Abstract
Ethyl methanesulfonate mutagenesis of the model legume Medicago truncatula has previously identified several genes required for early steps in nodulation. Here, we describe a new mutant that is defective in intermediate steps of nodule differentiation. The lin (lumpy infections) mutant is characterized by a 4-fold reduction in the number of infections, all of which arrest in the root epidermis, and by nodule primordia that initiate normally but fail to mature. Genetic analyses indicate that the symbiotic phenotype is conferred by a single gene that maps to the lower arm of linkage group 1. Transcriptional markers for early Nod factor responses (RIP1 and ENOD40) are induced in lin, as is another early nodulin, ENOD20, a gene expressed during the differentiation of nodule primordia. By contrast, other markers correlated with primordium differentiation (CCS52A), infection progression (MtN6), or nodule morphogenesis (ENOD2 and ENOD8) show reduced or no induction in homozygous lin individuals. Taken together, these results suggest that LIN functions in maintenance of rhizobial infections and differentiation of nodules from nodule primordia.
In response to specific soil bacteria called rhizobia, legume roots initiate a unique developmental program that culminates in the formation of nitrogen-fixing root nodules. The highly specialized microenvironment of the nodule provides conditions necessary for the conversion of atmospheric nitrogen to ammonium by the rhizobial nitrogenase enzyme. Nod factors, lipooligosaccharide signal molecules produced by the bacterial partner, are required for the parallel processes of bacterial infection and nodule morphogenesis (Geurts and Bisseling, 2002). In recent years, major advances have been made in defining the host's responses to Nod factors, culminating in the cloning of several plant genes required for Nod factor signal transduction (Schauser et al., 1999; Endre et al., 2002; Stracke et al., 2002; Limpens and Bisseling, 2003; Madsen et al., 2003; Radutoiu et al., 2003; Ané et al., 2004; Lévy et al., 2004). The characterization of these genes and their mutant phenotypes indicates that receptor-like kinases play multiple roles in the perception of Nod factor and that transduction of the signal proceeds via calcium-mediated responses and transcriptional regulation (Cullimore and Dénarié, 2003; Parniske and Downie, 2003; Riely et al., 2004).
Current understanding of the plant's contribution to symbiotic development subsequent to Nod factor perception, including rhizobial infection, primordium initiation, and nodule differentiation, comes largely from microscopic examination and characterization of plant genes that are differentially expressed during these processes. Early in the interaction, the bacteria gain access to the root through selective penetration of root hair cells. Invagination of the plant plasma membrane and deposition of cell wall material enable bacterial invasion via infection threads. Some, but not all, infection threads traverse the epidermal cells to invade root cortical cells that have been mitotically activated in response to Nod factor (Stougaard, 2000). Markers for infection and primordium initiation include RIP1, ENOD40, ENOD20, and MtN6. RIP1 (Cook et al., 1995), encoding a cell wall peroxidase, is induced prior to infection and may participate in infection initiation. ENOD40 is expressed in the dividing root cortical and pericycle cells and has been reported to have a role in cortical cell activation (Charon et al., 1997). Induction of both RIP1 and ENOD40 depends on the plant's perception of Nod factor (Crespi et al., 1994; Cook et al., 1995). ENOD20 is predicted to have a role in cell wall reorganization during infection thread growth and/or differentiation of the infected cells of the root nodules (Greene et al., 1998). MtN6 is expressed in root cortical cells, just in advance of infection thread penetration, and may have a role in preparing cells for infection (Mathis et al., 1999). The precise cellular functions of ENOD20 and MtN6 are not currently known.
Coincident with infection in the epidermis and outer cortical cells, cell divisions initiate in the root cortex, leading to the formation of a nodule primordium and ultimately to a differentiated nodule organ. In indeterminate nodules, such as those formed on alfalfa (Medicago sativa) and the model legume Medicago truncatula, some derivatives of the persistent meristem exit mitosis and enter a modified cell cycle characterized by endoreduplication, leading to an increase in ploidy and cell size (Cebolla et al., 1999). In M. truncatula nodules, expression of CCS52A, which codes for a WD-repeat protein, regulates endoreduplication of the cortical cells and is required for infection persistence (Vinardell et al., 2003). Other genes that are induced in specific tissue layers in nodules serve as developmental markers for progression through later symbiotic differentiation (e.g. Mitra and Long, 2004). For example, ENOD2, encoding a cell wall structural protein found in the nodule parenchyma surrounding the infected zone, and ENOD8, encoding a putative esterase, are associated with the development of nodule structures prior to the onset of nitrogen fixation (Dickstein et al., 1988, 1993; Pringle and Dickstein, 2004).
With the goal of identifying host genes that govern infection persistence, we screened ethyl methanesulfonate mutants of M. truncatula for mutants with an infection arrest phenotype. This study focuses on one such mutant, called lin (lumpy infections), that initiates but cannot sustain rhizobial infections. Although rhizobia elicit several Nod factor responses and nodule primordium formation on lin, the mutant is deficient in sustained meristematic activity. By investigating expression of genes that delineate stages of infection progression and primordium development, we demonstrate that lin is arrested prior to the onset of differentiation of nodule tissues. Genetic mapping has placed lin on linkage group 1 (LG1) of M. truncatula and fine mapping of the lin locus is under way. Thus, LIN defines a plant gene that is required for infection thread formation in root cortical cells and for formation of functional root nodules.
RESULTS
In an intensive screen for symbiotic mutants with altered nodulation phenotypes (Penmetsa and Cook, 2000), we identified a mutant line, C88, which developed small white bumps in response to rhizobia (Fig. 1, A–C). The phenotype of C88 was heritable in successive generations and was selected as a candidate for further analysis.
Figure 1.
Symbiotic phenotypes of wild-type and lin roots. Plants were inoculated with the wild-type Rhizobium strain ABS7M containing the chimeric hemA∷lacZ gene. A, Distribution of nodules (arrow) in the nodulation zone of a wild-type root (left) and primordia (arrowhead) in the corresponding zone of a lin root (right) at 18 dpi. B to G, Plant roots were harvested at different time points after inoculation and processed for β-galactosidase activity. B and C, Segments of lin root showing the recurrent occurrence of primordia (arrowheads in B) and infections (arrows in C) at 18 dpi. D and F, Wild-type infections. A root harvested at 1 dpi (in D) shows an infection thread (arrow) penetrating through the root hair and into the cortical cell layer, while F depicts a successful infection (arrow) ramifying within a developing primordium (np) at 3 dpi. E and G, Infections in the lin mutant. E, Infection (arrow) arrested within the root hair at 1 dpi. G, Arrested infection (arrow) that fails to penetrate the nodule primordium (np) at 3 dpi. Bars = 50 μm.
