Abstract
To investigate the legume-Rhizobium symbiosis, we isolated and studied a novel symbiotic mutant of the model legume Medicago truncatula, designated nip (numerous infections and polyphenolics). When grown on nitrogen-free media in the presence of the compatible bacterium Sinorhizobium meliloti, the nip mutant showed nitrogen deficiency symptoms. The mutant failed to form pink nitrogen-fixing nodules that occur in the wild-type symbiosis, but instead developed small bump-like nodules on its roots that were blocked at an early stage of development. Examination of the nip nodules by light microscopy after staining with X-Gal for S. meliloti expressing a constitutive GUS gene, by confocal microscopy following staining with SYTO-13, and by electron microscopy revealed that nip initiated symbiotic interactions and formed nodule primordia and infection threads. The infection threads in nip proliferated abnormally and very rarely deposited rhizobia into plant host cells; rhizobia failed to differentiate further in these cases. nip nodules contained autofluorescent cells and accumulated a brown pigment. Histochemical staining of nip nodules revealed this pigment to be polyphenolic accumulation. RNA blot analyses demonstrated that nip nodules expressed only a subset of genes associated with nodule organogenesis, as well as elevated expression of a host defense-associated phenylalanine ammonia lyase gene. nip plants were observed to have abnormal lateral roots. nip plant root growth and nodulation responded normally to ethylene inhibitors and precursors. Allelism tests showed that nip complements 14 other M. truncatula nodulation mutants but not latd, a mutant with a more severe nodulation phenotype as well as primary and lateral root defects. Thus, the nip mutant defines a new locus, NIP, required for appropriate infection thread development during invasion of the nascent nodule by rhizobia, normal lateral root elongation, and normal regulation of host defense-like responses during symbiotic interactions.
The symbiosis that develops between leguminous plants and soil rhizobia to form a nitrogen-fixing root nodule is a complex and unique interaction. The interaction begins with an exchange of signals between rhizobia and plants in the rhizosphere. Flavonoid compounds released by the host plant stimulate the expression of nod genes in an appropriate rhizobial species, resulting in the production of bacterial Nod factors. Nod factors, lipochitooligosaccharide molecules, are able to induce specific responses in the host plant, including root hair deformation and cortical cell division. Infection conduits called infection threads originate in deformed root hairs that curl to form a so-called shepherd's crook and facilitate entry of rhizobia into the root. Infection thread initiation and growth require living rhizobia that are synthesizing specific Nod factors (Ardourel et al., 1994; Limpens et al., 2003). Most infection threads abort in root epidermal cells, with only a small fraction of the infections proceeding to the interior of the root (Vasse et al., 1993; Penmetsa and Cook, 1997). For the infection threads that penetrate the interior of the root, the course by which they proceed is predetermined by cytoplasmic bridges or preinfection threads; these allow infection threads to pass through outer cortical cells toward the inside of the root (Kijne, 1992; van Brussel et al., 1992). When infection threads reach cells in the newly formed nodule primordium, they become confined to the intercellular space between cells (Kijne, 1992). As an infection thread ceases growth, irregular structures including unwalled droplets of infection thread matrix material containing rhizobia are formed. These are engulfed by the plant host plasma membrane, forming symbiosomes in a process resembling endocytosis. The peribacteroid membrane surrounding the symbiosome is initially formed by the plasma membrane of the host cell and expands by fusion with newly synthesized vesicles carrying peribacteroid membrane nodule-specific proteins (Brewin, 1998). Within symbiosomes, the rhizobia differentiate into bacteroids that are capable of fixing nitrogen. Reviews on legume root nodule development and the rhizobial infection process are available (Brewin, 1991; Hirsch, 1992; Kijne, 1992; Brewin, 1998; Gage and Margolin, 2000; Gage, 2004).
The means by which invading rhizobia are able to avoid triggering most of the legume root's host defense mechanisms are not understood. In a normal legume-rhizobia interaction, there are some detectable plant defense responses. Very localized hypersensitive-like reactions accompany aborted infections in epidermal cells and may be part of the plant host's autoregulation of nodule number (Vasse et al., 1993). When mutant rhizobia deficient in surface molecules, specifically exopolysaccharide (EPS) or lipopolysaccharide (LPS), interact with plant hosts, a larger and more generalized plant defense response is often activated (Niehaus et al., 1993; Perotto et al., 1994). The role of surface polysaccharides in the Rhizobium-legume symbiosis has recently been reviewed (Fraysse et al., 2003).
Genes and processes involved in lateral root formation may also be involved in nodule formation. Like nodule development, lateral root development is influenced by hormones as well as the nutritional status of the plant. Unlike nodules that develop from cortical cell divisions, lateral roots usually develop from divisions in the pericycle. Both types of divisions take place opposite a protoxylem pole, but lateral roots have a central arrangement of vascular bundles, whereas nodules have peripheral vascular bundles (Hirsch, 1992; Casimiro et al., 2003). When lateral roots emerge, tissues in the lateral root primordium become distinct, and the root cap, root meristem, and central stele are established.
In 1990, Medicago truncatula was proposed as a model legume, particularly for studying the Rhizobium-legume symbiosis, because of its small genome size (approximately 500 Mbp, n = 8), self-compatibility, relatively short generation time, ability to be transformed, and the well-characterized nature of its microsymbiont, Sinorhizobium meliloti (Barker et al., 1990). Since then, many genetic and genomic tools have been developed for M. truncatula (Cook, 1999); recently, detailed genetic maps have been produced (Thoquet et al., 2002; Choi et al., 2004), a genetically anchored physical map is emerging, and the euchromatic regions of M. truncatula's genome are being sequenced. M. truncatula produces the indeterminate type of nodule with a persistent meristem and, therefore, all stages of nodule development can be studied within a single mature nodule.
Here, we describe the isolation of a novel M. truncatula mutant, nip, and present data on its phenotype. nip plants respond to S. meliloti by producing abnormal nodules in which numerous aberrant infection threads are produced, with very rare rhizobial release into host plant cells. It has an abnormal defense-like response in root nodules as well as defects in lateral root development.
