Abstract
Embryonic regulators LEC2 (LEAFY COTYLEDON2) and FUS3 (FUSCA3) are involved in multiple aspects of Arabidopsis (Arabidopsis thaliana) seed development, including repression of leaf traits and premature germination and activation of seed storage protein genes. In this study, we show that gibberellin (GA) hormone biosynthesis is regulated by LEC2 and FUS3 pathways. The level of bioactive GAs is increased in immature seeds of lec2 and fus3 mutants relative to wild-type level. In addition, we show that the formation of ectopic trichome cells on lec2 and fus3 embryos is a GA-dependent process as in true leaves, suggesting that the GA pathway is misactivated in embryonic mutants. We next demonstrate that the GA-biosynthesis gene AtGA3ox2, which encodes the key enzyme AtGA3ox2 that catalyzes the conversion of inactive to bioactive GAs, is ectopically activated in embryos of the two mutants. Interestingly, both β-glucuronidase reporter gene expression and in situ hybridization indicate that FUS3 represses AtGA3ox2 expression mainly in epidermal cells of embryo axis, which is distinct from AtGA3ox2 pattern at germination. Finally, we show that the FUS3 protein physically interacts with two RY elements (CATGCATG) present in the AtGA3ox2 promoter. This work suggests that GA biosynthesis is directly controlled by embryonic regulators during Arabidopsis embryonic development.
Higher plant embryogenesis is divided into two major phases: embryo development (or morphogenesis) and seed maturation (West and Harada, 1993). During embryo development, early morphogenetic processes occur that give rise to embryonic cell types, tissues, and organs. During seed maturation, the fully developed embryo undergoes maturation, during which food reserves accumulate and dormancy and desiccation tolerance develop.
Seed development has been extensively studied in Arabidopsis (Arabidopsis thaliana) using mutants defective either in morphogenesis, such as GNOM (Mayer et al., 1991; Shevell et al., 1994) or KNOLLE (Mayer et al., 1991; Lukowitz et al., 1996), or in maturation, such as ABI (ABSCISIC ACID-INSENSITIVE) loci that were initially identified on the basis of the abscisic acid (ABA) hormone-resistant germination of mutants at these loci (Koornneef et al., 1984; Giraudat et al., 1992; Finkelstein et al., 1998; Finkelstein and Lynch, 2000). A particular set of mutants exhibiting the lec phenotype, which consists of a partial transformation of cotyledons into leaves, has allowed the identification of an important network of regulatory genes. The LEC1 (LEAFY COTYLEDON1; Meinke, 1992), LEC2 (Meinke et al., 1994), and FUS3 (FUSCA3; Keith et al., 1994; Meinke et al., 1994) genes, which are defined as the LEC genes hereafter, are the only known regulators required for normal development during both the morphogenesis and the maturation phases (Holdsworth et al., 1999; Harada, 2001). They are required, for instance, during the morphogenesis phase to specify cotyledon identity and during the maturation phase to inhibit precocious germination and anthocyanin accumulation. Furthermore, ectopic expression of LEC1 or LEC2 is sufficient to induce somatic embryogenesis from vegetative cells (Lotan et al., 1998; Stone et al., 2001), indicating that these genes are central regulators that act far upstream in the control of embryogenesis. The LEC1 protein shares significant identity with the HAP3 subunit of the CCAAT-binding transcription factors (CBFs, also called NF-Ys; Lotan et al., 1998; Lee and Schiefelbein, 2002). The LEC2 and FUS3 proteins belong to the same transcription factor family and share a conserved B3 domain, which is a DNA-binding motif unique to plants and is essential for the regulation of seed maturation genes (Giraudat et al., 1992; Luerssen et al., 1998; Stone et al., 2001). Both LEC2 and FUS3 proteins bind the so-called RY motif present in seed-specific promoters, as recently shown for the storage protein gene At2S3 (Reidt et al., 2000; Kroj et al., 2003).
The phenotypes of lec mutants are similar in several ways to abi mutant phenotype, especially mutants of ABI3, which has the most pleiotropic effects on seed maturation, regulating sensitivity to ABA inhibition of germination, desiccation tolerance, and dormancy. Genetic studies based on double mutant analysis indicate that LEC and ABI3 genes have complementary regulatory roles and act synergistically to control multiple processes during seed development (Baumlein et al., 1994; Keith et al., 1994; Meinke et al., 1994; West et al., 1994; Parcy et al., 1997; Nambara et al., 2000; Raz et al., 2001). The synergistic interaction is particularly well illustrated by ABA sensitivity at germination; while monogenic fus3 mutants are ABA sensitive and weak monogenic abi3 alleles are ABA resistant, fus3 abi3 double mutants are extremely resistant to exogenous ABA. While ABA establishes dormancy during seed development, the phytohormone gibberellic acids (GAs) break dormancy and induce germination of dry seed. The current theory of ABA/GAs antagonism in seed germination is based on experiments showing that ABA-deficient and ABA-insensitive mutants rescue the germination of the ga1 GA-deficient mutant and of seeds treated with GA biosynthetic inhibitor (Koornneef et al., 1982; Nambara et al., 1991; Leon-Kloosterziel et al., 1996). In maize (Zea mays), the ABA/GAs balance also governs seed development, as GA deficiency early in seed development suppresses precocious germination in ABA-deficient developing kernels and bioactive GA species accumulate prior to the peak in ABA content (White et al., 2000). While ABA biosynthesis is regulated both maternally and zygotically, the control of GA biosynthesis during seed development is largely unknown. Because seed development in Arabidopsis requires two processes—one that is regulated by LEC genes and a second that requires ABA (Raz et al., 2001)—and because trichome formation, a GA-dependent process on leaves (Perazza et al., 1998), is observed on lec embryos, we investigated whether the GA pathway was misregulated in lec mutants.
