Abstract
Thlaspi caerulescens is a heavy metal hyperaccumulator plant species that is able to accumulate extremely high levels of zinc (Zn) and cadmium (Cd) in its shoots (30,000 μg g−1 Zn and 10,000 μg g−1 Cd), and has been the subject of intense research as a model plant to gain a better understanding of the mechanisms of heavy metal hyperaccumulation and tolerance and as a source of genes for developing plant species better suited for the phytoremediation of metal-contaminated soils. In this study, we report on the results of a yeast (Saccharomyces cerevisae) complementation screen aimed at identifying candidate heavy metal tolerance genes in T. caerulescens. A number of Thlaspi genes that conferred Cd tolerance to yeast were identified, including possible metal-binding ligands from the metallothionein gene family, and a P-type ATPase that is a member of the P1B subfamily of purported heavy metal-translocating ATPases. A detailed characterization of the Thlaspi heavy metal ATPase, TcHMA4, demonstrated that it mediates yeast metal tolerance via active efflux of a number of different heavy metals (Cd, Zn, lead [Pb], and copper [Cu]) out of the cell. However, in T. caerulescens, based on differences in tissue-specific and metal-responsive expression of this transporter compared with its homolog in Arabidopsis (Arabidopsis thaliana), we suggest that it may not be involved in metal tolerance. Instead, we hypothesize that it may play a role in xylem loading of metals and thus could be a key player in the hyperaccumulation phenotype expressed in T. caerulescens. Additionally, evidence is presented showing that the C terminus of the TcHMA4 protein, which contains numerous possible heavy metal-binding His and Cys repeats residues, participates in heavy metal binding. When partial peptides from this C-terminal domain were expressed in yeast, they conferred an extremely high level of Cd tolerance and Cd hyperaccumulation. The possibilities for enhancing the metal tolerance and phytoremediation potential of higher plants via expression of these metal-binding peptides are also discussed.
There are a small number of terrestrial plant species that not only can tolerate high levels of toxic heavy metals in the soil but also can accumulate those metals to unusually high levels in their shoot biomass. These fascinating plant species, first coined hyperaccumulators by Brooks et al. (1977), are loosely categorized as plants that can accumulate metals in the shoot from 100- to 1,000-fold higher than normal, nonaccumulator plants (McGrath et al., 2002). Hyperaccumulating plant species have been identified for a number of heavy metals, including nickel (Ni), zinc (Zn), and cadmium (Cd), as well as for the metalloids selenium and arsenic. Probably the best known metal hyperaccumulator is Thlaspi caerulescens, a member of the Brassica family that has been the object of interest in the plant biology community for over a century, based on its ability to colonize calamine and serpentine soils containing naturally elevated levels of heavy metals such as Zn, Cd, Ni, and cobalt (Co). Certain ecotypes of T. caerulescens can accumulate Zn and Cd to extremely high levels in the shoot, with Zn reaching levels as high as 30,000 μg g−1 (Brown et al., 1995) and shoot Cd concentrations of 10,000 μg g−1 (Lombi et al., 2000). By comparison, shoot Zn concentrations in Zn-sufficient nonaccumulator plants are around 100 μg g−1, with 30 μg g−1 adequate and 300 to 500 μg g−1 toxic (Mengel and Kirkby, 1987); foliar Cd levels above 1 to 10 μg g−1 are usually toxic.
The growing awareness by the plant biology community regarding metal hyperaccumulating plants has, in part, spurred the considerable recent interest and research activity into phytoremediation as a “green” technology for the clean up of heavy metal-contaminated soils. A number of laboratories around the world are studying metal hyperaccumulators, such as T. caerulescens, as model plant systems to gain a better understanding of the mechanisms of heavy metal hyperaccumulation and tolerance, as well as a potential source of genes for developing high biomass plant species that are better suited for phytoremediation of metal-contaminated soils.
Our laboratory has been studying the physiology and molecular biology of heavy metal hyperaccumulation in T. caerulescens and has previously shown that altered metal ion transport plays an important role in the hyperaccumulation phenotype in this plant species (Lasat et al., 1996, 1998). In comparison with related nonaccumulating plant species, T. caerulescens mediates a much greater root heavy metal influx, much more rapid and efficient translocation of the absorbed metal from the root to the shoot in the xylem, and, of course, effective storage of the absorbed heavy metals in the shoot. In fact, one of the distinctive hallmarks of T. caerulescens and other metal hyperaccumulators is their ability to translocate most of the absorbed metal from the root to the shoot. A second hallmark for this hyperaccumulator is the extreme metal tolerance, which is exhibited both in roots and shoots. Mechanisms of metal tolerance can involve both ion transporters that transport the metal out of the cytoplasm (either into an internal compartment or out of the cell), and the synthesis of metal-binding ligands that can detoxify the metal in the cytoplasm (Clemens, 2001).
This study focused on the identification of candidate heavy metal tolerance genes from T. caerulescens via a yeast (Saccharomyces cerevisae)-functional complementation screen. A number of Thlaspi genes that conferred Cd tolerance to yeast were identified, including possible metal-binding ligands from the metallothionein gene family, and a P-type ATPase that is a member of the P1B subfamily of purported heavy metal-translocating ATPases (Axelsen and Palmgren, 1998). A detailed characterization of this Thlaspi heavy metal ATPase was conducted in order to investigate its possible role in metal hyperaccumulation. Here we present evidence that this ATPase facilitates a high degree of heavy metal tolerance in yeast by mediating the active efflux of heavy metals out of the cell. However in T. caerulescens, based on differences in tissue-specific and metal-responsive expression of this transporter compared with expression of its homolog in Arabidopsis (Arabidopsis thaliana; Mills et al., 2003; Hussain et al., 2004), we suggest that it may not be involved in metal tolerance in Thlaspi. Instead, we propose that it may play a role in xylem loading of metals and thus could be a key player in the hyperaccumulation phenotype expressed in T. caerulescens.