RESULTS
Isolation of the Recessive nip Mutant
Ethyl methane sulfonate-generated M. truncatula plants from the A17 genetic background were screened for nodulation phenotypes in the M2 generation by inoculating them with an S. meliloti strain containing the constitutive hemA∷lacZ construct (Boivin et al., 1990; Penmetsa and Cook, 1997, 2000). Putative mutants that had white nodule bumps instead of pink leghemoglobin-containing nitrogen-fixing nodules were screened at 10 d postinoculation (dpi) by staining roots with X-Gal for the presence of Sinorhizobium. Plants without rhizobia in their nodules and plants with lower than wild-type amounts of rhizobia were propagated and rescreened in the M3 and M4 generations for their nodulation phenotypes. nip was originally characterized from the C bulk as mutant C90, having small nodules/bumps on its roots with limited nodule invasion by rhizobia, short primary roots, and defective lateral roots. It was found to be proficient in the mycorrhizal symbiosis (M. Harrison, personal communication). Based on the phenotype (see below), the gene responsible for the mutant phenotype in C90 was named nip, for numerous infections and polyphenolics, in accordance with nomenclature rules for M. truncatula (VandenBosch and Frugoli, 2001), and registered with the M. truncatula gene nomenclature index (http://www.genome.clemson.edu/affiliated_cugi/medicago).
nip was back-crossed twice (BC2), utilizing the male-sterile tap mutation in the wild-type A17 background (Penmetsa and Cook, 2000) to ensure true crossing. nip, in the A17 M. truncatula genetic background, was also crossed into A20, a different polymorphic ecotype of M. truncatula, in order to facilitate future genetic-mapping studies. The A17/A20 pair forms the basis of one of the genetic maps of M. truncatula (Choi et al., 2004). Wild-type:nip segregated approximately 3:1 in F2 populations obtained in each back-cross to genotype A17 and in crosses to ecotype A20, consistent with a single-gene recessive mutation (Table I). Back-crossed nip plants were more robust in terms of overall growth than the original mutant. Characterization of the nip phenotype was carried out on plants BC2, except where noted.
Table I.
Segregation of nip phenotype in nip × wild-type crosses
| Cross, Progeny Tested | Wild-Type Nodules | Mutant Nodules | χ2 |
|---|---|---|---|
| nip × Mtap, (BC1) F2 | 80 | 24 | 0.21 |
| nip × Mtap, (BC2) F2 | 70 | 21 | 0.16 |
| nip × A20, F2 | 54 | 14 | 0.02 |
χ2 calculated for 3:1 ratio wild-type nodule:mutant nodule; P > 0.05 when χ2 < 3.84.
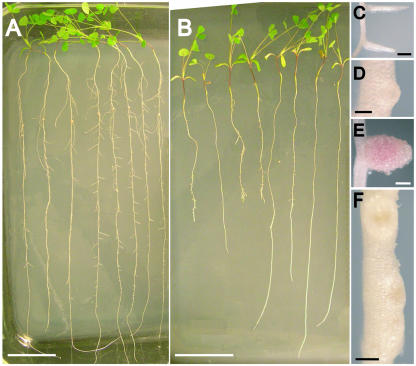
Representative wild-type and nip BC2 plants grown in an aeroponic chamber were photographed at 15 dpi (Fig. 1, A and B). nip roots showed variation in root length from plant to plant and were observed to have abnormal lateral roots. Unlike wild-type plants that had clearly emerged lateral roots (Fig. 1C), most nip plants had either no lateral roots, although lateral root primordia were visible (Fig. 1D), or had shorter lateral roots (Fig. 1B) than wild type (Fig. 1A). Instead of effective, pink, leghemoglobin-containing nodules as were found in wild type (Fig. 1E), nip developed straw-colored root nodules/bumps containing a brown pigment (Fig. 1F), suggesting an accumulation of polyphenolic compounds. Cosegregation of the lateral root defect with the nodulation defect was found in all nip F2 progeny tested in all crosses.
Figure 1.
Phenotype of nip and wild-type (A17) plants 15 dpi with S. meliloti. Plants were grown in aeroponic chambers as described in “Materials and Methods” and placed on an agar support for photography. A, Wild-type plants. Bar = 5.0 cm. B, nip plants. Bar = 5.0 cm. C, Wild-type primary root with emergent lateral roots. Bar = 1.0 mm. D, nip lateral root primordium. Bar = 0.25 mm. E, Wild-type nodule. Bar = 0.50 mm. F, nip nodules/bumps. Note the presence of a dark pigment at the distal ends of the nodules. Bar = 0.50 mm.
nip Nodules Accumulate Polyphenolic Compounds and Have Large, Proliferating Infection Threads That Rarely Release Rhizobia
When grown without nitrate and in the presence of compatible rhizobia, nip plants exhibited reduced vigor with chlorotic leaves that are characteristic of nitrogen deficiency. These symptoms were alleviated by supplementation with nitrate fertilizer and are indicative of a defect in the ability to form an effective nitrogen-fixing symbiosis. Growth of nip plants in the presence of nitrate fertilizer had no effect on the lateral root phenotype (data not shown).
To evaluate nip's nodulation phenotype, nip plants were grown in an aeroponic system and inoculated with S. meliloti carrying the hemA∷lacZ fusion. Staining with X-Gal at 15 dpi revealed a proliferation of S. meliloti inside numerous infection threads in the nodule primordia (Fig. 2, A and B). The nip nodule primordia emerged from the root only after prolonged nodule development times, at or after 25 dpi. In contrast, emerged elongated wild-type A17 nodules were filled with the blue-staining S. meliloti at 15 dpi.
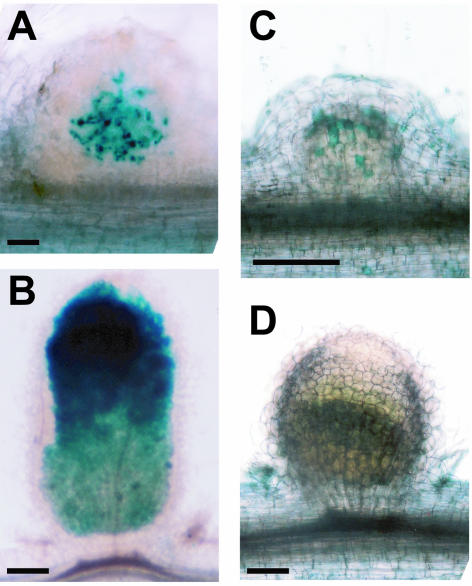
Figure 2.
Rhizobial infection (A and B) and polyphenolic accumulation (C and D) in nip and wild-type nodules. nip and A17 wild-type plants inoculated with S. meliloti carrying the hemA∷lacZ reporter were hand-sectioned and stained with X-Gal at 15 dpi. S. meliloti stain blue. A, nip nodule; B, A17 nodule. nip and A17 wild-type plants were inoculated with S. meliloti, fixed, and stained with potassium permanganate followed by methylene blue. Polyphenolics stain blue in this procedure. C, nip nodule; D, A17 nodule. Bars = 0.2 mm.
