Abstract
Xyloglucan endotransglucosylases/hydrolases (XTHs) that mediate cleavage and rejoining of the β (1-4)-xyloglucans of the primary cell wall are considered to play an important role in the construction and restructuring of xyloglucan cross-links. A novel rice (Oryza sativa) XTH-related gene, OsXTH8, was cloned and characterized after being identified by cDNA microarray analysis of gibberellin-induced changes in gene expression in rice seedlings. OsXTH8 was a single copy gene; its full-length cDNA was 1,298 bp encoding a predicted protein of 290 amino acids. Phylogenetic analysis revealed that OsXTH8 falls outside of the three established subfamilies of XTH-related genes. OsXTH8 was preferentially expressed in rice leaf sheath in response to gibberellic acid. In situ hybridization and OsXTH8 promoter GUS fusion analysis revealed that OsXTH8 was highly expressed in vascular bundles of leaf sheath and young nodal roots where the cells are actively undergoing elongation and differentiation. OsXTH8 gene expression was up-regulated by gibberellic acid and there was very little effect of other hormones. In two genetic mutants of rice with abnormal height, the expression of OsXTH8 positively correlated with the height of the mutants. Transgenic rice expressing an RNAi construct of OsXTH8 exhibited repressed growth. These results indicate that OsXTH8 is differentially expressed in rice leaf sheath in relation to gibberellin and potentially involved in cell elongation processes.
The plant primary cell wall is a complex and dynamic structure that plays an important role in controlling cell shape and plant morphology as a whole. Structural modification of the cell wall is important considering regulation of cell growth and differentiation. Flowering plants have type I wall in which the principal cellulose cross-linking glycans is xyloglucan and as much as 35% of the wall mass is pectin (Carpita and Gibeaut, 1993; Cosgrove, 1997). In the type II cell wall of the grasses and cereals, the predominant glycans that cross link the cellulose microfibrils are glucuronoarabinoxylan and (1,3)(1,4)-β-d-glucan (Buckeridge et al., 2004). Type II cell walls contain a relatively low amount of xyloglucan but could nevertheless be very important (Yokoyama et al., 2004). It is considered that structural changes in these networks are regulated by enzymatic modification, and therefore wall-modifying enzymes would be expected to play an important role in regulating the plasticity of the cell walls.
A class of enzymes known as xyloglucan endotransglucosylases/hydrolases (XTHs; Yokoyama and Nishitani, 2001) catalyzes the endo cleavage of xyloglucan polymers and the subsequent transfer of the newly generated reducing ends to other polymeric or oligomeric xyloglucan molecules (Fry et al., 1992; Nishitani and Tominaga, 1992). XTH action seems to achieve regulated wall loosening during turgor-driven expansion by rearranging load bearing xyloglucan cross-links between cellulose microfibrils. XTHs are also considered to catalyze molecular grafting reactions required to integrate nascent xyloglucan polysaccharide into the existing cell wall, maintaining cell wall thickness and integrity (Rose et al., 2002).
The advent of the genome sequencing projects has revealed the presence of multigene XTH families in various plant species, and XTH activity has been detected in a variety of plant tissues (Nishitani, 1997; Campbell and Braam, 1999a; Rose et al., 2002). In rice (Oryza sativa), a family of 29 XTH genes has been deduced from the rice genome sequence (Yokoyama et al., 2004). For any given XTH gene family, typically only a few genes have been demonstrated to encode true XTHs (Schroder et al., 1998; Campbell and Braam, 1999b), but the high homology among designated XTH genes and the presence of conserved key motifs among them strongly suggest that they encode proteins with XTH activity. In addition to its potential ability to alter and loosen the cell wall matrix, studies have shown a strong correlation between XTH expression and activity to cell elongation zones (Nishitani and Tominaga, 1991; Xu et al., 1996; Vissenberg et al., 2000). Similarly, GA treatment, which induces the elongation of leaves and stems in several plant species, increases XTH activity (Potter and Fry, 1994; Smith et al., 1996). Furthermore, specific XTH genes have shown to be up-regulated by the growth-promoting hormones like auxin, GA, and brassinosteroid (BR; Zurek and Clouse, 1994; Xu et al., 1996; Catala et al., 1997; Schunmann et al., 1997). However, XTH activity does not always correlate with growth rate, as activity has been detected in vegetative tissues that have ceased to elongate (Smith et al., 1996; Barrachina and Lorences, 1998) and in ripening fruit (Redgwell and Fry, 1993; Maclachlan and Brady, 1994). This indicates that various types of XTH genes are associated with wall reorganization during cellular differentiation and fruit ripening (Arrowsmith and de Silva, 1995; Saab and Sachs, 1996; Schroder et al., 1998).
Thus, the presence of XTHs with different tissue-specific expression, hormonal regulation, and/or potentially different enzymatic properties seems to be necessary for the metabolism of xyloglucan during various stages of plant growth and development. Therefore, the characterization of individual XTH genes within a single species is essential to understand their specific roles. In this study, a novel XTH gene, named OsXTH8, was identified by a cDNA microarray analysis of GA-regulated genes in rice. We describe OsXTH8 developmental and hormonal regulation and discuss its importance in growth processes of rice.
RESULTS
OsXTH8 Is a Novel XTH-Related Gene
A rice XTH gene named OsXTH8 was identified by a cDNA microarray analysis of GA-regulated genes in rice. In an effort to characterize the expression of OsXTH8 in rice, full-length OsXTH8 cDNA was amplified by RACE PCR and sequenced. The OsXTH8 full-length cDNA is 1,298 bp including a poly(A)+ tail, and it encodes a predicted protein of 290 amino acid residues. The deduced amino acid sequence indicated the presence of a putative signal peptide rich in hydrophobic amino acids in the N-terminal region (von Heijne, 1986). The deduced amino acid sequence also showed the presence of a potential site for N-linked glycosylation (N-X-T/S; the nucleotide sequence data reported will appear in the DNA Data Bank of Japan [DDBJ] under the accession no. AB110604).
As OsXTH is a multigene family in rice, to examine the precise expression pattern of OsXTH8 gene during development and in response to hormonal treatment, 3′ untranslated region (UTR) of OsXTH8 gene was used as a specific probe. To check the specificity of 3′ UTR OsXTH8, genomic DNA was digested with EcoRI, XbaI, and XhoI, respectively. Membrane probed with the 3′ UTR region of OsXTH8 produced a single band (Fig. 1), confirming the specificity of the 3′ UTR probe.
Figure 1.