RESULTS
Complementation Screening of Candidate Metal Tolerance Genes from T. caerulescens
Preliminary studies determined that when 90 μm Cd was included in solid modified synthetic dextrose (SD) media (see “Materials and Methods”), both the wild-type DY1457 yeast strain as well as DY1457 transformed with the empty pFL61 vector were unable to grow. Thus, this Cd level was used to select for Cd-tolerant transformants. Yeast cells were transformed with a T. caerulescens cDNA library constructed in the yeast expression vector, pFL61, and 35 yeast colonies were identified that were able to grow on this restrictive level of Cd. Yeast from each colony was grown up and replated on the same high Cd media to verify the Cd-tolerant phenotype. Subsequently, the plasmids bearing the T. caerulescens cDNA inserts were isolated, and the Thlaspi cDNAs were subjected to restriction digest analysis and DNA sequencing. Based on this analysis, seven unique Thlaspi genes were identified that conferred Cd tolerance to wild-type yeast. As depicted in Table I, which lists the identity of each of these putative metal tolerance genes, two had a close resemblance to metallothionein genes previously identified in Arabidopsis and Brassica juncea. Four of the other genes either had an unknown function based on DNA and protein sequence comparisons, or could play roles in processes such as photosynthesis, protein synthesis, or signal transduction. The final gene on the list in Table I was represented by two different length partial cDNA clones (423 and 1,152 bp in length) that had a strong similarity to the Arabidopsis heavy metal-transporting ATPase, AtHMA4. RACE-PCR techniques using the GeneRacer kit (Invitrogen Life Technologies, Carlsbad, CA) were employed to clone the full-length TcHMA4 cDNA from T. caerulescens. This technique targets only the 5′ end-capped mRNA, and thus allows for the cloning of only the full-length cDNA. Because of the possible importance of heavy metal ATPases to metal hyperaccumulation and tolerance in T. caerulescens, our subsequent research focused on a detailed characterization of TcHMA4.
Table I.
T. caerulescens genes that confer Cd tolerance in yeast
| Clone | Closest Match and Function | Accession No. | GenBank Hit and % Identity |
|---|---|---|---|
| 1 | B. juncea metallothionein-like protein | AY486002 | Y10849 (82%) |
| 2 | Arabidopsis metallothionein | AY486003 | L15389 (78%) |
| 3 | Arabidopsis integral membrane putative protein | AY486006 | NM_118925 (83%) |
| 4 | Sinapis alba subunit of oxygen evolving system of PSII | AY486007 | Y07498 (85%) |
| 5 | Arabidopsis light-regulated kinase | AY486008 | Z12120 (93%) |
| 6 | Arabidopsis chloroplast inner membrane putative protein | AY486009 | NM_116206 (79%) |
| 7 | Arabidopsis putative heavy metal P-type ATPase | AY486001 | AF412407 (71%) |
All the accession numbers provided are GenBank accession numbers.
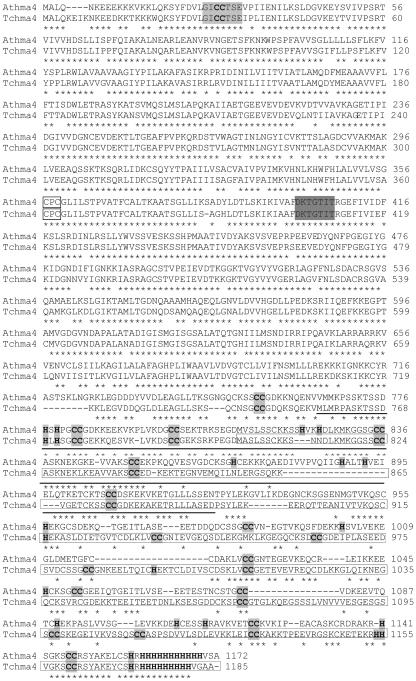
Both nucleotide and amino acid sequence comparisons between TcHMA4 and similar sequences from other organisms indicated that it is a member of the P-type ATPase superfamily and, more specifically, the P1B subfamily of ATPases that are purported to transport heavy metals. As mentioned above, the Arabidopsis P-type ATPase that it is most similar to is AtHMA4, which was recently shown to possibly be involved in Cd export and tolerance (Mills et al., 2003). The deduced amino acid sequence for TcHMA4 and its alignment with AtHMA4 are shown in Figure 1. The full-length TcHMA4 open reading frame is 3,561 bp in length and encodes a polypeptide of 1,186 amino acids. TcHMA4 is 71% identical to AtHMA4 and contains many of the same predicted motifs found in AtHMA4 and other heavy metal ATPases, including eight predicted membrane-spanning domains, and a large C-terminal tail that harbors a number of putative heavy metal-binding domains (see Fig. 1). These include a His repeat consisting of nine His residues, a number of Cys pairs, and multiple single His residues.
Figure 1.
Sequence alignment of TcHMA4 from T. caerulescens and the Arabidopsis homolog AtHMA4. Deduced amino acid sequences for TcHMA4 and AtHMA4 (accession no. 064474) are shown aligned using the ClustalW method. Asterisks indicate identical residues. A number of motifs common to P1B-type ATPases are indicated, including the E1-E2 ATPase phosphorylation site (shaded in dark gray), the highly conserved CPx motif (boxed), and a putative N-terminal heavy metal binding site (underlined). Also, the numerous His and Cys residues in the C terminus are also highlighted.
Expression of TcHMA4 in Yeast Confers Heavy Metal Tolerance
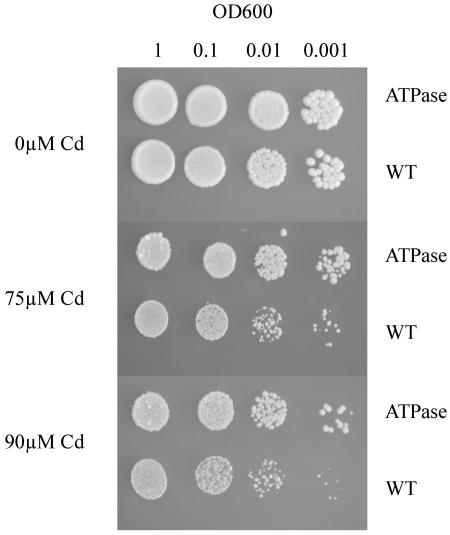
When wild-type yeast is transformed with the full-length TcHMA4, a significant level of Cd tolerance is conferred. As shown in Figure 2, growth of both the wild-type yeast expressing the empty pFL61 vector, and the untransformed wild-type strain (not shown) are strongly inhibited by inclusion of 75 and 90 μm Cd in solid SD media, while expression of TcHMA4 in yeast allows a significant increase in growth on both levels of Cd and also promotes growth compared with wild-type yeast on Cd concentrations as high as 120 μm (data not shown).