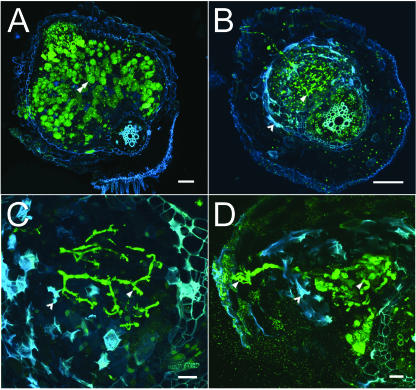
In order to more closely examine the extent of infection, nip and wild-type nodules were stained with nucleic acid-binding dye SYTO-13 and examined by laser-scanning confocal microscopy (LSCM). Stained rhizobia fluoresced in the green spectrum using this technique, while polyphenolics and cell walls fluoresced in the red spectrum, pseudocolored blue in the figure to enhance detail (Fig. 3). By 13 dpi, fully mature wild-type nodules had emerged from the surface of the primary root and infected cells were characterized by high rhizobial cellular occupancy (Fig. 3A). In contrast, despite an examination of over 160 nip nodules, we failed to find clear examples of infected host cells in the nip mutant. As was seen using the hemA∷lacZ marker and X-Gal staining, infection thread numbers were found to be substantially higher in nip (Fig. 3B), and LSCM revealed abnormal infection thread morphology in nip nodules. Infection threads, including the original infection thread that originates in the root epidermis, were thickened relative to wild-type threads with abnormal bulbous protrusions (Fig. 3C) that became more pronounced over time with an increase in diameter of the threads and the size of the protrusions (Fig. 3D). nip nodules were found to emerge from the root epidermis only late in development, and were also found to have large areas of autofluorescing cells, suggesting an induction of the plant defense response. The autofluorescing cells were localized near cells with infection threads, but did not themselves contain threads.
Figure 3.
Confocal light micrographs of M. truncatula wild-type (A17) and mutant (nip) nodules. A, Wild-type nodule 13 dpi. Nodule has emerged from the root and shows normal mature infected cells containing intracellular bacteroids (green). A typical infected cell is indicated by the double arrowhead. Bar = 100 μm. B, nip nodule 13 dpi demonstrating abnormal nodule formation. The nodule is infected with bacteria and there are an unusually high number of threads present (single arrowhead). Mutant nodules often did not emerge from the root and autofluorescence (blue) is observed in cells around the infection threads (barbed arrowhead). Bar = 100 μm. C, Higher magnification of nip mutant nodule 13 dpi showing abundant threads that terminate without release of bacteria into plant cell cytoplasm. Threads are thickened, exhibit abnormal bulges (arrowhead), and are flanked by cells exhibiting autofluorescence (blue). Bar = 20 μm. D, High magnification view of nip mutant nodule 29 dpi showing highly enlarged and distorted threads. Single arrowhead indicates an infection thread; barbed arrowhead indicates blue autofluorescence in nodule cells. Bar = 20 μm.
To determine if the autofluorescence and the accumulation of brown pigment represented an accumulation of polyphenolic compounds, nodules were stained with potassium permanganate using a method that results in blue precipitates at sites of polyphenols (Fig. 2, C and D). Individual cells in the nodule cortex of nip nodules stained heavily for polyphenolics, with other cell types displaying no staining (Fig. 2C). In contrast, in wild-type A17 nodules, no polyphenolic staining was detected in any of the cells in the nodule (Fig. 2D).
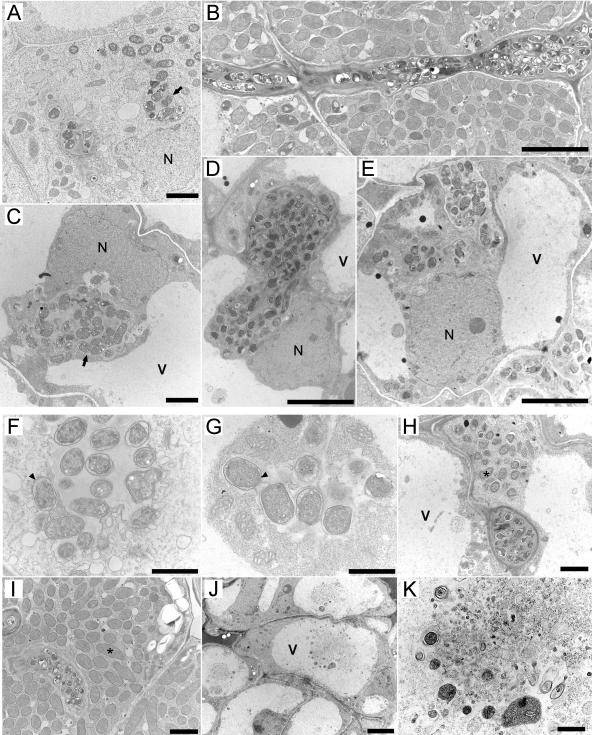
To further explore the nip nodule phenotype and examine the interface between the infection threads and plant host cells in detail, studies using transmission electron microscopy (TEM) on 15 and 21 dpi nip nodules were carried out. No differences between the 15 and 21 dpi nodules were observed and data from 15 dpi nodules are presented here. Wild-type nodules developed normally, and by 15 dpi had produced fully mature nodules exhibiting the typical developmental zones: meristem, prefixation zone (Fig. 4A), interzone, and nitrogen-fixation zone (Fig. 4, B and I). In contrast, nip nodules commonly remained unemerged at this stage and exhibited no distinct developmental zones; rather, the nodule interior most closely resembled an enlarged prefixation zone (Fig. 4, C and E). Infection droplets in the wild-type nodule were confined to a narrow zone below the meristem, were generally small, and exhibited normal release of bacteria (Fig. 4A), whereas droplets in nip were seen throughout the nodule interior and were often enlarged with excess matrix material (Fig. 4C). Wild-type infection threads were generally narrow and grew in a linear pattern through cells (Fig. 4B). In contrast, nip infection threads were much more numerous, often unusually wide and irregularly shaped (Fig. 4D), and commonly grew in a serpentine pattern (Fig. 4E). At high magnification, wild-type infection droplets (Fig. 4F) were similar in appearance to nip infection droplets, but nip infection droplets rarely released bacteria into host cells. In one instance, limited release of bacteria was seen in one cell of a nip nodule (Fig. 4H), but the bacteria were small and undifferentiated when compared to the mature fixation zone of a wild-type nodule (Fig. 4I). At this stage, nip plants were severely nitrogen stressed and it is unlikely that this limited infection could have progressed to a normally infected cell. In addition to abnormal threads, nip nodules also exhibited abnormal membrane profiles within vacuoles in the uninfected nodule periphery, several cell layers removed from the threads. Membrane profiles were seen in the vacuoles and were suggestive of cell death in these regions (Fig. 4, J and K). These cells correspond to the region in Figure 3 that contained autofluorescent compounds and to the region in Figure 2 that stained for polyphenolics.
Figure 4.