This study aimed to determine whether LEC2 and FUS3 are involved in the control of GA biosynthesis during Arabidopsis embryogenesis. We show here that levels of bioactive GAs are elevated in young developing seeds of lec2 and fus3 mutants relative to wild-type levels and that GA-dependent processes, such as trichome formation or expression of the GA target gene GL1 (GLABROUS1), occur in lec2 and fus3 embryos. We demonstrate that among a large set of GA biosynthesis genes, the AtGA3ox2 gene, which encodes the key enzyme AtGA3ox2 that catalyzes the ultimate step of bioactive GA biosynthesis, is transcriptionally up-regulated in embryonic mutants. Finally, we show that the FUS3 protein directly binds the two RY target sites present in the promoter of AtGA3ox2, suggesting a direct transcriptional regulation. This new implication of LEC2 and FUS3 genes in the control of GA biosynthesis sheds new light on their role during embryogenesis.
RESULTS
Levels of Bioactive GAs Are Altered in lec2-1 and fus3-8 Mutants
In Arabidopsis, bioactive GAs present in vegetative organs and at germination are GA1 and GA4, with GA4 being the most abundant and active form (Talon et al., 1990; Xu et al., 1999; Ogawa et al., 2003). In wild-type developing seeds, the levels of bioactive GAs had not been reported previously, presumably because the amount of material is limiting. To determine whether lec2 and fus3 mutants displayed abnormal bioactive GA levels, immature seeds were collected from wild-type and mutant developing siliques at 8 to 10 days after pollination (DAP), a stage at which both LEC2 and FUS3 are expressed (Kroj et al., 2003) and GA dosage was applied (Table I). In contrast to vegetative organs, the level of bioactive GA1 in wild-type immature seeds was similar to the level of bioactive GA4. In lec2-1 immature seeds, GA1 and GA4 levels were increased 4-fold and 1.4-fold, respectively, relative to wild-type levels. In fus3-8, the GA1 level was increased 1.4-fold while the GA4 level increased about 2-fold.
Table I.
GA levels in immature seeds of wild-type, lec2-1, and fus3-8 mutants
| Wild Type | lec2-1 | fus3-8 | |
|---|---|---|---|
| GA1 | 5.41 ± 0.35 | 25.70 ± 3.27 | 7.73 ± 0.41 |
| GA4 | 4.86 ± 0.44 | 7.21 ± 0.33 | 11.04 ± 0.30 |
Immature seeds were isolated from immature siliques at 8 to 10 DAP and levels of bioactive GA1 and GA4 were determined. Three independent assays from three biological samples collected independently were performed for each genotype. Small numbers represent sd. GA levels are in picograms per gram of fresh weight.
GAs Are Required for Ectopic Trichome Formation in lec2-1 and fus3-8 Mutants
Two genes are required for trichome initiation on true leaves: TTG1 (TRANSPARENT TESTA GLABRA1) and GL1, the latter being transcriptionally up-regulated by GAs (Perazza et al., 1998). Recently, it has been shown that FUS3 represses TTG1 expression in the epidermis of embryos (Tsuchiya et al., 2004). To determine whether the regulation of trichome development by LEC2 and FUS3 pathways is a GA-dependent process, we analyzed ectopic trichome formation on double mutants between the severe GA auxotroph mutant ga1-3 and lec mutants. Because GA deficiency impairs premature germination of double mutants (Raz et al., 2001), embryos were dissected out of immature green seeds and allowed to grow into seedlings. While single mutants had from 8 to 9 trichomes/cotyledon, double mutants with ga1-3 showed a complete suppression of ectopic trichome formation (Fig. 1A), suggesting that the GA pathway was abnormally activated during mutant embryogenesis. GL1, a myb gene required for trichome initiation, is a molecular marker of the pathway (Perazza et al., 1998). To determine whether GL1 was ectopically expressed during mutant embryogenesis, the β-glucuronidase (GUS) reporter gene under control of GL1 cis-regulatory elements (Larkin et al., 1993) was introduced into lec2-1 and fus3-8 mutant backgrounds. While wild-type embryos never showed GUS activity (Fig. 1B), developing embryos of lec2-1 and fus3-8 showed GUS staining of ectopic trichome primordia (Fig. 1B). Finally, real-time reverse transcription (RT)-PCR was used to show that GL1 transcript level was higher (50–60-fold) in both mutants (data not shown).
Figure 1.
GA-dependent trichome formation on mutant seedlings and PGL1∷GUS expression in mutant embryos. In A, immature seeds of single mutants at 11 to 13 DAP were isolated, sown in petri dishes on Murashige and Skoog medium, and grown for 10 d apart from wild type that was germinated from dry seeds. Immature embryos 11 to 13 DAP of double mutants and ga1-3 were dissected out of immature seeds, allowed to grow like single mutants, and the trichomes of 20 plants were counted for each genotype. Section B shows the GUS activity of the PGL1∷GUS construct in wild-type, lec2-1, and fus3-8 mutant embryos. Seeds were removed from immature siliques at 11 to 13 DAP, incubated with X-gluc, and embryos were dissected out of seeds.
We concluded that ectopic trichome formation is a GA-dependent process as in true leaves (Perazza et al., 1998), suggesting that the GA pathway is misactivated during embryogenesis of lec2 and fus3 mutants.