Figure 2.
Cd tolerance test for wild-type (transformed with empty pFL61 vector) and TcHMA4-transformed yeast cells. Yeast cells were grown to an OD600 of 1.0, serially diluted to an OD600 of 0.1, 0.01, and 0.001, and then 20-μL drops spotted on SD plates containing 0, 75, and 90 μm CdCl2. WT, Wild type.
Heavy Metal Accumulation in Wild-Type and TcHMA4-Transformed Yeast
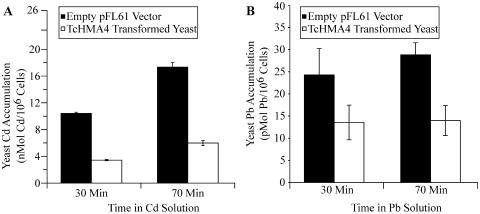
A plant transporter can confer metal tolerance when expressed in yeast either via mediating metal exclusion due to efflux across the plasma membrane or by sequestering the metal in an endomembrane compartment. The first scenario should result in reduced metal accumulation in yeast, while the second should cause enhanced metal accumulation. Thus, metal accumulation experiments were conducted with wild-type yeast expressing empty pFL61 and TcHMA4-transformed yeast; these experiments involved incubating both yeast genotypes in liquid SD media supplemented with either 20 μm CdCl2 or 10 μm PbCl2, ZnCl2, or CuCl2 and harvesting yeast cells after 30 and 70 min of metal accumulation. As shown in Figure 3, expression of TcHMA4 in yeast resulted in a large decrease in both Cd (Fig. 3A) and Pb (Fig. 3B) accumulation. After both 30 and 70 min of metal accumulation, TcHMA4-transformed cells accumulated approximately 70% less Cd and 50% less Pb. TcHMA4-transformed cells also accumulated less Zn and Cu than wild-type cells (data not shown). These findings are consistent with TcHMA4 operating at the yeast plasma membrane to pump metals out of the cell and having the ability to transport a number of different essential micronutrients and heavy metals.
Figure 3.
Cd and Pb accumulation by wild-type yeast cells (transformed with the empty pFL61 vector) and TcHMA4-transformed yeast cells. A, Yeast Cd accumulation for two time periods (30 and 70 min) in liquid SD media supplemented with 20 μm CdCl2. B, Yeast Pb accumulation for two time periods (30 and 70 min) in liquid SD media supplemented with 10 μm PbCl2. The error bars represent the mean of four replicate measurements ± the se of the mean. WT, Wild type.
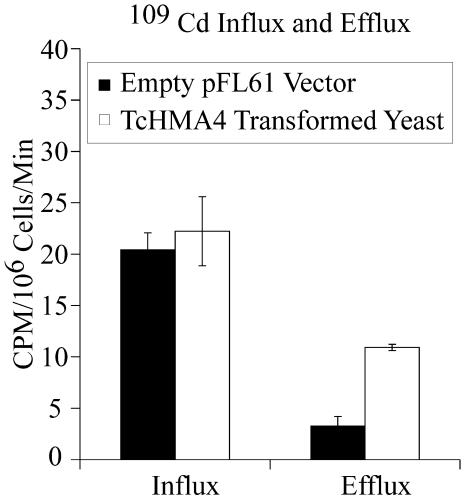
Quantitation of 109Cd Efflux and Influx
In order to more conclusively determine if TcHMA4 is functioning to pump metals across the yeast plasma membrane and out of the cell, 109Cd flux techniques were used to quantify Cd influx and efflux in control and TcHMA4-transformed yeast. As depicted in Figure 4, short-term 109Cd uptake experiments showed that there were no differences in Cd influx in control versus TcHMA4-transformed cells. However, when both yeast genotypes were loaded with 109Cd and allowed to efflux radiotracer into identical, unlabeled solution, the TcHMA4-transformed cells maintained a 3.5-fold higher rate of Cd efflux than did control cells not expressing the Thlaspi ATPase. These results further support the hypothesis that TcHMA4 is a metal efflux transporter operating at the plasma membrane. It is interesting to note than in these cells, the rates of Cd influx were 2- to 6-fold larger than Cd efflux, indicating that a net Cd uptake was occurring, which presumably is associated with rapidly growing and dividing cells in the mid-log phase of growth.
Figure 4.
Radiotracer (109Cd) Cd influx and efflux in wild-type yeast cells (transformed with the empty pFL61 vector) and TcHMA4-transformed yeast cells. The influx and efflux data are presented as 109Cd CPM/106 cells/ min. The error bars represent the mean of four replicate measurements ± the se of the mean.
Tissue-Specific Expression of TcHMA4 in T. caerulescens
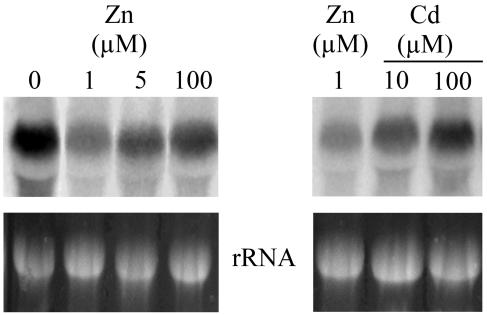
The expression of TcHMA4 in different plant tissues and in response to changes in plant mineral nutrient and heavy metal status were investigated via northern analysis in T. caerulescens seedlings. For this northern analysis, we used a 1,500-bp probe from the C-terminal region of TcHMA4 to avoid cross-hybridization with other HMAs (heavy metal associated). The C-terminal sequence is the most divergent region for the HMA subfamily of transporters. Southern analysis using this same 1,500-bp probe with T. caerulescens genomic DNA under the same stringency conditions used for the northern blots yielded results consistent with hybridization to a single copy of TcHMA4 in the genome (data not shown). These results suggest that the northern-analysis data are not complicated by hybridization of TcHMA4 to other members of the TcHMA subfamily.
