Abstract
The actin cytoskeleton mediates cellular processes through the dynamic regulation of the time, location, and extent of actin polymerization. Actin polymerization is controlled by several types of evolutionarily conserved proteins, including those comprising the ARP2/3 complex. In animal cells ARP2/3 activity is regulated by WAVE complexes that contain WAVE/SCAR proteins, PIR121, Nap125, and other proteins. The activity of the WAVE complex is regulated by Rho-GTPase-mediated signaling that leads to ARP2/3 activation by WAVE/SCAR proteins. We describe in this report Arabidopsis (Arabidopsis thaliana) genes encoding Nap and PIR proteins. Light-grown Atnap-1 and Atpir-1 mutant plants displayed altered leaf, inflorescence, silique, and seed set phenotypes. Dark-grown Atnap-1 and Atpir-1 seedlings also exhibited longer roots, enhanced skotomorphogenesis and Glc responses, and shorter thicker hypocotyls than those of wild type, showing that AtNAP and AtPIR participate in a variety of growth and developmental processes. Mutations in AtNAP and AtPIR caused cell morphology defects in cotyledon pavement cells and trichomes seen in mutants in ARP2/3 subunits and in plants expressing constitutively active Rop2 GTPase. The patterns and levels of actin polymerization observed in Atnap-1 and Atpir-1 mutant trichome cells and epidermal pavement cell morphology is consistent with Arabidopsis NAP and PIR proteins forming a WAVE complex that activates ARP2/3 activity. The multiple growth and developmental phenotypes of Atnap and Atpir mutants reveals these proteins are also required for a wider variety of cellular functions in addition to regulating trichome cell growth.
Reorganization of the actin cytoskeleton is required for many cell functions in a wide range of organisms. In animal cells extracellular signals often lead to dynamic changes in actin polymerization that alter cell morphology, movement, membrane dynamics, and other processes (Higgs and Pollard, 2001). Pharmacological and genetic studies have demonstrated that the actin cytoskeleton of plant cells plays an important role in a variety of cellular processes such as cell morphogenesis, stomatal closure, gravitropism, cell polarity and polarized cell growth, and cell division (Volkmann and Baluska, 1999; Vantard and Blanchoin, 2002; Deeks and Hussey, 2003).
The mechanisms regulating actin organization in plant cells are being revealed by genetic and cell biological analyses. Mutant alleles of ARP2/3 complex proteins, which nucleate the formation of networks of fine actin filaments, caused alterations in the expansion of epidermal pavement cell lobes and leaf trichomes and reduced coherence of the hypocotyl epidermis during cell expansion (Li et al., 2001; Le et al., 2003; Mathur et al., 2003a; Mathur et al., 2003b). These changes were associated with reduced diffuse cortical F-actin levels in rapidly expanding regions of cells undergoing diffuse growth and the accumulation of F-actin bundles in mature cells. Overexpression of profilin, which facilitates actin polymerization, leads to altered polarized growth and cell shape in Arabidopsis (Arabidopsis thaliana; Ramachandran et al., 2000). Reduced levels of an actin interacting protein in Arabidopsis cause a distorted actin cytoskeleton, altered cell shape, and developmental defects (Ketelaar et al., 2004). Formin proteins also nucleate actin filament formation, and overexpression of the Arabidopsis forming protein AFH1 leads to supernumerary actin cable formation and growth arrest in pollen tubes (Cheung and Wu, 2004).
In animal cells actin polymerization is regulated via extracellular receptors that transduce signals to members of the Rho GTPase family such as Rac and cdc42 (Etienne-Manneville and Hall, 2002). These proteins mediate the activity of a variety of effector proteins, including members of the WASP/WAVE/SCAR family (Mullins, 2000), which subsequently modulate activity of the ARP2/3 complex. The activity of WAVE and SCAR proteins is regulated by a complex of proteins comprising Nap125, PIR121, NCK, and HSPC300 in response to Rac-mediated signals (Smith and Li, 2004; Stradal et al., 2004). Disruption of animal Nap and PIR genes causes morphological defects. The Drosophila Kette gene encodes a homolog of Nap1, and disruption of Kette function leads to formation of excess F-actin and loss of axonal growth and path finding (Hummel et al., 2000). These neuronal defects were suppressed by reducing the copy number of the scar/wave gene, suggesting that Kette represses scar/wave activity (Hummel et al., 2000; Bogdan and Klambt, 2003). In Caenorhabditis elegans the GEX-2 and GEX-3 proteins, which are highly similar to PIR121 and Nap125, respectively, interact in a yeast two-hybrid system and associate with cell boundaries (Soto et al., 2002). Disruption of either gene leads to failure of embryonic tissues to organize correctly due to aberrant migration, resulting in embryo arrest. These cellular phenotypes in both C. elegans and Drosophila were shared with mutants affecting Rac signaling systems, and the kette mutation was partially rescued by expression of an activated DRAC1 protein (Bogdan and Klambt, 2003). The maize (Zea mays) BRICK1 gene encodes a protein that is highly similar to the human HSPC300 protein (Frank and Smith, 2002) that is a component of the human WAVE complex (Smith and Li, 2004; Stradal et al., 2004). Disruption of the maize BRICK1 gene caused decreased cell lobing in epidermal cell (Frank and Smith, 2002; Frank et al., 2003). In wild-type cells a diffuse form of cortical F-actin accumulated at sites of incipient lobing, and this was reduced in brick1 mutant cells. Cortical actin patterns are also altered in epidermal cells of Arabidopsis lines expressing constitutively active (CA-rop2) and dominant-negative (DN-rop2) forms of Rop GTPase, suggesting Rop activity may regulate cortical actin formation (Fu et al., 2002). Mutations in the Arabidopsis SPIKE1 gene, which encodes a member of the CDMS (CED-5, DOCK180, MBC) family of membrane proteins that associate with Rac, lead to randomized cortical actin filaments and epidermal cell shape defects (Qiu et al., 2002). Together these data suggest that actin cytoskeleton dynamics in plant cells may be regulated by signals from Rop proteins that activate ARP2/3, perhaps through the activity of BRICK/HSPC300-like proteins. Therefore, plant proteins performing analogous functions to NAP, PIR, and WASP/WAVE/SCAR proteins may be involved in transducing signals to the actin cytoskeleton in plants (Smith and Li, 2004).
Here we characterize two Arabidopsis genes encoding proteins similar to human Nap125 and PIR121 proteins. Analysis of insertion mutations in AtNAP and AtPIR genes revealed cell morphology defects in epidermal pavement cells and trichomes, and levels of F-actin filaments were reduced in trichome cells. These defects are consistent with AtNAP and AtPIR regulating ARP2/3 activity (Eden et al., 2002; Blagg et al., 2003; Kunda et al., 2003; Rogers et al., 2003). Mutations in AtNAP and AtPIR also caused a range of growth and developmental phenotypes, suggesting an Arabidopsis WAVE complex may regulate other processes in addition to cell shape.