The GA-Biosynthetic Gene AtGA3ox2 Is Ectopically Expressed in lec2-1 and fus3-8 Mutants
The above analysis suggested that GA biosynthesis was abnormally induced during embryogenesis of lec2-1 and fus3-8 mutants. In Arabidopsis, the biosynthesis pathway of GAs converts the geranylgeranyl diphosphate to C20- and C19-GA molecules (Fig. 2A). The genes encoding most of the enzymes involved in GA biosynthesis have been isolated (Hedden and Phillips, 2000; Olszewski et al., 2002; O'Neill and Ross, 2002; Ogawa et al., 2003). We analyzed the expression levels of 12 GA-biosynthetic genes in siliques of wild-type, lec2-1, and fus3-8 mutants using real-time RT-PCR (see “Materials and Methods” for a complete list). The majority of these genes displayed no significant modification of their transcript level, as shown for AtCPS, AtGA20ox1, and AtGA3ox1, three genes playing a role in major biosynthesis steps and whose developmental regulations are well known (Silverstone et al., 1997; Meier et al., 2001; Yamaguchi et al., 2001; Fig. 2B). In contrast, the AtGA3ox2 gene showed a transcript level that was strongly up-regulated in mutants relative to wild type (Fig. 2B). AtGA3ox2 encodes AtGA3ox2, a key enzyme that catalyses the ultimate step of GA biosynthesis leading to the production of bioactive GA1 and GA4 molecules (Fig. 2A). AtGA3ox2 is not expressed in wild-type siliques and is not subjected to feedback regulation by bioactive GAs, unlike AtGA3ox1 or AtGA20ox1 (Yamaguchi et al., 1998, 2001). In lec2-1 and fus3-8 developing seeds, AtGA3ox2 transcript level was increased between 10- to 40-fold, with fus3-8 showing the highest levels of derepression.
Figure 2.
The major GA biosynthesis pathway in Arabidopsis and expression of four well-known biosynthetic genes in wild-type, lec2-1, and fus3-8 mutant siliques. Section A describes the main steps of GA biosynthesis. The first committed step in the GA pathway is the biosynthesis of ent-kaurene by AtCPS and AtKS (not represented). AtGA20oxidases catalyze the biosynthesis of inactive substrates GA9 and GA20 (C19-GAs) from GA12 molecule (C20-GA). AtGA3ox1 and AtGA3ox2 catalyze the biosynthesis of the two bioactive forms in Arabidopsis, GA1, and GA4. Multiple arrows indicate multiple biosynthesis steps while single arrows indicate direct synthesis. Section B shows the transcript levels of AtCPS, AtGA20ox1, AtGA3ox1, and AtGA3ox2 in wild-type (white bars), lec2-1 (black bars), and fus3-8 (dashed bars) mutants. Real-time RT-PCR was performed from total RNAs isolated from 10 to 11 DAP siliques. Because AtGA20ox1 is weakly expressed in all backgrounds, the threshold cycle was close to the ultimate cycle (Ct close to 40) in all genetic backgrounds. We considered the putative activation of AtGA20ox1 not reliable in these experimental conditions. Error bars represent sd. Three independent replicates were done from three set of seeds collected independently.
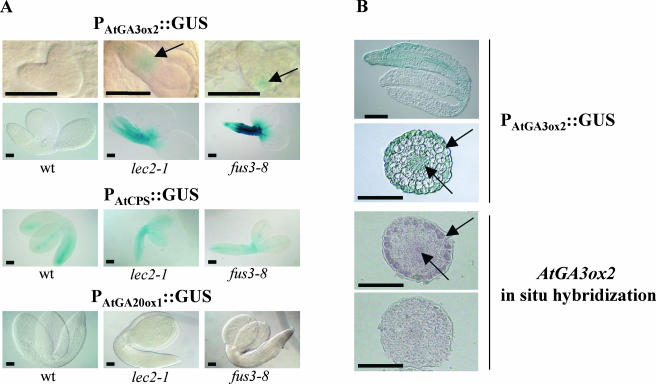
To study the misexpression of AtGA3ox2 during mutant embryogenesis in more detail, an AtGA3ox2∷GUS reporter line was crossed to lec2-1 and fus3-8 mutants. In wild type, this translational fusion is expressed only in germinating seeds and shows a GUS expression pattern in the cortex and endodermis of the embryo axis (Yamaguchi et al., 1998, 2001). As shown in Figure 3A, no GUS activity was observed in developing wild-type seeds or in embryos at any stage of development. In contrast, GUS activity was observed in the lec2-1 and fus3-8 mutant embryos as early as the torpedo-stage embryo in lec2-1 and the heart-stage embryo in fus3-8 (Fig. 3A). The staining was localized predominantly at the basal pole of the embryo in a region that corresponds to the precursor to the root cortex initials and the central region of the root cap (West and Harada, 1993). Later in development, the staining was visible along the axis of lec2-1 and fus3-8 embryos and persisted until the end of seed development (Fig. 3A). In both mutants, the staining often reached the base of cotyledons, a region where trichomes are initiated in true leaves (Lloyd et al., 1994; Szymanski et al., 2000). As reported recently for the At2S3 gene expression, the pattern and extent of staining varied between embryos from the same siliques (Kroj et al., 2003).
Figure 3.
Expression patterns of three biosynthetic genes in wild-type, lec2-1, and fus3-8 mutant embryos. Section A shows the GUS reporter gene expression patterns in wild-type, lec2-1, and fus3-8 mutants of (first row) AtGA3ox2 in heart-stage or torpedo-stage embryos, (second row) AtGA3ox2 in mature embryos, (third row) AtCPS in mature embryos, and (fourth row) AtGA20ox1 in mature embryos. Section B shows the GUS reporter gene and in situ hybridization of AtGA3ox2 expression in sections of fus3-8 mutant embryos. Two upper images, longitudinal section (top image), and transverse section of the embryo axis (bottom image) of PAtGA3ox2∷GUS fus3-8 embryos. Arrows on the transverse section indicate staining in epidermal cells and vascular tissues. Two lower images, transverse sections of fus3-8 embryo axis hybridized with the antisense (top image) and sense (bottom image) AtGA3ox2 probe. Arrows on the top image indicate hybridization signal in epidermal cells and vascular tissues.