As depicted in Figure 5, TcHMA4 is expressed strongly in roots, and we also found a very low level of expression in aboveground plant tissues and organs (data not shown). This finding suggests that TcHMA4 is primarily a root-associated metal transporter. With regards to the relationship of TcHMA4 expression and plant Zn status, both Zn deficiency as well as exposure of plants to high Zn levels induced a significant increase in TcHMA4 transcript abundance (Fig. 5). Furthermore, when plants were challenged with high levels of Cd in the nutrient solution, this also induced a strong increase in TcHMA4 expression (Fig. 5). This response to increasing plant Cd status in T. caerulescens is quite different than what has been reported for its homolog in Arabidopsis, where it has been shown that root expression of AtHMA4 is down-regulated by plant exposure to Cd (Mills et al., 2003).
Figure 5.
Northern analysis of TcHMA4 in roots of T. caerulescens plants grown under Zn-deficient, -sufficient, and high-Zn conditions, or under high-Cd conditions. Seedlings were grown on hydroponic media containing 0, 1, 5, 10, and 100 μm ZnCl2, or 10 and 100 μm CdCl2. The 25S ribosomal bands are shown as loading controls. Each specific northern was repeated at least twice, with the same results.
Analysis of Partial TcHMA4 Clones
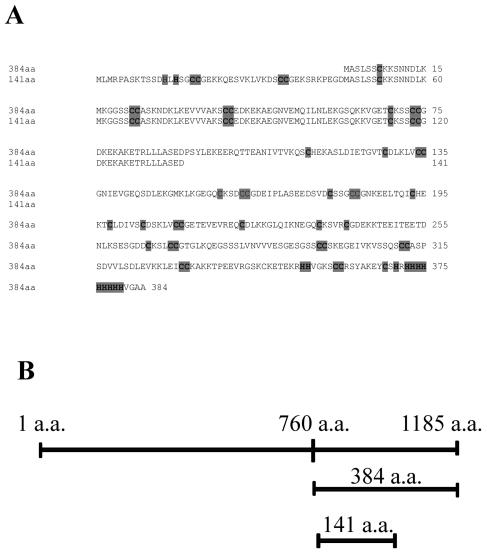
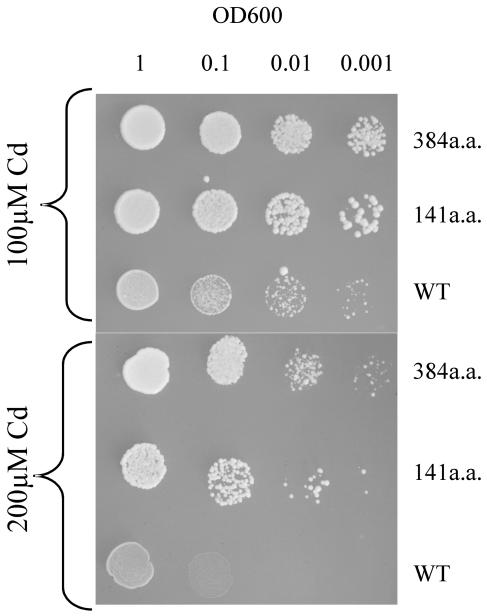
As described above for the initial yeast complementation screen for Thlaspi Cd tolerance genes, two different partial TcHMA4 clones conferred a significant degree of Cd tolerance when expressed in yeast. These two clones encode peptides predicted to be 384 and 141 amino acids in length that encompass most (for the 384-amino acid peptide) or a portion (for the 141-amino acid peptide) of the large C-terminal cytoplasmic tail of TcHMA4 that harbors a number of putative heavy metal-binding domains (Fig. 6). These domains include a number of Cys pairs, a His-9 repeat, and numerous single His residues. As neither of these peptides contain any predicted membrane-spanning domains, it is clear that they are not functioning as metal transporters. Thus, the possibility that they confer heavy metal tolerance in yeast by acting as metal-binding peptide ligands in the yeast cytoplasm was investigated. As shown in Figure 7, both the 141- and 341-amino acid peptides conferred a very high degree of Cd tolerance, resulting in significant yeast growth even in solid media supplemented with 200 μm CdCl2. Thus these peptides confer a considerably higher degree of tolerance than does the full TcHMA4 protein, as yeast expressing the full TcHMA4 protein would not grow on media containing Cd at concentrations higher than 120 μm. It is interesting to note that the longer TcHMA4 peptide confers a greater degree of Cd tolerance than does the shorter, 141-amino acid peptide (see Fig. 7). The longer peptide contains both the His-9 concatamer as well as the numerous Cys pairs and single His residues, while the shorter peptide lacks the His-9 repeat. Thus, it might be expected that the longer peptide, which covers most of the C-terminus cytoplasmic tail with its numerous potentially metal-binding amino acids, might have a greater capacity to bind Cd and afford a greater degree of metal detoxification in the cytoplasm. It should also be pointed out that the His concatamer as well as the vicinal Cys residues also can effectively interact with other micronutrients and heavy metals; therefore, it is also possible in the full-length ATPase that this region of the peptide could also be involved in the binding and/or sensing of a range of metals in addition to Cd, including Zn, Co, and Ni.
Figure 6.
A, Amino acid sequence alignment of the 384- and 141-amino acid partial peptides from the C terminus of TcHMA4 that were identified from the initial yeast complementation screen. The conserved His stretch, numerous Cys repeats, and single His residues in the C-terminal region of the peptide are indicated in light gray. B, Schematic representation of location of two partial peptides sequences relative to each other and to the full-length TcHMA4 protein.
Figure 7.
Cd tolerance test for yeast expressing the partial TcHMA4 peptides. Yeast cells were transformed either with the empty pFL61 vector (WT) or with the 384- or 141-amino acid peptides (see Fig. 6 for their sequence). The plates were set up as described in the legend for Figure 2. Metal tolerance was assayed by visualizing growth on SD plates containing 100 and 200 μm CdCl2.