TEMs of ultrathin nodule sections from 15 dpi. A, Invasion zone of a wild-type nodule. Arrow indicates an infection droplet releasing bacteria into the plant cell cytoplasm. N, Nucleus. Bar = 2 μm. B, Remnant infection thread in the fixation zone of a wild-type nodule. Note that the thread is straight and relatively narrow. Bar = 5 μm. C, Cells of the extensive invasion zone in nip nodules. Arrow indicates an enlarged infection droplet-like structure in a nip nodule cell. V, Vacuole. Bar = 5 μm. D, Enlarged and irregular infection thread in nip nodule. Bar = 5 μm. E, Meandering infection thread in nip nodule. Bar = 5 μm. F, Wild-type infection droplet. Arrow indicates bacterium being released. Bar = 1 μm. G, Infection droplet-like structure in nip nodule. Arrow indicates a comparable bacterium to that shown in F. Bar = 1 μm. H, Rare instance of limited bacterial release in a nip nodule. *, Released bacterium. Bar = 2 μm. I, Large, mature bacteroids in fixation zone of wild-type nodule. *, Bacteroid. Bar = 2 μm. J, Cells at the nip nodule periphery with membrane profiles in the vacuoles. Bar = 5 μm. K, Higher magnification image of the vacuolar membrane profiles in nip cells. Bar = 1 μm.
nip Plants Have Normal Shoots and Abnormal Lateral Roots
Growth characteristics were investigated by growing F3 plants derived from the BC2 generation and from crosses into ecotype A20, using the aeroponic system with nitrate-free media in the presence of rhizobia. The results are shown in Tables II and III. In plants propagated from BC2, nip primary shoots were found to be approximately the same size as wild type at 5 and 10 dpi, and averaged 75% of wild type at 15 dpi, presumably reflecting a nitrogen deficiency (Table II).
Table II.
Shoot growth characteristics of nip plants compared to wild type
| dpi | A17 Shoot Length | nip Shoot Length |
|---|---|---|
| cm | cm | |
| 5 | 2.0 ± 0.4 | 2.1 ± 0.4 |
| 10 | 5.2 ± 0.4 | 5.0 ± 0.8 |
| 15 | 6.4 ± 1.1 | 4.7 ± 1.0 |
Plants were grown in aeroponic chambers as described in “Materials and Methods.” Four or five plants were harvested and measured for each time point. Averages ± sds are given.
Table III.
Root system characteristics of individual nip × A20 F2 nip nodule phenotype plants' F3 progeny
| Plant(s) | Number Plants Examined | Number of Plants with Lateral Roots | Number of Lateral Roots per Plant (When Present) | Root Lengths |
|---|---|---|---|---|
| cm | ||||
| A17 | 9 | 9 | 4.7 ± 3.6 | 19.7 ± 1.7 |
| A20 | 10 | 10 | 3.3 ± 1.5 | 18.6 ± 3.5 |
| nip × A20, nip no. 1 | 7 | 2 | 1, 3 | 16.7 ± 1.5 |
| nip × A20, nip no. 2 | 14 | 5 | 1, 1, 2, 3, 3 | 22.3 ± 3.8 |
| nip × A20, nip no. 3 | 6 | 2 | 2, 7 | 20.9 ± 4.0 |
| nip × A20, nip no. 4 | 13 | 2 | 1, 1 | 15.7 ± 4.1 |
| nip × A20, nip no. 5 | 7 | 0 | 16.3 ± 2.9 |
All nip plants measured were progeny of nip × A20 F2 plants having lateral roots and wild-type length primary roots. Plants were grown in aeroponic chambers as described in “Materials and Methods.” The nip and control plants' characteristics were measured at 10 dpi.
As was noted above (see Fig. 1), in F3 plants derived from the BC2 population, nip plants showed root abnormalities. To further investigate the nip root phenotype, nip plants with the most normal-looking root systems were crossed to ecotype A20. Several F2 plants resulting from this cross were observed that had primary roots of similar length to wild type and possessed lateral roots. To determine the correlation between altered lateral root development with symbiotic phenotype, 47 individual F3 progeny from these F2 plants were examined for their root phenotypes (Table III). The F3 progeny had roots that averaged a similar length as wild type, 18.4 ± 4.4 cm, compared to 19.7 ± 1.7 cm for A17, and 18.6 ± 3.5 cm for A20. However, only 11, or approximately 25% of the 47 individual F3 plants, had lateral roots, although, as before, lateral root primordia were evident. When present, the lateral roots occurred at a lower frequency than wild type (Table III) and were shorter in length compared to wild type (data not shown). This strongly suggests that the lateral root defect in nip is incompletely penetrant.
Ethylene Responsiveness Is Similar in nip and Wild Type
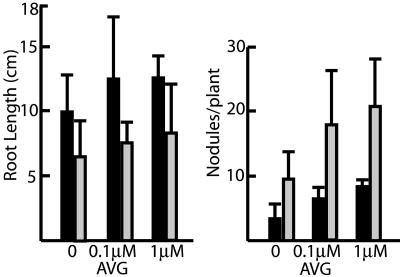
Previous studies have shown that the plant hormone ethylene plays a central role in regulation of successful infections as well as root growth in M. truncatula (Penmetsa and Cook, 1997, 2000; Oldroyd et al., 2001; Penmetsa et al., 2003). To determine if the nip mutant was altered in ethylene responses, nip root length and nodule number were measured in the presence of various concentrations of the ethylene inhibitor l-α-(2-aminoethoxyvinyl)-Gly (AVG) and in the presence of the ethylene precursor 1-aminocyclopropane carboxylic acid (ACC). The results, shown in Figure 5, demonstrate a similar percentage increase in root length and nodule number in nip BC2 plants as compared to wild type (A17) in the presence of increasing AVG concentration. When nip plants were grown in the presence of ACC, a similar decrease in root length and nodule number in nip plants, as compared to wild-type A17, was observed (data not shown). Together, the AVG and ACC data suggest that the nip mutant phenotype is unlikely to be mediated by altered ethylene biosynthesis or perception.
Figure 5.
Root length and nodule number in plants grown in the presence of AVG. Wild-type A17 (black bars) and nip plants (gray bars) were grown on solid media in petri dishes containing the indicated concentrations of AVG with their roots shielded from light. Four or five plants were measured for each concentration of AVG tested.
nip Nodule Gene Expression Studies Indicate a Block to Nodule Differentiation and Elevated Levels of a Transcript Associated with Plant Defense
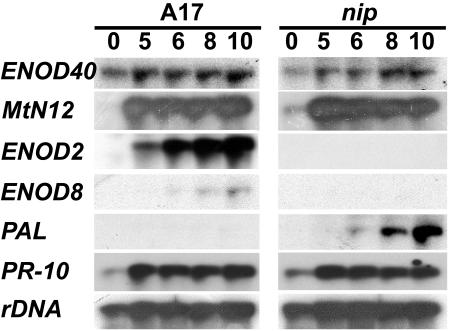
To evaluate the progression of nip nodule development at the transcriptional level, the expression of four genes associated with nodule development and two genes associated with plant host defense were investigated in nip and wild-type nodulating roots. The ENOD40 gene is correlated with formation of nodule and lateral root primordia (Crespi et al., 1994; Fang and Hirsch, 1998). The MtN12 gene encodes a nodule-specific extensin associated with the IT matrix (Gamas et al., 1996; Rathbun et al., 2002). The ENOD2 gene is expressed in the nodule parenchyma (van de Wiel et al., 1990) and is a marker for early stages of nodule organogenesis. ENOD8 is expressed in infected nodule cells in M. truncatula relatively early in nodule development (Dickstein et al., 1993, 2002) and is a marker for infected cell formation (L. Coque, K. Wilson, and R. Dickstein, unpublished data). As shown in Figure 6, ENOD40 and MtN12 are expressed at comparable levels in nip and wild-type nodulating roots, while expression of ENOD2 and ENOD8 are not detectable in nip nodulating roots. These data are consistent with nip roots responding to rhizobial signals by producing nodule primordia that become invaded by rhizobia in infection threads. The absence of ENOD2 and ENOD8 expression, each of which is associated with specific aspects of nodule organogenesis, is consistent with nip nodules being halted at a very early stage of nodule differentiation.