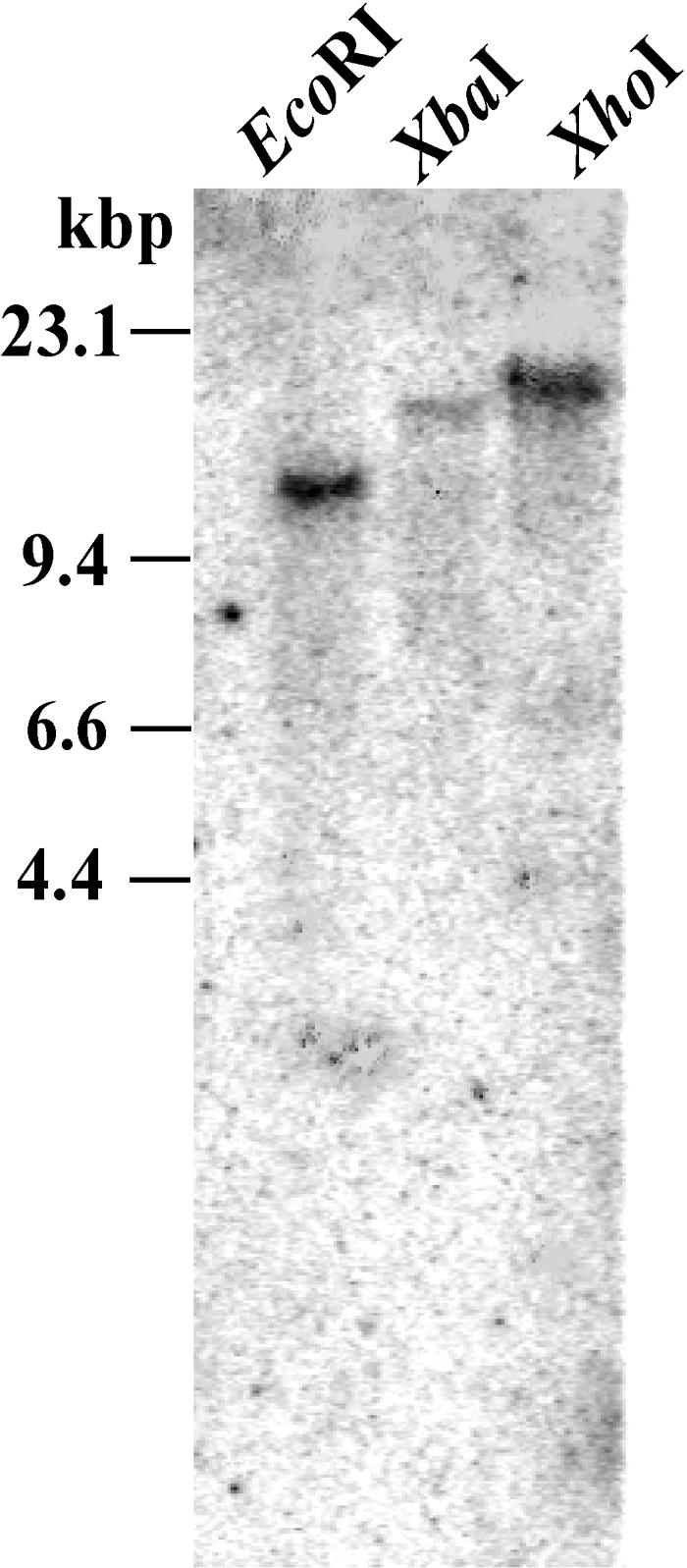
Genomic Southern-blot analysis of OsXTH8. Rice genomic DNA was digested completely with EcoRI, XbaI, and XhoI, respectively, separated by agarose gel electrophoresis, and then blotted onto positively charged nylon membrane. The blot was hybridized to 3′ UTR of OsXTH8. Molecular size markers are indicated on the left.
The deduced amino acid sequence of OsXTH8 showed sequence homology and the presence of a functional motif in comparison with other rice XTHs sequences (Fig. 2). OsXTH8 showed a sequence identity of 43%, 46%, 33%, and 38% to OsXTR1, OsXTR2, OsXTR3, and OsXTR4 (Uozu et al., 2000), respectively. XTHs contain a conserved sequence (DEIDFEFLG) that matches the bacterial endo-β-1, 3-1, 4-glucanase (Borriss et al., 1990) and has been suggested as the catalytic center for the XTH family. The corresponding sequence in OsXTH8 has two amino acid substitutions (DEIDIEFMG) compared to the consensus sequence.
Figure 2.
Amino acid sequence alignment of OsXTH8 and other members of the rice XTH gene family. The deduced amino acid sequence of OsXTH8 is aligned with other members of rice XTH gene family by Genetyx-WIN. DEIDFEFLG motif (marked by line) indicates a possible conserved catalytic region shared with the Bacillus β-glucanase. The possible N-linked glycosylation residues are indicated by asterisks. Conserved Cys residues are marked by arrowheads.
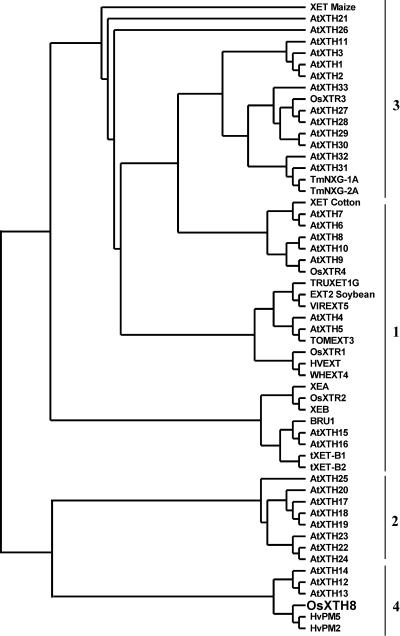
To find the evolutionary relationships of OsXTH8 with XTHs of other plant species, a phylogenic tree was generated using full-length protein sequences. The analysis revealed that XTHs could be loosely grouped into four distinct groups (Fig. 3), as reported by other authors (Nishitani, 1997; Campbell and Braam, 1999a; Catala et al., 2001). Group 1 contains genes that share a high level of sequence identity among different species and that are expressed in young developing tissues (Catala et al., 1997; Shimizu et al., 1997; Akamatsu et al., 1999; Takano et al., 1999). Group 2 comprises XTH genes from several species showing diverse patterns of expression and response to hormonal or mechanical stimuli, including touch-inducible, flooding-response, BR-inducible, and fruit ripening-related XTHs (Catala et al., 2001). Group 3 represents a divergent group of XTHs, including NXG1 from nasturtium (Tropaeolum majus) that can act as xyloglucan hydrolase and transglycosylase (de Silva et al., 1993). OsXTH8 has the highest homology to sequences in Group 4, consisting of well-characterized barley (Hordeum vulgare) genes, HvPM2 and HvPM5, which are up-regulated by GA in barley leaf sheaths and leaves (Smith et al., 1996). Monocot members of group 4 revealed to have two substitutions (DEIDIEFMG) compared to the consensus sequence (DEIDFEFLG). The presence of different amino acid residues in the putative catalytic region may attribute to unique enzymatic activity of OsXTH8.
Figure 3.
Phylogenic-alignment of the OsXTH8-deduced amino acid sequence with other plant XTHs. OsXTH8 was aligned with 54 full-length deduced amino acid sequences using the ClustalW and tree View software. Details and GenBank accession numbers are: AtXTH1, At4g13080; AtXTH2, At4g13090; AtXTH3, At3g25050; AtXTH4, At2g06850; AtXTH5, At5g13870; AtXTH6, At5g65730; AtXTH7, At4g37800; AtXTH8, At1g11545; AtXTH9, At4g03210; AtXTH10, At2g14620; AtXTH11, At3g48580; AtXTH12, At5g57530; AtXTH13, At5g57540; AtXTH14, At4g25820; AtXTH15, At4g14130; AtXTH16, At3g23730; AtXTH,17, At1g65310; AtXTH18, At4g30280; AtXTH19, At30290; AtXTH20, At5g48070; AtXTH21, At2g18800; AtXTH22, At5g57580; AtXTH23, At4g25810; AtXTH24, At4g30270; AtXTH25, At5g57550; AtXTH26, At4g28850; AtXTH27, At2g01850; AtXTH28, At1g14720; AtXTH29, At4g18990; AtXTH30, At1g32170; AtXTH31, At3g44990; AtXTH32, At2g36870; AtXTH33, At1g10550; TmNXG-1A, X68254; TmNXG-2A, X68255; TRUXET1G, L43094; VIREXT5, D16458; EXT2 (soybean), D16455; HVEXT, X91659; WHEXT4, D16457; TOMEXT3, D16456; XET (cotton), D88413; tXET-B1, X82685; tXET-B2, X82684; XEB X93175; XEA, X93174; XET (maize), U15781; HvPM2, X91660; HvPM5, X93173; and OsXTH8, AB110604. OsXTR1, OsXTR2, OsXTR3, and OsXTR4 sequences were noted down from Uozu et al. (2000).
Alternative Organ- and Cell Type-Expression Profiles of OsXTH8
It has been shown that different members of XTH gene family are specifically regulated by various physiological and environmental stimuli (Xu et al., 1996). In this study, OsXTH8 was characterized for temporal and spatial expression patterns and its response to different hormones.