We verified that this misexpression was specific for AtGA3ox2 reporter by monitoring the expression of other GUS constructs. Transgenic reporter lines that carry the GUS gene under control of AtCPS or AtGA20ox1 cis-regulatory elements were crossed to lec2-1 and fus3-8 mutants. AtCPS (GA1 gene) is expressed mainly in the provasculature of the wild-type embryo, as shown on Figure 3A (Silverstone et al., 1997; Yamaguchi et al., 2001). A GUS staining pattern similar to wild type was observed in all mutants and wild type (Fig. 3A). AtGA20ox1 is not expressed during wild-type embryogenesis as shown on Figure 3A (Xu et al., 1999; Meier et al., 2001). In lec2-1 and fus3-8 mutants, the activity of the AtGA20ox1 transcriptional GUS fusion was also undetectable (Fig. 3A), in agreement with our real-time RT-PCR data above.
We analyzed in more detail the expression pattern of AtGA3ox2 in sections of fus3-8 embryos (Fig. 3B). Surprisingly, the GUS staining in the embryo axis of PAtGA3ox2∷GUS fus3-8 lines was observed mainly in the epidermis and vascular tissues but not in the cortex and endodermis as reported at germination (Yamaguchi et al., 1998, 2001). To confirm this observation, in situ hybridization was performed on fus3-8 embryos using a specific AtGA3ox2 antisense probe (Yamaguchi et al., 2001). The observed signal was restricted essentially to the epidermis and to a lesser extent to vascular tissues, confirming GUS observations.
Taken together, these results show that AtGA3ox2 is transcriptionally regulated by LEC2 and FUS3 pathways.
The FUS3 Protein Physically Interacts with RY Motifs Present in the AtGA3ox2 Promoter
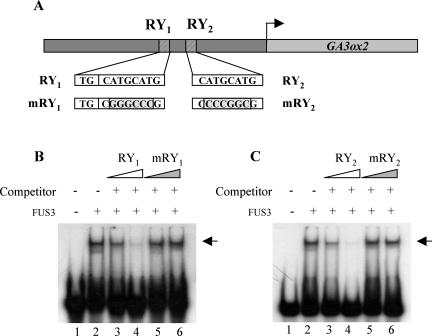
The FUS3 protein binds to the RY element CATGCATG present in promoters of several seed maturation genes (Reidt et al., 2000; Kroj et al., 2003). Two perfectly conserved RY sites, RY1 (TGCATGCATG) and RY2 (CATGCATG), are present in the promoter of the AtGA3ox2 gene (Fig. 4A). To determine whether FUS3 physically interacts with these two putative target sites, we performed electromobility shift assay with radiolabeled probes carrying RY1 and RY2 (Fig. 4, B and C, respectively). FUS3 protein present in bacterial extracts was able to shift radiolabeled wild-type probes (Fig. 4, B [lane 2] and C [lane 2]). The binding was competed away with large amounts of unlabeled wild-type probes (Fig. 4, B [lanes 3 and 4] and C [lanes 3 and 4]) but not with large amounts of probes carrying mutated RY sites (Fig. 4, B [lanes 5 and 6] and C [lanes 5 and 6]). In the same conditions, no gel retardation of the wild-type radiolabeled probes was observed with the LEC2 protein as described previously with the At2S3 promoter (data not shown; Kroj et al., 2003).
Figure 4.
Direct interaction of the FUS3 protein with the two RY elements present in the AtGA3ox2 promoter. Section A is a schematic representation of the two RY sites in the AtGA3ox2 promoter. The sequence of the two wild-type sites (RY1 and RY2) is indicated together with the two mutated versions (mRY1 and mRY2). Section B shows an electrophoretic mobility shift assay experiment with RY1 site. A radiolabeled probe carrying the RY1 site (TGCATGCATG) was incubated with the FUS3 protein present in bacterial extracts. Lane 1, radiolabeled probe only; lane 2, rabiolabeled probe incubated with FUS3 extracts; lanes 3 and 4, radiolabeled probe incubated with FUS3 extracts after incubation with an excess of 100- and 500-fold, respectively, of unlabeled probe; lanes 5 and 6, same as in lanes 3 and 4 except that the unlabeled probe carried a mutated RY1 site (mRY1). Section C is similar to section B except that the radiolabeled probe (lane 1) carried the RY2 site (CATGCATG). The probe is retarded by FUS3 extracts (lane 2) and competition experiments were performed with an unlabeled probe carrying the native (lanes 3 and 4) or mutated (lanes 5 and 6) RY2 site (mRY2). The arrows indicate FUS3-RY complexes.
We concluded that FUS3 specifically interacts with RY sites present in the AtGA3ox2 promoter, suggesting a direct transcriptional regulation of AtGA3ox2 by FUS3.
DISCUSSION
Embryonic regulators LEC2 and FUS3 are both involved in the regulation of higher plant embryogenesis. Our biochemical and molecular analysis of GA biosynthesis in immature seeds indicates that GA biosynthesis is misactivated in lec2 and fus3 mutants relative to wild type.
Bioactive GAs during Seed Development
Our biochemical data indicate that the level of bioactive GA1 is similar to or higher than the level of bioactive GA4 during wild-type embryogenesis at 8 to 10 DAP. In mature wild-type seeds (12–13 DAP), we have also consistently detected a GA1:GA4 ratio of 10:1 (data not shown) in contrast to vegetative development or at germination where this ratio is about 1:10 (Talon et al., 1990; Xu et al., 1999; Ogawa et al., 2003). This is the first report, to our knowledge, of a specific developmental process where GA1 is the predominant bioactive GA in Arabidopsis, although we cannot exclude that GA4 is more active than GA1 during embryogenesis as it is during vegetative development. Ogawa et al. (2003) have recently shown that GA4, but not GA1, is the major bioactive GA during germination. The switch from one pathway to the other is not known. Nevertheless, our data suggest that this switch occurs at some point between late seed development and germination.