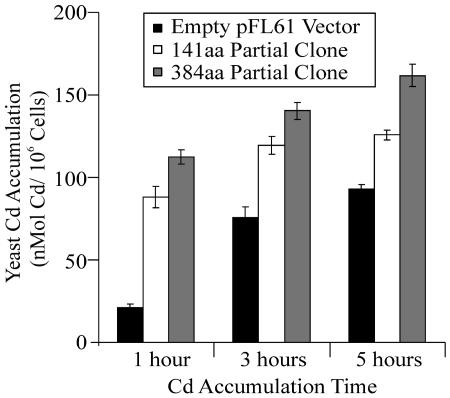
It can be predicted for a heavy metal tolerance mechanism, based on the production of a metal-binding ligand, that this tolerance mechanism would be associated with a larger cellular metal accumulation compared with wild-type yeast cells. As shown in Figure 8, this is what occurs; yeast cells transformed with the partial clones exhibit a 4- to 6-fold greater Cd accumulation than wild-type yeast expressing the empty pFL61 vector after 1 h of accumulation. Furthermore, Cd accumulation in yeast cells expressing the longer peptide was always greater than in yeast cells expressing the shorter peptide at all three time points where Cd accumulation was measured (Fig. 8). Again, this correlates nicely with the differing degree of Cd tolerance conferred by these two clones and with the relative metal-binding capacity of these two peptides as predicted from their amino acid sequences. It is interesting to note that the net Cd accumulation in yeast expressing these partial peptides was approximately 10-fold higher than the rates of Cd accumulation in yeast expressing the full-length transporter. This is consistent with Cd tolerance via the partial clones associated with increased symplasmic binding and accumulation of Cd, while for the full length transporter, tolerance is conferred by the pumping of the absorbed metal back out of the cell, which would result in a lower rate of net accumulation.
Figure 8.
Cd accumulation by yeast expressing the empty pFL61 vector (WT), compared with yeast expressing the 384- and 141-amino acid partial peptides from TcHMA4. Cd accumulation was conducted in liquid SD media supplemented with 20 μm CdCl2. The error bars represent the mean of four replicate measurements ± the se of the mean.
DISCUSSION
In this study, we identified a number of genes that may be involved in the mechanisms of metal tolerance and hyperaccumulation in the heavy metal hyperaccumulating plant species, T. caerulescens (Table I). Some of the genes, such as those encoding metallothioneins and heavy metal ATPases, have previously been suggested to play a role in metal tolerance (see, for example, Clemens, 2001; Cobbett and Goldsbrough, 2002). However, for most of the other genes in Table I, it is more difficult to ascribe a function in metal tolerance mechanisms based on sequence similarity to previously characterized genes. In should be noted that, because the yeast complementation screen used in this study will identify genes that can confer tolerance at the single-cell level, it is possible that some of these genes may not participate in metal tolerance in T. caerulescens, particularly if more complex metal tolerance mechanisms based on the operation and interplay of multiple cell types, tissues, and/or organs is functioning in Thlaspi.
TcHMA4 Confers Metal Tolerance in Yeast Via Metal Efflux Out of the Cell
From the yeast complementation screen, we identified a P1B-type heavy metal ATPase that is 71% identical on a protein level to the Arabidopsis heavy metal ATPase, AtHMA4 (Mills et al., 2003). The P1B subfamily of ATPases is believed to be involved in the transport of heavy metals such as Zn, Cu, Co, Cd, Pb, and silver that are either essential micronutrients or noessential toxic metals, although only a few members of this subfamily have been well characterized. Genome sequencing efforts as well as isolation and analysis of individual members of this subgroup from different organisms indicate these heavy metal ATPases are expressed in a wide range of organisms, ranging from the archaea and bacteria to eukaryotic organisms including Arabidopsis (see, for example, Rensing et al., 1997; Axelsen and Palmgren, 1998, 2001; Tong et al., 2002; Mills et al., 2003).
TcHMA4 shares many of the conserved motifs found in other P1B-type ATPases, including eight putative membrane-spanning domains, and conserved CPx (amino acids 357–359) and HP (amino acids 441–442) motifs. Also, TcHMA4 shares certain unique structural features with its Arabidopsis homolog, in that both AtHMA4 and TcHMA4 have a long polar C-terminus region that is predicted to reside in the cytoplasm and contains numerous metal-binding amino acids, including a long (9-amino acid) His stretch at the very end of the C terminus, 13 Cys pairs, and a large number of single His residues (Figs. 1 and 6) and might be expected to participate in the binding of a number of heavy metals, although an involvement of this putative metal-binding region in transport has not been demonstrated.
As shown in Figure 2, expression of TcHMA4 in wild-type yeast conferred a significant increase in heavy metal (Cd) tolerance. Metal tolerance at the single cell level can be achieved in a number of different ways. These include: (1) metal efflux, where the metal is first transported into the cell and then actively pumped back out via the action of an efflux transporter; (2) true tolerance, where the synthesis of metal-binding ligands bind and detoxify the metal in the cytoplasm; and (3) internal sequestration, where an endomembrane-localized metal transporter sequesters the metal in an internal cellular compartment (e.g. the vacuole). Yeast accumulation experiments indicated that expression of TcHMA4 in yeast resulted in reduced metal accumulation, and that this occurred rapidly, within minutes. Furthermore, TcHMA4 can mediate this reduced accumulation for a number of heavy metals and micronutrients, including Cd and Pb (Fig. 3) as well as Zn and Cu (data not shown). This TcHMA4-mediated reduction in metal accumulation is consistent with the TcHMA4 functioning to mediate metal efflux across the yeast plasma membrane. This possibility was investigated in more detail using 109Cd radiotracer flux techniques, and these experiments clearly showed that TcHMA4 was associated with metal efflux out of the cell (Fig. 4). Thus, it is quite likely that in yeast and plants, TcHMA4 operates as a plasma membrane-localized metal ATPase to pump Cd and possibly other heavy metals and micronutrients across the plasma membrane and out of the cell.
Possible Function of TcHMA4 in Heavy Metal Hyperaccumulation
Heavy metal hyperaccumulation in T. caerulescens is associated with several traits, including: (1) the ability to tolerate high levels of metals both in the soil and within the plant; (2) an enhanced ability to absorb metals from the soil; (3) the ability to efficiently and rapidly translocate the absorbed metals from the root to the shoot; and (4) the ability to store very high levels of the metals in leaf epidermal cells (Lasat et al., 1996, 1998; Küpper et al., 1999). One of the most distinctive features that differentiate metal hyperaccumulator plants from nonaccumulators is their ability to translocate most of the absorbed metal from the root to the shoot. Nonaccumulator plants tend to sequester heavy metals in the roots, while hyperaccumulators, and especially T. caerulescens, rapidly translocate the bulk of the heavy metal to the shoot for storage in the leaf epidermis (Lasat et al., 1996, 1998).