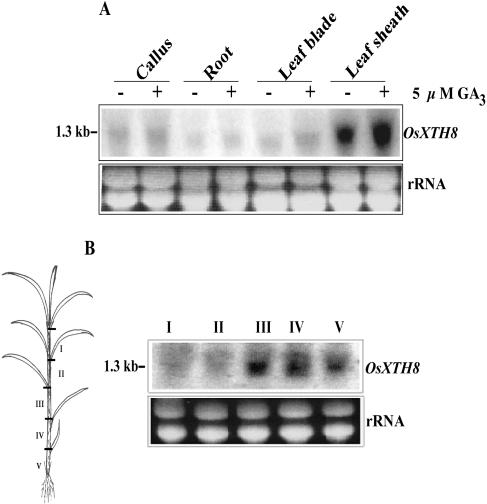
To examine the tissue specificity of OsXTH8, total RNAs from rice callus, root, leaf blade, and leaf sheath were hybridized with OsXTH8-specific DNA probe. A strong signal was detected in leaf sheaths but weak or no signal was observed in leaf blades, roots, and calli (Fig. 4A). These expressions were enhanced by GA3 treatment (Fig. 4A). When 1-month-old seedlings were used to characterize the expression in leaf sheath, the expression was mainly found in the three basal parts of leaf sheath (Fig. 4B). Enhanced expression of OsXTH8 in the third part of 1-month-old rice seedling, which corresponded to the second internode of leaf sheath, compared to two basal parts of leaf sheath showed that OsXTH8 is differentially expressed in leaf sheath.
Figure 4.
Tissue-specific expression of OsXTH8. A, Expression of OsXTH8 in different tissues. Two-week-old seedlings were treated with 5 μm GA3 for 24 h. For tissue-specific expression, total RNAs were extracted from roots, leaf blades, leaf sheaths, and calli. B, Expression of OsXTH8 in five different sections of leaf sheaths using 1-month-old rice seedlings. The rice leaf sheath was divided in five sections: leafy section (I), second and third leaf section (II), first and second leaf section (III), coleoptile and first leaf section (IV), and coleoptile section (V) as shown in the figure drawing. Total RNA (20 μg each) transferred onto membrane was probed with PCR- amplified 3′ UTR of OsXTH8 cDNA clone. rRNA stained with ethidium bromide was used as a loading control.
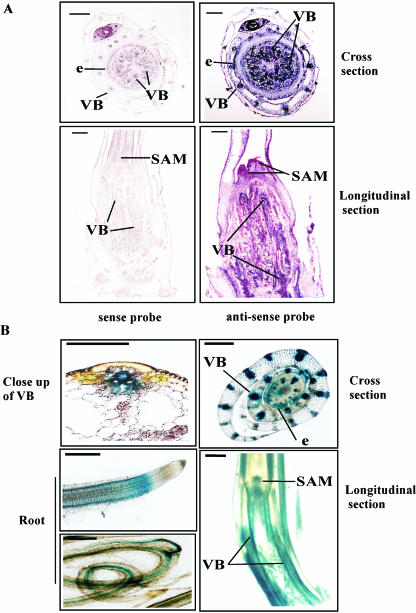
In situ hybridization was performed using the basal part (culm) of 2-week-old rice seedlings to learn more about the expression pattern. On hybridization with gene-specific OsXTH8 antisense probe, the cross and longitudinal sections of culm tissue revealed an accumulation of OsXTH8 mRNA in shoot apex meristem, vascular tissues, and young leaves (Fig. 5A). Although the expression did not seem to be delimited to specific cell types, significant hybridization was observed in large and small vascular bundles of leaf sheath and peripheral cylinder of the vascular bundles and fibers in the nodal region (Fig. 5A). No significant signal was visible when sense probe was used.
Figure 5.
In situ hybridization of OsXTH8 and GUS expression driven by OsXTH8 promoter in different tissues of rice. A, In situ localization of OsXTH8 mRNA. Sections of rice leaf sheaths were hybridized with Dig-labeled antisense RNA prepared from OsXTH8 expressed sequence tag (A, right). Dig-labeled sense RNA was used as a negative control (A, left). B, GUS expression in different tissues of transgenic rice carrying OsXTH8∷GUS construct. Cross-section and longitudinal section of leaf sheath and intact roots were used for histochemical localization of GUS activity. VB, Vascular bundle; e, peripheral cylinder of vascular tissues; SAM, shoot apical meristem. Bar represents 500 μm.
To further characterize the spatial distribution patterns of OsXTH8 gene expression, 2,325 bp of the OsXTH8 upstream region from the proposed first translational start site was fused to the β-glucuronidase (GUS) reporter gene. This fusion gene was introduced into rice cells, and transgenic plants were regenerated. Putative OsXTH8∷GUS transgenic plants were screened by PCR. Only PCR-confirmed, transgenic lines were used for GUS staining and GUS assay. To assess whether the GUS staining patterns were consistent with the result of in situ hybridization, similar tissue sections of leaf sheath were used. Figure 5B shows the GUS expression pattern driven by OsXTH8 promoter in the basal part of leaf sheath and young nodal roots of 2-week-old rice seedlings. In the case of leaf sheath, strong GUS staining was observed in shoot apex meristem and vascular bundles, very much similar to the result of in situ hybridization (Fig. 5, A and B). Microscopic observation of vascular bundles revealed GUS expression in vascular bundle sheath and mesotomic sheath surrounding xylem and phloem. Stele or vascular cylinder region in young nodal roots of the coleoptile node and roots arising from nodal roots also showed GUS staining (Fig. 5B). Weak GUS staining was found in the sclerenchyma cells lining the epidermis of young leaves (Fig. 5B).
Hormonal Regulation of OsXTH8 Expression
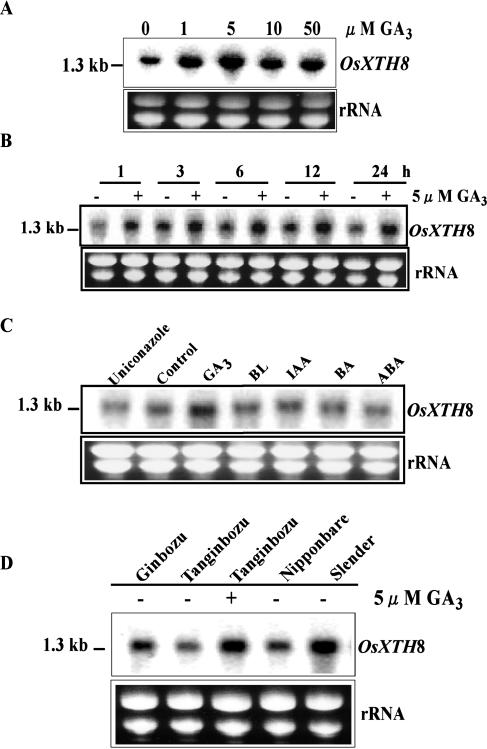
GA dose-dependent expression patterns of OsXTH8 mRNA were determined in rice leaf sheaths using different concentration of GA3 (1, 5, 10, and 50 μm). OsXTH8 expression was found to be up-regulated with the increase in GA3 concentration as there was no inhibitory effect of increase in GA3 concentration up to 50 μm; however, 5 μm GA3 induced maximum expression of OsXTH8 (Fig. 6A). To determine temporal expression patterns of OsXTH8 mRNA, leaf sheaths were treated for 1, 3, 6, 12, and 24 h. OsXTH8 mRNA accumulation in leaf sheath showed that during treatment of leaf sheath fragments with GA3, OsXTH8 expression was up-regulated and continued to increase throughout the 24-h incubation period (Fig. 6B). As 2-week-old rice seedlings were used for time-course experiment, because of the growth of leaf sheaths, increase in OsXTH8 expression in control could also be detected.
Figure 6.
Hormonal regulation of OsXTH8 expression. A, Dose-dependent effect of GA3. Rice leaf sheaths were treated with 0, 1, 5, 10, and 50 μm GA3 for 24 h. B, Time-course changes in the expression of OsXTH8. Rice leaf sheaths were treated with 5 μm GA3 for 1, 3, 6, 12, and 24 h for time-course experiment. C, Effects of different phytohormones on the expression of OsXTH8. Rice leaf sheaths were treated with 1 μm BL, 10 μm uniconazole, and 5 μm each GA3, IAA, BA, and ABA for 24 h. D, OsXTH8 expression in different GA mutants exhibiting abnormal heights. Ginbozu and Nipponbare are the wild type of Tanginbozu and Slr1 mutant, respectively. Total RNA (20 μg each) was extracted from leaf sheaths of 2-week-old seedlings and probed with 3′ UTR of OsXTH8. Experiments were replicated three times.