Elevated GA1 levels were observed in lec2-1, while elevated GA4 levels were observed in fus3-8. As the two forms differ by a 13-β-hydroxylation, the simplest hypothesis to explain this observation is that the 13-β-hydroxylase that is encoded by an unknown gene is differentially regulated by LEC2 and FUS3 pathways.
Premature Germination versus Germination
Premature germination can be seen as a simple temporal shift of germination events during seed development. Our data indicate that this is not true for GA biosynthesic genes. In wild-type germination, the expression of several GA biosynthetic genes such as AtGA3ox1, AtGA3ox2, or AtGA20ox1 is detected, while genes involved in GA catabolism are not expressed (Ogawa et al., 2003). In contrast, the regulation of only AtGA3ox2 is affected in lec2 and fus3 mutants, and we have observed that the AtGA2ox3 gene that encodes a deactivating enzyme responsible for bioactive GA catabolism (Thomas et al., 1999) is strongly induced in 12 to 13 DAP seeds of lec2 and fus3 mutants (data not shown). Furthermore, we show that AtGA3ox2 expression pattern in fus3-8 does not overlap at all with the expression pattern at germination. Finally, premature germination of the lec1 mutant has been shown to be GA independent (Raz et al., 2001), and we have also observed that ectopic trichome formation on lec1-3 ga1-3 double mutants is not abolished (data not shown). As far as GA biosynthesis is concerned, this suggests that premature germination is distinct from germination and that LEC and FUS3 pathway act specifically on AtGA3ox2.
Strong synergistic interactions between LEC genes and ABI loci have been described, particularly in response to ABA at germination (Baumlein et al., 1994; Keith et al., 1994; Meinke et al., 1994; West et al., 1994; Parcy et al., 1997; Nambara et al., 2000; Brocard-Gifford et al., 2003). This study strongly suggests that this synergistic interaction is due to the disruption of the GA/ABA balance during embryogenesis with the GA pathway controlled by LEC genes and the ABA pathway controlled by ABI genes. Similarly, the PKL (PICKLE) gene has been proposed to be a component of a GA-modulated developmental switch that functions during germination to prevent reexpression of the embryonic developmental state (Ogas et al., 1999; Dean Rider et al., 2003). The penetrance of the pkl phenotype that consists of primary root meristems retaining characteristics of embryonic tissue is strongly enhanced by low concentrations of inhibitors of gibberellin biosynthesis. Expression of this aberrant differentiation state is suppressed by exogenous GAs (Ogas et al., 1997). PKL, which encodes a chromatin-remodeling factor, is necessary for repression of LEC1, LEC2, and FUS3 (Ogas et al., 1999; Dean Rider et al., 2003). Our results suggest that pkl mutants have lower GA levels relative to wild-type levels because of an increased repression of GA biosynthesis by LEC genes.
Regulation of AtGA3ox2 Expression by FUS3
The derepression of AtGA3ox2 is observed in both fus3-8 mutant and lec2-1 mutants. Derepression in lec2 might be a consequence of FUS3 down-regulation in lec2 as recently described (Kroj et al., 2003). Therefore, it is possible that AtGA3ox2 repression is a primary action of FUS3. Our data support the possibility that this action is direct, as we show that FUS3 binds specifically to RY elements present in the promoter of the AtGA3ox2 gene. Alternatively, we cannot exclude the fact that both FUS3 and LEC2 are needed to repress AtGA3ox2 gene expression. In both cases, AtGA3ox2 would be the first direct target gene of B3-domain proteins to be described, although several genes are known to be down-regulated by ABI3 and FUS3 (Nambara et al., 2000). It is intriguing that AtGA3ox2 is derepressed specifically in epidermal cells of fus3 embryo. In contrast, AtGA3ox2 is not expressed in epidermal cells at germination but in cortex and endodermis (Yamaguchi et al., 2001). Therefore, it is possible that distinct cis-regulatory elements within the promoter control the regulation of AtGA3ox2 during embryogenesis and at germination. It has recently been shown that FUS3 is expressed specifically in epidermal cells of young embryos, where it represses the expression of TTG1 (Tsuchiya et al., 2004). Moreover, expression of FUS3 under control of the epidermis-specific AtML1 promoter is sufficient to fully suppress fus3 mutant phenotype, indicating a noncell autonomous action of FUS3 during embryogenesis (Tsuchiya et al., 2004). It will be of interest to place the AtGA3ox2 gene under control of the AtML1 promoter to determine to what extent GA biosynthesis participates to the fus3 phenotype.
In conclusion, we have shown that LEC2 and FUS3 pathways repress AtGA3ox2 expression during embryogenesis. While previous work has shown that GAs are required for plant embryogenesis (Singh et al., 2002), this study shows the necessity for plants to down-regulate GA biosynthesis in embryos. The fine-tuning of GA biosynthesis regulation is likely to involve cross-talk between other plant hormone pathways such as ABA, ethylene, or auxin that also play a role in embryogenesis. Future work will be required to understand the precise molecular mechanisms underlying these complex signaling networks during embryogenesis.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) ecotype Landsberg erecta (Ler) was used as the wild type. The ga1-3 mutant (Koornneef et al., 1983) is in the Ler background. lec2-1 and fus3-8 were initially in the Wassilewskija and Columbia backgrounds, respectively, and were backcrossed to Ler as described (Raz et al., 2001). Wild-type seeds were planted on soil or surface sterilized and grown in petri dishes on Murashige and Skoog for Arabidopsis (MSAR) medium (Koncz et al., 1990).
lec2-1 and fus3-8 mutants were maintained by isolating immature seeds out of immature siliques at 10 to 13 DAP and sowing in petri dishes on MSAR medium and later on soil. Plants were grown at 22°C under a photoperiod of 16 h of light/8 h of dark.
