants, make this a potentially valuable tool in the elucidation of mechanisms of thylakoid biogenesis.
MATERIALS AND METHODS
Plant Growth and Maintenance
Arabidopsis (Arabidopsis thaliana) seeds were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH). Plants were maintained and seeds were collected and treated, essentially as described (Weigel and Glazebrook, 2002). Seeds were sown in soil and plants were maintained in a growth chamber at 23°C. Lighting conditions were kept at 16-h-light/8-h-dark cycles unless indicated otherwise.
Rosette growth was determined by measuring at the maximum diameter. Ten plants of each genotype were measured daily and were no longer measured after they started to bolt.
Thf1 Sequence Analysis
The Arabidopsis Thf1 cDNA was amplified via RT-PCR from total RNA of Arabidopsis (ecotype Columbia) using an oligo(dT) primer and M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA) for first-strand synthesis. Amplification was carried out with specific primers (forward primer, 5′-ctcacttcaaggtaccgatggctgc-3′; reverse primer, 5′-catatacacgtctagaatgttcttag-3′) based on the GenBank accession AY062799. Products of the RT-PCR were cloned into a plasmid vector, and identification of the cloned cDNA was confirmed via DNA sequencing.
The predicted Thf1 protein was analyzed for the presence of a putative chloroplast transit signal using the ChloroP program (Emanuelsson et al., 1999) and for coiled-coil regions with the COILSCAN program (Lupas et al., 1991) contained within the GCG sequence analysis package.
The predicted amino acid sequence encoded by Arabidopsis Thf1 was aligned with similar full-length sequences, identified in a BLAST search, using the Clustal program within the GCG sequence analysis package. Amino acid sequence distances were calculated using Protdist and an unrooted tree was constructed via the unweighted pair group method with arithmetic mean (UPGMA) method in the NEIGHBOR program; both programs are contained in PHYLIP version 3.5c (http://evolution.genetics.washington.edu/phylip/html).
Thf1 RNA and Protein Analyses
Total RNA was isolated from leaves frozen in liquid nitrogen and extracted with TriReagent (Molecular Research Center, Cincinnati) following the manufacturer's directions. RNA was separated on 1% agarose formaldehyde gels, transferred to nylon membranes, and hybridized with 32P-labeled Thf1 cDNA as a probe using established methods (Sambrook et al., 1989).
For in vitro chloroplast import assays, the Thf1 cDNA was first cloned into the pGEM4z plasmid vector for protein expression. Linearized DNA was used in an in vitro transcription/wheat germ translation system in the presence of 35S-labeled Met to produce radiolabeled protein. Import of radiolabeled proteins into isolated pea (Pisum sativum) chloroplasts, subfractionation of chloroplasts, gel electrophoresis, and radiography were carried out as described (Henry et al., 1994). The labeling and import of cab80 (LHCP; Cashmore, 1984) and p36 (Schnell et al., 1990) were performed alongside the Thf1 studies.
The open reading frame of AtThf1 (At2g20890) was first cloned into the entry vector pENTR/SD/-TOPO (Invitrogen). For expression of the GFP-tagged Thf1 in plant cells, the open reading frame of AtThf1 was recombined into Gateway destination vectors pGWB5 (Research Institute of Molecular Genetics, Shimane, Japan). Stable transformed suspension cells and transgenic plants (Ferrando et al., 2000; Bechtold et al., 1993) were generated for Thf1-GFP subcellular localization. Protoplasts were isolated according to the protocol described by Sheen (2001). Fluorescence images were captured using an Olympus IX81 platform (Tokyo) equipped with a Roper cascade II camera.
T-DNA-Tagged Line
The T-DNA-tagged mutant of Arabidopsis (Alonso et al., 2003) was selected via a sequence-based search of the Salk Institute Genomic Analysis Laboratory database. Seed from lines SALK_094925 and SALK_094926 were obtained and planted. Individual plants were screened via a PCR method (Siebert et al., 1995) to test for T-DNA insertion in the Thf1 gene, using the LBb1 primer (5′-gcgtggaccgcttgctgcaact-3′) and a Thf1-specific primer (5′-ctcacttcaaggtaccgatggctgc-3′). Presence of a PCR product of the expected size was used as preliminary evidence of T-DNA insertion at the desired site. Confirmation of T-DNA insertion was obtained via DNA sequencing of PCR products and through DNA hybridization. DNA electrophoresis, blotting, hybridization, and random-primer labeling of DNA were all carried out as described (Sambrook et al., 1989).
Plant Transformation
For plant transformation, the Arabidopsis Thf1 cDNA was synthesized via RT-PCR from total RNA as described above. The sense fragment used for overexpression was amplified with the forward and reverse primers described above for cDNA amplification. The inverted Thf1 fragment used in an antisense construct was amplified with a forward primer (5′-ctcacttctagatagcgatggctgc-3′; introduced XbaI site in bold) and a reverse primer (5′-ggttatatagggtacctcccag-3′; introduced KpnI site in bold). The sense and antisense constructs in a binary vector were generated by directional ligation of the amplified inserts into a modified pCAMBIA2300 vector (GenBank accession no. AF234315) containing a 35S promoter of cauliflower mosaic virus (provided by Y. Yang, University of Arkansas, Fayetteville, AR). The resulting constructs were introduced into Agrobacterium tumefaciens strain GV3010 via electroporation. Plants were transformed using a floral dip method (Clough and Bent, 1998).