Observations made on stained whole mounts of fixed roots or root sections of mutant and wild-type plants showed aberrant development of infection threads in C88. Infection threads were first visible in the curled root hairs of both wild-type and mutant plants by 1 d postinoculation (dpi). In wild-type plants, the infection threads invaded the cortical layer by 3 dpi (Fig. 1, D and F). However, in C88, all infections arrested within root hairs, typically after very limited progression into the infected cell.
Similar to wild-type plants, C88 plants displayed foci of cell divisions in the root cortex that formed the nascent nodule primordia. These developing primordia were not invaded by the infection threads and thus showed an arrest in differentiation (Fig. 1G) in comparison to wild-type nodules (Fig. 1F). As demonstrated in Figure 2, cellular differentiation was substantially reduced within C88 nodule primordia, although prevascular strands were sometimes noted to connect to the vascular cylinder of the root.
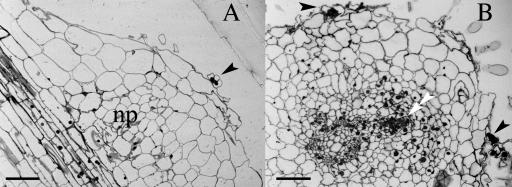
Figure 2.
Cytological characterization of infection arrest and nodule primordia of lin. Sinorhizobium-infected lin roots were fixed and stained for β-galactosidase activity. Semithin sections of infected roots were stained with methylene blue plus azure II. A, A longitudinal section through a lin root at 21 dpi showing a nodule primordium (np) with an infection (arrowhead) arrested in the root hair. B, A cross-section of a lin root at 21 dpi shows two arrested infections (arrowheads), one opposite a xylem pole and one opposite a phloem pole. A developing vascular strand (double arrowhead) is found within the nodule primordium (np) adjacent to a xylem pole. Bars = 50 μm.
Infections in C88 occurred at less than one-fourth the frequency of infections in wild-type plants (Table I), and most infections occurred in coincidence with subtending nodule primordia. Interestingly, infection and formation of nodule primordia in C88 recurred continually during prolonged growth in the presence of rhizobium (Fig. 1, A–C). This phenotype is in contrast to wild-type plants, where initial infection and nodulation are transient, resulting in a narrow zone of primary nodule formation.
Table I.
Comparison of nodulation phenotype between A17 and lin
| Phenotype | A17 | lin |
|---|---|---|
| No. of infection per root at 3 dpia | 116.5 ± 20.0 | 25.0 ± 6.5 |
| No. of primordia at 18 dpi | 0.0 ± 0.0 | 18.0 ± 2.0 |
| No. of nodules at 18 dpi | 8.0 ± 2.0 | 0.0 ± 0.0 |
| Nitrogenase activity at 18 dpi (C2H4 nmol/plant/h)b | 8.00 ± 2.00 | 0.01 ± 0.00 |
Values represent mean ± sd.
Infection count was made using plants inoculated with wild-type Rhizobium strain ABS7M containing the chimeric hemA∷lacZ gene, processed for β-galactosidase activity observed by light microscopy.
Nitrogenase activity was assayed by measuring acetylene reduction on a gas chromatograph.
The Mutant Phenotype in C88 Is Conditioned by a Monogenic, Recessive Mutation That Maps to LG1 of the M. truncatula Map
For purposes of genetic characterization, C88 plants were crossed with the wild-type genotype A17. As shown in Table II, the frequency of nod+ and nod− phenotypes in the F2 generation was consistent with segregation as a single recessive allele. To determine whether C88 was allelic to other nodulation mutants, complementation analysis was performed with mutant lines C19 and C96, which showed an infection arrest phenotype, the production of small white nodules, or both. The F1 progeny from these crosses revealed that none of the lines tested was allelic to C88. C88, therefore, identifies a new complementation group. The gene conditioning the mutant phenotype in C88 was designated lin-1, for lumpy infections allele 1, in accordance with nomenclature rules for M. truncatula (VandenBosch and Frugoli, 2001). The gene name has been registered with the Medicago gene nomenclature index (http://www.genome.clemson.edu/affiliated_cugi/medicago).
Table II.
Genetic analysis of the M. truncatula nodulation mutant lin
| Parental Lines
|
F2 Progeny Analyzed
|
No. of Plants
|
Observed
|
||
|---|---|---|---|---|---|
| Nod+ | Nod− | χ2 Value | P Value | ||
| lin × A17 | 122 | 99 | 23 | 2.459 | >0.100 |
| lin × A20 | 125 | 93 | 32 | 0.024 | >0.25 |
Mapping of lin was initiated to allow eventual map-based cloning of this gene. A mapping population was constructed by crossing lin in the background of the male sterile mutant tap with ecotype A20 (Penmetsa and Cook, 2000). The F1 progeny from the cross between lin and A20 showed a wild-type phenotype for nodulation, consistent with the recessive nature of the mutation. One hundred twenty-five F2 individuals were phenotyped for nodulation, with a segregation pattern consistent with the recessive nature of lin-1 (Table II).