Identification of Mutants with GUS Reporter Genes
Homozygous lec2-1 and fus3-8 mutants were crossed to AtCPS, AtGA20ox1, AtGA3ox2, or GL1 GUS homozygous reporter lines. The PAtGA3ox2∷GUS reporter line used in this study is the TL line described in Yamaguchi et al. (2001). F3 families that segregated for the lec phenotype were surface sterilized and grown in petri dishes on MSAR medium supplemented with kanamycin (50 μg mL−1) to identify lines homozygous for the reporter gene construct. Immature F4 seeds with the lec phenotype were identified and allowed to germinate precociously. F5 seeds were incubated for 8 to 10 h with GUS substrate in GUS buffer (0.1 m phosphate sodium, pH 7.5, 2 mm X-Gluc, 10 mm EDTA, 10 mm, 0.2% [v/v] Triton X-100, 0.1 mm potassium ferricyanide) and destained as described (Gallagher, 1992; Kroj et al., 2003). Staining of the PAtGA3ox2∷GUS reporter line in both mutant and wild-type backgrounds was shorter (6 h) as compared to other GUS reporter lines (10 h) because the expression of AtGA3ox2 is strong in lec2-1 and fus3-8 backgrounds.
Reverse Transcription and Real-Time PCR
Total RNA was isolated from immature siliques at 11 to 13 DAP using the RNeasy plant kit (Qiagen, Valencia, CA). RT was performed using random hexamers and reverse transcriptase (Applied Biosystem, Sunnyvale, CA) and PCRs were performed according to the manufacturer on the ABI 5700-SDS (Applied Biosystem). Relative transcript levels of biosynthesis were calculated with the ΔΔCt method (Applied Biosystem) using the APT1 (adenine phosphoribosyltransferase) gene (Cowling et al., 1998) or the EF-1α (Elongation Factor-1α) gene as the reference. Activation of a given gene was considered to be reliable when at least a 10-fold modification of transcript level was detected. Oligonucleotides were designed with the OligoExpress 1.5 software (Perkin Elmer, Foster City, CA) and are as shown in Table II.
Table II.
Primers used in real-time RT-PCR experiments
| Gene Name (Accession No.) | Forward Primer | Reverse Primer |
|---|---|---|
| Reference Genes | ||
| APT1 (At1g27450) | 5′-TGCAATCCGACTACTTGAACGA-3′ | 5′-CAAGCACATTCAACAATCTTCACTC-3′ |
| EF-1α (At5g60390) | 5′-CCCAGGCTGATTGTGCTGT-3′ | 5′-GGGTAGTGGCATCCATCTTGTT |
| Unique GA Biosynthetic Genes | ||
| AtCPS (At4g02780) | 5′-GCGGAAATCATCAATCGAATC-3′ | 5′-CCTTGCCTTTAAGTATTGGCGA-3′ |
| AtKS (At1g79460) | 5′-TGTCTTACGATCCGCTAAAACC-3′ | 5′-CGATTATTGCTTGCTCATGGC-3′ |
| AtKO1 (At5g25900) | 5′-TGAACGGTCTTTTGGGTGCTA-3′ | 5′-CTCTGTAATGTCTTTTTCGTTTCTGTG-3′ |
| AtKAO1 (At1g05160) | 5′-TCAATATTCCTGGATTTGCTTATCAT-3′ | 5′-GTGTTTTCCTCGCCTTGAGTG-3′ |
| AtKAO2 (At2g32440) | 5′-TCCTCAAATACCGGGTGGAA-3′ | 5′-CAAGAACATCACCGGACATCC-3′ |
| GA20oxidase Family | ||
| AtGA20ox1 (At4g25420) | 5′-CTTCCATCAACGTTCTCGAGC-3′ | 5′-GGTTTTGAAGGTCGATGAGAGG-3′ |
| AtGA20ox2 (At5g51810) | 5′-AGAAACCTTCCATTGACATTCCA-3′ | 5′-AGAGATCGATGAACGGGACG-3′ |
| AtGA20ox3 (At5g07200) | 5′-ACTCGTCTCAAAGGCTGCAAC-3′ | 5′-GAGGCTCTCATCGACACCATG-3′ |
| GA3oxidase Family | ||
| AtGA3ox1 (At1g15550) | 5′-GATCTCCTCTTCTCCGCTGCT-3′ | 5′-GAGGGATGTTTTCACCGGTG-3′ |
| AtGA3ox2 (At1g80340) | 5′-CTGCCGCTCATCGACCTC-3′ | 5′-AGCATGGCCCACAAGAGTG-3′ |
| AtGA3ox3 (At1g80330) | 5′-GATCACACCAAGTACTGCGGTATAA-3′ | 5′-TTCCATTTCGTCCACGTATTCTT-3′ |
| AtGA3ox4 (At4g21690) | 5′-CGCTACACTCTTATGGCCCG-3′ | 5′-TCCATCACATTGCAGAACTCG-3′ |
| GA2oxidase Family | ||
| AtGA2ox1 (At1g78440) | 5′-CCAAGTCTTCTCAAAAGCCCG-3′ | 5′-GTACTCTTCCAATGCGTTTCTGAA-3′ |
| AtGA2ox2 (At1g30040) | 5′-GGTTCCGGTTCTCACTTCCC-3′ | 5′-GGATCGGCTAGGTTGACGAC-3′ |
| AtGA2ox3 (At2g34555) | 5′-AGCCAGCCAGTTTTGATAGCA-3′ | 5′-GCGGTTTGCATTTTGGATTAAC-3′ |
| AtGA2ox4 (At1g02400) | 5′-GATCCTTTCAAGTTCAGCTCGG-3′ | 5′-TCTAACCGTGCGTATGTAATCATTC-3′ |
| GL1 (At3g27920) | 5′-AGCTCCTCGGCAATAGATGGT-3′ | 5′-TGTGGCGGCAGTGATGAA-3′ |
Measurement of GA Levels in Immature Seeds
Samples of immature seeds (20 mg) and 1 mL of extraction medium (80% MeOH) with 2H2-GAs (Prof. L. Mander, Canberra, Australia) as internal standards, were added to an Eppendorf tube. The extraction was performed using an MM 301 Vibration mill (Retsch GmbH & Co. KG, Haan, Germany) at a frequency of 30 Hz s−1 for 3 min after adding 3 mm tungsten carbide beads (Retsch GmbH & Co. KG) to each tube to increase the extraction efficiency. After extraction the samples were placed at 4°C for 2 h and then centrifuged in an Eppendorf centrifuge for 3 min at 14,000 rpm. The supernatants were evaporated to dryness in a speed-vac concentrator (Savant Instrument, Framingdale, NY). The samples were dissolved in 500 μL of water, pH 3.0, and applied to a preequilibrated 2-mL C8 500-mg Oasis (Waters, Milford, MA) SPE-column. After elution with 2 mL 80% MeOH, the samples were evaporated to dryness, methylated with diazomethane, purified further with HPLC, and finally analyzed by gas chromatography/mass spectrometry-selected reaction monitoring (JMS-MStation 700, JEOL, Tokyo) as described earlier (Peng et al., 1999). The sensitivity was in the range of 100 fg. The amount of internal standard added to the extract was in the range of 50 to 100 pg.