Figure 6.
RNA-blot analysis of gene expression during nip nodule development. Gel blots were prepared from 20 μg of total RNA extracted from A17 or nip roots inoculated with S. meliloti at the indicated times after inoculation (dpi). Blots were hybridized sequentially with radiolabeled ENOD40, MtN12, ENOD2, ENOD8, PAL, and PR-10 probes, with rDNA serving as the loading control.
Cytological evaluation demonstrated an unusual plant defense-like response in nip nodules (Figs. 2C, 3, and 4, J and K). Therefore, expression of two genes associated with plant host defense was studied in nodulating nip and wild-type roots. Elevated levels of pathogenesis-related protein-10 (PR-10) and Phe ammonia lyase (PAL) are both associated with plant host defense and plant stress (Hammond-Kosack and Jones, 2000). PR-10 mRNA accumulation in nip nodulating roots was found to be comparable to wild-type nodulating roots. In contrast, PAL expression was found to increase dramatically during nip nodule development compared to wild type. Because PAL encodes the enzyme catalyzing the first step in phenylpropanoid metabolism, the elevated levels of PAL mRNA are consistent with the accumulation of polyphenolics detected in nip nodules.
Allelism Tests
Although nip has a different phenotype from other legume mutants studied by several groups in the Medicago scientific community, it is possible that it is allelic to another mutant. To test this possibility, nip was crossed to other mutants and two to six independent F1 progeny from each cross were scored for nodulation phenotype. Of particular interest were mutants that are able to initiate nodulation but are defective in rhizobial invasion or later steps in the symbiosis. Genetic crosses of the nip mutant to each of several M. truncatula mutants, lin, rit, bit, TE7 (Mtsym1), dnf1, dnf2, dnf3, dnf4, dnf5, dnf6, or dnf7 (Benaben, 1994, 1995; Mitra and Long, 2004; Kuppusamy et al., 2004), were carried out. All F1 progeny from each cross were found to have wild-type nodulation, indicating that nip was not allelic to any of these loci. Additionally, nip complemented dmi1, dmi2, and nsp1, which are defective in Nod factor signal transduction (Catoira et al., 2000). Notably, nip did not complement latd. latd is a M. truncatula mutant with phenotypes somewhat similar to, but much more severe than, nip. (J. Harris, personal communication and unpublished observations).
DISCUSSION
We identified a novel symbiotic mutant in M. truncatula called nip. The monogenic and recessive nip mutant is able to initiate nodule development, but is not competent to attain functional nitrogen-fixing nodules when grown in the presence of the compatible Rhizobium sp., S. meliloti. As our studies using light, confocal and electron microscopy show, the nip mutant is capable of developing nodule primordia and initiating rhizobial invasion through plant-derived infection threads. However, nip infection threads are thicker than wild type and characterized by abnormal bulbous protrusions and unusual branching. The block to nodule development is most likely at release of rhizobia from infection threads and endocytosis into the host cytoplasm. The nip mutant is very slightly leaky, rarely allowing release of rhizobia into host cells, but neither the rhizobia nor the host cells appeared to differentiate in response to release. Our data do not allow us to distinguish whether the infection thread characteristics observed are a consequence of the failure to release rhizobia from the threads or if infection thread characteristics, including size, proliferation, or biochemical constitution, cause the rhizobia not to be released.
nip nodules show evidence of an abnormal plant host defense-like response. We noted that the cells with the defense-like response, those that stain for polyphenolics, are autofluorescent and with a vacuolar accumulation of membrane fragments, are only a subset of cells within the nodule and are generally adjacent to cells not undergoing a defense-like response. This feature of defense-like response in nip resembles the hypersensitive response, where cells undergoing programmed cell death are interspersed among living cells (Hammond-Kosack and Jones, 2000). The cells with features of defense response were found several cell layers from the abnormal infection threads. Moreover, the nip-specific induction of PAL, a transcript frequently associated with host responses to pathogens, provided additional circumstantial evidence of a host defense response. It is unclear whether the defense-like response is a primary effect of the nip mutation or is a secondary response to the block to nodule development or to the abnormal infection threads.
Ethylene is a plant hormone that has been implicated in regulation of plant defense, rhizobial infections, and root growth (Penmetsa and Cook, 1997; Oldroyd et al., 2001; Penmetsa et al., 2003). Growth of nip plants on media containing ethylene inhibitors or precursors revealed similar effects as on wild-type plants. Thus, it is unlikely that ethylene metabolism or perception is compromised in the nip mutant.
nip plants showed an approximate 2-fold increase in nodule number as compared to wild-type plants (e.g. see Fig. 5). Similar increases in nodule number have been noted in studies with other legume and rhizobial mutants that form ineffective nodules (Dickstein et al., 1988; Benaben et al., 1995; Kuppusamy et al., 2004). Similar to other symbiotic mutants, the lack of mature nodule meristems in nip may lead to recurrent nodule initiation, a downstream effect of the absence of nitrogen-fixing nodules, rather than a specific effect of the nip mutation.
Nodule-specific marker genes for nodule primodia (ENOD40) and infection thread formation (MtN12) were expressed in nodulating nip root systems. Genes that are associated with nodule organogenesis (ENOD2, ENOD8) were not. Lack of ENOD2 expression in nip nodules is consistent with recent studies that showed nodule-like structures formed in M. truncatula in response to S. meliloti exo mutants are associated with an absence of ENOD2 expression (Mitra and Long, 2004). The studies presented here demonstrate that, in M. truncatula, formation of nodule primordia invaded by rhizobia within infection threads is not sufficient for ENOD2 or ENOD8 expression.