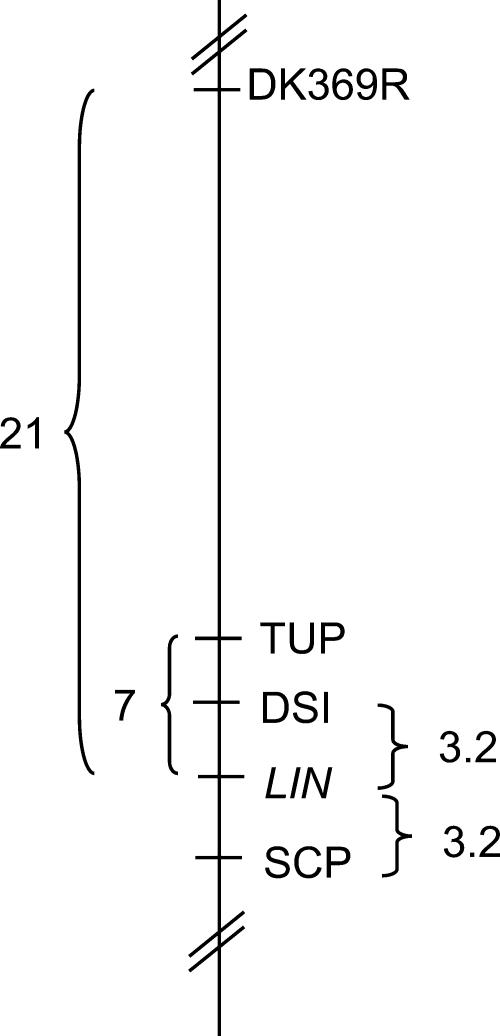
Several molecular markers distributed on the eight linkage groups of the M. truncatula genetic map (Choi et al., 2004) were selected to determine the linkage relationship of each to lin (see “Materials and Methods” for list). As a first screen, the genotypes of these markers were determined in 16 F2 lin mutant plants (plants homozygous for the lin parental allele). This analysis revealed a presumptive linkage between lin and the TUP marker of LG1. Analysis of the expanded population of 125 individuals, as well as other molecular markers linked to TUP, confirmed the location of lin on the lower arm of LG1, positioned between markers DSI and SCP (Fig. 3). Four of the eight recombination events were in the lin homozygous background, permitting location of the recombinants relative to lin and the codominant markers DSI and SCP based on F2 data alone. The remaining four recombinants occurred in phenotypically wild-type plants. Mapping these recombinants relative to lin was accomplished by phenotyping of F3 progeny, placing lin equidistant between DSI and SCP (Fig. 3).
Figure 3.
Genetic mapping of lin to LG1 of M. truncatula. Localization of the LIN locus on LG1 of the M. truncatula genetic map. Markers DK369R, TUP, DSI, and SCP from the genetic map of LG1 (Choi et al., 2004) were found to be linked to the LIN locus. The recombination frequency between the two flanking markers DSI and SCP is 6.4 based on the lin mapping population.
The nodule-expressed gene MtN6 (Mathis et al., 1999) was also mapped to the lower arm of LG1 of M. truncatula (T. Huguet and P. Gamas, personal communication) based on a mapping population derived from the mapping parents Jemalong and DZA315 (Thoquet et al., 2002). Both the map location and the fact that MtN6 expression occurs spatially in advance of growing infection threads (Mathis et al., 1999) suggested that MtN6 and LIN could be the same gene. To test this hypothesis, we mapped MtN6 in the lin mapping population to examine the extent of cosegregation between the two loci. Eight plants were identified with recombinations between lin and MtN6, thus demonstrating that lin and MtN6 are distinct loci. MtN6 was determined to map north of lin on LG1 to a position most closely linked to the genetic marker DK369R.
lin Nodule Development Is Blocked before the Stage of Nodule Differentiation
The morphological phenotype of lin suggested that LIN is necessary for nodule differentiation. To determine the nature of the developmental block in lin, we used RNA blots to examine the expression of four nodulin genes as markers of various stages of nodulation. The early nodulins RIP1 and ENOD40 were selected as markers of early responses to Nod factor. As shown in Figure 4, the onset of ENOD40 induction was similar between wild-type and mutant plants, with increased transcript abundance evident at 1 dpi. RIP1 was also induced in lin and wild-type plants, with transcript first detectable in both genotypes at 6 h postinoculation. ENOD2 and ENOD8 were chosen as markers of nodule differentiation. Observations on RNA blots revealed that, in wild-type plants, ENOD2 expression commenced about 4 dpi, and ENOD8 transcript accumulated by 6 dpi. Transcripts corresponding to these two nodulins could not be detected in lin (Fig. 4). A duplicate blot, with RNA from independent biological replicates, verified the expression patterns of these nodulins (data not shown).
Figure 4.
Northern-blot analysis of expression of early nodulins in nodulating roots of lin and wild-type plants. Total RNA from the main root zone (see “Materials and Methods”) of plants before and after inoculation was electrophoresed, blotted, and probed sequentially with cDNA probes. The rDNA probe is a loading control.
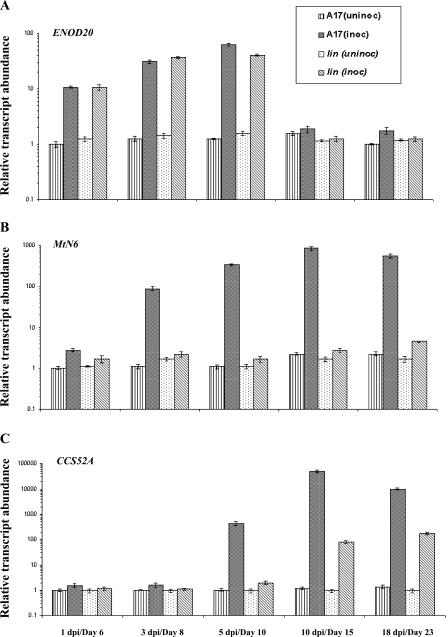
Together, these results indicated that the early responses of the plant to rhizobia were present in lin. By contrast, cytological events related to nodule organogenesis, including nodule differentiation, were partially or completely blocked in lin. We therefore chose three additional markers of early primordium formation and infection thread penetration (ENOD20, MtN6, and CCS52A) to examine the molecular phenotype of lin. Real-time PCR analysis indicated that both genotypes showed an increasing expression of ENOD20 in inoculated roots through 5 dpi (Fig. 5A), with slightly higher expression in wild type relative to lin at 5 dpi. By contrast, expression of MtN6 in lin was significantly lower at all time points compared to wild type. MtN6 induction in lin was 1.5-fold at 1 dpi and reached a maximum of 2.5-fold at 18 dpi (Fig. 5B), as compared to the wild-type roots in which a dramatic increase of MtN6 occurred between 1 dpi (2.7-fold) and 10 dpi (380-fold). For both genes, the differences in basal levels of expression in roots between the two genotypes were slight, and transcript abundance in uninoculated roots varied little over time.
Figure 5.
Relative transcript abundance of marker genes for primordium development in wild-type and lin roots. A to C, Relative transcript abundance of ENOD20 (A), MtN6 (B), and CCS52A (C) in root samples of A17 and lin over a time course (x axes) with and without inoculation with rhizobia. Lined bars represent uninoculated root tissue of A17; black hatched bars represent inoculated root samples of A17; spotted bars represent uninfected samples of lin; and gray hatched bars represent infected root samples of lin. Transcript abundance (y axes) for all samples was estimated relative to A17 uninoculated roots on day 6. Each point represents the mean of three or four replicates with error bars representing the sd. Note that the y axis is in logarithmic scale and the scale is different for each gene. Uninoculated samples are indicated in terms of their physiological age in days and infected samples are indicated in dpi; for each time point, uninoculated and inoculated samples were harvested at the same age.