It has been shown that XTH genes are regulated by various hormones. For example, BRU1, a soybean XTH, is regulated by BR (Zurek and Clouse, 1994), and TCH4, an Arabidopsis (Arabidopsis thaliana) XTH gene, is up-regulated by auxin and BR (Xu et al., 1995). To characterize hormonal regulation of OsXTH8 expression, the effect of several plant hormones was examined on OsXTH8 mRNA abundance (Fig. 6C). When leaf sheaths were treated with GA3, brassinolide (BL), 6-bezyladenine (BA), indole-3-acetic acid (IAA), and abscisic acid (ABA), GA3 up-regulated the expression of OsXTH8 and there was very little effect of other hormones. Uniconazole, which is a potent GA biosynthesis inhibitor, had an inhibitory effect on the OsXTH8 mRNA accumulation (Fig. 6C).
To understand the physiological functions of OsXTH8, its expression in rice mutants with abnormal heights was investigated. Tanginbozu is a GA-deficient semidwarf mutant and a single recessive gene controls the semidwarfism of Tanginbozu. Mutation in Tanginbozu blocks the three oxidative steps whereby ent-kaurene is converted to ent-kaurenoic acid resulting in less accumulation of active GA (Ogawa et al., 1996). Northern-blot analysis showed that the level of OsXTH8 mRNA in the mutant was lower than that in its wild-type cv Ginbozu (Fig. 6D). The expression of OsXTH8 in the mutant was induced to exceed wild-type level following treatment with GA3 for 24 h. Slender rice1 (slr1), a GA-insensitive mutant, shows a constitutive GA-response phenotype (Itoh et al., 2002). Stem of the slr1 mutant grows 2 to 3 times more than the stem of wild-type cv Nipponbare. Northern-blot analysis confirmed that the level of OsXTH8 expression was higher in slr1 mutant than that of its wild type (Fig. 6D).
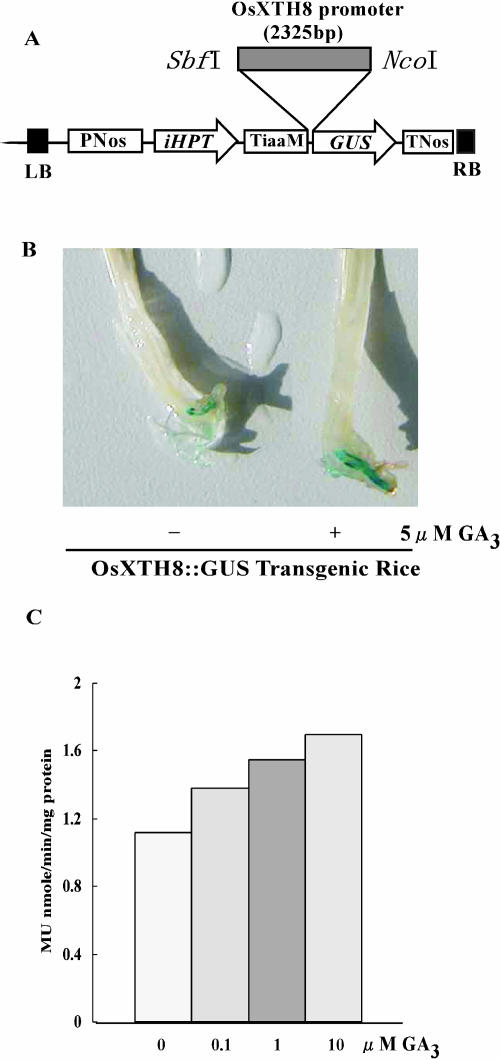
To analyze whether the 2,325-bp 5′-promoter region of the OsXTH8 locus is sufficient for the induction of its expression, an independent transgenic line (Fig. 7B), which was transformed with 2,325 bp of OsXTH8 promoter fused to the GUS reporter gene, was treated with 5 μm GA3. It was observed that GA3 treatment enhanced the expression of the GUS reporter gene compared to untreated (mock) OsXTH8∷GUS transgenic rice (Fig. 7B). This indicated that the 2,325-bp promoter region of OsXTH8 was sufficient for hormone-induced OsXTH8 expression. This observation was also confirmed by GUS assay using methyl-umbelliferylglucuronide as a substrate (Fig. 7C).
Figure 7.
GUS activity in OsXTH8∷GUS transgenic rice seedlings in response to GA treatment. A, Binary vector pSMAHdN627 harboring GUS gene under the control of the 2,325-bp promoter region of OsXTH8. B, Histochemical localization of GUS activity in transgenic rice treated with or without GA3. C, Ten-day-old seedlings were treated with 0, 0.1, 1, and 10 μm GA3 for 24 h. GUS activity was measured by fluorescent method as described in “Methods and Materials.” Values are mean of triplicate experiment.
RNAi OsXTH8 Transgenic Plants Exhibited Altered Development
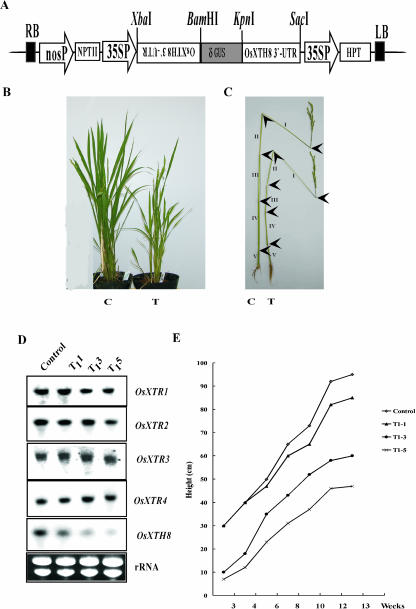
To assess the effects of loss of function of OsXTH8 on rice growth and development, a 360-bp 3′ UTR fragment of OsXTH8 was cloned into the pIG121-Hm vector in both sense and antisense orientations (Fig. 8A) for subsequent generation of rice RNAi transgenic plants. RNAi OsXTH8 was introduced into rice using Agrobacterium-mediated transformation. Rice plants transformed with only pIG121-Hm vector were used as control. At regeneration stage, the regeneration efficiency of the RNAi OsXTH8 transformed callus was only about 37% when compared to control transformed callus. After transformation, 29 transformants were generated, out of which 13 plants appeared like wild type and 16 exhibited altered vegetative growth. RNAi transformed plants showed various degrees of repressed growth and were 20% to 50% shorter than the control plants when they reached maturity (Fig. 8B). Three different RNAi transgenic lines (T1-1, T1-3, and T1-5) were selected for further analysis. RNAi transgenic line T1-1 appeared like control in phenotype, while the other two transgenic lines T1-3 and T1-5 had almost a 40% to 50% reduced height when compared to the control (Fig. 8E). To determine whether a relationship existed between the OsXTH8 RNA levels and the phenotype, RNA was extracted from the three transgenic lines to analyze their transcript levels compared with that in the control. The results are shown in Figure 8D, with ethidium bromide-stained rRNA to show that equal amounts of RNA were used. Two lines, T1-3 and T1-5, in which OsXTH8 transcript accumulated in very little amount, had a 40% and 50% reduction in height when compared to the control. Transgenic line T1-1 that appeared like control in phenotype had slightly reduced OsXTH8 mRNA accumulation. To assess whether the phenotype of the RNAi transgenic plants was caused by the knockout of OsXTH8 or other OsXTHs genes were also affected, these lines were tested for the expression of OsXTR1, OsXTR2, OsXTR3, and OsXTR4. It is clear from Figure 8D that there was no significant reduction in the transcript level of OsXTH genes tested. The internodes of RNAi OsXTH8 transgenic elongated almost normally (T in Fig. 8C), but three internodes (II, III, and IV) seemed to be shorter than the control plants. The apparent repressed growth of transgenic rice was attributed to the reduced elongation of these three internodes.
Figure 8.