The nip nodule phenotype is similar in some respects to defective nodules elicited by other mutations in either the bacterial or legume symbiotic partners. Both S. meliloti EPS I-deficient mutants and LPS-deficient R. leguminosarum mutants induce nodule structures on alfalfa and pea, respectively, showing signs of a host defense reaction. Thickened infection thread-like structures containing rhizobia were observed in intercellular spaces in the nodule structures, with some rhizobia able to form symbiosomes, although at a higher frequency than was observed in the nip mutant (Niehaus et al., 1993; Perotto et al., 1994). S. meliloti LPS mutants form Fix− nodules on M. truncatula with enlarged infection threads that released rhizobia, but failed to form symbiosomes properly, and with a host defense reaction (Niehaus et al., 1998). Given the similarity of the nip nodules to nodules elicited by EPSI- or LPS-deficient Rhizobium sp., it is tempting to speculate that the defect in nip is in a receptor or part of a signal transduction pathway for molecules on the surface of the Rhizobium sp. However, alternative explanations remain a possibility because auxotrophic rhizobial mutants, like the S. meliloti hemA mutant controlling the first step of heme biosynthesis (Dickstein et al., 1991), as well as a rhizobial mutant defective in the synthesis of signal peptides (Muller et al., 1995), also produce nodules containing infection threads without rhizobial endocytosis into host plant cells.
The nip nodule phenotype is also similar to that of other M. truncatula symbiotic mutants, especially the lin and Mtsym1 (TE7) mutants. lin forms nodule primordia in which infection thread development halts at the root epidermis, apparently before the blockage in nip. Similar to nip, nodulating lin root systems express genes associated with nodule primordium formation but not the ENOD2 and ENOD8 genes associated with nodule differentiation (Kuppusamy et al., 2004). By contrast, nodule development in Mtsym1 appears to progress further than nip as Mtsym1 nodules are invaded by rhizobia that senesce upon release (Benaben et al., 1995). Interestingly, Mtsym1 nodules exhibit evidence of defense-like responses, including deposition of polyphenolic substances, similar to the current observation of polyphenolic accumulation in nip nodules. Benaben (1994) observed ENOD2, but not ENOD8, expression in certain MtSym1 nodules, while a more recent study of Mtsym1 nodules concluded that ENOD2 was not expressed (Mitra and Long, 2004). The difference between the two studies may be the result from different growth conditions. Based on developmental phenotypes, we propose that the NIP gene acts after LIN and before MtSYM1. The M. truncatula dnf mutants have all been found to express ENOD2 (Mitra and Long, 2004); thus, NIP acts before the DNF genes.
Because of the diverse effects of the nip mutation on infection thread morphology, nodule differentiation, marker gene expression, polyphenolic accumulation, and lateral root growth, we speculate that NIP may have a regulatory role in root and nodule development. NIP is also required to suppress a defense-like response. It is unclear whether NIP is a direct suppressor of the defense-like response or whether the infection thread proliferation and failure to release rhizobia resulting from NIP loss of function triggers the response. The molecular identification of NIP should further elucidate the intersection between symbiotic nodule and lateral root development and may yield insight into aspects of the balance between symbiosis and host defense.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Medicago truncatula A17 wild-type plants were germinated and grown in aeroponic growth chambers misted with an inorganic nutrient media lacking NH4NO3 (Lullien et al., 1987) as described previously (Dickstein et al., 2002; Catalano et al., 2004). Briefly, seeds greater than 3 months old were scarified with concentrated H2SO4 for 8 min, rinsed in sterile water, surface sterilized with 6% sodium hypochlorite for 1 min, and rinsed with sterile water. nip plants were treated similarly, but the sodium hypochlorite treatment was omitted. Seeds were imbibed with sterile water with gentle rotation for 5 to 7 h at ambient temperature, then stored in water at 4°C for 16 to 24 h, plated on petri dishes, and then left in the dark at room temperature for 14 to 15 h for germination. Freshly harvested seeds (1–2 weeks after pod maturation) were left in sterile deionized water for 24 h on a rotating shaker and then germinated in the dark at room temperature for 14 to 15 h. For vernalization, plants were maintained in a moist chamber for 2 weeks at 4°C. Plants were placed into aeroponic chambers when their roots were at least 0.5 cm long and misted with a nitrogen-free nutrient solution (Lullien et al., 1987). Plants were maintained in a growth room at 22°C on a 16-h-light and 8-h-dark schedule under Phillips Agro-Lite bulbs (Phillips, Somerset, NJ) at 60 μmol m−2 s−1. For nodulation studies, plants were inoculated with Sinorhizobium meliloti strain ABS7 (Bekki et al., 1987), strain Rm2011 (Rosenberg et al., 1981), or S. meliloti/pXLDG4 (Boivin et al., 1990; Penmetsa and Cook, 1997) 5 d after germination. Infected roots and nodules were harvested at the indicated times after inoculation. Plants used for confocal microscopy and TEM studies were grown as above with some modifications. A17 seeds were scarified for 6 min and surface sterilized for 3 min with 6% sodium hypochlorite and rinsed thoroughly with sterile water after each step. Both nip and wild-type A17 seeds were imbibed in sterile water at ambient temperature for 5 h, stored at 4°C overnight, and rinsed with sterile water over a 6-h period prior to spreading on a petri dish. Seeds were sown immediately after germination on aeroponic chambers with Lullien solution containing NH4NO3, changed to Lullien solution without NH4NO3 after 5 d, and inoculated 9 d postsowing with S. meliloti Rm2011.
For the ethylene inhibitor and precursor studies, plant growth and nodulation studies were performed on agar plates. The same media were used with the appropriate AVG or ACC concentrations and solidified with 1% w/v phytagar (Invitrogen, Carlsbad, CA). The root portion was shaded by covering the bottom half of plates with aluminum foil. Growth conditions were the same as for aeroponic growth.
Histochemical Staining
S. meliloti/pXLDG4 containing the hemA∷lacZ reporter was visualized on plant roots as described (Boivin et al., 1990). Briefly, whole roots were vacuum infiltrated for three cycles of 30 s with 2.5% glutaraldehyde in 0.1 m PIPES (pH 7.2), fixed for 1 h, and rinsed in 0.1 m PIPES (pH 7.2) twice. Samples were then incubated in staining solution containing 50 mm potassium ferricyanide, 50 mm potassium ferrocyanide, 0.08% X-Gal, and 0.1 m PIPES (pH 7.2) for 16 h at room temperature. Samples were rinsed in 0.1 m PIPES (pH 7.2), cleared with 2.4% sodium hypochloride for 5 min, sectioned on a vibratome (model 1000; Vibratome, St. Louis), sectioned by hand or left whole, mounted on glass slides with coverslips, and observed under an Olympus BX50 microscope (Olympus, Melville, NY) using bright field.
For LSCM studies, nodulated roots were prepared and imaged as described (Haynes et al., 2004). Briefly, nodulated roots were harvested into 80 mm PIPES (pH 7.0) and nodules were hand-sectioned longitudinally. Nodule halves were stained with 1 μL mL−1 SYTO-13 (Molecular Probes, Eugene, OR) in PIPES buffer for 15 m, transferred to a Lab-Tech chambered no. 1.5 coverglass system (Nalge/Nunc, Naperville, IL), and imaged on an inverted Zeiss LSM 510 NLO laser-scanning microscope (Carl Zeiss, Jena, Germany).