During the maturation of indeterminate nodules, selected cells within the primordium switch from mitotic cycles into endoreduplicating cycles. This is mediated by the cell cycle-switching gene CCS52A. We analyzed the expression of CCS52A in lin compared to that of wild-type plants. As shown in Figure 5C, transcripts increased in both wild-type and lin genotypes, although the response was both earlier and of substantially greater magnitude in wild type compared to lin (i.e. 50- to 500-fold greater induction in wild type).
DISCUSSION
A Symbiotic Mutant, lin, Identifies a Gene in M. truncatula Required for Persistence of Rhizobial Infections and Differentiation of Nitrogen-Fixing Nodules
In an effort to identify plant genes required for infection maintenance and nodule organogenesis in the legume-Rhizobium symbiosis, we identified a symbiotic mutant in M. truncatula called lin that is blocked in development prior to invasion of nodule primordia by infection threads. In lin, the number of infections is reduced by 5-fold compared to wild type. Infections that initiate in lin are of abnormal structure and are arrested within the root epidermis, failing to penetrate cells of the root cortex. Genetic analysis of lin showed that the mutation is conferred by a single recessive allele, and genetic mapping has placed LIN in the lower arm of LG1.
The lin phenotype supports the hypothesis that progressive invasion of the infection thread is necessary for complete nodule organogenesis (Dénarié et al., 1992). Previous findings have indicated that invading bacteria can induce nodule initiation in the absence of a persistent infection (Dénarié et al., 1992). However, the developing nodule meristem persists to produce a nodule with differentiated internal tissues only when the growing infection thread invades the primordium. This could be due to the production of unknown growth-stimulating substances by the invading bacteria (Truchet et al., 1980; Yang et al., 1992), or by a mechanism that couples the continuous differentiation of nodule meristems to the invasion of the meristem derivatives by rhizobial infection threads.
The lin mutation blocks the penetration of the infection thread into the cortex but permits mitotic activation of cortical cells and the initial stages of primordium formation. The lin phenotype could be due to one or more factors. Failure to transduce the Nod factor or other signals properly could result in lack of continued symbiotic development. Conversely, deployment of an inappropriate defense response could result in an active infection arrest. The former hypothesis was addressed in further phenotypic characterization of the mutant.
LIN Is Required for Appropriate Regulation of Nodule Initiation
In M. truncatula, the host plant regulates the number of persistent rhizobial infections and nodules by at least two distinct mechanisms. Ethylene insensitivity, as caused by the skl mutation (Penmetsa and Cook, 1997), causes a large increase in infection persistence, resulting in hypernodulation in the susceptible zone of the mutant root. The mutation in the sunn locus causes an independent hypernodulation phenotype that is normally sensitive to ethylene, although sunn root growth is insensitive to ethylene (Penmetsa et al., 2003). In lin, the continuous recurrence of primordia and arrested infections in the nodulation zone of roots during long exposure to rhizobia could indicate that there is another level of unexplored regulation imposed by the host plant in controlling nodule numbers. Analogous to mechanisms proposed for autoregulation of nodule formation (Caetano-Anollés et al., 1991; Wopereis et al., 2000), mature nodule meristems may trigger suppression of new nodulation events in the adjacent nodulation zone. The absence of effective nodules in lin could result in absence of suppression, leading to a recurrent initiation of new foci of cell divisions. This phenomenon is likely to be a downstream effect of the absence of mature nodules rather than from a specific effect of the lin gene.
Developmental Markers for Nodule Differentiation Are Misregulated in lin
While recent successes in map-based cloning have advanced understanding of plant genetic control of Nod factor perception, knowledge of genes regulating stages of nodule development and infection is still rudimentary. This intermediate stage encompasses a range of activities, including cortical cell activation leading to formation of primordia, infection thread formation and penetration into the primordium, release of bacteria as membrane-bound organelles within cells of the primordium, and endoreduplication and subsequent differentiation of these cells. In comprehensive genetic and phenotypic analyses of pea (Pisum sativum) mutants, Tsyganov et al. (2002) identified two pea genes, sym37 and sym38, that developed nodule primordia arrested at the stage of nodule meristem development, as in lin. However, both sym37 and sym38 showed very different infection thread morphology as compared to what is seen in lin. In sym37, infection threads were arrested immediately after they began to grow in contrast to lin, where infection threads fail to elongate. In sym38, the percentage of infection threads was 10% to 30% higher than in the wild type and, although all infections arrested in root hair cells, the degree of progression within the root hair was highly variable. lin was further distinguished from sym37 and sym38 because the early arrest of infection thread growth in lin was accompanied by radial expansion of the infection threads (causing a bulbous appearance) and continued bacterial cell division. Arrested development was also described for a Lotus japonicus mutant, called crinkle, defining the Ljsym79 locus (Tansengco et al., 2003), and a M. truncatula mutant, called Mtsym1 (Bénaben et al., 1995). In addition to forming white bump-like structures as seen in lin, both Mtsym1 and Ljsym79 formed nodule structures containing some bacteroid-infected cells, suggesting that the developmental block in these mutants is at a stage subsequent to where lin acts.
We used northern-blot and real-time PCR analysis to analyze expression of various nodulin genes according to the lin mutation, and by comparison to wild type we were able to infer additional aspects of the lin phenotype. As depicted in Table III, the composite of the current results with those from previous studies allows the ordering of both gene expression and the action of genetic loci within the context of a pathway for nodule development and gene transcription. Three genes (RIP1, ENOD20, and ENOD40) were induced in inoculated lin roots at levels nearly comparable to wild type. The induction of RIP1 and ENOD40 is consistent with the ability to perceive and transduce the Nod factor signal by lin, as in wild type (Crespi et al., 1994; Cook et al., 1995). LIN is thus predicted to act downstream of the Nod factor signal transduction genes, including NFP, DMI1, DMI2, and DMI3, which are required for induction of RIP1, ENOD40, and another early nodulin, ENOD11 (Catoira et al., 2000; Ben Amor et al., 2003). ENOD20 expression appeared to be similar between the wild type and lin, indicating that its induction does not require infection thread penetration of the root cortex.
Table III.