RESULTS
Identification and Genetic Characterization of Arabidopsis AtNAP and AtPIR Genes Encoding Nap125- and PIR121-Like Proteins
The identification of the maize BRICK1 gene, encoding a plant protein related to animal HSPC300, suggested the existence of a WAVE-like regulatory complex in plants (Frank and Smith, 2002). To identify potential components of a WAVE complex in plants and understand their functions, we conducted a search for Arabidopsis proteins with significant similarity to known components of the human WAVE regulatory complex. Two single-copy genes encoding Nap125 and PIR121-like proteins from the Arabidopsis genome sequence were identified and named AtNAP (Arabidopsis Nap125-like protein; At2g35110) and AtPIR (Arabidopsis PIR121 like-protein; At5g18410). Full-length cDNAs were synthesized and sequenced to establish exon-intron boundaries. The predicted AtNAP has 34.8%, 32.9%, and 32.5% amino acid similarity with related proteins in human (Nap1), C. elegans (GEX-3), and Drosophila melanogaster (HEM-2), respectively (Fig. 1A). The predicted (AtPIR) has 47.7%, 47.8%, and 47.4% overall amino acid similarity with related proteins in human (PIR121), C. elegans (GEX-2), and D. melanogaster (CYFIP), respectively (Fig. 1B). One gene (At2g22640) encoding an HSPC300-like protein from the Arabidopsis genome sequence was also found (data not shown).
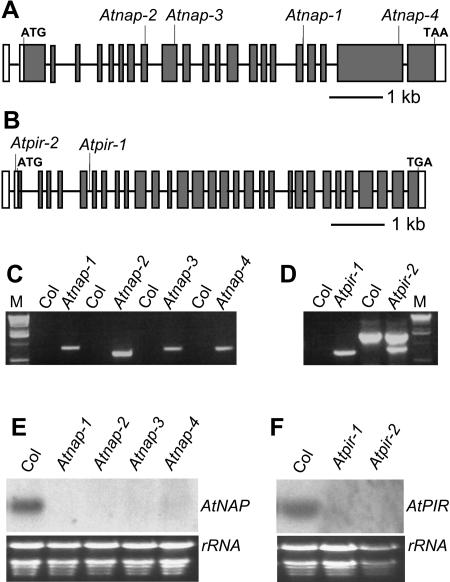
Figure 1.
Alignment of Arabidopsis NAP and PIR sequences with related proteins in Homo sapiens, D. melanogaster, and C. elegans. A, The amino acid sequence of Arabidopsis NAP aligned with H. sapiens (NAP1), D. melanogaster (HEM-2), and C. elegans (GEX-3) NAP125 proteins. B, The amino acid sequence of Arabidopsis PIR aligned with H. sapiens (PIR121), D. melanogaster (CYFIP), and C. elegans (GEX-2) PIR121 proteins. An asterisk indicates identical amino acid residues in the alignment. A colon indicates conserved substitutions in the alignment. A period indicates semiconserved substitutions in the alignment.
To investigate the function of AtNAP and AtPIR, we obtained several insertion alleles from the SALK and GABI-Kat Arabidopsis T-DNA mutant collection (Fig. 2, A and B). Atnap-1 (SALK_038799), Atnap-2 (SALK_014298), Atnap-3 (SALK_135634), and Atnap-4 (SALK_009695) were isolated with a T-DNA insertion in the 18th intron, the 9th exon, the 10th exon, and the 21st exon, respectively (Fig. 2, A and C). Atpir-1 (GABI-Kat 313F03) and Atpir-2 (SALK_106757) were identified with T-DNA insertions in 6th intron and 5′ untranslated region, respectively (Fig. 2, B and D). T-DNA insertions were confirmed by PCR using T-DNA specific and flanking primers and sequencing PCR products (Fig. 2, C and D). Northern-blot analysis revealed that neither the four Atnap mutant lines nor the two Atpir mutant lines had detectable transcripts of their respective genes (Fig. 2, E and F). These were therefore considered to be null mutant alleles.
Figure 2.
Molecular characterization of Atnap and Atpir mutants. A, Location of T-DNA insertions in Atnap-1, Atnap-2, Atnap-3, and Atnap-4 mutant lines are shown on the exon map of the AtNAP gene. The position of exons and introns are indicated by black rectangles and lines, respectively. B, Location of the T-DNA insertions in Atpir-1 and Atpir-2 mutant lines are shown on the exon map of AtPIR gene. The position of exons and introns are indicated by black rectangles and lines, respectively. The untranslated regions are shown by white rectangles. C, PCR identification of T-DNA insertion in Atnap mutants with T-DNA specific primers and flanking primers. Atnap lines yielded PCR products and Col wild-type lines did not amplify. M, One-kilobase DNA ladder. D, PCR identification of T-DNA insertion in Atpir mutants with T-DNA specific primers and flanking primers. Compared with Col wild type, Atpir mutant lines had specific PCR products. M, One-kilobase DNA ladder. E, Northern-blot analysis of AtNAP transcript levels in Col wild-type, Atnap-1, Atnap-2, Atnap-3, and Atnap-4 seedlings. F, Northern-blot analysis of AtPIR transcript levels in wild-type, Atpir-1, and Atpir-2 seedlings.
All the Atnap and Atpir mutants exhibited similar defects in trichomes, epidermal cells, and seedling development (see below). The respective T-DNA insertions cosegregated with the defective trichome phenotypes in their F2 populations, demonstrating that the disruptions in AtNAP and AtPIR caused the mutant phenotypes observed. According to trichome phenotypes we observed that all the heterozygotic lines had the wild-type phenotype and the F2 population showed a segregation ratio of three wild type to one mutant, indicating that Atnap-1 and Atpir-1 are single recessive mutants (Table I). Progeny of crosses of the two lines with insertions in the AtPIR gene demonstrated the insertions were allelic, and crosses of the four lines with insertions in the AtNAP gene showed all these insertions were also allelic with respect to trichome phenotypes. This further confirmed that the insertions in AtNAP and AtPIR caused the trichome phenotypes.
Table I.
Genetic analysis of Atnap-1 and Atpir-1 trichome phenotype
| F2 Population
|
||||||
|---|---|---|---|---|---|---|
| Mutants | Heterozygotes | Wild Type | Mutant | Total | x2 (3:1) | P Value |
| Atnap-1 | 12 | 286 | 89 | 375 | 0.32 | 0.5–0.1 |
| Atpir-1 | 16 | 302 | 87 | 389 | 1.08 | 0.5–0.1 |
Mutations in AtNAP and AtPIR Affect Growth and Development
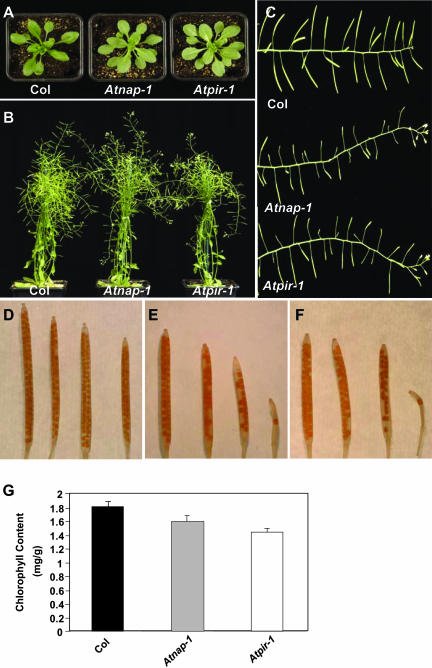
The effect of mutations in AtNAP and AtPIR on seedling development was investigated. No obvious differences in the strength of phenotype between Atnap-1, Atnap-2, Atnap-3, and Atnap-4 or between Atpir-1 and Atpir-2 were observed; therefore, we chose the Atnap-1 and Atpir-1 mutant lines for further characterization. Soil-grown Atnap-1 and Atpir-1 plants exhibited a variety of growth defects. The leaves of Atnap-1 and Atpir-1 mutants were paler green than those of wild type (Fig. 3A). This was due to reduced chlorophyll content (Fig. 3G). Leaf morphogenesis was also slightly altered compared to wild type. Wild-type rosette leaves, before bolting, were slightly epinastic, whereas the rosette leaves of Atnap-1 and Atpir-1 plants were flatter (Fig. 3A). The growth and development of siliques, seeds, and inflorescences was abnormal in Atnap-1 and Atpir-1 plants. Early developing siliques in mutant plants were straight and had a similar length to wild type. However, later-developing siliques were occasionally very short and contained fewer seeds (Fig. 3, C–F). The siliques in Atnap-1 and Atpir-1 contained slightly larger seed than wild type (data not shown). The inflorescences of Atnap-1 and Atpir-1 mutants continued indeterminate growth until senescence, in contrast to most wild-type inflorescences, which ceased flowering and elongation before senescence (Fig. 3, B and C).