When one examines the metal transport and gene expression data for TcHMA4 presented here in comparison with recently published expression data for AtHMA4 in Arabidopsis, the findings lead us to speculate about an intriguing scenario regarding the role of TcHMA4 in metal hyperaccumulation in Thlaspi. In Arabidopsis, it has been reported that the homolog of TcHMA4 is expressed throughout the plant, with somewhat stronger expression in roots than in other plant organs and tissues (Mills et al., 2003). In this paper it was also reported that AtHMA4 expression was down-regulated by plant exposure to high levels of Cd. In a more recent article, Hussain et al. (2004) showed in Arabidopsis that the tissue-specific expression of AtAtHMA4 is localized primarily to the vascular tissue in roots, leaves, and stems. They also showed that when AtHMA4 is knocked out, the mutant accumulates significantly less Zn in the shoots compared with wild-type plants. These findings suggest that at least in the Arabidopsis root, HMA4 may be involved in Zn translocation to the shoot. As seen in Figure 5, TcHMA4 is expressed strongly and almost exclusively in the Thlaspi root, and its expression is up-regulated by seedling exposure to high levels of Cd and Zn, as well as by Zn deficiency. Given the tissue-specific localization of AtHMA4 expression, it is reasonable to assume that TcHMA4 follows a similar cell-specific expression pattern in Thlaspi roots. The Thlaspi and Arabidopsis HMA4 expression data, along with the information concerning the function of the Arabidopsis homolog and our findings in yeast that are consistent with TcHHA4 operating at the root-cell plasma membrane to pump heavy metals and micronutrients out of the cell, have led us to hypothesize that TcHMA4 does not directly play a role in heavy metal tolerance in T. caerulescens. Instead, we propose that it functions in metal xylem loading by mediating heavy metal and micronutrient efflux from xylem parenchyma into xylem vessels in the Thlaspi root, and thus plays a key role in the mechanisms underlying metal hyperaccumulation in T. caerulescens. This would be consistent with the observed up-regulation of expression of this transporter by heavy metal exposure. It is interesting to also note that this transporter is also induced by Zn deficiency. Thus, TcHMA4 may be involved not only in heavy metal hyperaccumulation but also in Thlaspi Zn nutrition; that is, under Zn deficiency expression of this transporter is increased in an attempt to maintain shoot Zn status for reproduction and the completion of the plant's life cycle. The possible role of TcHMA4 in Thlaspi heavy metal hyperaccumulation and Zn homeostasis will be the subject of future research.
The C Terminus as a Metal-Binding Peptide. A Possible Role in Phytoremediation?
During our initial yeast screen for heavy metal tolerance genes, the two heavy metal ATPase clones that conferred Cd tolerance in wild-type yeast were not the full-length ATPase, but instead were partial TcHMA4 clones. As seen in Figure 6, these two clones are 141 and 384 amino acids in length and both are from the C-terminal region of the TcHMA4 protein, which is predicted to reside in the cytoplasm and contains numerous His and Cys residues that have high affinities for heavy metals. From their predicted amino acid sequence, it is clear that the partial clones lack any membrane-spanning domains and are too small to function as a metal transporter. The only explanation for the increased Cd tolerance conferred by these partial clones is that they are functioning as Cd-binding ligands in the yeast cytoplasm. Our yeast metal tolerance and accumulation experiments with these partial clones verified this hypothesis, as expression of both clones conferred a high degree of Cd tolerance (considerably greater than the Cd tolerance associated with the full-length TcHMA4) and Cd hyperaccumulation (4- to 6-fold increase in yeast Cd accumulation after 1 h). The longer 384-amino acid clone harbors both the long, His-9 concatamer as well as the numerous Cys pairs and single His residues of the TcHMA4 C terminus, while the shorter 141-amino acid peptide lacks the poly-His tail as well as several of the Cys-Cys repeats. There was a clear correlation between the number of metal-binding motifs and the degree of metal tolerance and accumulation conferred by the two peptides, with the longer peptide conferring significantly higher Cd tolerance and Cd hyperaccumulation in yeast.
It has previously been suggested that the Cys dipeptides and His-rich domains in heavy metal ATPases may be involved in heavy metal-binding (Solioz and Vulpe, 1996; Williams et al., 2000), and the findings presented here confirm these speculations for the TcHMA4 protein. Based on the association constants for the binding of heavy metals to di-Cys and di-His residues (Motekaitis et al., 1997), the His and Cys repeats, particularly in the longer peptide, should provide a large number of high-affinity binding sites for a range of heavy metals and micronutrients, including Cd, Pb, mercury, Zn, Co, Ni, and Cu. Therefore, the findings in yeast suggest that these two plant peptides, when expressed to a high level in plants, may confer significant increases in tolerance and accumulation for a range of heavy metals and thus may have useful applications with regard to enhancing the phytoremediation potential of plants via biotechnology. We are currently expressing these partial TcHMA4 clones in Arabidopsis in order to study their potential environmental applications, including the creation of plants with increased heavy metal tolerance and possibly enhanced metal hyperaccumulation.
MATERIALS AND METHODS
Plant Growth Conditions
Thlaspi caerulescens (ecotype Prayon) seedlings were grown on a modified Johnson's solution that had a macronutrient composition of 1.2 mm KNO3, 0.8 mm Ca(NO3)2, 0.2 mm NH4H2PO4, and 0.2 mm MgSO4 and a micronutrient composition of 50 μm KCl, 12.5 μm H3BO4, 1 μm MnSO4, 1 μm ZnSO4, 0.5 μm CuSO4, 0.1 μm Na2MoO4, 0.1 μm NiSO4, and 7.5 μm Fe-EDDHA (N,N′-ethylenediamine-di(O-hydroxyphenylacetic acid)). The solution was buffered at a pH of 5.5 with 1 mm MES (2-[N-morpholino]-ethanesulfonic acid) buffer. Thlaspi seeds were placed in a drop of 0.7% (w/v) low-temperature gelling agarose on nylon mesh circles (1-mm mesh openings), which, in turn, were positioned on a coarser mesh support sealed to the bottom of black plastic cups. The cups and seeds were fitted into holes cut into black plastic lids covering 5-L black plastic pots. Seedlings were grown in a growth chamber at 25/15°C (light:dark, 16:8 h) under a light intensity of 300 μmol photons m−2 s−1 for 2 weeks, and then seedlings were transferred into identical growth containers containing modified Johnson's solution supplemented with specific concentrations of Zn (0, 1, 5, or 100 μm) or Cd (10 or 100 μm). Tissue samples were harvested after 20 d growth in the Zn- and Cd-supplemented medium.