Electron Microscopy
Fresh material from rosette leaves was prepared and viewed via electron microscopy as described by Kim and Fulton (1984). Briefly, samples were fixed in a solution of 2% glutaraldehyde/2% paraformaldehyde and further fixed in 1% OsO4. Tissues were stained with uranyl acetate, dehydrated in ethanol, and embedded in Spurr's medium prior to thin sectioning. Samples were stained again and examined with a JEOL (Tokyo) 100 CX electron microscope.
Chloroplast Isolation and Immunoblots
Fully expanded rosette leaf tissue (5g) was homogenized using polytron (Brinkman model PT 10-35) in ice-cold extraction buffer (20 mL) containing 0.35 m sorbitol, 50 mm Tris-HCl, pH 8.0, 5 mm EDTA, 0.1% BSA (w/v), and 0.1% β-mercaptoethanol (v/v). All steps were carried out at 4°C. Cheesecloth-filtered homogenate was centrifuged at 2,000g for 20 min. The pellet was gently resuspended in 2 mL of extraction buffer and centrifuged again at 2,000g for 15 min. The final pellet was mixed with 2 mL of lysis buffer (50 mm Tris-HCl, pH 8.0, 5 mm EDTA, 20 μL of a 100× protease inhibitor cocktail). The chloroplast extract was treated via sonication and insoluble debris was removed by centrifugation at 10,000g for 15 min. Protein concentration was determined using the Bio-Rad (Hercules, CA) protein assay. All SDS-PAGE and immunoblots were carried out as described (Harlow and Lane, 1988) and membranes were probed with rabbit antiserum raised against a conserved region of the potato form of Thf1 (R. Sullivan and K. Korth, unpublished data).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY087394.
Acknowledgments
We thank Sandy Goeke for acquiring the TEM images. We thank Yinong Yang for providing the binary vector, K.S. Kim for help with interpreting electron microscopy images, and the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants. The LhcA1 clone was kindly provided by Dr. N.E. Hoffman.
This work was supported in part by the Arkansas Rice Research and Promotion Board (grants to K.L.K.), by the National Institutes of Health COBRE Program of the National Center for Research Resources (grant no. P20–RR15569 to R.L.H.), by the Department of Energy (grant no. DE–FG02–01ER15161 to R.L.H.), by the National Institute of General Medical Sciences (grant no. GM65989–01 to A.M.J.), and by the National Science Foundation (grant no. MCB–0209711 to A.M.J.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.049841.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aluru MR, Bae H, Wu D, Rodermel SR (2001) The Arabidopsis immutans mutation affects plastid differentiation and the morphogenesis of white and green sectors in variegated plants. Plant Physiol 127: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JM (1986) Photoregulation of the composition, function, and structure of thylakoid membranes. Annu Rev Plant Physiol 37: 93–136 [Google Scholar]
- Andersson J, Wentworth M, Walters RG, Howard CA, Ruban AV, Horton P, Jansson S (2003) Absence of the Lhcb1 and Lhcb2 proteins of the light-harvesting complex of photosystem II—effects on photosynthesis, grana stacking and fitness. Plant J 35: 350–361 [DOI] [PubMed] [Google Scholar]
- Andersson MX, Sandelius AS (2004) A chloroplast-localized vesicular transport system: a bio-informatics approach. BMC Genomics 5: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Ser III Life Sci 316: 1194–1199 [Google Scholar]
- Bock JB, Matern HT, Peden AA, Scheller RH (2001) A genomic perspective on membrane compartment organization. Nature 409: 839–841 [DOI] [PubMed] [Google Scholar]
- Budziszewski GJ, Lewis SP, Glover LW, Reineke J, Jones G, Ziemnik LS, Lonowski J, Nyfeler B, Aux G, Zhou Q, et al (2001) Arabidopsis genes essential for seedling viability: isolation of insertional mutants and molecular cloning. Genetics 159: 1765–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard P, Stetefeld J, Strelkov S (2001) Coiled coils: a highly versatile protein folding motif. Trends Cell Biol 11: 82–88 [DOI] [PubMed] [Google Scholar]
- Carde JP, Joyard J, Douce R (1982) Electron microscopic studies of envelope membranes from spinach plastids. Biol Cell 44: 315–324 [Google Scholar]
- Cashmore AR (1984) Structure and expression of a pea nuclear gene encoding a chlorophyll a/b binding polypeptide (Pisum sativum var. Progress 9). Proc Natl Acad Sci USA 81: 2960–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Choi Y, Voytas DF, Rodermel S (2000) Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. Plant J 22: 303–313 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- De Santis-Maciossek G, Kofer W, Bock A, Schoch S, Maier RM, Wanner G, Rüdiger W, Koop H-U, Herrmann RG (1999) Targeted disruption of the plastid RNA polymerase genes rpoA, B and C1: molecular biology, biochemistry and ultrastructure. Plant J 18: 477–489 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando A, Farras R, Jasik J, Schell J, Koncz C (2000) Intron-tagged epitope: a tool for facile detection and purification of proteins expressed in Agrobacterium-transformed plant cells. Plant J 22: 553–560 [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Henry RL, Kapazoglou A, McCaffery M, Cline K (1994) Differences between lumen targeting domains of chloroplast transit peptides determine pathway specificity for thylakoid transport. J Biol Chem 269: 10189–10192 [PubMed] [Google Scholar]