Figure 3.
Phenotypes of Atnap-1 and Atpir-1 plants. A, Two-week-old seedlings of Col wild type, Atnap-1, and Atpir-1. Mutant leaves are epinastic and a paler green color. B, Mature Col wild-type, Atnap-1, and Atpir-1 plants. Note the extended growth of mutant inflorescence stems. C, Inflorescence stems of Col wild type, Atnap-1 and Atpir-1. D, Siliques of Col wild type. E, Siliques of Atnap-1. A significant proportion of these are smaller with reduced seed set. F, Siliques of Atpir-1. A significant proportion of these are smaller with reduced seed set. G, Chlorophyll content analysis of Col wild type, Atnap-1, and Atpir-1. Error bars represent se (n = 4).
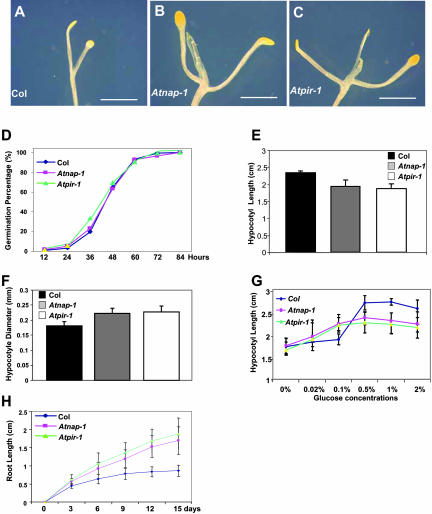
Atnap-1 and Atpir-1 seedlings grown on vertical plates in the dark exhibited increased shoot development (Fig. 4, A–C). Wild-type seedlings grown for 15 d in the dark had partially expanded cotyledonary petioles, and true leaves had just started to develop (Fig. 4A). Dark-grown Atnap-1 and Atpir-1 plants had fully expanded cotyledonary petioles and the first true leaves had formed (Fig. 4, B and C). This increased dark development or skotomorphogenesis in mutants was not a result of earlier germination, since the germination of Atnap-1 and Atpir-1 seeds was the same as wild-type and mutant seed germinated with similar kinetics to wild-type seed (Fig. 4D). Furthermore, dark-grown Atnap-1 and Atpir-1 seedlings had shorter and thicker hypocotyls than wild type (Fig. 4, E and F), suggesting that AtNAP and AtPIR may regulate cell size and radial expansion of hypocotyls. Interestingly, hypocotyl elongation of Atnap-1 and Atpir-1 mutants showed enhanced responses to sugar in the dark. Wild-type Columbia (Col) seedlings do not exhibit significant hypocotyl elongation in response until Glc levels are between 0.1% and 0.5%, and hypocotyl elongation is only inhibited at Glc concentrations above 3% (data not shown). In contrast, both Atnap-1 and Atpir-1 seedlings had longer hypocotyls than wild type at low Glc levels, and at higher Glc concentrations hypocotyls were shorter than wild type (Fig. 4G). This suggested the mutants seedlings displayed Glc-hypersensitive hypocotyl elongation. Finally, roots of dark-grown Atnap-1 and Atpir-1 were substantially longer than that of wild type (Fig. 4H).
Figure 4.
Characterization of dark-grown Atnap-1 and Atpir-1 mutant phenotypes. A to C, Fifteen-day-old dark-grown seedlings of Col wild type (A), Atnap-1 (B), and Atpir-1 (C) show partially developed cotyledonary petioles and expansion of the cotyledon in wild type (A), and the fully developed cotyledonary petioles, cotyledons, and the first pair of true leaves in Atnap-1 (B) and Atpir-1 (C). The scale bars represent 2 mm in A to C. D, Germination of wild-type, Atnap-1, and Atpir-1 seedlings. Seeds were stratified for 6 d on Murashige and Skoog medium supplemented with 1% Glc. Germination was scored as the percentage of seeds showing radicle emergence at 20°C in constant light. E, Hypocotyl length of 15-d-old dark grown seedlings of wild type, Atnap-1, and Atpir-1 grown on vertical plates. Error bars represent sds (n > 50). F, Hypocotyl diameter of 15-d-old dark-grown seedlings of wild type, Atnap-1, and Atpir-1 grown on vertical plates. Error bars represent sds (n > 50). G, Hypocotyl length of 7-d-old dark-grown seedlings of wild type, Atnap-1, and Atpir-1 grown on vertical plates at different Glc concentrations. Error bars represent sds (n > 30). H, Root length of dark-grown seedlings of wild-type, Atnap-1, and Atpir-1 seedlings grown on vertical plates. Error bars represent sds (n > 50).