Phenotypes of transgenic rice constitutively overexpressing RNAi OsXTH8. A, A binary vector pIG121-Hm harboring RNAi OsXTH8 under the control of CaMV 35S promoter. B, Transgenic rice 2 months after transferred to soil in the isolating green house. C, Elongation of the upper five internodes of the control (C) and RNAi OsXTH8 transgenic rice (T). D, Expression levels of OsXTR1, OsXTR2, OsXTR3, OsXTR4, and OsXTH8 mRNA in RNAi OsXTH8 transgenic lines. E, Growth curves of the three RNAi OsXTH8 transgenic plants and a vector control transgenic plant.
DISCUSSION
The existence of a family of 29 XTH genes in rice suggests that individual XTHs may exhibit distinct patterns of expression in terms of tissue specificity and responses to hormonal and environmental stimuli. The individual XTH enzymes encoded by the XTH gene family are thought to have varying kinetic properties as well as different catalytic functions, including transferase and hydrolase activities. Therefore, it is predicted that combinatorial expression of multiple XTHs is critical for a broad spectrum of plant developmental processes (Rose et al., 2002). In this study, we characterized the new XTH-related gene, named OsXTH8, which is highly expressed in rice leaf sheath and is up-regulated by GA3. Southern-blot analysis and database search revealed that OsXTH8 is a novel gene. To study the functional role of OsXTH8 in rice, the enzymatic activity profile of OsXTH8 must be examined.
The deduced amino acid sequence of OsXTH8 shares high homology, and many conserved sequences, with other XTH family members. OsXTH8 also has amino acid residues peculiar to monocotyledons within the presumed catalytic region. Most XTH family members in rice and other plants have the conserved amino acid sequence DEIDFEFLG (Fig. 2), whereas the corresponding sequence in OsXTH8 is DEIDIEFMG. This conserved sequence is also found in the catalytic region of bacterial endo-β-1, 3-1-4-glucanases (Borriss et al., 1990). Site-directed mutagenesis of Bacillus licheniformis endo-β-1, 3-1, 4-glucanases indicated that the second Glu (E) is critical for catalytic activity, while other studies demonstrated the importance of the first Glu (E) and the second Asp (D) (Planas et al., 1992; Juncosa et al., 1994). The conservation of these critical amino acids in the catalytic domain of OsXTH8 argues that its enzymatic activity will act on xyloglucan-β-1, 4-glycosyl linkages in plants.
Most sequence differences among XTH proteins are found in the carboxyl terminal and, based on the relatedness of carboxyl-terminal regions, the XTHs from many plant species can be loosely organized into four groups (Campbell and Braam, 1999a). The three main subfamilies (1, 2, and 3 in Fig. 3) are found in a wide range of flowering plants, including both dicots and monocots. The diversification of the structure of XTH genes among these three subfamilies may reflect a unique functional assignment for each subfamily that is essential throughout flowering plants. An obvious consideration is that this phylogenetic divergence reflects the evolution of XTH subgroups with different biochemical mechanisms of action, such as transglucosylation versus hydrolysis (Rose et al., 2002). The OsXTH8 gene isolated in this study grouped into the fourth subfamily (Fig. 3), suggesting that it may have some peculiar functional features. Other related XTH genes that do not fit within the three main subfamilies have been reported in barley (Schunmann et al., 1997) and rice (Uozu et al., 2000) and show a unique expression pattern during leaf sheath and stem development.
An insight into the physiological role of XTHs can be gained from their enzymatic activity, the expression pattern of XTH genes, and the activity of promoter-GUS fusions for individual genes at various developmental stages in various parts of plants. OsXTH8 is expressed in rice leaf sheath, and, less abundantly, in root, leaf blade, and callus (Fig. 4). Similarly, OsXTR1, OsXTR2, OsXTR3, and OsXTR4 were shown to express mainly in rice culm (stem), and their expression patterns in culm seem to overlap with that of OsXTH8. OsXTR1 is expressed in both the elongation and division zones of internodes, with a higher level of expression in the former. In contrast, OsXTR4 is expressed in the division zone of internodes and nodes. OsXTR2 is expressed in most organs, with the exception of well-developed leaf blades and roots. OsXTR3 expression showed a strict organ specificity in elongating stems, being notably higher in the internode elongation zone (Uozu et al., 2000). OsXTH8 may play a cooperative role with the above-mentioned genes and others that are expressed in overlapping regions. Further detailed expression studies, as well as biochemical analysis, will provide insight into the physiological function of OsXTH8. mRNA localization by in situ hybridization and histochemical localization of GUS activity in OsXTH8∷GUS transgenics indicated that OsXTH8 was expressed in growing regions such as shoot apical meristem, vascular bundles of leaf sheath, and young crown roots developing from nodes (Fig. 5); thus, OsXTH8 exhibited a unique expression pattern in terms of organ and stage specificity during leaf sheath elongation and young nodal root development. The expression of OsXTH8 in the shoot apex meristem, vascular bundles in leaf sheath, and young nodal roots supports its role in cell wall modification processes during active growth.
OsXTH8 expression increased in a dose- and time-dependent manner with GA3 treatment. A significant increase in OsXTH8 mRNA was observed in leaf sheath segments or intact plants with an increase in GA3 concentration, and there was no inhibitory effect associated with further increase in GA3 concentration up to 50 μm (Fig. 6A). OsXTH8 also showed a temporal increase in transcript levels when leaf sheaths were treated with GA3. Relative to controls, the earliest increase in OsXTH8 mRNA level and leaf sheath growth was detectable after 1 h of incubation with GA3, and it continued to increase up to 24 h of incubation (Fig. 6B). Transcriptional up-regulation of XTH gene expression by GA has been demonstrated in several instances (Uozu et al., 2000), but the mechanism has not yet been established. GAs regulate tissue elongation in several plants, and an effect of GA on wall extensibility, including promotion of wall loosening reactions, has been proposed (Cosgrove and Sovonick-Dunford, 1989). It has been reported that GA has a large effect on internodal elongation, especially activating cell division and cell elongation (Kamijima, 1981). Furthermore, it has been proposed that XTH activity and mRNA levels of XTH-related genes are regulated by GA to induce leaf elongation (Smith et al., 1996; Schunmann et al., 1997). In agreement with the above reports, GA also induced the expression of OsXTH8 and acted as a candidate marker of rice leaf sheath growth. This suggests that OsXTH8 acts to alter the structure of cell wall in response to this hormone.
Tanginbozu is a GA-deficient semidwarf mutant of rice, the phenotype of which is controlled by a single recessive gene. The mutation in Tanginbozu blocks the three oxidative steps whereby ent-kaurene is converted to ent-kaurenoic acid, resulting in less accumulation of active GA (Ogawa et al., 1996). The level of OsXTH8 mRNA in Tanginbozu indicated that reduced accumulation of active GA was accompanied by preferential suppression of OsXTH8 expression, and OsXTH8 expression increased when exogenous GA3 was applied to the semidwarf mutant (Fig. 6D). This result confirms the up-regulation of OsXTH8 by GA and the correlation of this gene's expression with leaf sheath elongation. The tall mutant Slr1 is thought to have a defect in a suppressive gene in the GA signal transduction pathway, which causes a constitutive GA response without application of exogenous GA (Itoh et al., 2002). This finding that OsXTH8 expression is up-regulated in slr1 mutant compared to the wild-type Nipponbare further supports the regulation of OsXTH8 by GA.
GUS activity and histochemical localization of GUS activity showed that a 2,325-bp fragment 5′ upstream of the OsXTH8-coding region was sufficient to drive and up-regulate GUS reporter gene expression in response to GA3 (Fig. 7, B and C). Computer analysis using the PLACE signal scan program, a database of plant cis-acting regulatory DNA elements (Higo et al., 1999), revealed many potential cis-elements in the 2,325-bp sequence of OsXTH8. In the putative promoter region of OsXTH8, three elements of a GA-response complex were found, including a pyrimidine box CCTTTT (Skriver et al., 1991), a GA-response element (GARE) TAACGTAG (Gubler and Jacobsen, 1992), and a CAACTC regulatory element (CARE) CAACTCAA (Sutoh and Yamauchi, 2003), at 228, 154, and 311 bp upstream of the translation initiation site, respectively. It will be interesting to dissect further the OsXTH8 promoter region to find out whether it contains other known or novel cis regulatory elements that function to confer responsiveness to different stimuli or hormones. Although many auxin-regulated genes have been shown to be transcriptionally controlled, regulation of BRU1 by BRs is thought to occur posttranscriptionally (Zurek and Clouse, 1994; Xu et al., 1995). It is possible that OsXTH8 is regulated both transcriptionally and posttranscriptionally, depending on the type of hormone or stimulus. Identification of trans-acting factors will help elucidate the signal transduction pathways by which environmental stimuli or different hormones lead to regulation of OsXTH8.