Detection of polyphenolics was accomplished by staining with potassium permanganate to visualize polyphenols as described (Vasse et al., 1993). Briefly, whole roots were fixed in 2.5% glutaraldehyde, 0.01 m PIPES (pH 7.2), and stained in 0.04% potassium permanganate for 1 h, rinsed in 0.01 m PIPES (pH 7.2), then stained with 0.01% aqueous solution of methylene blue. The roots were cleared with 2.4% sodium hypochlorite for 3 min and visualized as above.
TEM
Nodules were produced aeroponically as described and harvested at 5, 10, 15, and 21 dpi. Nodulated roots were harvested into a solution of 4% formaldehyde and 1% glutaraldehyde in 80 mm PIPES (pH 7.0). Wild-type nodules were dissected from the root system and cut longitudinally to aid in infiltration. nip nodules were generally unemerged and formed in clusters, so nodulated portions of the root were cut into small segments for fixation. Nodules were vacuum infiltrated in fixative 4 × 2 min and fixed overnight at 4°C with rotation. Nodules were rinsed 3 × 15 min with distilled water, stained 4 h with 1% OsO4 (aq), rinsed 3 × 15 min with distilled water, dehydrated in a graded series of acetone, and infiltrated with a graded series of Epon Araldite resin (Mollenhauer, 1964). Samples were polymerized in Epon Araldite resin for 48 h at 60°C. Nodules were sectioned with glass knives and then 70-nm sections were collected using a Diatome diamond knife (Diatome-U.S., Fort Washington, PA) on a Reichert Ultracut E ultramicrotome. Sections were collected onto hexagonal gold grids and were counterstained with alkaline lead citrate (Reynolds, 1963) for 7 min and 0.5% uranyl acetate (aq) for 12 min. Samples were visualized and documented on a Zeiss CEM 902 TEM.
RNA Extraction and Blots
RNA was extracted and northern blots prepared with 20 μg RNA for each sample as previously described (Dickstein et al., 2002). DNA was labeled for hybridization by random priming (Feinberg and Vogelstein, 1983). The probes used were ENOD40 (Crespi et al., 1994); ENOD2, GenBank accession number X12580 (Dickstein et al., 1988); ENOD8, GenBank accession number AF064775 (Liu et al., 1998); MtN12 (Gamas et al. 1996); PAL (GenBank accession no. BF635112); PR-10 (GenBank accession no. AW587229), with rDNA (Dickstein et al., 1991) as loading control.
Genetic Analysis
Mutants were crossed into the male sterile M. truncatula A17 mutant (MtAp) as previously described (Penmetsa and Cook, 2000). Male-sterile, nodule-deficient F2 mutants that resulted from this cross were used as female parents in crosses with A17 and other mutants. Plants were evaluated for phenotype 10 dpi by examining their roots for the presence of white bumps or pink nitrogen-fixing nodules by eye or with the aid of a dissecting microscope.
Acknowledgments
We gratefully acknowledge Kate VandenBosch, in whose lab some mutant screening was performed; Colby Starker, Giles Oldroyd, Sharon Long, Kate VandenBosch, Thierry Huguet, and Jeanne Harris for seed of various M. truncatula nodulation and nitrogen-fixation mutants; Jannon Fuchs and Harry Schwark for help with the vibratome; Kate VandenBosch, Joe Clouse, and Pascal Gamas for cDNA clones; Jeanne Harris for insightful discussions; and Kirk Czymmek and Carol Carlson for assistance with confocal microscopy.
This work was supported by University of North Texas Faculty Research Funds (to R.D.), the University of Delaware Research Foundation, and a National Institutes of Health BRIN (no. RR16472–02) to the Delaware Biotechnology Institute (to D.J.S).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.049064.
References
- Ardourel M, Demont N, Debelle F, Maillet F, de Billy F, Prome JC, Denarie J, Truchet G (1994) Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6: 1357–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DG, Bianchi S, London F, Dattee Y, Duc G, Essad S, Flament P, Gallusci P, Genier G, Muel X, Tourneur J, Denarie J, et al (1990) Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol Biol Rep 8: 40–49 [Google Scholar]
- Bekki A, Trinchant J-C, Rigaud J (1987) Nitrogen fixation (C2H2 reduction) by Medicago nodules and bacteroids under sodium chloride stress. Physiol Plant 71: 61–67 [Google Scholar]
- Benaben V (1994) Caracterisation cytologique et moleculaire du mutant symbiotique TE7 de Medicago truncatula. PhD thesis. Universite Paul Sabatier, Toulouse, France
- Benaben V, Duc G, Lefebvre V, Huguet T (1995) TE7, an inefficient symbiotic mutant of Medicago truncatula Gaertn. cv Jemalong. Plant Physiol 107: 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin C, Camut S, Malpica CA, Truchet G, Rosenberg C (1990) Rhizobium meliloti genes encoding catabolism of trigonelline are induced under symbiotic conditions. Plant Cell 2: 1157–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewin NJ (1991) Development of the legume root nodule. Annu Rev Cell Biol 7: 191–226 [DOI] [PubMed] [Google Scholar]
- Brewin NJ (1998) Tissue and cell invasion by Rhizobium: the structure and development of infection threads and symbiosomes. In HP Spaink, A Kondorosi, PJJ Hooykaas, eds, The Rhizobiaceae, Molecular Biology of Model Plant-Associated Bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 417–429
- Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ (2003) Dissecting Arabidopsis lateral root development. Trends Plant Sci 8: 165–171 [DOI] [PubMed] [Google Scholar]
- Catalano C, Lane WS, Sherrier DJ (2004) Biochemical characterization of symbiosome membrane proteins from Medicago truncatula root nodules. Electrophoresis 25: 519–531 [DOI] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Denarie J (2000) Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell 12: 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H-K, Kim D, Uhm T, Limpens E, Lim H, Kalo P, Penmetsa VR, Seres A, Kulikova O, Bisseling T, Kiss G, Cook DR (2004) A sequence-based genetic map of Medicago truncatula and comparison of marker co-linearity with Medicago sativa. Genetics 166: 1463–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DR (1999) Medicago truncatula—a model in the making! Curr Opin Plant Biol 2: 301–304 [DOI] [PubMed] [Google Scholar]
- Crespi MD, Jurkevitch E, Poiret M, d'Aubenton-Carafa Y, Petrovics G, Kondorosi E, Kondorosi A (1994) enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J 13: 5099–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickstein R, Bisseling T, Reinhold VN, Ausubel FM (1988) Expression of nodule-specific genes in alfalfa root nodules blocked at an early stage of development. Genes Dev 2: 677–687 [DOI] [PubMed] [Google Scholar]
- Dickstein R, Hu X, Yang J, Ba L, Coque L, Kim D-J, Cook DR, Yeung AT (2002) Differential expression of tandemly duplicated Enod8 gene in Medicago. Plant Sci 163: 333–343 [Google Scholar]