Atnap-1 and Atpir-1 Plants Exhibit Defects in Epidermal Cell Morphogenesis
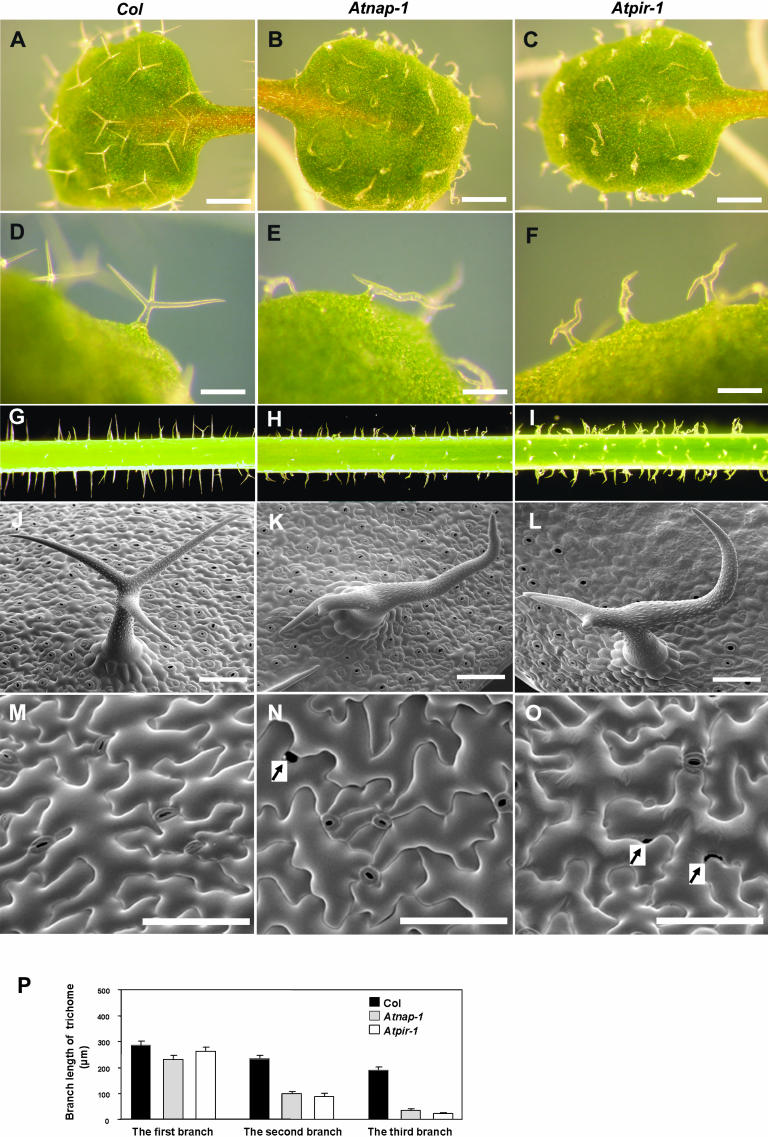
The effect of the Atnap-1 and Atpir-1 mutations on trichome development was examined because mutations in the ARP2/3 complex of Arabidopsis exhibit distorted trichomes. Wild-type Arabidopsis trichomes generally form three branches, the position, shape, and length of which are tightly regulated during trichome development (Huelskamp et al., 1994; Szymanski et al., 1999). At maturity, wild-type trichome branches are characteristically straight or very slightly curved. Wild-type branches were between 190 μm and 285 μm in length and each of the three trichome branches was progressively shorter (Fig. 5P). To analyze branch length and position in the mutants, the longest branch was named the first branch, and the second and third branches were the progressively shorter ones. The Atnap-1 and Atpir-1 mutations also caused similar severe trichome defects on leaves and stems (Fig. 5, A–J). As shown in Figure 5, J to L, the first and second trichome branches of Atnap-1 and Atpir-1 mutants were wavy and slightly distorted and the third branch was very short compared to wild type, and the trichome stalk was often swollen. Quantitative analysis showed that the length of the first branch in Atnap-1 and Atpir-1 mutants was slightly shorter than wild type, the length of the second branch was decreased to 42.0% and 37.5% that of wild type, and the shortest branch was 17.2% and 11.3% that of wild type, respectively (Fig. 5P). We also observed that trichome branch position was altered; the first and second branches were much farther apart compared with wild type (Fig. 5, A–L). Finally, papillae on the tip of aborted branches in Atnap-1 and Atpir-1 were often absent (data not shown).
Figure 5.
Trichome and epidermal cell defects of Atnap-1 and Atpir-1 mutants. A and D, Col wild-type leaf trichomes. B and E, Atnap-1 leaf trichomes. C, and F, Atpir-1 leaf trichomes. G, Col wild-type stem trichomes. H, Atnap-1 stem trichomes. I, Atpir-1 stem trichomes. J, Scanning electron microscope image of trichomes on leaves from Col wild-type plants. K, Scanning electron microscope image of trichomes on leaves from Atnap-1 plants. L, Scanning electron microscope images of trichomes on leaves from Atpir-1 plants. M, Scanning electron microscope image of the upper surface of Col wild-type cotyledon pavement cells. N, Scanning electron microscope image of the upper surface of Atnap-1 cotyledon pavement cells. O, Scanning electron microscope image of the upper surface of Atpir-1 cotyledon pavement cells. P, Branch length of 9-d-old leaf trichomes. The branches of each trichome were classified into the longest branch, the second longest branch, and the shortest branch. Arrows indicate gaps between adjacent pavement cells. The scale bars represent 500 μm in A to C, 200 μm in D to F, and 100 μm in J to O.
Since the maize brick and Arabidopsis arp2/3 subunit mutants have defects in epidermal cell lobing, we examined pavement cell shape and size in the Atnap-1 and Atpir-1 mutants. Mature pavement cells on the adaxial surface of wild-type cotyledons have regular lobes that interlock tightly with adjacent cells. In Atnap-1 and Atpir-1 mutants lobe shape was indistinguishable from wild type (Fig. 5, M–O). However, some cotyledon pavement cells failed to tessellate normally, resulting in gaps between adjacent cells (Fig. 5, M–O). These gaps were not observed in true leaves.
Aberrant Actin Organization in Atnap-1 and Atpir-1 Mutants
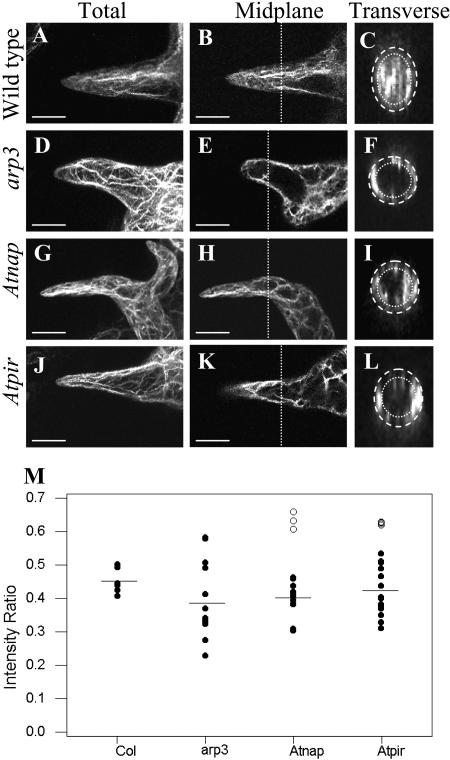
As the Atnap-1 and Atpir-1 mutants affected trichome development, and because related genes are involved in regulating the actin cytoskeleton in animal cells, we visualized the actin cytoskeleton of Atnap-1 and Atpir-1 trichomes using fluorochrome-conjugated phalloidin. We compared the actin cytoskeleton in the Atnap-1 and Atpir-1 mutants with an arp3 mutant to determine any similarities between these mutants. During stage 2 trichome development, the trichome stalks elongate, and during stage 3 rudimentary branches develop (Szymanski et al., 1999). At these stages the pattern of actin localization in trichomes of Atnap-1 and Atpir-1 mutants was similar to wild type (data not shown). The first differences in actin architecture between wild-type, Atnap-1, and Atpir-1 mutant trichomes were detected in stages 4 and 5, when the trichome branches and stalk were expanding rapidly (Szymanski et al., 1999). In the branches of wild-type trichomes, F-actin cables and bundles were long and generally aligned with the direction of branch growth (Fig. 6, A and B). Mutant arp3, Atnap-1, and Atpir-1 trichomes had more disorganized bundles of F-actin in stage 4/5 trichomes (Fig. 6, D–K). Compared to wild type, more presumptive vacuoles (observed as dark regions) were found in the branches of stage 4/5 trichomes of arp3, Atnap-1, and Atpir-1 mutants (Fig. 6, D–K). In stage 4/5 wild-type trichomes more actin was observed in the core of trichome branches compared to arp3, Atnap-1, and Atpir-1 mutants (Fig. 6, C, F, I, and L).
Figure 6.
Actin localization and quantification in wild-type, arp3, Atnap-1, and Atpir-1 trichomes. Whole mount stage 4/5 trichome branches stained with Alexafluor phalloidin. A, D, G, and J are maximum projections. B, E, H, and K are midplane sections and the dotted line indicates the midpoint of branch length where transverse section was taken. C, F, I, and L are transverse sections of branches, which have been enlarged for clearer viewing. Core actin was within a 2.5-μm perimeter from the cell surface, indicated by the dotted lines. M, Intensity of core actin to total actin in stage 4/5 trichomes. Horizontal lines indicate average intensity ratio of black circle data points. Compared to wild type, a distinct subset of Atnap-1 and Atpir-1 trichomes did not have a reduced intensity ratio (white circles).