- Hugueney P, Bouvier F, Badillo A, D'Harlingue A, Kuntz M, Camara B (1995) Identification of a plastid protein involved in vesicle fusion and/or membrane protein translocation. Proc Natl Acad Sci USA 92: 5630–5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, Chen L-J, Li H-M, Peto CA, Fankhauser C, Chory J (1998) An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 282: 100–103 [DOI] [PubMed] [Google Scholar]
- Keegstra K, Cline K (1999) Protein import and routing systems of chloroplasts. Plant Cell 11: 557–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS, Fulton RW (1984) Ultrastructure of Datura stramonium infected with an Euphorbia virus suggestive of a whitefly-transmitted Gemini virus. Phytopathology 74: 236–241 [Google Scholar]
- Kim SJ, Jansson S, Hoffman NE, Robinson C, Mant A (1999) Distinct “assisted” and “spontaneous” mechanisms for the insertion of polytopic chlorophyll-binding proteins into the thylakoid membrane. J Biol Chem 274: 4715–4721 [DOI] [PubMed] [Google Scholar]
- Kroll D, Meierhoff K, Bechtold N, Kinoshita M, Westphal S, Vothknecht UC, Soll J, Westhoff P (2001) VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc Natl Acad Sci USA 98: 4238–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J (1991) Predicting coiled-coils from protein sequences. Science 252: 1162–1164 [DOI] [PubMed] [Google Scholar]
- Martin W, Stoebe B, Goremykin V, Hansmann S, Hasegawa M, Kowallik KV (1998) Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393: 162–165 [DOI] [PubMed] [Google Scholar]
- Morré DJ, Selldén G, Sundqvist C, Sandelius AS (1991) Stromal low temperature compartment derived from the inner membrane of the chloroplast envelope. Plant Physiol 97: 1558–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Cho JH, Kang SG, Jang HJ, Pih KT, Piao HL, Cho MJ, Hwang I (1998) A dynamin-like protein in Arabidopsis thaliana is involved in biogenesis of thylakoid membranes. EMBO J 17: 859–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilgrim ML, van Wijk K-J, Parry DH, Sy DAC, Hoffman NE (1998) Expression of a dominant negative form of cpSRP54 inhibits chloroplast biogenesis in Arabidopsis. Plant J 13: 177–186 [DOI] [PubMed] [Google Scholar]
- Praefcke GJK, McMahon HT (2004) The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol 5: 133–147 [DOI] [PubMed] [Google Scholar]
- Rizo J, Südhof TC (2002) SNARES and MUNC18 in synaptic vesicle fusion. Nat Rev Neurosci 3: 641–653 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schnell DJ, Blobel G, Pain D (1990) The chloroplast import receptor is an integral membrane protein of chloroplast envelope contact sites. J Cell Biol 111: 1825–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127: 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Siebert PD, Chenchik A, Kellogg DE, Kukyanova KA, Lukyano SA (1995) An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res 23: 1087–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel J, Wiley D (1998) Coiled coils in both intracellular vesicle and viral membrane fusion. Cell 95: 871–874 [DOI] [PubMed] [Google Scholar]
- Ungermann C, Nichols BJ, Pelham HR, Wickner W (1998) A vacuolar v-t-SNARE complex, the predominant form in vivo and on isolated vacuoles, is disassembled and activated for docking and fusion. J Cell Biol 140: 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten J (2003) Unusual life style of giant chlorella viruses. Annu Rev Genet 37: 153–195 [DOI] [PubMed] [Google Scholar]
- Vothknecht UC, Westhoff P (2001) Biogenesis and origin of thylakoid membranes. Biochim Biophys Acta 1541: 91–101 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Westphal S, Soll J, Vothknecht UC (2001) A vesicle transport system inside chloroplasts. FEBS Lett 506: 257–261 [DOI] [PubMed] [Google Scholar]
- Westphal S, Soll J, Vothknecht UC (2003) Evolution of chloroplast vesicle transport. Plant Cell Physiol 44: 217–222 [DOI] [PubMed] [Google Scholar]
- Wu D, Wright DA, Wetzel C, Voytas DF, Rodermel S (1999) The IMMUTANS variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell 11: 43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak E, Norling B, Maitra R, Huang F, Andersson B, Pakrasi HB (2001) The initial steps of biogenesis of cyanobacterial photosystems occur in plasma membranes. Proc Natl Acad Sci USA 98: 13443–13448 [DOI] [PMC free article] [PubMed] [Google Scholar]