One way to examine the role of XTHs in plant growth and development is to increase or decrease endogenous XTHs' mRNA content by applying transgenic methodology. RNAi OsXTH8 expressed under the control of constitutive cauliflower mosaic virus (CaMV) 35S promoter produced plants with altered growth (Fig. 8B). The repression of growth in a subpopulation of RNAi transgenic rice was attributable to the reduced second, third, and fourth internodal elongation (Fig. 8C) and is consistent with the observation that in 1-month-old wild-type rice seedlings, OsXTH8 mRNA accumulation was only observed in the first three parts of the leaf sheath (Fig. 4B), which at this stage of development correspond to the third and fourth internode. As OsXTH8 is a member of a multigene family, it is possible that silencing of OsXTH8 could affect the expression of other OsXTH genes. For this reason, we selected four OsXTR genes, where OsXTR1, OsXTR2, and OsXTR4 are fairly closely related genes and OsXTR3 is distantly related gene to OsXTH8 (Yokoyama et al., 2004). As the expressions of these genes were not affected by expression of RNAi OsXTH8 (Fig. 8D), it could be argued that the silencing of OsXTH8 was quite specific.
Whether the repressed internode elongation in the transgenic plants is because of some discernible alteration in cell expansion or development awaits further investigation. Sakamoto et al. (2003) modified the level of GA by overproduction of a GA catabolic enzyme, GA 2-oxidase, and the first (uppermost) internode was more severely affected than the other internodes. In contrast, all internodes were severely reduced in severe transformants. Transgenic Arabidopsis plants with altered expression of AtXTH24 showed developmental defects (Verica and Medford, 1997), but one cannot conclude specifically that these defects were due to the alterations in AtXTH24 expression. Transgenic tobacco plants with reduced XET activity were shown to accumulate xyloglucan with a Mr at least 20% greater than that of wild-type plants (Herbers et al., 2001). The consequences of these alterations for wall properties and plant fitness are not yet clear (Rose et al., 2002). In spite of the low amount of xyloglucan in rice and the possibility that other wall loosening agents also mediate growth, our results support that XTHs have a role in cell wall modification and growth promoting functions in rice.
In conclusion, this study shows that OsXTH8 is differentially expressed in rice leaf sheath in relation to GA. Based on the specific accumulation of OsXTH8 transcripts in young rice leaf sheath and the altered phenotype of RNAi OsXTH8, it is suggested that OsXTH8 is involved in cell wall modification processes during rice growth and development.
MATERIALS AND METHODS
Plant Materials and Treatments
Wild-type rice (Oryza sativa L. cv Nipponbare or cv Ginbozu) and two rice mutants, Tanginbozu and Slr1, were grown under white fluorescent light (about 200 μmol m−2 s−1, 12-h light period/d) at 25°C and 75% relative humidity in a growth chamber. Leaf sheath segments of 2-week-old seedlings were floated on 10 mL of distilled water in 60 mm × 15 mm petri dishes containing BL (Fuji Chemical, Toyama, Japan), IAA, GA3, ABA, and BA uniconazole (Wako Pure Chemical, Osaka) for various times.
cDNA Microarray Analysis
A cDNA microarray containing 4,000 cDNA clones randomly selected from the rice cDNA library was used. The mRNAs for microarray probes preparation were purified with an Oligotex-dT-30 mRNA purification kit (Takara, Kyoto) according to the manufacturer's instructions. mRNA samples (1 μg) prepared from leaf sheath treated with 5 μm GA3 for 24 h or water as a control, were reverse transcribed in 20-μL volume containing 1 mm Cy3 or Cy5 dCTP (Amersham Biosciences, Piscataway, NJ), anchored oligo(dT)25, random nonamer, dithiothreitol, dNTPs, and SuperScript II (Invitrogen, Carlsbad, CA). After incubation at 42°C for 2 h, the reaction was stopped, and RNA was degraded by first heating at 94°C for 3 min and then treated with NaOH at 37°C for 15 min. Fluorescently labeled probes were purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany). Probe hybridization and scanning of the hybridized microarray slide were done according to the method of Yazaki et al. (2000). Hybridization was done twice and data were analyzed with Array Gauge version 1.21 (Fujifilm, Tokyo). Only those clones that showed the same changes in two experiments were selected as the candidates.
cDNA Cloning and Sequencing
RACE-PCR was used to clone the full-length sequence of OsXTH8 using Gene Racer kit (Invitrogen). Oligo(dT) was used for first strand synthesis. PCR was performed by using Gene Racer 5′ Primer and a gene-specific primer (5′-CACACCGCCCAACTGTGCAAGATGAACT-3′). The PCR product was purified and cloned into pCR 4Blunt-TOPO vector (Zero Blunt TOPO PCR Cloning Kit for Sequencing; Invitrogen). Sequencing of the full-length cDNA was accomplished for both strands using dye-labeled terminations (PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit; Applied Biosystems, Foster City, CA) and an automated DNA sequencer (model 373A; Applied Biosystems). DNA sequence was analyzed with Genetyx-WIN Version 5.1 (Software Development, Tokyo).
Genomic Southern Hybridization
Rice genomic DNA was digested with EcoRI, XbaI, and XhoI, respectively, separated by 0.8% agarose gel electrophoresis, and then transferred onto Hybond+ membranes under alkaline conditions. A 3′ UTR of OsXTH8, which was PCR amplified using gene-specific primers, purified from agarose gel (QIAEXII Gel Extraction Kit; Qiagen), and radiolabeled using [alpha-32P] dCTP (Amersham Biosciences) random prime labeling system (Rediprime II; Amersham Biosciences), was used as probe for Southern-blot analysis. Hybridization was performed at 42°C in an ultrasensitive hybridization buffer (ULTRAhyb, Ambion, Austin, TX) overnight. The blot was washed twice first in 2× SSC, 0.1% × SDS at 42°C for 5 min and in 0.1× SSC, 0.1% SDS at 68°C for 15 min. It was finally analyzed by the phosphor image program with the Typhoon 8600k variable imager (Amersham Biosciences).
RNA Extraction and Northern-Blot Analysis
Tissue samples were quick-frozen in liquid nitrogen. Samples (0.5 g) were ground to powder by mortar and pestle, and total RNAs were isolated according to the procedure of Chomczynski and Sacchi (1987). For northern-blot analysis, 20 μg of total RNA was separated on 1.2% agarose containing 6% formaldehyde and transferred on to a Hybond-N+ nylon membrane (Amersham Biosciences). Loading of equal amounts of total RNA for northern blots was determined by visualization of ethidium bromide-stained rRNA bands. Probe for northern blots was the 3′ UTR of OsXTH8, which was cut from pBluescript SK+ or SK− plasmids, purified from agarose gels (QIAEXII Gel Extraction kit; Qiagen), and radio labeled using [alpha-32P]dCTP (Amersham Biosciences) random prime labeling system (Rediprime II; Amersham Biosciences). Hybridization was performed at 42°C in the ultrasensitive hybridization buffer (ULTRAhyb) overnight. The blots were washed twice first in 2× SSC, 0.1% × SDS at 42°C for 5 min and in 0.1× SSC, 0.1% SDS at 68°C for 15 min. They were finally detected with x-ray film (Kodak, Rochester, NY) or analyzed by the phosphor image program with the Typhoon 8600k variable imager (Amersham Biosciences).