To quantify these observations we measured the ratio of subcortical (core) actin to total actin abundance in trichome branches in wild-type Col, arp3, Atnap-1, and Atpir-1 mutants. In stage 4/5 trichome branches the average ratio of core to total actin in arp3 mutants was significantly reduced (0.39 ± se 0.03), compared to wild type (0.46 ± se 0.02; Fig. 6M). In Atnap-1 and Atpir-1 stage 4/5 branches the average ratio of core to total actin including all data points was 0.45 ± se 0.03 and 0.47 ± se 0.02, respectively (Fig. 6M). Although this was similar to wild type we found that actin distribution in the trichomes of Atnap-1 and Atpir-1 formed two distinct groups. Compared to wild type approximately two-thirds of Atnap-1 (n = 14) and Atpir-1 (n = 17) branches (Fig. 6M, black circles) have reduced relative amounts of core actin, 0.40 ± se 0.01 and 0.42 ± se 0.02, respectively. The remaining Atnap-1 (n = 5) and Atpir-1 (n = 6) branches (Fig. 6M, white circles) have on average slightly increased relative amounts of core actin, 0.63 ± se 0.02 and 0.62 ± se 0.00, respectively.
AtNAP and AtPIR Have Similar Expression Patterns
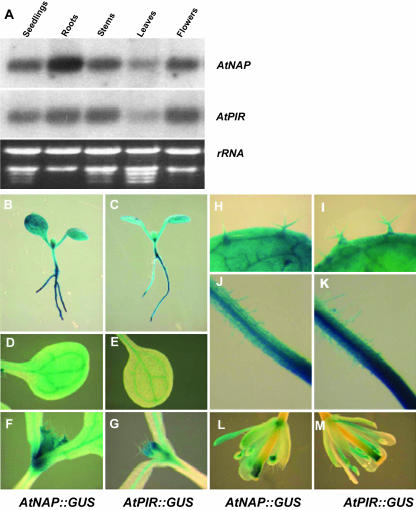
Levels of AtNAP and AtPIR mRNA were analyzed in various tissues by RNA gel-blot analysis. Figure 7A shows that AtNAP and AtPIR transcripts were detected in all tissues examined, including young seedlings, roots, stems, rosette leaves, and flowers. AtNAP mRNA levels were highest in roots. The spatial expression patterns of AtNAP and AtPIR were revealed by histochemical assays of β-glucuronidase (GUS) activity of transgenic plants containing AtNAP promoter:GUS fusions (AtNAP∷GUS) or AtPIR promoter:GUS fusions (AtPIR∷GUS). Histochemical staining shows AtNAP and AtPIR gene expression throughout the plant, including roots, root hairs, hypocotyls, cotyledons, true leaves, trichomes, and flowers (Figure 7, B–M). The highest GUS activity was detected in roots (Fig. 7, B and C), consistent with northern-blot analysis. In addition, relatively high levels of GUS activity were detected in vascular tissues and the shoot apex of AtNAP∷GUS and AtPIR∷GUS lines (Fig. 7, D–G). Northern gel blot and GUS histochemical staining also indicated that AtNAP and AtPIR had similar expression patterns.
Figure 7.
Expression patterns of AtNAP and AtPIR genes. A, Northern-blot analysis of AtNAP and AtPIR gene expression. Total RNA was isolated from seedlings, roots, stems, leaves, and flowers. B, GUS activity in AtNAP∷GUS seedlings. C, GUS activity in AtPIR∷GUS seedlings. D, GUS activity in AtNAP∷GUS cotyledons. E, GUS activity in AtPIR∷GUS cotyledons. F, GUS activity in AtNAP∷GUS shoot apex. G, GUS activity in AtPIR∷GUS shoot apex. H, GUS activity in AtNAP∷GUS line in trichomes. I, GUS activity in AtPIR∷GUS line in trichomes. J, GUS activity in AtNAP∷GUS line in root hairs. K, GUS activity in AtPIR∷GUS line in root hairs. L, GUS activity in AtNAP∷GUS line in flowers. M, GUS activity in AtPIR∷GUS line in flowers.
DISCUSSION
A survey of the Arabidopsis genome for proteins that may regulate ARP2/3 activity identified two proteins encoded by At2g35110 and At5g18410 with significant overall similarity to human, Drosophila, and C. elegans Nap125 and PIR121 proteins, respectively. The preservation of identity and frequent conserved substitutions throughout the Arabidopsis, human, Drosophila, and C. elegans proteins suggested these Arabidopsis proteins may perform similar cellular functions to their animal counterparts. After submission of this study, several related studies describing the identification and initial characterization AtNAP and AtPIR were also published (Basu et al., 2004; Brembu et al., 2004; Deeks et al., 2004; El-Assal Sel et al., 2004). These studies report similar effects of mutations in AtNAP and AtPIR on the actin cytoskeleton to those described here, and also showed interactions between AtNAP and AtPIR and between AtPIR and AtROP2 (Basu et al., 2004). Furthermore, Arabidopsis proteins similar to SCAR proteins were also reported (Brembu et al., 2004; Deeks et al., 2004). The C-terminal VCA motif of these proteins, which is common to both WAVE/SCAR and WASP proteins, drives conformational changes in the ARP2/3 complex that promote actin nucleation and the VCA domain of the Arabidopsis SCAR protein encoded by At2g34150 was shown to bind actin in vitro (Deeks et al., 2004). These analyses, together with the characterization of BRICK/HSPC300 proteins (Frank and Smith, 2002) and SPIKE1 (Qiu et al., 2002) that are related to other known components of animal WAVE complexes, provide initial evidence for Arabidopsis WAVE complexes that regulate actin cytoskeleton dynamics. Trichome development was strongly affected in the Atnap and Atpir mutants; they exhibited frequent swollen trunks bearing second and third branches much reduced in length. Related distorted trichome phenotypes result from overexpression of CA-rop2 in Arabidopsis (Fu et al., 2002). Mutations of subunits of the ARP2/3 complex also lead to characteristic distorted trichomes with short distended branches and a bulbous base (Mathur and Chua, 2000; Le et al., 2003; S. Li et al., 2003; Mathur et al., 2003a). Among the distorted trichome class of mutants, no fewer than four encode subunits of the ARP2/3 complex (Deeks and Hussey, 2003; Smith and Li, 2004), indicating that proteins regulating the actin cytoskeleton cause distorted trichomes. Recently the gnarled locus was shown to be allelic to Atnap mutants (Brembu et al., 2004; El-Assal Sel et al., 2004), and Atpir was reported to be allelic to klunker (Mutondo et al., 2004). However, this finding is currently inconsistent with another report claiming Atpir mutants are not allelic to klunker (Basu et al., 2004). Despite this apparent difference it is likely that other distorted mutants may also encode cytoskeletal regulators.