Electrophoretic Mobility Shift Assay
pGEX expression vectors carrying FUS3 and LEC2 cDNAs were kindly provided by Dr. Jesus Vincente Carbajosa (Madrid). Protein extracts were prepared as follow. Bacteria were grown in 20 mL of Luria-Bertani medium supplemented with ampicilin (100 μg mL−1) to an O.D.600 of 0.8 to 1. FUS3 and LEC2 protein synthesis was induced by 0.5 mm of isopropylthio-β-galactoside during 3 h at 30°C. Cells were washed twice with 10 mL of washing buffer (100 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, and 10% [v/v] glycerol) and resuspended in 1.5 mL of binding buffer (50 mm KCl, 10 mm HEPES, pH 7.9, 1 mm dithiothreitol, and 1 mm MgCl2). Fractions of 0.2 mL were sonicated 5 × 10 s and centrifuged 20 min at 12,000g. Aliquots of the supernatant were stored at −80°C. The labeling of the probes was performed using oligonucleotides encompassing the AtGA3ox2 RY sites as already described (Kroj et al., 2003). Oligonucleotide sequences are available upon request. The binding reaction was performed in 20 μL at room temperature for 20 min in binding buffer with 1 nm of radiolabeled probe, 0.5 to 1 μL of bacterial supernatant, and 1 to 5 μg of poly (dA.dT) (Pharmacia, Piscataway, NJ) as nonspecific competitor. Loading and migration was performed as already described (Kroj et al., 2003).
In Situ Hybridization
AtGA3ox2 probe has been described previously (Yamaguchi et al., 1998; Yamaguchi et al., 2001). The probe was amplified by PCR from Arabidopsis genomic DNA and cloned into the pSPT19 vector (Roche Diagnostics, Meylan, France). To synthesize digoxygenin labeled probe, in vitro transcription reactions were carried out as recommended by the manufacturer (Roche). Embryos were fixed in 4% paraformaldehyde solution for 10 min at room temperature, transferred in 40% saccharose for 1 h, and frozen in Polyfreeze solution (Polysciences, Warrington, PA) in liquid nitrogen. Sectioning (8 μm) was done at −20°C. Hybridization was done in hybridization buffer according to the manufacturer's protocol (Roche). Localization of the digoxygenin-labeled probe was immunologically detected using alkaline phosphatase-conjugated antidigoxygenin antisera (Roche).
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all parts of the material. Obtaining any permission will be the responsibility of the requestor.
Acknowledgments
We thank Dr. Tai-ping Sun (Duke University) for sending AtGA3ox2 and AtCPS reporter lines and Dr. John Mundy (Copenhagen) for sending the AtGA20ox1 reporter line. We thank Dr. Jesus Vincente Carbajosa (Madrid) for the gift of pGEX-FUS3 and pGEX-LEC2 expression vectors and Ingabritt Carlsson for technical help with the GA analysis. We also thank Jean-Marc Bonneville for critical reading of the manuscript. We thank Nicole Potier and Eliane Charpentier for their technical assistance.
This work was supported by the Région Rhône-Alpes (through the Programme Thématiques Prioritaires [2000–2003] and a doctoral fellowship to J.C.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.047266.