- Dickstein R, Prusty R, Peng T, Ngo W, Smith ME (1993) ENOD8, a novel early nodule-specific gene, is expressed in empty alfalfa nodules. Mol Plant Microbe Interact 6: 715–721 [DOI] [PubMed] [Google Scholar]
- Dickstein R, Scheirer DC, Fowle WH, Ausubel FM (1991) Nodules elicited by Rhizobium meliloti heme mutants are arrested at an early stage of development. Mol Gen Genet 230: 423–432 [DOI] [PubMed] [Google Scholar]
- Fang Y, Hirsch AM (1998) Studying early nodulin gene ENOD40 expression and induction by Nodulation factor and cytokinin in transgenic alfalfa. Plant Physiol 116: 53–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132: 6–13 [DOI] [PubMed] [Google Scholar]
- Fraysse N, Couderc F, Poinsot V (2003) Surface polysaccharide involvement in establishing the rhizobium-legume symbiosis. Eur J Biochem 270: 1365–1380 [DOI] [PubMed] [Google Scholar]
- Gage DJ (2004) Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev 68: 280–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage DJ, Margolin W (2000) Hanging by a thread: invasion of legume plants by rhizobia. Curr Opin Microbiol 3: 613–617 [DOI] [PubMed] [Google Scholar]
- Gamas P, de Carvalho-Niebel F, Lescure N, Cullimore J (1996) Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. Mol Plant Microbe Interact 9: 223–242 [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack K, Jones JDG (2000) Responses to plant pathogens. In BB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Biologists, Rockville, MD, pp 1102–1165
- Haynes JG, Cyzmmek KJ, Carlson CA, Veereshlingam H, Dickstein R, Sherrier DJ (2004) A novel method for rapid analysis of legume root nodule development using confocal microscopy. New Phytol 163: 661–668 [DOI] [PubMed] [Google Scholar]
- Hirsch AM (1992) Developmental biology of legume nodulation. New Phytol 122: 211–237 [DOI] [PubMed] [Google Scholar]
- Kijne JW (1992) The Rhizobium infection process. In G Stacey, HJ Evans, RH Burris, eds, Biological Nitrogen Fixation. Chapman and Hall, London, pp 349–398
- Kuppusamy KT, Endre G, Prabhu R, Penmetsa RV, Veereshlingam H, Cook DR, Dickstein R, VandenBosch KA (2004) LIN, a Medicago truncatula gene required for nodule differentiation and persistence of rhizobial infections. Plant Physiol 136: 3682–3691 [DOI] [PMC free article] [PubMed]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633 [DOI] [PubMed] [Google Scholar]
- Liu C, Yeung AT, Dickstein R (1998) The cDNA sequence of Medicago truncatula cv. Jemalong Enod8, a gene associated with nitrogen fixing root nodule organogenesis. Plant Physiol 117: 1127 [Google Scholar]
- Lullien V, Barker DG, de Lajudie P, Huguet T (1987) Plant gene expression in effective and ineffective root nodules of alfalfa (Medicago sativa). Plant Mol Biol 9: 469–478 [DOI] [PubMed] [Google Scholar]
- Mitra RM, Long SR (2004) Plant and bacterial symbiotic mutants define three transcriptionally distinct stages in the development of the Medicago truncatula/Sinorhizobium meliloti symbiosis. Plant Physiol 134: 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollenhauer H (1964) Plastic embedding mixtures for use in electron microscopy. Stain Technol 39: 111–114 [PubMed] [Google Scholar]
- Muller P, Klaucke A, Wegel E (1995) TnphoA-induced symbiotic mutants of Bradyrhizobium japonicum that impair cell and tissue differentiation in Glycine max nodules. Planta 197: 163–175 [Google Scholar]
- Niehaus K, Kapp D, Puhler A (1993) Plant defence and delayed infection of alfalfa pseudonodules induced by an exopolysaccharide (EPSI)-deficient Rhizobium meliloti mutant. Planta 190: 415–425 [Google Scholar]
- Niehaus K, Lagares A, Puhler A (1998) A Sinorhizobium meliloti lipopolysaccharide mutant induces effective nodules on the host plant Medicago sativa (alfalfa) but fails to establish a symbiosis with Medicago truncatula. Mol Plant Microbe Interact 11: 906–914 [Google Scholar]
- Oldroyd GED, Engstrom EM, Long SR (2001) Ethylene inhibits the Nod factor signal transduction pathway of Medicago truncatula. Plant Cell 13: 1835–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa RV, Cook DR (1997) A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275: 527–530 [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Cook DR (2000) Production and characterization of diverse developmental mutants of Medicago truncatula. Plant Physiol 123: 1387–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa RV, Frugoli JA, Smith LS, Long SR, Cook DR (2003) Dual genetic pathways controlling nodule number in Medicago truncatula. Plant Physiol 131: 998–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perotto S, Brewin NJ, Kannenberg EL (1994) Cytological evidence for a host defense response that reduces cell and tissue invasion in pea nodules by lipopolysaccharide-defective mutants of Rhizobium leguminosarum strain 3841. Mol Plant Microbe Interact 7: 99–112 [Google Scholar]
- Rathbun E, Naldrett MJ, Brewin NJ (2002) Identification of a family of extensin-like glycoproteins in the lumen of Rhizobium-induced infection threads in pea root nodules. Mol Plant Microbe Interact 15: 350–359 [DOI] [PubMed] [Google Scholar]
- Reynolds ES (1963) The use of lead citrate at high pH as an electon opaque stain in electron microscopy. J Cell Biol 17: 208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg C, Boistard P, Denarie J, Casse-Delbart F (1981) Genes controlling early and late functions in symbiosis are located on a megaplasmid in Rhizobium meliloti. Mol Gen Genet 184: 326–333 [DOI] [PubMed] [Google Scholar]
- Thoquet P, Gherardi M, Journet E-P, Kereszt A, Ane J-M, Prosperi J-M, Huguet T (2002) The molecular genetic linkage map of the model legume Medicago truncatula: an essential tool for comparative legume genomics and the isolation of agronomically important genes. BMC Plant Biol 2: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brussel AAN, Bakhuizen R, van Spronsen PC, Spaink HP, Tak T, Lugtenberg BJJ, Kijne JW (1992) Induction of pre-infection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of Rhizobium. Science 257: 70–72 [DOI] [PubMed] [Google Scholar]
- van de Wiel C, Norris JH, Bochenek B, Dickstein R, Bisseling T, Hirsch AM (1990) Nodulin gene expression and ENOD2 localization in effective, nitrogen-fixing and ineffective, bacteria-free nodules of alfalfa. Plant Cell 2: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBosch KA, Frugoli J (2001) Standards and guidelines for genetic nomenclature for the model legume Medicago truncatula. Mol Plant Microbe Interact 14: 1364–1367 [DOI] [PubMed] [Google Scholar]
- Vasse J, Billy F, Truchet G (1993) Abortion of infection during the Rhizobium meliloti-alfalfa symbiotic interaction is accompanied by a hypersensitive reaction. Plant J 4: 555–566 [Google Scholar]