Reduced cell polarity in ARP2/3 mutants has been proposed to underlie these changes in trichome cell shape, suggesting that cell polarity may be altered in the Atnap and Atpir mutants. This is supported by the observation of reduced tessellation and gaps between epidermal pavement cells in Atnap and Atpir cotyledons. This is also seen in ARP2/3 mutants and is thought to be due to reduced cell lobing. Similar observations have also been made recently for Atpir mutants (Basu et al., 2004). Loss of BRICK1 function in maize also led to reduced leaf epidermal lobing (Frank and Smith, 2002), and this was postulated to be due to reduced activity of a putative WAVE complex containing BRICK1 that may activate ARP2/3 activity (Smith and Li, 2004). These data suggest a WAVE complex regulates ARP2/3 activity during epidermal cell growth. In animal cells the activity of the WAVE complex is regulated by Rac-mediated signaling (Stradal et al., 2004). The hypocotyls of CA-rop2, Atnap, and Atpir plants also exhibited significantly increased diameters, and this phenotype has been shown to be due to increased radial cell expansion in CA-rop2 (Fu et al., 2002). These phenotypes suggest Rac-mediated signals may regulate a putative WAVE complex that promotes hypocotyl cell expansion in the dark.
The severity of trichome branch distortion in arp3, Atnap, and Atpir mutant plants correlated with the degree of disruption in F-actin cable organization. In more disrupted trichomes, actin cables were randomly arranged into a mesh-like structure in all three mutants compared to wild-type trichomes. Measurement of the distribution of F-actin showed that most trichome branches of the arp3 mutant had reduced core F-actin in proportion to total actin filaments. About two-thirds of stage 4/5 Atnap-1 and Atpir-1 mutant trichome branches also showed a reduction in core F-actin. The remaining one-third of Atnap-1 and Atpir-1 trichome branches had slightly elevated ratios of core actin filaments to total actin levels that were not clearly related to the degree of trichome branch distortion. The reduced levels of core actin filaments seen in the majority of mutant trichomes was also seen in related studies (Basu et al., 2004; Deeks et al., 2004; El-Assal Sel et al., 2004) and is most consistent with the putative Arabidopsis WAVE complex functioning as a positive regulator of ARP2/3 activity with respect to trichome cell expansion.
Current evidence obtained from studies in human, Dictyostelium, and Drosophila cells supports two distinct mechanisms of WAVE function (Stradal et al., 2004). RNAi experiments in Drosophila and mammalian cells (Kunda et al., 2003; Rogers et al., 2003) showed that reduced expression of WAVE, Nap1, PIR121, and Abi1 reduced Rac-mediated actin remodeling and lamellipodia formation. This is consistent with current data on actin levels in Arabidopsis trichomes in Atnap-1 and Atpir-1 mutants that suggest the putative Arabidopsis WAVE complex is a positive regulator of ARP2/3 mediated actin filament formation (Basu et al., 2004; Deeks et al., 2004; El-Assal Sel et al., 2004). In contrast, biochemical studies in human cells show that constitutively active WAVE/SCAR proteins can be held in an inactive form in a complex with proteins including Nap125, PIR121, Abi2, and HSPC300 proteins (Eden et al., 2002). Activation of this complex by GTP-Rac leads to release of a WAVE/HSPC300 complex that subsequently activates ARP2/3. This model of Nap125 and PIR121 as negative regulators of ARP2/3 activity is supported by genetic evidence in Dictyostelium (Blagg et al., 2003). Deletion of the PirA gene, encoding PIR121, resulted in unusually large cells displaying reduced motility and excessive actin polymerization. Disruption of the SCAR gene in Dictyostelium resulted in small cells exhibiting reduced actin polymerization and movement defects. Double pir121/scar mutants showed the same phenotype as scar- mutants, suggesting PIR121 requires SCAR in vivo. Further analysis of Arabidopsis mutants, such as double mutant analysis and biochemical analyses of actin polymerization, will help define the mechanisms of ARP2/3 regulation in plants and contribute to an evolutionarily broader definition of WAVE complex function.
We have also identified a wider range of developmental and growth defects in the Atnap and Atpir mutants. These include low chlorophyll levels, leaf epinasty, reduced seed set, deformed siliques, and enhanced responses to Glc, root growth, and skotomorphogenesis. This range of phenotypes is consistent with the widespread expression patterns of the AtNAP and AtPIR genes. In contrast, mutations in ARP2/3 subunits are reported to be restricted to altered morphogenesis of trichome and epidermal cells (Le et al., 2003; S. Li et al., 2003; Mathur et al., 2003a). Several phenotypes seen in Atnap-1 and Atpir-1 plants are also exhibited by plants expressing CA-rop2, such as increased seed size, deformed siliques and increased seed abortion, increased radial hypocotyl growth, and increased skotomorphogenesis (Li et al., 2001; Fu et al., 2002). We speculate that Rac-mediated signaling to a putative Arabidopsis WAVE complex may contribute to some of these common phenotypes. The enhanced skotomorphogenesis and Glc response phenotypes are also shared with ARP2/3 mutants (K. Sorefan, G. Hemmann, R. Holman, M. Baier, and M.W. Bevan, unpublished data), suggesting that ARP2/3 mediated changes in the actin cytoskeleton may contribute to these phenotypes and that AtNAP and AtPIR are positive regulators of ARP2/3 with respect to these phenotypes. Cell shape or cell polarity defects may contribute to these phenotypes, but it is also possible that other cellular processes regulated by the actin cytoskeleton (Wasteneys and Galway, 2003) could also contribute to these phenotypes. For example, vacuole integrity is altered in ARP2/3 mutants (Mathur et al., 2003a), and alterations in vacuolar ATPase function in the det3 mutant affects sugar and starch levels (Schumacher et al., 1999), suggesting a potential link with altered carbohydrate responses. Further work aims to identify other processes regulated by ARP2/3 and WAVE complex activity that may lead to a wider understanding of the cellular functions of the Arabidopsis WAVE complex.
MATERIALS AND METHODS
Database Search and Bioinformatics
To identify Arabidopsis (Arabidopsis thaliana) proteins related to members of the WAVE complex, human, Drosophila, and Dictyostelium WAVE complex members were used as queries for BLASTP and TBLASTN searches of the National Center for Biotechnology Information database (www.ncbi.nlm.nil.gov) and the Arabidopsis Information Resource database (www.arabdopsis.org). Amino acid alignment was conducted using ClustalW (www.ebi.ac.uk/clustalw) and Bioedit.
Identification of AtNAP and AtPIR cDNA Sequences
To identify cDNA sequences of AtNAP and AtPIR, we performed reverse transcription (RT)-PCR using AtNAP and AtPIR specific primers and sequenced RT-PCR products. Total RNA was extracted from Arabidopsis seedlings using an RNeasy Plant Mini kit (Qiagen, West Sussex, UK) according to the kit manual. RT-PCR analysis was performed as described (Y. Li et al., 2003). Briefly, first stand cDNA was transcribed reversibly from total RNA with oligo(dT) as the primer and used as the template to amplify the transcripts with a program (40 cycles) of 94°C for 15 s, 58°C for 15 s, and 72°C for 1 min. The AtNAP gene primers used for RT-PCR are: SNAPS1 (F 5′-CGACTTCTGTGAGATCAAGGG-3′ and R 5′-ACGAGATATCCAAGAAGCACC-3′); SNAPS2 (F 5′-TTTTCTATCTGCAGATACGAGG-3′ and R 5′-TCGAGATGTTGCACAATACGC-3′); SNAPS3 (F 5′-CTTTCTTCATCTGCTGGAAGG-3′ and R 5′-CTCTGCCAAGTGAACTATGC-3′); SNAPS4 (F 5′-GAATGCATCCTTGGGAACTTC-3′and R 5′- CATTCAGAGTCGTGATCACC-3′); and SNAPS5 (F 5′-CTCGATGATCTCTGGCATCG-3′ and R 5′-TGTCTCTTTTGTAGAATGTGGG-3′). The AtPIR gene primers used for RT-PCR are as follows: SPIRS1 (F 5′-TCCTGTAGAGGAAGCAATCGC-3′ and R 5′-GAGAAGAATATACCGCTCAGG-3′); SPIRS2 (F 5′-CCTGCACGTTGAGATGTTCC-3′ and R 5′-TTCTTTCGGAAGGTGGTACG-3′); SPIRS3 (F 5′-GAGTGTAGGATCGATGCTGC-3′ and R 5′-TGAGACTGAAAGGGTCTATGG-3′); SPIRS4 (F 5′-TGAAAAGTTCTCCATCCAGCC-3′ and R 5′-ACCTGGACTTGATACAACTGC-3′); and SPIRS5 (F 5′-TTCATGCAAACAGCTCCATGG-3′ and R 5′-CAGACACGGTATTCTCAAACC-3′). The available expressed sequence tag sequences used to confirm AtNAP and AtPIR cDNA sequences were T88379, AA394638, AV545854, AV546555, AV547935, AV547490, AV548158, AV548139, AV554904, AV554853, AV554801, and AV556813 for the AtNAP gene and AI992622, N96263, AV540814, and AV781909 for the AtPIR gene.