References
- Baumlein H, Misera S, Luerssen H, Kolle K, Horstmann C, Wobus U, Muller AJ (1994) The FUS3 gene of Arabidopsis thaliana is a regulator of gene expression during late embryogenesis. Plant J 6: 379–387 [Google Scholar]
- Brocard-Gifford IM, Lynch TJ, Finkelstein RR (2003) Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol 131: 78–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling RJ, Kamiya Y, Seto H, Harberd NP (1998) Gibberellin dose-response regulation of GA4 gene transcript levels in Arabidopsis. Plant Physiol 117: 1195–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean Rider S Jr, Henderson JT, Jerome RE, Edenberg HJ, Romero-Severson J, Ogas J (2003) Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J 35: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher SR (1992) Quantitation of GUS activity by fluorometry. In SR Gallagher, ed, GUS Protocols Using the GUS Gene as a Reporter of Gene Expression. Academic Press, San Diego, pp 47–59
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada JJ (2001) Role of Arabidopsis LEAFY COTYLEDON genes in seed development. J Plant Physiol 158: 405–409 [Google Scholar]
- Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5: 523–530 [DOI] [PubMed] [Google Scholar]
- Holdsworth M, Kurup S, McKibbin R (1999) Molecular and genetic mechanisms regulating the transition from embryo development to germination. Trends Plant Sci 4: 275–280 [Google Scholar]
- Keith K, Kraml M, Dengler NG, McCourt P (1994) fusca3: a heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell 6: 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Mayerhofer R, Koncz-Kalman Z, Nawrath C, Reiss B, Rédei GP, Schell J (1990) Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. EMBO J 9: 1337–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Jorna ML, Brinkhorst-van der Swan DLC, Karssen CM (1982) The isolation of abscisic acid (aba)-deficient mutants by selection of induced revertants in non-germinating gibberellin-sensitive lines of Arabodopsis L. Heynh. Theor Appl Genet 61: 385–393 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant 61: 377–383 [Google Scholar]
- Koornneef M, Van Eden J, Hanhart CJ, Jongh AMM (1983) Genetic fine-structure of the GA1 locus in the higher plant Arabidopsis thaliana (L.) Heynh. Genet Res Camb 41: 57–68 [Google Scholar]
- Kroj T, Savino G, Valon C, Giraudat J, Parcy F (2003) Regulation of storage protein gene expression in Arabidopsis. Development 130: 6065–6073 [DOI] [PubMed] [Google Scholar]
- Larkin JC, Oppenheimer DG, Pollock S, Marks MD (1993) Arabidopsis GLABROUS1 gene requires dowstream sequence for function. Plant Cell 5: 1739–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J (2002) Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 14: 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10: 655–661 [DOI] [PubMed] [Google Scholar]
- Lloyd AM, Schena M, Walbot V, Davis RW (1994) Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid-inducible regulator. Science 266: 436–439 [DOI] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Luerssen H, Kirik V, Herrmann P, Misera S (1998) FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J 15: 755–764 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jurgens G (1996) Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84: 61–71 [DOI] [PubMed] [Google Scholar]
- Mayer U, Torres Ruiz RA, Berleth T, Misera S, Jürgens G (1991) Mutations affecting body organization in the Arabidopsis embryo. Nature 353: 402–407 [Google Scholar]
- Meier C, Bouquin T, Nielsen ME, Raventos D, Mattsson O, Rocher A, Schomburg F, Amasino RM, Mundy J (2001) Gibberellin response mutants identified by luciferase imaging. Plant J 25: 509–519 [DOI] [PubMed] [Google Scholar]
- Meinke DW (1992) A homeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 258: 1647–1650 [DOI] [PubMed] [Google Scholar]
- Meinke DW, Franzmann L, Nickle TC, Yeung EC (1994) Leafy cotyledon mutants of Arabidopsis. Plant Cell 6: 1049–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Akazawa T, McCourt P (1991) Effects of the gibberellin biosynthetic inhibitor uniconazol on mutants of Arabidopsis. Plant Physiol 97: 736–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Hayama R, Tsuchiya Y, Nishimura M, Kawaide H, Kamiya Y, Naito S (2000) The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Dev Biol 220: 412–423 [DOI] [PubMed] [Google Scholar]
- Ogas J, Cheng JC, Sung ZR, Somerville C (1997) Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277: 91–94 [DOI] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C (1999) PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S (2003) Gibberellin biosynthesis and response during arabidopsis seed germination. Plant Cell 15: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N, Sun T-p, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell (Suppl) 14: S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill DP, Ross JJ (2002) Auxin regulation of the gibberellin pathway in pea. Plant Physiol 130: 1974–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Valon C, Kohara A, Misera S, Giraudat J (1997) The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell 9: 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, et al (1999) ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261 [DOI] [PubMed] [Google Scholar]
- Perazza D, Vachon G, Herzog M (1998) Gibberellins promote trichome formation by up-regulating GLABROUS1 in arabidopsis. Plant Physiol 117: 375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz V, Bergervoet JH, Koornneef M (2001) Sequential steps for developmental arrest in Arabidopsis seeds. Development 128: 243–252 [DOI] [PubMed] [Google Scholar]
- Reidt W, Wohlfarth T, Ellerstrom M, Czihal A, Tewes A, Ezcurra I, Rask L, Baumlein H (2000) Gene regulation during late embryogenesis: the RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J 21: 401–408 [DOI] [PubMed] [Google Scholar]
- Shevell DE, Leu WM, Gillmor CS, Xia G, Feldmann KA, Chua NH (1994) EMB30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell 77: 1051–1062 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Chang C, Krol E, Sun TP (1997) Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J 12: 9–19 [DOI] [PubMed] [Google Scholar]
- Singh DP, Jermakow AM, Swain SM (2002) Gibberellins are required for seed development and pollen tube growth in Arabidopsis. Plant Cell 14: 3133–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski DB, Lloyd AM, Marks MD (2000) Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci 5: 214–219 [DOI] [PubMed] [Google Scholar]
- Talon M, Koornneef M, Zeevaart JAD (1990) Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of the semidwarf ga4 and ga5 mutants. Proc Natl Acad Sci USA 87: 7983–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SG, Phillips AL, Hedden P (1999) Molecular cloning and functional expression of gibberellin 2- oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA 96: 4698–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, Nambara E, Naito S, McCourt P (2004) The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis. Plant J 37: 73–81 [DOI] [PubMed] [Google Scholar]
- West M, Harada JJ (1993) Embryogenesis in higher plants: an overview. Plant Cell 5: 1361–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M, Yee KM, Danao J, Zimmerman JL, Fischer RL, Goldberg RB, Harada JJ (1994) LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6: 1731–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CN, Proebsting WM, Hedden P, Rivin CJ (2000) Gibberellins and seed development in maize. I. Evidence that gibberellin/abscisic acid balance governs germination versus maturation pathways. Plant Physiol 122: 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y-L, Li L, Gage D, Zeevaart J (1999) Feedback regulation of GA5 expression and metabolic engineering of gibberellin levels in Arabidopsis. Plant Cell 11: 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Kamiya Y, Sun T (2001) Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J 28: 443–453 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RG, Kamiya Y, Sun T (1998) Phytochrome regulation and differential expression of gibberellin 3beta-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10: 2115–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]