Promoter Analysis and GUS Localization
To amplify OsXTH8 promoter fragment, rice genomic DNA was extracted from 1-week-old seedlings, grown on Murashige and Skoog medium (Murashige and Skoog, 1962) using DNeasy Plant Mini kit (Qiagen). The expected OsXTH8 promoter fragment was amplified using primer pairs of 5′-ATGCCCTGCAGGGAGGGAGTAGTAGCTAGCTTGAG-3′ (5′ side, SbfI site is underlined in the adaptor sequence) and 5′-TTCGCCATGGCTACTGTACTTGCTTG-3′ (3′-side, NcoI site is underlined in the adopter sequence). The 3′-side primer was designed in a way that it gave the initiation codon of OsXTH8 on cutting with NcoI restriction enzyme. The OsXTH8 promoter fragment was amplified using KOD plus (Toyobo, Osaka) using the PCR conditions: 94°C for 2 min (1 cycle), 94°C for 15 s, 63°C for 30 s, 68°C for 2 min (30 cycles), and 68°C for 7 min (1 cycle). The amplified fragment was gel purified using Wizard SV gel and PCR clean up system (Promega, Madison, WI). The purified product was incubated with dATP and Ex Taq (Takara) at 72°C for 5 min and purified again using Wizard SV gel and PCR clean up system. The purified fragment was cloned into pGEM-T easy of pGEM-T easy vector system (Promega). OsXTH8 promoter fragment was confirmed by sequencing using ABI 3100. OsXTH8 promoter fragment was released by digesting with SbfI and NcoI and cloned into a binary vector pSMAHdN627 (H. Nakamura, unpublished data; Fig. 7A). The resulting plasmid carrying OsXTH8 promter∷GUS fusion was transformed into rice via Agrobacterium-mediated transformation (Tanaka et al., 2001).
GUS assay was conducted according to Jefferson (1987). For histochemical analysis, leaf, stem, and root segments were sectioned into 30-μm-thick pieces by a microslicer and incubated in 50 mm sodium phosphate buffer, pH 7.2, containing 1.0 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (Wako Pure Chemical) and 5% methanol at 37°C for 2 to 24 h. The reaction was stopped by adding ethanol.
GUS activity was measured using the fluorogenic substrate 4-methyl-umbelliferylglucuronide (Sigma, St. Louis) and measured with an MTP-100F microplate reader (Corona Electric, Ibaraki, Japan). Each assay was carried out with crude extracts containing 5 μg proteins. The relative GUS activity was calculated as the mean of triplicate experiment.
In Situ Hybridization
Nodal part tissues taken from 2-week-old seedlings were fixed in 4% paraformaldehyde and 0.25% glutaraldehyde under vacuum. Fixed samples were then dehydrated through a graded ethanol series followed by a t-butanol series, and finally embedded in paraplast. Microtome sections (10 μm thick) were mounted on silicon-coated glass slides (Matsunami, Hamamatsu, Japan). Paraplast was removed through a graded ethanol series. Probes for in situ hybridization were labeled with digoxigenin11-UTP (Roche Diagnostics, Manheim, Germany). OsXTH8 expressed sequence tag pBluescript SK plasmid was either treated with XhoI and transcribed with T7 RNA polymerase (Stratagene, La Jolla, CA; antisense probe) or digested with EcoRI and transcribed with T3 RNA polymerase (Stratagene; sense probe). Immunological detection was done with an anti-digoxygenin-AP conjugate and 4-nitrobluetetrazolium (Roche Diagnostics; Kouchi and Hata, 1993).
Construction of RNAi OsXTH8 Transgenic Rice
For construction of RNAi transgenic plants, OsXTH8 cDNA in the pBluescript SK+ vector was amplified by PCR in both sense and antisense directions. Sense RNAi fragment was amplified using primer pairs of 5′-GGGGTACCTTTTGAACTCGATCGATTCAAA-3′ (5′ side, KpnI site underlined as a linker) and 5′-GCGAGCTCTGTTCATACCTGAGAGCATAAG-3′ (3′-side, ScaI site underlined as a linker). Anti-sense RNAi fragment was amplified using primer pairs of 5′-CGTCTAGATGTTCATACCTGAGAGCATAAG-3′(5′ side, XbaI site underlined as a linker) CGGGATCCTTTTGAACTCGATCGATTCAAA-3′ (3′ side, BamHI site underlined as a linker). The resulting PCR fragments were ligated between the CaMV 35S promoter and nopaline synthase terminator in the binary vector pIG121-Hm (Ohta et al., 1990) in a position sandwiching 700 bp of partial GUS coding region (Fig. 8A). The pIG121-Hm/RNAi OsXTH8 construct was confirmed by restriction mapping and sequencing. The pIG121-Hm/RNAi OsXTH8 plasmid and the control vector pIG121-Hm were then transferred into Agrobacterium tumefaciens strain EHA 105 (Hood et al., 1986) and transformed into rice as described (Toki, 1997). Transgenic plants were selected on medium containing hygromycin. Hyygromycin-resistant plants were transferred to soil and grown to maturity at 30°C in 16-h light/8-h dark cycle in closed greenhouse.
Total RNAs were extracted from leaves of 2-month-old control and transgenic plants and subjected to northern-blot analysis. The membrane was hybridized with the same probe and condition as described in the previous northern-blot analysis section.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AB110604.
Supplementary Material
Acknowledgments
We are grateful to Dr. T. Murakami and Dr. Y. Ohashi of the National Institute of Agrobiological Sciences for their technical help. We are also grateful to Dr. K. Nakamura of Nagoya University for providing pIG121-Hm vector for rice transformation, and Dr. E.E. Hood of ProdiGene for providing Agrobacterium strain EHA105.
This work was supported in part by a grant from the Program for Promotion of Basic Research Activities for Innovative Biosciences, Japan.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.052274.
References
- Akamatsu T, Hanzawa H, Ohtake Y, Takahashi T, Nishitani K, Komeda Y (1999) Expression of endoxyloglucan transferase genes in acaulis mutants of Arabidopsis. Plant Physiol 121: 715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrowsmith DA, de Silva J (1995) Characterization of two tomato fruit-expressed cDNAs encoding xyloglucan endotransglucosylases. Plant Mol Biol 28: 391–403 [DOI] [PubMed] [Google Scholar]
- Barrachina C, Lorences EP (1998) Xyloglucan endotransglucosylase activity in pine hypocotyls: intracellular localization and relationship with endogenous growth. Physiol Plant 102: 55–60 [DOI] [PubMed] [Google Scholar]
- Borriss R, Buettner K, Maentsaelae P (1990) Structure of the β-1,3-1,4-glucanase gene of Bacillus macerans: homologies to other β-glucanases. Mol Gen Genet 222: 278–283 [DOI] [PubMed] [Google Scholar]
- Buckeridge MS, Rayon C, Urbanowics B, Tine Aurelio MAS, Carpita NC (2004) Mixed linkage (1–>3), (1–>4) β-D-glucans of grasses. Cereal Chem 18: 115–127 [Google Scholar]
- Campbell P, Braam J (1999. a) Xyloglucan endotransglucosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci 4: 361–366 [DOI] [PubMed] [Google Scholar]
- Campbell P, Braam J (1999. b) In vitro activities of four xyloglucan endotransglucosylases from Arabidopsis. Plant J 18: 371–382 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Catala C, Rose JKC, Bennett AB (1997) Auxin regulation and spatial localization of an endo-1, 4-β-D-glucanase and a xyloglucan endotransglucosylase in expanding tomato hypocotyls. Plant J 12: 417–426 [DOI] [PubMed] [Google Scholar]
- Catala C, Rose JKC, York WS, Albersheim P, Darvill AG, Bennett AB (2001) Characterization of a tomato xyloglucan endotransglucosylase gene that is down-regulated by auxin in etiolated hypocotyls. Plant Physiol 127: 1180–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (1997) Assembly and enlargement of the primary cell wall in plants. Annu Rev Cell Dev Biol 13: 171–201 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Sovonick-Dunford SA (1989) Mechanism of gibberellin-dependent stem elongation in peas. Plant Physiol 89: 184–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva J, Jarman CD, Arrowsmith DA, Stronach MS, Chengappa S, Sidebottom C, Reid JS (1993) Molecular characterization of a xyloglucan-specific endo-(1, 4)-β-D-glucanase (xyloglucan endotransglucosylase) from nasturtium seeds. Plant J 3: 701–711 [PubMed] [Google Scholar]
- Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ (1992) Xyloglucan endotransglucosylase, a new wall-loosening enzyme activity from plants. Biochem J 282: 821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Jacobsen JV (1992) Gibberellin-responsive elements in the promoter of a barley high-pI alpha-amylase gene. Plant Cell 4: 1435–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbers K, Lorences EP, Barrachina C, Sonnewald U (2001) Functional characterization of Nicotiana tabacum xyloglucan endotransglucosylase (NtXET-1): generation of transgenic tobacco plants and changes in cell wall xyloglucan. Planta 212: 279–287 [DOI] [PubMed] [Google Scholar]
- Hood EE, Helmer GL, Fraley RT, Chilton MD (1986) The hyper virulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T-DNA. J Bacteriol 168: 1291–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M (2002) The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14: 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 [Google Scholar]
- Juncosa M, Pons J, Dot T, Querol E, Planas A (1994) Identification of active site carboxylic residues on Bacillus licheniformis 1,3-1,4-β-D-glucan 4-glucanylhydrolase by site-directed mutagenesis. J Biol Chem 269: 14530–14535 [PubMed] [Google Scholar]
- Kamijima O (1981) Consideration on the mechanism of expression of dwarf genes in rice plants. II. The actions of dwarf genes on cell division and cell elongation in parenchyma of internode. Jpn J Breed 31: 302–315 [Google Scholar]
- Kouchi H, Hata S (1993) Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol Gen Genet 238: 106–119 [DOI] [PubMed] [Google Scholar]
- Maclachlan G, Brady C (1994) Endo-1, 4-β-glucanase, xyloglucanase and xyloglucan endotransglucosylase activities versus potential substrates in ripening tomatoes. Plant Physiol 105: 965–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nishitani K (1997) The role of endoxyloglucan transferase in the organization of plant cells. Int Rev Cytol 173: 157–205 [DOI] [PubMed] [Google Scholar]
- Nishitani K, Tominaga R (1991) In vitro molecular weight increase in xyloglucan by an apoplastic enzyme preparation from epicotyls of Vigna angularis. Physiol Plant 82: 490–497 [Google Scholar]
- Nishitani K, Tominaga R (1992) Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J Biol Chem 267: 21058–21064 [PubMed] [Google Scholar]
- Ogawa S, Toyomasu T, Yamane H, Murofushi N, Ikeda R, Mormoto Y, Nishimura Y, Omori T (1996) A step in the biosynthesis of gibberellins that is controlled by the mutation in the semi-dwarf rice cultivar Tan-Ginbozu. Plant Cell Physiol 37: 363–368 [Google Scholar]
- Ohta S, Mita S, Hattori T, Nakamura K (1990) Construction and expression in tobacco of a β-glucronidase (GUS) reporter gene containing an intron within the coding sequence. Plant Cell Physiol 31: 805–813 [Google Scholar]
- Planas A, Juncosa M, Lloberas J, Querol E (1992) Essential catalytic role of Glu134 in endo-β-1, 3-1,4-D-glucan 4-glucanohydrolase from B. licheniformis as determined by site-directed mutagenesis. FEBS Lett 308: 141–145 [DOI] [PubMed] [Google Scholar]
- Potter I, Fry SC (1994) Changes in xyloglucan endotransglucosylase (XET) activity during hormone-induced growth in lettuce and cucumber hypocotyls and spinach cell suspension cultures. J Exp Bot 45: 1703–1710 [Google Scholar]
- Redgwell RJ, Fry SC (1993) Xyloglucan endotransglucosylase activity increases during kiwifruit (Actinidia deliciosa) ripening. Plant Physiol 100: 1318–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43: 1421–1435 [DOI] [PubMed] [Google Scholar]
- Saab I, Sachs M (1996) A flooding-induced xyloglucan endotransglucosylase homologue in maize is responsive to ethylene and associated with aerenchyma. Plant Physiol 112: 385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Morinaka Y, Ishiyama K, Kobayashi M, Itoh H, Kayano T, Iwahori S, Matsuoka M, Tanaka H (2003) Genetic manipulation of gibberellin metabolism in transgenic rice. Nat Biotechnol 21: 909–913 [DOI] [PubMed] [Google Scholar]
- Schroder R, Atkinson RG, Langenkamper G, Redgwell RJ (1998) Biochemical and molecular characterization of xyloglucan endotransglucosylase from ripe kiwifruit. Planta 204: 242–251 [DOI] [PubMed] [Google Scholar]
- Schunmann PHD, Smith RC, Lang V, Matthews PR, Chandler PM (1997) Expression of XET-related genes and its relation to elongation in leaves of barley (Hordeum vulgare L.). Plant Cell Environ 20: 1439–1450 [Google Scholar]
- Shimizu Y, Aotsuka S, Hasegawa O, Kawada T, Sakuno T, Sakai F, Hayashi T (1997) Changes in levels of mRNAs for cell wall-related enzymes in growing cotton fiber cells. Plant Cell Physiol 38: 375–378 [DOI] [PubMed] [Google Scholar]
- Skriver K, Olsen FL, Rogers JC, Mundy J (1991) cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc Natl Acad Sci USA 88: 7266–7270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RC, Matthews PR, Schunmann PHD, Chandler PM (1996) The regulation of leaf elongation and xyloglucan endotransglucosylase by gibberellin in “Himalaya” barley (Hordeum vulgare L.). J Exp Bot 47: 1395–1404 [Google Scholar]
- Sutoh K, Yamauchi D (2003) Two cis-acting elements necessary and sufficient for gibberellin-upregulated proteinase expression in rice. Plant J 34: 35–45 [DOI] [PubMed] [Google Scholar]
- Takano M, Fuji N, Higashitani A, Nishitani K, Hirasawa T, Takahashi H (1999) Endoxyloglucan transferase cDNA isolated from pea roots and its fluctuating expression in hydrotropically responding roots. Plant Cell Physiol 40: 135–142 [DOI] [PubMed] [Google Scholar]
- Tanaka H, Kayano T, Ugaki M, Shiobara F, Onodera H, Ono K, Tagiri A, Nishizawa Y, Shibuya N (2001) Transformation technique for monocotyledons. Patent Cooperation Treaty Application No. WO 01/06844 A1 (December 22, 2000)
- Toki S (1997) Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Mol Biol Rep 15: 16–21 [Google Scholar]
- Uozu S, Tanaka-Ueguchi M, Kitano H, Hattori K, Matsuoka M (2000) Characterization of XET-related genes of rice. Plant Physiol 122: 853–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verica JA, Medford JI (1997) Modified MER15 expression alters cell expansion in transgenic Arabidopsis plants. Plant Sci 125: 201–210 [Google Scholar]
- Vissenberg K, Martinez-Vilchez IM, Verbelen JP, Miller JG, Fry SC (2000) In vivo colocalization of xyloglucan endotransglycosylase activity and its donor substrate in the elongation zone of Arabidopsis roots. Plant Cell 12: 1229–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G (1986) A new method for predicting signal sequence cleavage sites. Nucleic Acids Res 14: 4683–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Campbell P, Vargheese AK, Braam J (1996) The Arabidopsis XET-related gene family: environmental and hormonal regulation of expression. Plant J 9: 879–889 [DOI] [PubMed] [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J (1995) Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglucosylase. Plant Cell 7: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki J, Kishimoto N, Nakamura K, Fujii F, Shimbo K, Otsuka Y, Wu J, Yamamoto K, Sakata K, Sasaki T, et al (2000) Embarking on rice functional genomics via cDNA microarray: use of 3′ UTR probes for specific gene expression analysis. DNA Res 7: 367–370 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K (2001) A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol 42: 1025–1033 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Rose JK, Nishitani K (2004) A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiol 134: 1088–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek DM, Clouse SD (1994) Molecular cloning and characterization of a brassinosteroid-regulated gene from elongating soybean (Glycine max L.) epicotyls. Plant Physiol 104: 161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.