Plant Material and Growth Conditions
All experiments described in this study involve Arabidopsis ecotype Col-0. Atnap-1 (SALK_038799), Atnap-2 (SALK_014298), Atnap-3 (SALK_135634), Atnap-4 (SALK_009695), Atpir-2 (SALK_106757), and Atpir-1 (GABI-Kat 313F03) lines were identified in the AtIDB database (www.atidb.org) and obtained from the Nottingham Arabidopsis Stock Centre or GABI-Kat. Seeds were surface-sterilized with 100% isopropanol and 20% (v/v) household bleach, washed at least five times with sterile water, stratified at 4°C for 6 d in the dark, and germinated on Murashige and Skoog medium (Duchefa Biochemie BV, Haarlem, The Netherlands) supplemented with 0.9% agar and 1% Glc. Seedlings were grown in media under continuous light at 22°C and grown in soil under 16-h light periods at 20°C to 25°C.
Atnap and Atpir Allele Characterization
Arabidopsis genomic DNA preparation was performed as described (Qian et al., 2001). SALK_038799, SALK_014298, SALK_135634, and SALK_009695 T-DNA insertions in the AtNAP gene were confirmed by PCR and sequencing, by using primers SALK_038799LP (TTTTCCAGATAGGATTCGAGCA), SALK_038799RP (GACCACCCAACGAGCACCATA), SALK_014298LP (TGATCATTTTCTCAACTTGTTTTGC), SALK_014298RP (CCTGGAGGCTGTAGGACCCA), SALK_135634LP (TGGGAAAGTTTAGGGAAGGGAA), SALK_135634RP (TCCTCCAAGGATTCTATCTTGAAAA), SALK_009695LP (GGATGCTGTTGAGAGCGGTGT), SALK_009695RP (CCGGGAATTGATTGAAGCAGC), and T-DNA specific primer LBa1 (TGGTTCACGTAGTGGGCCATCG). SALK_106757 T-DNA insertions in AtPIR were identified by primers SALK_106757 LP (CGATGGGACTATCGGTTGCTG), SALK_106757 RP (TCCCAAAATAAGAAATGGTTGAGGA), and LBa1. The GABI-Kat 313F03 T-DNA insertion in AtPIR was confirmed by primers 313F03LP (TGATCCAGCTGAACCAAGACG) and T-DNA specific primer O08760 (GGGCTACACTGAATTGGTAGCTC).
RNA Gel-Blot Analysis
Total RNA was extracted from Arabidopsis seedlings, roots, stems, leaves, and flowers using an RNeasy Plant Mini kit (Qiagen) according to the manual. RNA gel-blot analysis was performed as described (Rook et al., 2001). Gene specific probes were digoxigenin-labeled (DIG-dUTP) by RT-PCR with PCR DIG Labeling Mix (Roche Diagnostics, Lewes, UK) and gene specific primers. The gene specific primers used for RTPCR are as follows: NAP-F (CTCGATGATCTCTGGCATCG), NAP-R (TGGATGAGCGATACGGATGG), PIR-F (TGCAGTTGTATCAAGTCCAGG), and PIR-R (CAGACACGGTATTCTCAAACC). Hybridization was as described (Rook et al., 2001); washes were with 0.2× SSC, 0.1% SDS at 65°C, twice for 15 min. Detection used antidigoxigenin-A, Fab fragments, and the chemiluminescent substrate CSPD, according to the manufacturer's instructions (Roche, Mannheim, Germany).
Scanning Electron Microscopy
Seedlings grown for 9 d in constant light were frozen in nitrogen slush at −190°C. Ice was sublimed at −90°C, and the specimen was sputter coated and examined on an XL 30 FEG (Philips, Eindhoven, The Netherlands) cryoscanning electron microscope fitted with a cold stage.
Visualization of F-Actin and Confocal Microscopy
To visualize actin in trichomes, young leaves were incubated in 2% formaldehyde in PEM buffer (100 mm PIPES, 5 mm EGTA, 4 mm MgCl2, 100 mm mannitol, and 0.01% IGEPAL) for 30 min. Tissue was washed three times in PEM buffer before incubation overnight in PEM buffer and 0.8 units Alexa Fluor 488-phalloidin (Molecular Probes, Leiden, The Netherlands). F-actin was visualized using a Leica (Wetzlar, Germany) SP confocal microscope and images were analyzed with Leica Confocal Software. Stage 4/5 branches were between 16 μm and 50 μm. Measurements were taken only from trichomes where all branches were less than 50 μm. The average branch length of col, arp3, Atnap-1, and Atpir-1 was 25 μm, 32 μm, 35 μm and 33 μm respectively. ImageJ software was used for measuring integrated fluorescence intensity of transverse sections taken at the midpoint of the branch. The core fluorescence was within a region 2.5 μm from the cell surface. Seven branches for wild type, 12 branches for arp3, 14 branches for Atnap-1, and 18 branches for Atpir-1 were measured.
Transgenic Lines and GUS Histochemistry
The AtNAP promoter-GUS construct (AtNAP∷GUS) and the AtPIR promoter-GUS construct (AtPIR∷GUS) were made using a PCR-based Gateway system. The promoter specific primers for the AtNAP gene were NAPP-F (5′-CACCAGCCGAGTACAAAGAAGAAGC-3′) and NAPP-R (5′-TAATTCAGTACAATAATCTCTACAATA-3′) and for the AtPIR gene were PIRP-F (5′-CACCCATCAGCCTTGCCCGTATAGC-3′) and PIRP-R (5′-TGAGTCACCTGGAAAGATCAG-3′). PCR products were subcloned into pENTR/D-TOPO using TOPO enzyme and sequenced. Then the AtNAP and AtPIR promoters were further subcloned into Gateway Binary Vector (pGWB3) containing the GUS reporter gene. Arabidopsis transformation was made by dipping method using Agrobacterium strain GV3101. Transformants were selected on kanamycin (50 μg/mL) medium. Seedlings were stained in a solution of 1 mm X-gluc, 50 mm NaPO4 buffer, 0.4 mm each K3Fe(CN)6/K4Fe(CN)6, 0.1% (v/v) Triton X-100 and incubated at 37°C for 10 to 24 h. After GUS staining chlorophyll was removed using 70% ethanol.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY787211 and AY787212.