Expression of nodulins in M. truncatula mutants defective in infection or nitrogen fixation
| Expressed Genes
|
Host Genotypes
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Inf−, Nod-Mutants
|
Infection Arrest Mutants
|
Fix-Mutants
|
Wild Type
|
||||||||
| nfpa | dmi1, dmi2, dmi3b | nsp1b | nspc | hcld | line | bit, rit, Mtsym1f | dnf 1, dnf5f | dnf2, dnf3, dnf4, dnf6, dnf7f | A17 | ||
| Early Nodulins | RIP 1 | − | − | ± | ± | + | + | + | |||
| ENOD11 | − | − | ± | − | + | + | |||||
| ENOD20 | + | + | |||||||||
| ENOD40 | − | − | − | + | + | ||||||
| CCS52A | ± | + | |||||||||
| Late Early Nodulins | MtN6 | − | + | ||||||||
| ENOD2 | − | − | + | + | + | ||||||
| ENOD8 | − | + | |||||||||
| LEC4 | − | + | + | + | |||||||
| Late Nodulins | MtN31 | − | − | + | + | ||||||
| MtCAM1 | − | − | + | + | |||||||
| MtLB | − | − | + | + | |||||||
−, Nodulin tested was not induced in roots inoculated with rhizobia or treated with Nod factor or in nodules; +, rhizobial induction; and ±, induction at markedly reduced levels in comparison to wild type. No entry indicates that assay has not been done.
Oldroyd and Long (2004).
This study.
The M. truncatula mutants hcl, nsp1, and nsp2 also show modest-to-normal induction of one or more early nodulins that are induced in response to Nod factor, but otherwise have different phenotypes from lin (Catoira et al., 2000, 2001; Oldroyd and Long, 2003). Mutants at these three loci exhibit root hair deformation, but do not become infected. Also, cell divisions are induced in the inner root cortex of hcl by rhizobia, but do not persist to generate a nodule primordium. We hypothesized that a class of molecular markers for infection initiation and primordium formation would differentiate lin from the other two mutants. The cell cycle switch gene CCS52A is a suitable marker for the transitional stages between early Nod factor perception and nodule differentiation. CCS52A was induced around the same time in lin and wild type, although its transcript levels were much lower in the mutant. This suggests that LIN is required for full expression of CCS52A, but not for its induction in response to Rhizobium. Reduced expression of CCS52A in lin could indicate that lin nodules undergo only a modest increase in ploidy. The infection arrest phenotype of lin supports the prediction from Vinardell et al. (2003) that posits that ramification of infection threads is necessary for full activation of CCS52A. In addition to results from monitoring CCS52A expression presented here, a more in-depth search for genes involved in primordium formation is currently under way using comparative cDNA microarray analysis of lin and wild type. A similar approach, based on identification of genes that are misregulated in lin, should also identify genes that function in primordium differentiation and infection thread growth.
Three genes (MtN6, ENOD8, and ENOD2) were identified that fail to be induced in lin. Consistent with previous findings, levels of MtN6 increased in wild-type plants during infection progression and nodule development. In contrast, lin roots showed minimal induction of MtN6, suggesting that preinfection events related to primordium differentiation are not activated in lin. The similarity of the lin-arrested infections and nodule structures to those of empty alfalfa nodules induced by Rhizobium meliloti EPSI (exo−) mutants (Yang et al., 1992) led us to investigate the expression of ENOD2 and ENOD8 in lin. ENOD2 and ENOD8 are expressed in empty alfalfa nodules induced by exo− mutants (Dickstein et al., 1988, 1993). Surprisingly, we found that neither ENOD2 nor ENOD8 are expressed in lin nodule structures. This demonstrated that lin nodule structures are not the equivalent of the previously studied empty alfalfa nodules and are probably blocked at an earlier stage. This is consistent with recent findings of Mitra and Long (2004) that the nodule-like structures formed in M. truncatula in response to R. meliloti exo− mutants are not morphologically similar to those observed in alfalfa, and that ENOD2 fails to be induced in M. truncatula symbioses that are blocked at the stage of infection penetration of the root inner cortex. Together with MtLEC4, a nodule-specific lectin that is misregulated in infection arrest mutants (Mitra and Long, 2004), ENOD2, ENOD8, and MtN6 genes could be considered late early nodulins, due to the apparent requirement for infection progression for their induction (Table III).
In summary, the phenotypic analyses of lin presented here suggest that LIN functions downstream of initial events in Nod factor signal transduction, but is required for both infection persistence and nodule differentiation. We hypothesize that LIN may also be required to suppress a host defense response, allowing some infections to proceed. Further work to investigate this hypothesis is in progress. The map-based cloning and molecular characterization of LIN, initiated here with genetic mapping of the lin mutation to LG1 of M. truncatula, should provide further insight into LIN function.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Medicago truncatula cv Jemalong genotype A17 was used as the wild-type control for phenotypic and genotypic analysis. The plants were grown as previously described by Cook et al. (1995). For nodulation studies, seeds were scarified (Penmetsa and Cook, 2000) and seedlings were transplanted to aeroponic chambers 1 d after germination and inoculated 5 d later with a late log phase culture of Sinorhizobium meliloti. The day of rhizobial inoculation was considered as the zero time point for rhizobial studies. The bacterial strain ABS7M (pXLGD4) of S. meliloti was used for all studies of inoculated seedlings. It carries a constitutively expressed lacZ construct, which makes it an efficient system for detection of infection in the plant root (Penmetsa and Cook, 1997). The mutant line C88 was isolated in the process of screening for putative mutants in seed-bulk C that formed a part of an ethyl methanesulfonate-mutagenized population of A17 seedlings as previously described (Penmetsa and Cook, 2000). The mutant was back-crossed to the wild-type parent (A17) that was homozygous for the tap mutation. The tap mutation renders flowers male-sterile and female-fertile with no apparent effects on nodulation phenotype, and thus facilitates unambiguous crossing (Penmetsa and Cook, 2000). The F1 progeny were allowed to self-pollinate to obtain lines homozygous for the C88 mutation. C88 homozygotes with a wild-type flower phenotype were allowed to self-pollinate, and seeds from the F3− or F4− generation plants were used for all analyses of the mutant phenotype.
Light Microscopic Analysis of Roots and Nodules
To visualize infection of roots and nodules by rhizobia, tissues inoculated with the rhizobia bearing the lacZ construct were stained for β-galactosidase activity (Penmetsa et al., 2003) and examined by bright-field microscopy under Nikon Eclipse E800 photo microscope (Frank Fryer, Huntly, IL).