Yeast Growth Conditions
The wild-type yeast strain DY1457 (MATa ade6 can1 his3 trp1 ura3) was transformed either with the empty yeast expression vector pFL61 (Minet et al., 1992) or pFL61 containing T. caerulescens cDNA. Transformed yeast strains were grown on SD-minimal medium (Rose et al., 1990) supplemented with 0.1% casamino acids, adenine sulfate (20 mg/mL), l-trypthophan (20 mg/mL), l-His (20 mg/mL), and l-Leu (30 mg/mL). This supplemented media will be referred to as SD media in the manuscript. Solid media consisted of the same SD media supplemented with granulated agar (Difco Laboratories, Sparks, MD) at a concentration of 20 g/L. To test the heavy metal tolerance of control and Thlaspi-transformed yeast, growth was conducted on high Cd plates consisting of SD solid media supplemented with 90 μm CdCl2. Plates were inoculated with an aliquot of liquid media yeast culture and incubated at 30°C for 3 d before plates were photographed to document the relative Cd tolerance of the different yeast strains.
Functional Complementation Assay for Heavy Metal Tolerance
A cDNA library was constructed with combined polyA+ RNA from roots and shoots of T. caerulescens seedlings grown on both Zn-deficient and Zn-replete nutrient solution. The cDNA was synthesized using the Superscript Choice System (Life Technologies/Gibco-BRL, Cleveland), then ligated using BstXI/EcoRI adapters into the bifunctional yeast/Escherichia coli expression plasmid vector pFL61 (Pence et al., 2000). This vector contains a yeast phosphoglycerate kinase promoter and a uracil selection marker. Preliminary experiments determined that the yeast DY1457 wild-type stain was unable to grow on solid SD media containing 90 μm Cd. Subsequently, wild-type yeast was transformed with T. caerulescens cDNA and plated on high Cd (90 μm) solid SD media. After growth for 3 d at 30°C on Cd-containing media, 35 individual yeast colonies were identified that could grow under the restrictive, high-Cd conditions. Each yeast colony was replated on high Cd plates to verify the metal tolerance phenotype, and then DNA was extracted from each colony and the nucleotide sequence determined (Applied Biosystems Automated 3730 DNA Analyzer; Cornell University).
Quantification of Yeast Metal Accumulation
Wild-type DY1457 yeast strain containing the empty pFL61 vector and TcHMA4-transformed yeast were grown on liquid SD medium. At the mid-log phase of growth, the metal accumulation experiment was initiated by adding either CdCl2 or PbCl2 to a final concentration of 20 or 10 μm, respectively. After 5, 30, and 70 min of metal accumulation, aliquots of yeast cells were taken, centrifuged at 10,000g for 2 min to separate yeast cells from the metal-containing media, washed once with 5 mm CaCl2 to desorb Cd2+ or Pb2+ from the yeast cell walls, and then centrifuged again at 10,000g for 2 min. The heavy metal content of the yeast cell pellet was analyzed using an inductively coupled, plasma trace analyzer emission spectrometer (Model ICAP 61E, Thermo-Jarrell Ash, Waltham, MA).
The same procedure was carried out when determining Cd accumulation in yeast transformed with the partial TcHMA4 clones, except that Cd accumulation was determined after 1, 3, and 5 h of yeast Cd accumulation. All the metal accumulation values are the average ± the se determined from four replicate experiments.
Determination of 109Cd2+ Influx and Efflux in Yeast
Radiotracer (109Cd) flux methodologies were used to determine Cd2+ influx and efflux in control and transformed yeast cells (transformed with either full-length or partial TcHMA4 clones). For the Cd2+ influx experiments, the pFL61- and TcHMA4-transformed yeast were grown on liquid SD medium until they reached the mid-log phase of growth. At this time, the Cd2+ influx experiment was initiated by adding 109CdCl2 to a final [Cd] of 10 μm. Yeast was sampled at 30 s (which was an approximation of Cd bound to cell walls), 3, 5, 10, and 15 min. The yeast aliquots for each time point were transferred into 1.5 mL plastic microfuge tubes containing a 200-μL silicon oil/dinonyl phthalate pad on top of 2 μL of 40% perchloric acid. Tubes were centrifuged at 7,000g for 1 min to separate the yeast from the radiolabeled uptake solution, and the supernatant was removed. The microfuge tube with the remaining cell pellet was placed in a scintillation vial and the amount of 109Cd accumulation determined using a Perkin Elmer WIZARD 3 1480 Automatic Gamma Counter (Perkin Elmer Applied Biosystems, Foster City, CA). The amount of 109Cd associated with yeast cells after 30 s was determined in preliminary experiments to be a measure primarily of Cd associated with the yeast cell wall, and this value was subtracted from each time point to obtain a measure of Cd accumulation in the yeast cell symplasm. Preliminary experiments determined that Cd2+ accumulation was linear between 30 s and 15 min, and these values were used to calculate Cd2+ influx values.
For the Cd efflux experiments, the control and TcHMA4-transformed yeast cells were grown on liquid SD medium and again, at the mid-log phase of growth, 109CdCl2 was added to a final concentration of 10 μm. The yeast cells were allowed to accumulate 109Cd2+ for 90 min, centrifuged at 7,000g for 2 min, washed briefly in 5 mm CaCl2, and then centrifuged again. The washed yeast cells were then transferred to liquid SD medium containing nonradioactive CdCl2 (10 μm) to initiate the Cd efflux, and yeast aliquots were sampled at 30, 60, 90, and 120 min. The yeast cells were treated as described above for the influx experiments to determine the amount of 109Cd appearing in the efflux solution, and Cd efflux was calculated based on the rate of arrival of 109Cd into the efflux solution. We also found in separate experiments that the same rates of Cd efflux were obtained when the efflux was calculated based on the rate of disappearance of 109Cd from yeast cells. All of the Cd influx and efflux values are the average ± the se determined from four replicate experiments.