Acknowledgments
We thank Kim Findlay for assistance with scanning electron microscopy, the Nottingham Arabidopsis Stock center (NASC) for mutant lines, and members of the Bevan group for advice.
This work was supported by the Biotechnology and Biological Sciences Research Council (grant no. 208/EGM16126), by Syngenta (grant no. PMC19), and by the John Innes Centre Core Strategic Grant (to M.W.B.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.053173.
References
- Basu D, El-Assal Sel D, Le J, Mallery EL, Szymanski DB (2004) Interchangeable functions of Arabidopsis PIROGI and the human WAVE complex subunit SRA1 during leaf epidermal development. Development 131: 4345–4355 [DOI] [PubMed] [Google Scholar]
- Blagg SL, Stewart M, Sambles C, Insall RH (2003) PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Curr Biol 13: 1480–1487 [DOI] [PubMed] [Google Scholar]
- Bogdan S, Klambt C (2003) Kette regulates actin dynamics and genetically interacts with Wave and Wasp. Development 130: 4427–4437 [DOI] [PubMed] [Google Scholar]
- Brembu T, Winge P, Seem M, Bones AM (2004) NAPP and PIRP encode subunits of a putative wave regulatory protein complex involved in plant cell morphogenesis. Plant Cell 16: 2335–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, Wu HM (2004) Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell 16: 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks MJ, Hussey PJ (2003) Arp2/3 and ‘The Shape of things to come’. Curr Opin Plant Biol 6: 561–567 [DOI] [PubMed] [Google Scholar]
- Deeks MJ, Kaloriti D, Davies B, Malho R, Hussey PJ (2004) Arabidopsis NAP1 is essential for Arp2/3-dependent trichome morphogenesis. Curr Biol 14: 1410–1414 [DOI] [PubMed] [Google Scholar]
- Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW (2002) Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418: 790–793 [DOI] [PubMed] [Google Scholar]
- El-Assal Sel D, Le J, Basu D, Mallery EL, Szymanski DB (2004) Arabidopsis GNARLED encodes a NAP125 homolog that positively regulates ARP2/3. Curr Biol 14: 1405–1409 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A (2002) Rho GTPases in cell biology. Nature 420: 629–635 [DOI] [PubMed] [Google Scholar]
- Frank MJ, Cartwright HN, Smith LG (2003) Three Brick genes have distinct functions in a common pathway promoting polarized cell division and cell morphogenesis in the maize leaf epidermis. Development 130: 753–762 [DOI] [PubMed] [Google Scholar]
- Frank MJ, Smith LG (2002) A small, novel protein highly conserved in plants and animals promotes the polarized growth and division of maize leaf epidermal cells. Curr Biol 12: 849–853 [DOI] [PubMed] [Google Scholar]
- Fu Y, Li H, Yang Z (2002) The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14: 777–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs HN, Pollard TD (2001) Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu Rev Biochem 70: 649–676 [DOI] [PubMed] [Google Scholar]
- Huelskamp M, Misera S, Juergens G (1994) Genetic dissection of trichome cell development in Arabidopsis. Cell 76: 555–566 [DOI] [PubMed] [Google Scholar]
- Hummel T, Leifker K, Klambt C (2000) The Drosophila HEM-2/NAP1 homolog KETTE controls axonal pathfinding and cytoskeletal organization. Genes Dev 14: 863–873 [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T, Allwood EG, Anthony R, Voigt B, Menzel D, Hussey PJ (2004) The actin-interacting protein AIP1 is essential for actin organization and plant development. Curr Biol 14: 145–149 [DOI] [PubMed] [Google Scholar]
- Kunda P, Craig G, Dominguez V, Baum B (2003) Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr Biol 13: 1867–1875 [DOI] [PubMed] [Google Scholar]
- Le J, El-Assal Sel D, Basu D, Saad ME, Szymanski DB (2003) Requirements for Arabidopsis ATARP2 and ATARP3 during epidermal development. Curr Biol 13: 1341–1347 [DOI] [PubMed] [Google Scholar]
- Li H, Shen JJ, Zheng ZL, Lin Y, Yang Z (2001) The Rop GTPase switch controls multiple developmental processes in Arabidopsis. Plant Physiol 126: 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Blanchoin L, Yang Z, Lord EM (2003) The putative Arabidopsis arp2/3 complex controls leaf cell morphogenesis. Plant Physiol 132: 2034–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Qian Q, Zhou Y, Yan M, Sun L, Zhang M, Fu Z, Wang Y, Han B, Pang X, et al (2003) BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15: 2020–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Chua NH (2000) Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant Cell 12: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kernebeck B, Hulskamp M (2003. a) Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell 15: 1632–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kirik V, Kernebeck B, Srinivas BP, Hulskamp M (2003. b) Arabidopsis CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region specific fine F-actin formation. Development 130: 3137–3146 [DOI] [PubMed] [Google Scholar]
- Mullins RD (2000) How WASP-family proteins and the Arp2/3 complex convert intracellular signals into cytoskeletal structures. Curr Opin Cell Biol 12: 91–96 [DOI] [PubMed] [Google Scholar]
- Mutondo M, Zimmerman I, Saedler H, Huelskamp M (2004) The Arabidopsis KLUNKER gene encodes a putative regulator of the ARP2/3 complex (abstract no. T03-039). In 15th International Conference Arabidopsis Research, July 11–14, 2004, Berlin
- Qian Q, Li YH, Zeng D, Teng S, Wang Z, Li X, Dong Z, Dai N, Sun L, Li J (2001) Isolation and genetic characterization of a fragile plant mutant rice (Oryza sativa L.). Chin Sci Bull 46: 2082–2085 [Google Scholar]
- Qiu JL, Jilk R, Marks MD, Szymanski DB (2002) The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell 14: 101–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Christensen HE, Ishimaru Y, Dong CH, Chao-Ming W, Cleary AL, Chua NH (2000) Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol 124: 1637–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Wiedemann U, Stuurman N, Vale RD (2003) Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J Cell Biol 162: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW (2001) Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J 26: 421–433 [DOI] [PubMed] [Google Scholar]
- Schumacher K, Vafeados D, McCarthy M, Sze H, Wilkins T, Chory J (1999) The Arabidopsis det3 mutant reveals a central role for the vacuolar H(+)-ATPase in plant growth and development. Genes Dev 13: 3259–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LG, Li R (2004) Actin polymerization: riding the wave. Curr Biol 14: R109–R111 [PubMed] [Google Scholar]
- Soto MC, Qadota H, Kasuya K, Inoue M, Tsuboi D, Mello CC, Kaibuchi K (2002) The GEX-2 and GEX-3 proteins are required for tissue morphogenesis and cell migrations in C. elegans. Genes Dev 16: 620–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stradal TE, Rottner K, Disanza A, Confalonieri S, Innocenti M, Scita G (2004) Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol 14: 303–311 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Marks MD, Wick SM (1999) Organized F-actin is essential for normal trichome morphogenesis in Arabidopsis. Plant Cell 11: 2331–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vantard M, Blanchoin L (2002) Actin polymerization processes in plant cells. Curr Opin Plant Biol 5: 502–506 [DOI] [PubMed] [Google Scholar]
- Volkmann D, Baluska F (1999) Actin cytoskeleton in plants: from transport networks to signaling networks. Microsc Res Tech 47: 135–154 [DOI] [PubMed] [Google Scholar]
- Wasteneys GO, Galway ME (2003) Remodeling the cytoskeleton for growth and form: an overview with some new views. Annu Rev Plant Biol 54: 691–722 [DOI] [PubMed] [Google Scholar]