To facilitate the examination of the internal structure of the root, segments containing infections were dissected from the main root, dehydrated in an ethanol series, and embedded in LR White acrylic resin as described previously (Sherrier and VandenBosch, 1994). Series of semithin sections were cut on glass knives using an Ultracut microtome (Leica, Deerfield, IL) and placed on poly-L Lys-coated slides. Sections were stained with 0.05% methylene blue and 0.05% azure II in 0.1% sodium metaborate and observed by bright-field optics.
Acetylene Reduction Assay
Nitrogenase activity of inoculated roots was determined by assaying acetylene reduction, using roots harvested from an aeroponic chamber 21 dpi (Somasegaran and Hoben, 1994). Excised nodulation zones from wild-type plants and the entire root system of the mutant plants were assayed for nitrogenase activity. For each genotype, samples consisting of three roots each were measured for acetylene reduction in 2-mL sealed vials in the presence of 10% acetylene. After 1.5 h of incubation in acetylene, a sample (5–10 μL) was injected into a Photovac 10S Plus gas chromatograph (Photovac, Markham, Canada) that used a photo-ionizing detector. Samples were compared to a standard curve generated against a 5 ppm ethylene standard, and nitrogenase activity was expressed as moles of C2H4 evolved (per minute per gram fresh weight).
Genetic Analysis and Mapping of lin
For allelism tests of lin with other nodulation mutants, either pollen from plants homozygous for other nodulation mutants was applied to the stigmatic surface of the male-sterile line of lin to obtain F1 seeds, or vice versa. The nodulation phenotype of the F1 progeny for each cross was assessed to determine whether the mutant was allelic to lin.
For mapping studies, a mapping population was constructed by crossing lin in the tap genetic background (that does not affect nodulation) with ecotype A20. F1 seeds were collected and seedlings were tested for nodulation phenotype, as described above. F2 seeds were obtained by self-pollinating the F1 seeds. A population of 125 F2 individuals was visually screened twice for the presence or absence of nodules at 10 and 30 dpi. A trifoliate leaf from each of the F2 plants was used to extract genomic DNA for PCR as described (Williams and Ronald, 1994). The following PCR-based codominant molecular markers well distributed along the eight linkage groups of the M. truncatula genetic map (Choi et al., 2004; www.medicago.org) were chosen to test for possible linkage to the trait: LG1: MDH2, TUP; LG2: EST158, PGKI, DK293R, DK445R; LG3: DK501R, DK417L, EST400, DK313L; LG4: UNK3, APX; LG5: DK018R, MtU4, DK006R; LG6: CrS, DK321L, TE013; LG7: DK322L, DK296L; LG8: DK455L, PPDK, FIS1. Additional markers from LG1: DK369R, DSI, SCP. PCR and restriction digestion experiments for all of the above markers were carried out according to Choi et al. (2004). To generate a genetic marker for MtN6 in our mapping population, a pair of intron-targeted primers was designed based on the sequence information available for MtN6 at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov). The GenBank accession number for the genomic sequence is AJ133118 and the mRNA sequence is Y18225. The primer sequences are as follows: MtN6F (5′-CAAATGGAACTTCTTGGACTC-3′) and MtN6R (5′-AGCCAAGCCAAAATCAAGGT-3′). The PCR products generated using these primers from A17 and A20 parents were sequenced. The sequences were analyzed using the Vector NTI suite 7.1 software. One nucleotide difference between the two sequences affected the cleavage site of BstBI restriction enzyme and was used to generate a cleaved-amplified polymorphic sequence marker specific for MtN6. The method of color mapping was used to analyze the genotype results for linkage relations between each marker and the gene of interest (Kiss et al., 1998).
Northern-Blot Analysis
M. truncatula plants were germinated, transferred to an aeroponic chamber, grown in nitrogen-free media for 5 d, and inoculated with S. meliloti strain ABS7M. At various times before or after inoculation, plants were harvested and the bottom 2 cm of the root tip were removed to eliminate the root meristem tissue, as in Peng et al. (1996). The remaining main root zone (minus the root tips), containing the region of the root where nodulation occurs, was immediately frozen in liquid nitrogen and stored at −80°C. RNA from the main root zone was isolated and hybridized as previously described (Dickstein et al., 1993). Blots were prepared as in Peng et al. (1996), so that each lane contained RNA from approximately 20 roots, based on average yields of RNA isolated. Amounts of RNA loaded were the same for both lin and wild-type (A17) root systems at any given time point. Probes for ENOD2 (Dickstein et al., 1988), ENOD8 (Dickstein et al., 2002), RIP1 (Cook et al., 1995), ENOD40 (Crespi et al., 1994), and rDNA (Dickstein et al., 1991) were cDNAs.