Comparative Determination of Cd Tolerance for Different Yeast Genotypes
To determine the relative Cd tolerance of control and TcHMA4-transformed cells (transformed with either full or partial clones), the different yeast genotypes were grown on regular liquid SD medium until they achieved an optical density (OD) of 1.0. At this point, the cells were harvested and serial dilutions were made to achieve ODs of 0.1, 0.01, and 0.001. SD plates were made containing the appropriate concentration of CdCl2, and a 20-μL aliquot for each cell dilution was spotted on to the plates, as described by Lee et al. (2003). The plates were placed in a 30°C incubator for 3 d, after which pictures were taken.
Northern Analysis in T. caerulescens
T. caerulescens plants were grown in modified Johnson's nutrient solution as described above that was supplemented with different Zn2+ and Cd2+ concentrations. Total RNA was isolated from roots and shoots, denaturated, separated with denaturating agarose gel electrophoresis, and transferred to a nylon membrane (Hybond N+; Amersham Pharmacia; Piscataway, NJ). Probes were labeled with [α-32P]dCTP by random hexamer primers and hybridized to the membrane overnight. Next, 20 μg of RNA was loaded in each line and equal loading was ensured by ethidium bromide staining of ribosomal subunits. Following hybridization at 65°C, the nylon membranes were washed twice for 15 min at 65°C in a low-stringency wash solution (2× SSC, 0.1% SDS). Each northern experiment was repeated at least twice.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY486001, AY486002, AY486003, AY486006, AY486008, and AY486009.
This work was supported by the National Science Foundation (grant no. IBN–0129844) and a National Research Initiative Competitive Grant from the U.S. Department of Agriculture (grant no. 2002–35100–12487 to L.V.K.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.044503.
References
- Axelsen KB, Palmgren MG (1998) Evolution of substrate specificities in the P-type ATPase superfamily. J Mol Evol 46: 84–101 [DOI] [PubMed] [Google Scholar]
- Axelsen K, Palmgren MG (2001) Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol 126: 696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks RR, Lee J, Reeves RD, Jaffre T (1977) Detection of metalliferous rocks by analysis of herbarium specimens of indicator plants. J Geochem Explor 7: 49–77 [Google Scholar]
- Brown S, Chaney R, Angle J, Baker A (1995) Zinc and cadmium uptake of T. caerulescens grown in nutrient solution. Soil Sci Soc Am J 59: 125–133 [Google Scholar]
- Clemens S (2001) Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212: 457–486 [DOI] [PubMed] [Google Scholar]
- Cobbett C, Goldsborough P (2002) Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu Rev Plant Biol 53: 159–182 [DOI] [PubMed] [Google Scholar]
- Hussain D, Haydon M, Wang Y, Wong E, Sherson S, Young J, Camakaris J, Harper J, Cobbett C (2004) P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 16: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küpper H, Zhao FJ, McGrath SP (1999) Cellular compartmentation of zinc in leaves of the hyperaccumulator Thlaspi caerulescens. Plant Physiol 119: 305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasat MM, Baker AJM, Kochian LV (1996) Physiological characterization of root Zn2+ absorption and translocation to shoots in Zn hyperaccumulator and nonaccumulator species of Thlaspi. Plant Physiol 112: 1715–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasat MM, Baker AJM, Kochian LV (1998) Altered zinc compartmentation in the root symplasm and stimulated Zn2+ absorption into the leaf as mechanisms involved in zinc hyperaccumulation in Thlaspi caerulescens. Plant Physiol 118: 875–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Bae H, Jeong J, Lee J, Yang Y, Hwang I, Martinoia E, Lee Y (2003) Functional expression of bacterial heavy metal transporter in Arabidopsis enhances resistance to and decreases uptake of heavy metals. Plant Physiol 133: 589–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombi E, Zhao F, Dunham S, McGrath S (2000) Cadmium accumulation in populations of Thlaspi caerulescens and Thlaspi goesingense. New Phytol 145: 11–20 [Google Scholar]
- McGrath SP, Zhao FJ, Lombi E (2002) Phytoremediation of metals, metalloids and radionuclides. Adv Agron 75: 1–56 [Google Scholar]
- Mengel K, Kirkby E (1987) Principals of Plant Nutrition, Ed 4. International Potash Institute, Bern, Switzerland
- Mills R, Krijger G, Baccarini P, Hall JL, Williams L (2003) Functional expression of AtHMA4, a P1B-type ATPase of the Zn/Co/Cd/Pb subclass. Plant J 35: 164–176 [DOI] [PubMed] [Google Scholar]
- Minet M, Dufour ME, Lacroute F (1992) Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J 2: 417–422 [DOI] [PubMed] [Google Scholar]
- Motekaitis RJ, Smith RM, Martell AE (1997) National Institute of Standards and Technology Metal Complexes Database. U.S. Department of Commerce, Gaithersburg, MD
- Pence N, Larsen P, Ebbs S, Letham D, Lasat M, Garvin D, Eide D, Kochian V (2000) The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens. Proc Natl Acad Sci USA 97: 4956–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing C, Mitra B, Rosen B (1997) The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc Natl Acad Sci USA 94: 14326–14331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P (1990) Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 178–179
- Solioz M, Vulpe C (1996) CPx-type ATPases: a class of P-type ATPases that pump heavy metals. Trends Biochem Sci 21: 237–241 [PubMed] [Google Scholar]
- Tong L, Nakashima S, Shibasaka M, Katsuhara M, Kasamo K (2002) A novel histidine-rich CPx-ATPase from the filamentous cyanobacterium Oscillatoria brevis related to multiple-heavy-metal cotolerance. J Bacteriol 184: 5027–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Pittman J, Hall J (2000) Emerging mechanisms for heavy metal transport in plants. Biochim Biophys Acta 1465: 104–126 [DOI] [PubMed] [Google Scholar]