Real-Time PCR Analysis
Root tissues were collected from infected plants at 1, 3, 5, 10, and 18 dpi and also from uninoculated, uninfected plants of equivalent age. For harvests at early time points, the root tips were excised from the rest of the root before freezing samples for RNA extraction. In contrast, for plants grown for 10 and 18 dpi and from their equivalent uninfected plants, approximately 10 cm of the root tissues targeting the nodulation competent zone were harvested. Total RNA was extracted using the RNeasy plant kits (Qiagen, Valencia, CA) according to the manufacturer's instructions. RNA samples were harvested for each time point from three independent biological replicates that were then pooled in equal amounts to average out biological variability. For cDNA synthesis, DNase-treated total RNA (Ambion, Austin, TX) was used and the reverse transcription was carried out with the oligo(dT) primers (Invitrogen, Carlsbad, CA) using the C. Therm. polymerase reverse transcription kit (Roche Applied Sciences, Indianapolis) following the manufacturer's directions. Gene-specific primers were designed based on the sequence information available at NCBI (http://www.ncbi.nlm.nih.gov) and the Institute for Genomic Research MtGI, version 7.0 (http://www.tigr.org/tdb/tgi/mtgi) databases. To allow detection of amplification from contaminant genomic DNA, primers were located within exons and selected to span one or more introns. The specific primers were RT1F (5′-TTGGCTGTTGGTTTGGGTAAC-3′) and RT1R (5′-TACTCATCTTGCTGTTGGAAC-3′) for CCS52A (TC37234; GenBank accession no. AF134835); RT2F (5′-CAAATGGAACTTCTTGGACTC-3′) and RT2R (5′-CCAACTATCTCACCAGCAACT-3′) for MtN6 (TC88012; GenBank accession no. AJ133118); RT3F (5′-CTCTTATTCCACATCTATC-3′) and RT3R (5′-ACCCGTTG GGCCTCTAACT-3′) for ENOD20 (TC77148; GenBank accession no. X99467). Transcripts detected from two housekeeping genes (actin [TC85697] and an O-linked N-acetyl glucosamine transferase [TC77416], otherwise called secret agent [Hartweck et al., 2002]) were used to confirm the quality of the cDNAs of the unknown samples. The primer sequences used for actin were actF (5′-ATGTTGCTATTCAGGCCG-3′) and actR (5′-GCTCATAGTCAAGGGCAAT-3′) and for secret agent were secF (5′-GGCAGGTCTGCCTATGGTTA-3′) and secR (5′-GGTCAGACGCACAGATTTGA-3′). For preparation of external standards, specific amplification of either selected cDNA clones (if available) or genomic DNA was carried out. The PCR products were purified using the Microcon-PCR centrifugal filter devices (Millipore, Bedford, MA) and were quantified based on the A260. Serial dilutions of the standards were prepared starting with an initial concentration of 1 ng μL−1 and were further used for the LightCycler reactions. Real-time PCR using LightCycler (Roche Applied Sciences, Indianapolis) and LightCycler FastStart DNA Master SYBR Green I kit (Roche Applied Sciences) was carried out with the gene-specific primers at a final concentration of 3.75 pM each and 2 μL (the equivalent of 30 ng total RNA) of cDNA as template. PCR cycling conditions comprised an initial denaturation step at 95°C for 10 min, followed by 45 to 50 cycles at 95°C for 0 s, 50°C for 7 s, and 72°C for 30 s. To eliminate the detection of nonspecific products, fluorescence was acquired during each cycle at 81°C, which was above the melting temperature of the primer dimers. Background subtraction and determination of the maximum second derivative (Rasmussen, 2001) for each amplification curve was performed using the LightCycler 3.5 analysis software (Roche Applied Sciences). The cycle numbers defining the second maximum derivative (Rasmussen, 2001) of the standards were plotted against their logarithmic concentrations to derive a standard curve for each of the selected genes. Transcript abundance corresponding to specific genes in the unknown samples was interpolated from this standard curve. Transcript abundance of the unknown samples relative to the uninfected wild-type sample (t = day 6) was determined and their values in logarithmic scale were represented as the relative transcript abundance.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AJ133118, AF134835, and X99467.
Acknowledgments
We thank Thierry Huguet and Pascal Gamas for sharing information on the map location of MtN6 prior to publication. We gratefully acknowledge Jeff Esch, Michelle Graham, and M. David Marks for their technical assistance in the real-time PCR analysis. We also thank Lynn Hartweck for sharing information on secret agent as a constitutively expressed gene prior to publication.
This work was supported by a grant from the U.S. Department of Agriculture-National Research Initiative (award no. 98–35305–6686 to K.VdB.), by funding from the University of Minnesota (to K.VdB.), and by funding from the University of North Texas (to R.D.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.045575.
References
- Ané JM, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GE, Ayax C, Lévy J, Debellé F, Baek JM, Kalo P, et al (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367 [DOI] [PubMed] [Google Scholar]
- Ben Amor B, Shaw SL, Oldroyd GED, Maillet F, Penmetsa RV, Cook D, Long SR, Dénarié J, Gough C (2003) The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J 34: 1–12 [DOI] [PubMed] [Google Scholar]
- Bénaben V, Duc G, Lefebvre V, Huguet T (1995) TE7, an inefficient symbiotic mutant of Medicago truncatula Gaertn. cv Jemalong. Plant Physiol 107: 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano-Anollés G, Paparozzi ET, Gresshoff PM (1991) Mature nodules and root tips control nodulation in soybean. J Plant Physiol 137: 389–396 [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Dénarié J (2000) Four genes of Medicago truncatula controlling components of a Nod factor signal transduction pathway. Plant Cell 12: 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Timmers ACJ, Maillet F, Galera C, Penmetsa RV, Cook D, Dénarié J, Gough C (2001) The HCL gene of Medicago truncatula controls Rhizobium induced root hair curling. Development 128: 1507–1518 [DOI] [PubMed] [Google Scholar]
- Cebolla A, Vinardell JM, Kiss E, Olah B, Roudier F, Kondorosi A, Kondorosi É (1999) The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargements in plants. EMBO J 18: 4476–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon C, Johansson C, Kondorosi E, Kondorosi A, Crespi M (1997) enod40 induces dedifferentiation and division of root cortical cell in legumes. Proc Natl Acad Sci USA 94: 8901–8906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HK, Kim DJ, Uhm T, Limpens E, Lim H, Kalo P, Penmetsa RV, Seres A, Kulikova O, Bisseling T, et al (2004) A sequence-based genetic map of Medicago truncatula and comparison of marker colinearity with Medicago sativa. Genetics 166: 1463–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D, Dreyer D, Bonnet D, Howell M, Nony E, VandenBosch K (1995) Transient induction of a peroxidase gene in Medicago truncatula precedes infection by Rhizobium meliloti. Plant Cell 7: 43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi MD, Jurkevitch E, Poiret M, d'Aubenton-Carafa Y, Petrovics G, Kondorosi E, Kondorosi A (1994) enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J 13: 5099–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullimore J, Dénarié J (2003) How legumes select their sweet talking symbionts. Science 302: 575–578 [DOI] [PubMed] [Google Scholar]
- Dénarié J, Debellé F, Rosenberg C (1992) Signaling and host range variation in nodulation. Annu Rev Microbiol 46: 497–531 [DOI] [PubMed] [Google Scholar]
- Dickstein R, Bisseling T, Reinhold VN, Ausubel FM (1988) Expression of nodule-specific genes in alfalfa root nodules blocked at an early stage of development. Genes Dev 2: 677–687 [DOI] [PubMed] [Google Scholar]
- Dickstein R, Hu X, Yang J, Ba L, Coque L, Kim DJ, Cook DR, Yeung AT (2002) Differential expression of tandemly duplicated Enod8 genes in Medicago. Plant Sci 163: 333–343 [Google Scholar]
- Dickstein R, Prusty R, Peng T, Ngo W, Smith ME (1993) ENOD8, a novel early nodule-specific gene, is expressed in empty alfalfa nodules. Mol Plant Microbe Interact 6: 715–721 [DOI] [PubMed] [Google Scholar]
- Dickstein R, Scheirer DC, Fowle WH, Ausubel FM (1991) Nodules elicited by Rhizobium meliloti heme mutants are arrested at an early stage of development. Mol Gen Genet 230: 423–432 [DOI] [PubMed] [Google Scholar]
- Endre G, Kerestz A, Kevei Z, Mihacea S, Kalo P, Kiss BG (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Geurts R, Bisseling T (2002) Rhizobium Nod factor perception and signaling. Plant Cell (Suppl) 14: S239–S249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene AE, Erard M, Dedieu A, Barker DG (1998) MtENOD16 and 20 are members of a family of phytocyanin-related early nodulins. Plant Mol Biol 36: 775–783 [DOI] [PubMed] [Google Scholar]
- Hartweck LM, Scott CL, Olszewski NE (2002) Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics 161: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss GB, Kereszt A, Kiss P, Endre G (1998) Color mapping: a nonmathematical procedure for genetic mapping. Acta Biol Hung 49: 47–64 [Google Scholar]
- Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ané JM, Lauber E, Bisseling T, et al (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1363 [DOI] [PubMed] [Google Scholar]
- Limpens E, Bisseling T (2003) Signaling in symbiosis. Curr Opin Plant Biol 6: 343–350 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szcyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Mathis R, Grosjean C, de Billy F, Huguet T, Gamas P (1999) The early nodulin gene MtN6 is a novel marker for events preceding infection of Medicago truncatula roots by Sinorhizobium meliloti. Mol Plant Microbe Interact 6: 544–555 [DOI] [PubMed] [Google Scholar]
- Mitra R, Long SR (2004) Plant and bacterial symbiotic mutants define three transcriptionally distinct stages in the development of the Medicago truncatula/Sinorhizobium meliloti symbiosis. Plant Physiol 134: 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED, Long SR (2003) Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in nod factor signaling. Plant Physiol 131: 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske M, Downie AJ (2003) Locks, keys and symbioses. Nature 425: 569–570 [DOI] [PubMed] [Google Scholar]
- Peng HM, Dreyer DA, VandenBosch KA, Cook D (1996) Gene structure and differential regulation of the Rhizobium-induced peroxidase gene rip1. Plant Physiol 112: 1437–1446 [DOI] [PMC free article] [PubMed]
- Penmetsa RV, Cook DR (1997) A legume ethylene insensitive mutant hyper-infected by its rhizobial symbiont. Science 275: 527–530 [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Cook DR (2000) Production and characterization of diverse developmental mutants of Medicago truncatula. Plant Physiol 123: 1387–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa RV, Frugoli JA, Smith LS, Long SL, Cook DR (2003) Dual genetic pathways controlling nodule number in Medicago truncatula. Plant Physiol 131: 998–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle D, Dickstein R (2004) Purification of ENOD8 proteins from Medicago sativa root nodules and their characterization as esterases. Plant Physiol Biochem 42: 73–79 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Gronlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 569–570 [DOI] [PubMed] [Google Scholar]
- Rasmussen R (2001) Quantification on the light cycler. In S Meuer, C Wittwer, K Nakagawara, eds, Rapid Cycle Real-Time PCR. Springer-Verlag, New York, pp 21–34
- Riely BK, Ané JM, Penmesta RV, Cook DR (2004) Genetic and genomic analysis in model legumes brings Nod-factor signaling to center stage. Curr Opin Plant Biol 7: 408–413 [DOI] [PubMed] [Google Scholar]
- Schauser L, Rousiss A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Sherrier DJ, VandenBosch KA (1994) Secretion of cell wall polysaccharides in Vicia root hairs. Plant J 5: 185–195 [Google Scholar]
- Somasegaran P, Hoben HJ (1994) Handbook for Rhizobia. Methods in Legume-Rhizobium Technology. Springer-Verlag, New York, pp 392–398
- Stougaard J (2000) Regulators and regulation of legume root nodule development. Plant Physiol 125: 531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, et al (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Tansengco ML, Hayashi M, Kawaguchi M, Anraku IH, Murooka Y (2003) crinkle, a novel symbiotic mutant that affects the infection thread growth and alters the root hair, trichome, and seed development in Lotus japonicus. Plant Physiol 131: 1054–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoquet P, Ghérardi M, Journet EP, Kereszt A, Ané JM, Prosperi JM, Huguet T (2002) The molecular genetic linkage map of the model legume Medicago truncatula: an essential tool for comparative legume genomics and the isolation of agronomically important genes. BMC Plant Biol 2: 1 (http://www.biomedcentral.com/1471.2229/2/1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truchet G, Michel M, Dénarié J (1980) Sequential analysis of the organogenesis of lucerne (Medicago sativa) root nodules using symbiotically defective mutants of Rhizobium meliloti. Differentiation 16: 163–172 [Google Scholar]
- Tsyganov VE, Voroshilova VA, Priefer UB, Borisov AY, Tikhonovich IA (2002) Genetic dissection of the initiation of the infection process and nodule tissue development in the Rhizobium-pea (Pisum sativum L.) symbiosis. Ann Bot (Lond) 89: 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBosch KA, Frugoli J (2001) Standards and guidelines for genetic nomenclature for the model legume Medicago truncatula. Mol Plant Microbe Interact 14: 1364–1367 [DOI] [PubMed] [Google Scholar]
- Vinardell JM, Fedorova E, Cebolla A, Kevei Z, Horvath G, Kelemen Z, Tarayre S, Roudier F, Mergaert P, Kondorosi A, et al (2003) Endoreduplication mediated by the anaphase-promoting complex activator CCS52A is required for symbiotic cell differentiation in Medicago truncatula nodules. Plant Cell 15: 2093–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CE, Ronald PC (1994) PCR template-DNA isolated quickly from monocot and dicot leaves without tissue homogenization. Nucleic Acids Res 22: 1917–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wopereis J, Pajuelo E, Dazzo FB, Jiang D, Gresshoff PM, de Bruijn FJ, Stougaard J, Szczyglowski K (2000) Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. Plant J 23: 97–114 [DOI] [PubMed] [Google Scholar]
- Yang C, Signer ER, Hirsch AM (1992) Nodules initiated by Rhizobium meliloti exopolysaccharide mutants lack a discrete, persistent nodule meristem. Plant Physiol 98: 143–151 [DOI] [PMC free article] [PubMed] [Google Scholar]